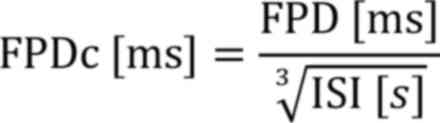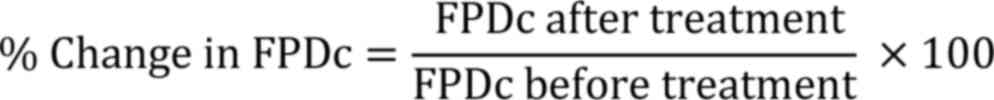Introduction
Drug-induced arrhythmias are new or exacerbated
existing arrhythmias in patients during or after drug treatment and
are the most common clinical adverse cardiovascular events
(1). Drug-induced TdP is serious
and fatal and multiple compounds were recalled in the 1990s and
early 2000s due to TdP (2). As
such, drug-related arrhythmia is one of the main causes of drug
withdrawal and drug recalls and how to predict drug-induced TdP is
a key problem in drug research and development (3).
Human ether-a-go-go related gene (hERG) block and QT
prolongation are thought to be the main causes of TdP (4). Consequently, the International
Council on Harmonization of Technical Requirements for Registration
of Pharmaceuticals for Human Use issued guidelines S7B and E14 in
2005(5). S7B recommends in
vitro assays to evaluate whether compounds and their
metabolites block hERG currents, while E14 focuses on the overall
clinical QT monitoring to determine whether the drugs prolong QT
(6). These two are currently the
main guidelines for the safety evaluation of drug-induced
arrhythmias.
The implementation of these two guidelines
effectively reduces cardiotoxicity risk in the later drug
development stage (7). However,
hERG block and QT prolongation are sensitive but not specific to
TdP risk identification (8).
Cardiomyocyte repolarization is a complex process resulting in
multiple time- and voltage-dependent ion flows across membrane
(9). In addition, hERG is an
important outward potassium current during repolarization and hERG
block can lead to prolonged repolarization and prolonged QT
interval (4). Nevertheless, it
does not induce QT prolongation if a compound, for instance
verapamil, blocks both outward potassium and inward calcium
currents (10). Therefore,
drug-induced hERG block cannot be definitively proven to be
exclusively associated with QT prolongation or TdP (11).
The Food and Drug Administration (FDA) launched the
CiPA program in 2013 that proposed a new mechanism and model
information approach to evaluate cardiac safety of new drugs
(12). The initiative focuses on
the effects of drugs on multiple ion channels using computer models
to simultaneously grade the risk of arrhythmias along with a
validation assessment using hiPSC-CMs and evaluates the expected
risk of drugs in phase I of clinical study (13). A key CiPA principle is that
ventricular repolarization and TdP depend on changes in the
equilibrium of ion flows in and outside the cell membrane and is
not solely on hERG blockade (14).
Moreover, hiPSC-CMs are more suitable for the evaluation of
arrhythmias compared with the noncardiac myocytes stably
transfected with the human hERG gene. hiPSC-CMs express
cardiocontractile proteins and functional ion channels, which
ensure that the cells simulate the function of the human conduction
system (15,16). Thus, it has become the preferred
model to predict TdP (17).
Previous studies of TdP prediction models have
mainly been based on the electrophysiological effects of compounds
on hiPSC-CMs, commonly employing regression- and machine
learning-based methods (18). Ando
et al (17) established a
regression-based risk prediction model based on field potential
duration (FPD) prolongation and early afterdepolarization, which
has been used for risk score prediction of TdP risk. In addition,
da Rocha et al (19)
established a regression-based TdP risk model based on the Voltage
Sensitive Dye Assay and used only a single predictor, action
potential (AP). Raphel et al (20) constructed an algorithm based on the
modelling dictionary and greedy optimization to predict the risk of
proarrhythmia using hiPSC-CMs and used FPD prolongation as a model
predictor. The prediction model published in the 2018 FDA report
included seven predictors but only used one algorithm, LR model
(21). Single predictor models are
prone to false positive and false negative results in the
application and a single algorithm, especially logical regression,
which may not be able to identify some significant non-linear
relations and detect the correlation between predictive variables,
is prone to result in a false-negative practical application
(22). Machine learning techniques
overcome the shortcomings of traditional regression-based methods
and have excellent performance in predictive models (23). In addition, machine learning can
predict accurately using multiple predictors following nonlinear
links and interaction between multiple variables (24).
In the present study, the effects of compounds on
the electrophysiology of synergistically beating 2D cardiomyocytes
on a label-free cardiac safety screening device were examined. A
total of 28 CiPA compounds (eight high, 11 intermediate and nine
low TdP risk) were used as training set. Overall, six endpoints of
electrophysiological responses were used as model predictors and
seven models were selected. Subsequently, two models [LR and
Adaptive Boosting (AdaBoost)] with high accuracy and good stability
were selected as sub-models, establishing a new TdP prediction
model with higher accuracy and sensitivity. This model is predicted
to reduce the impact of differences in species and reduce the
evaluation shortcomings caused by the use of hERG inhibition as a
single indicator. This model is expected to be used for preclinical
drug-induced TdP prediction and also for the re-evaluation of
post-marketing drugs.
Materials and methods
hiPSC-CMs culture
hiPSC-CMs (cat. no. CA2201106; Beijing Cellapy
Biotechnology Co., Ltd.) were cryopreserved in liquid nitrogen
(approx. -196˚C) for ~30 days after differentiation. The
cardiomyocytes used in the present study consist mainly of
ventricular myocytes with autonomous electrophysiological activity,
but also contain a small proportion of atrial myocytes and
sinoatrial node-like cells. Cell culture was conducted following
the manufacturer's protocols and four batches of hiPSC-CMs were
used. Briefly, 5 µl fibronectin (50 µg/ml, cat. no. F2006;
Sigma-Aldrich; Merck KGaA) was added to the central electrode of
the CardioExcyte 96 sensor plates (cat. no. 020730; Nanion
Technologies GmbH), then the plate was incubated at 37˚C for 1-1.5
h. The cells were then thawed in a 37˚C water bath and centrifuged
at 200 x g for 5 min at room temperature. The supernatant was later
discarded and the cell concentration was adjusted to
4-5x104 cells/ml. Fibronectin coating solution from the
sensor plate was discarded and 5 µl of cell suspension was added
into each well before being incubated for 1 h at 37˚C, 5%
CO2. Lastly, 200 µl of medium (cat. no. CA2015002;
Cellapy; Beijing Saibei Biotechnology Co., Ltd.) was added to each
well and the plate was cultured for 14-21 days at 37˚C, 5%
CO2. The medium was replaced 48 h after plating and
medium changing was performed every other day.
Test compounds
In total, 35 compounds were used (17 were purchased
from Sigma-Aldrich, Merck KGaA; 16 were made by medicinal chemists
at the National Institute for Food and Drug Control; one from
Selleck Chemicals; and one from MedChemExpress). All compounds were
dissolved in DMSO or H2O2 to prepare the
stock solutions, which were at least 1,000 times more concentrated
compared with the highest concentration used experimentally
(Table SI).
Myocardial cell activity, beat
frequency and field potential detection
The sensor plate was placed on the CardioExcyte 96
and incubated at 37˚C for 1-2 days before drug administration. The
medium was changed on the day of drug administration. Different
compounds (100 µl) were added 3-4 h after culture medium change and
0.1% DMSO was set as the negative control for each plate. The data
collection interval was 30 min within 2 h, the interval then
changed to 2 h. Thus, the data collection period was >24 h. For
each experiment, three to four concentrations were set for each
compound and three duplicates were set for each concentration. Each
experiment was repeated in duplicate or quadruplicate. Data for
nicotinamide and mannitol were from a single experiment and eight
replicates were set for each concentration. The main indices were
amplitude (Ampl, the representative amplitude traces were shown in
Fig. S1), beats per minute (BPM)
and FPD. FPD was corrected to FPDc by Fridericia's formula:
FPD, Field potential duration; ISI, Inter-spike
interval.
Recording of arrhythmia-like
waveforms
Asakura et al (25) recorded the compound-induced
arrhythmia waveforms; an arrhythmia-like waveform was observed and
recorded as 1 and without arrhythmia-like waveforms was recorded as
0.
Data processing
CardioExcyTecontrol (version 1.4.5.6; Nanion
Technologies GmbH) software was used to process the data. The
negative control, 0.1% DMSO, was used as the baseline to calibrate
the online analysis parameters. Ampl, BPM and FPDc (Corrected FPD)
data were then normalized to preadministration data. The FPDc
change was calculated according to the following formula:
Model establishment and
evaluation
In total, 28 CiPA compounds were used as the
training set. In addition, a prediction model of TdP risk was
constructed following the physiological effects of these compounds
on hiPSC-CMs. effective therapeutic plasma concentration (ETPC),
arrhythmia-like events, drug concentration (folds over ETPC when
drug-induced arrhythmias were first observed), type of FPDc change,
the degree of change in FPDc and drug concentration (folds over
ETPC) at which the change of FPDc was observed (Table I) were used as model predictors.
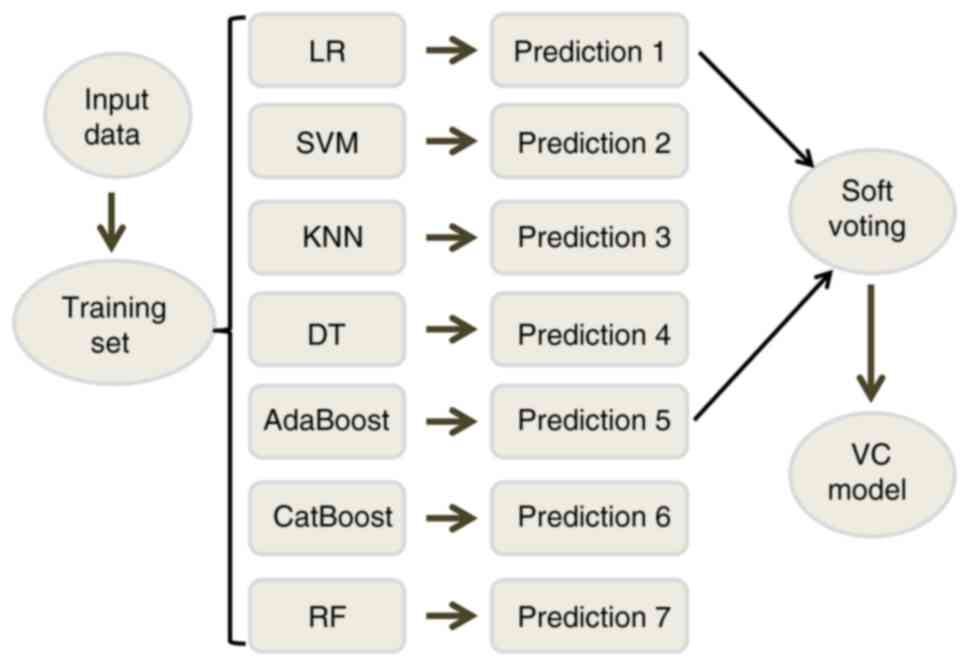
Based on these six predictors, seven models were established in the
training set for high- and intermediate-risk compounds versus
low-risk compounds. The seven models were LR, Support Vector
Machine (SVM), k-nearest neighbor algorithm (KNN), decision
tree (DT), AdaBoost, CatBoost and Random Forest (RF).
 | Table IModel predictors for all compounds
tested. |
Table I
Model predictors for all compounds
tested.
| TdP risk | Compounds | ETPC, µM | ETPC1 | Arrhythmia-like
Events | ETPC2 | FPDc changes | %change in
FPDc |
|---|
| H | Azimilide |
7.0x10-2 |
7.1x10-1 | 1 |
7.1x10-1 | 1 | 35 |
| H | Bepridil |
3.2x10-2 |
3.1x102 | 1 |
9.4x10-1 | 3 | -11 |
| H | Disopyramide |
7.0x10-1 |
1.4x10-1 | 1 |
1.4x10-1 | 1 | 65 |
| H | Dofetilide |
2.0x10-3 | - | 0 |
1.5x101 | 1 | 30 |
| H | Ibutilide |
1.0x10-1 |
1.0x10-1 | 1 |
1.0x10-1 | 1 | 77 |
| H | Quinidine | 3.0x10 |
3.0x10-2 | 1 |
3.0x10-2 | 1 | 33 |
| H | Sotalol |
1.5x101 | 2.0x10 | 1 |
7.0x10-2 | 1 | 59 |
| H | Vandetanib |
3.0x10-1 |
3.0x10-2 | 1 |
3.3x101 | 1 | 53 |
| I | Astemizole |
3.0x10-4 |
3.3x102 | 1 | 3.3x10 | 1 | 50 |
| I | Chlorpromazine |
3.5x10-2 | - | 0 |
8.7x101 | 1 | 57 |
| I | Cisapride |
2.6x10-3 | - | 0 |
1.2x103 | 2 | 10 |
| I | Clarithromycin | 1.2x10 | - | 0 |
8.0x10-2 | 3 | -43 |
| I | Clozapine |
7.1x10-2 |
1.4x102 | 1 |
4.2x10-1 | 1 | 65 |
| I | Domperidone |
2.0x10-2 | 1.5x10 | 1 |
1.5x101 | 1 | 45 |
| I | Droperidol |
1.6x10-2 |
1.9x101 | 1 | 6.3x10 | 1 | 38 |
| I | Ondansetron |
3.7x10-1 |
8.1x10-1 | 1 | 8.1x10 | 2 | 9 |
| I | Pimozide |
4.3x10-4 | 2.3x10 | 1 | 2.3x10 | 1 | 49 |
| I | Risperidone |
1.8x10-3 |
5.6x102 | 1 |
5.6x101 | 1 | 35 |
| I | Terfenadine |
2.9x10-4 | - | 0 |
3.5x103 | 2 | 10 |
| L | Diltiazem |
1.3x10-1 | - | 0 |
2.3x10-1 | 3 | -16 |
| L | Loratadine |
4.5x10-4 | - | 0 |
6.7x101 | 3 | -19 |
| L | Ranolazine | 1.9x10 |
1.5x101 | 1 |
5.1x10-1 | 1 | 27 |
| L | Metoprolol | 1.8x10 | - | 0 |
6.0x10-2 | 1 | 60 |
| L | Mexiletine | 2.5x10 | - | 0 | 1.2x10 | 2 | 10 |
| L | Nifedipine |
7.7x10-3 | - | 0 |
1.3x102 | 3 | -10 |
| L | Nitrendipine |
3.0x10-3 |
3.3x102 | 1 |
9.9x10-1 | 3 | -51 |
| L | Tamoxifen |
2.1x10-2 |
1.4x102 | 1 |
1.4x101 | 3 | -46 |
| L | Verapamil |
4.5x10-2 | - | 0 | - | 0 | 0 |
| Non | Amiodarone |
7.0x10-4 | - | 0 |
1.4x104 | 1 | 25 |
| Non | Flecainide |
7.5x10-1 | 1.3x10 | 1 |
1.3x10-1 | 1 | 41 |
| Non | Moxifloxacin | 3.6x10 | - | 0 |
8.4x101 | 1 | 40 |
| Non | E4031 |
8.4x10-3 | - | 0 |
1.2x10-1 | 3 | -33 |
| Non | Methadone |
9.9x10-1 | - | 0 |
2.0x10-1 | 1 | 51 |
| Non | Nicotinamide |
1.2x103 | - | 0 |
4.0x10-2 | 1 | 37 |
| Non | Mannitol |
2.4x103 | - | 0 |
2.0x10-5 | 1 | 33 |
Accuracy, recall rate or sensitivity and AUC were
used to evaluate the ability to distinguish capacity, where the
larger the index value is, the higher the prediction ability of the
model is (26,27). Seven compounds with known TdP risk
were used as the test set to evaluate the calibration
capacity-defined as the consistency between predicted and observed
results (28). The models with
excellent comprehensive performance and good stability in the
training and test data sets were selected and the voting classifier
strategy was then used for model training. Each sub-model was
assigned a similar weight and the new voting classifier (VC)
prediction model was established through soft voting; the weight
here refers to the relative importance of the indicator in the
overall evaluation. The VC model is the combination of two
classifiers that combined the predicted probabilities for each
classifier and the highest probability is then voted (Fig. 1).
Statistical analysis
Data are expressed as the mean ± standard deviation.
SPSS 26.0 (IBM Corp) was used to perform Pearson's and Spearman's
correlation analysis, P<0.05 was considered to indicate a
statistically significant difference. Python 3.8.0 (python.org/) and Prism 8.0 (GraphPad Software, Inc.)
were used to establish prediction models and plot data.
Results
Effects of compounds on myocardial
cell activity and beating frequency
Ampl is an impedance parameter, which is a key
parameter for the estimation of cell viability (29). In total, four out of eight high TdP
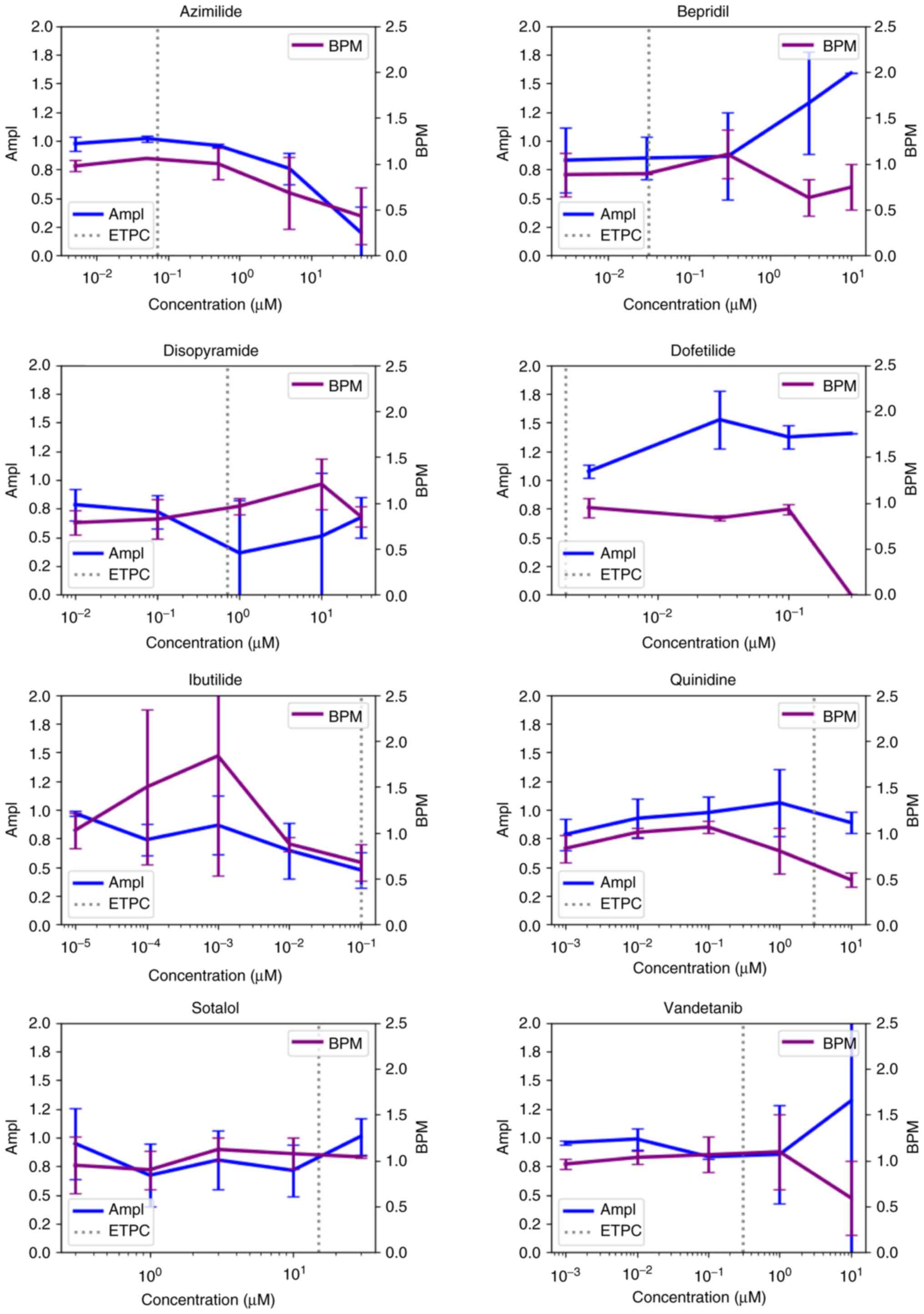
risk compounds resulted in decreased cell viability (Fig. 2), among which, disopyramide,
ibutilide and quinidine could cause decreased cell activity at
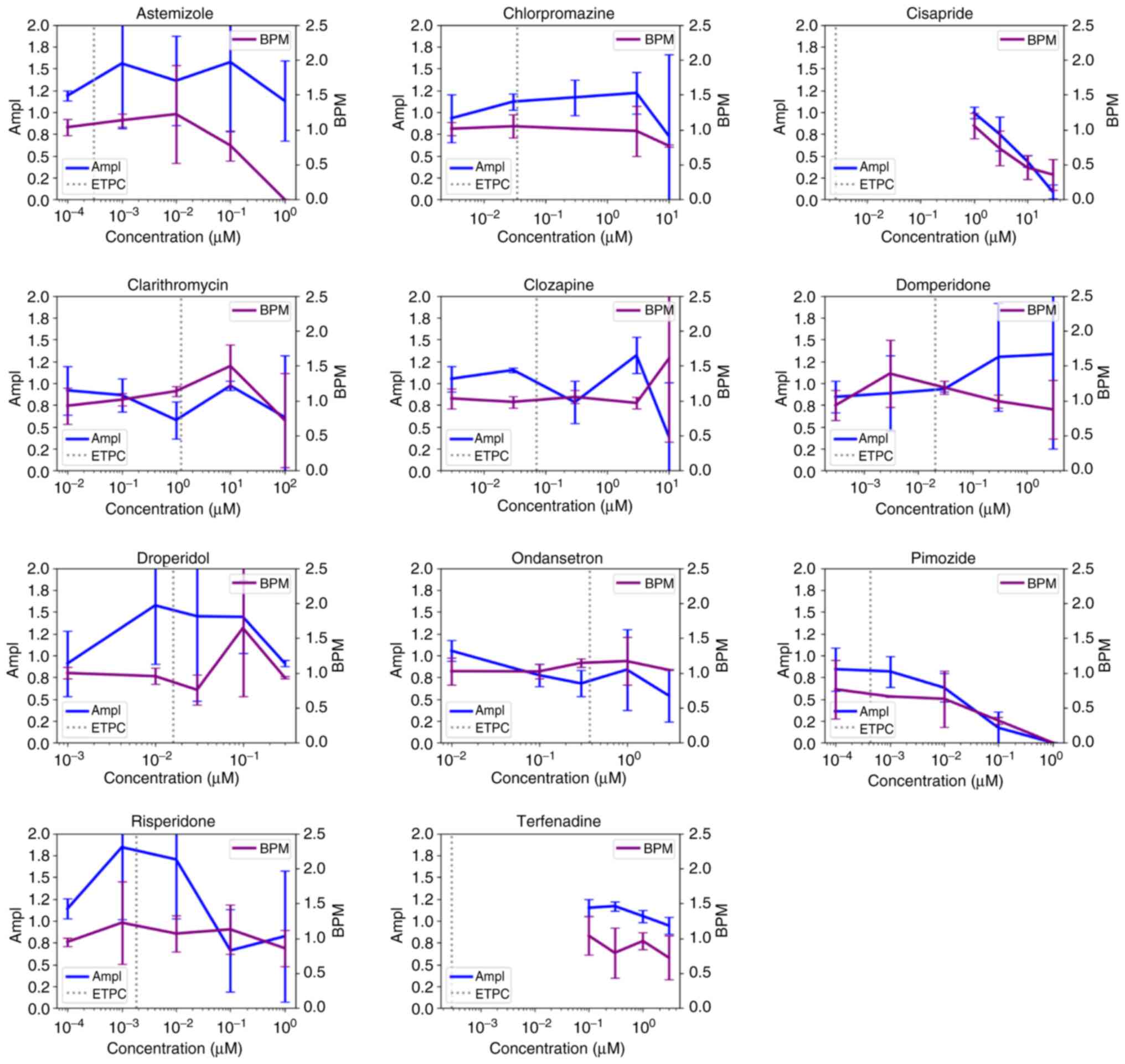
concentrations less than ETPC. Moreover, nine (Astemizole,
Chlorpromazine, Cisapride, Clarithromycin, Droperidol, Ondansetron,
Pimozide, Risperidone and Terfenadine) out of eleven and eight
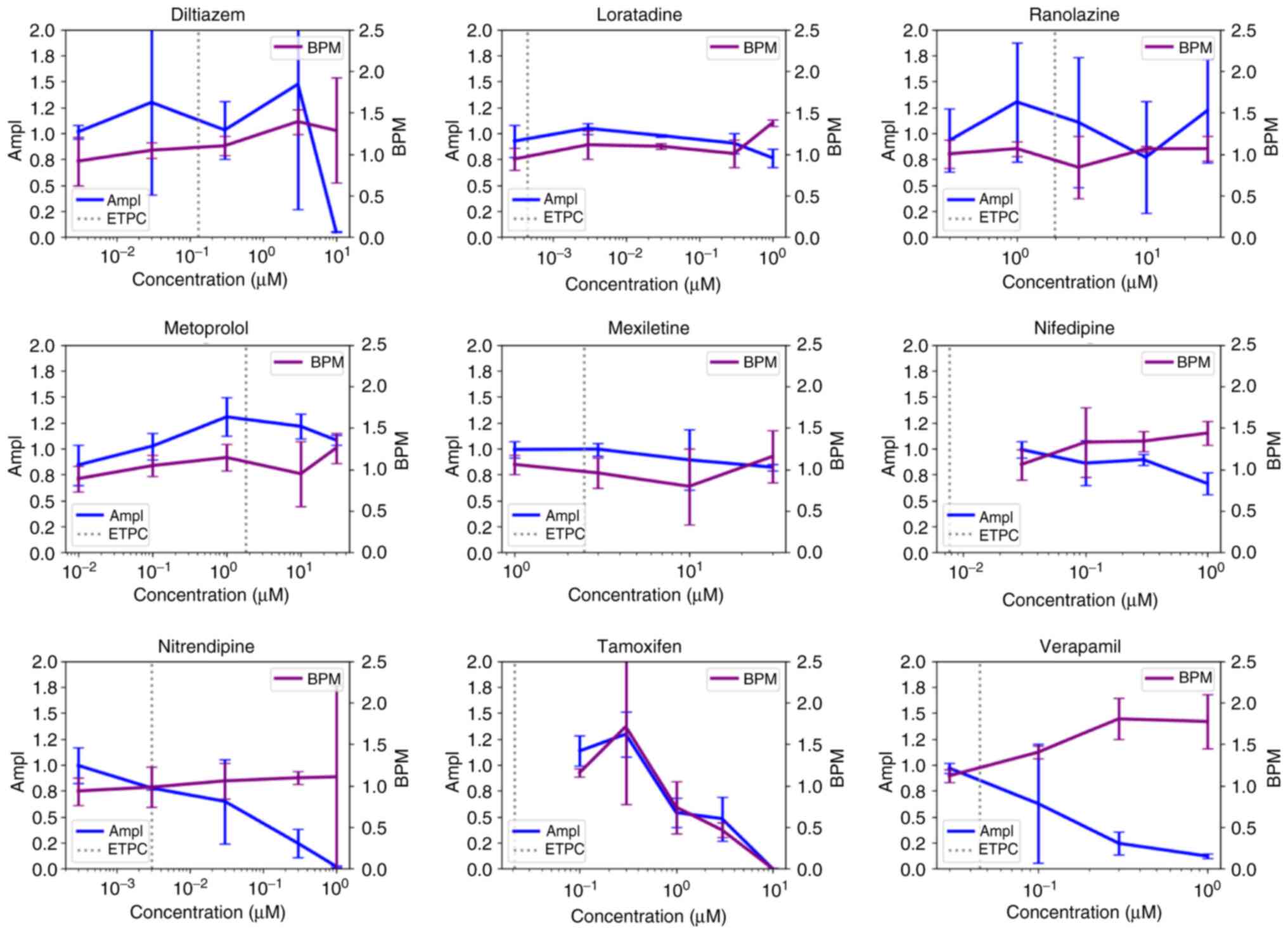
(Diltiazem, Loratadine, Metoprolol, Mexiletine, Nifedipine,
Nitrendipine, Tamoxifen and Verapamil) out of nine intermediate and
low TdP risk compounds, respectively, decreased the activity of
cardiomyocytes at a concentration higher than ETPC (Figs. 3 and 4, Table
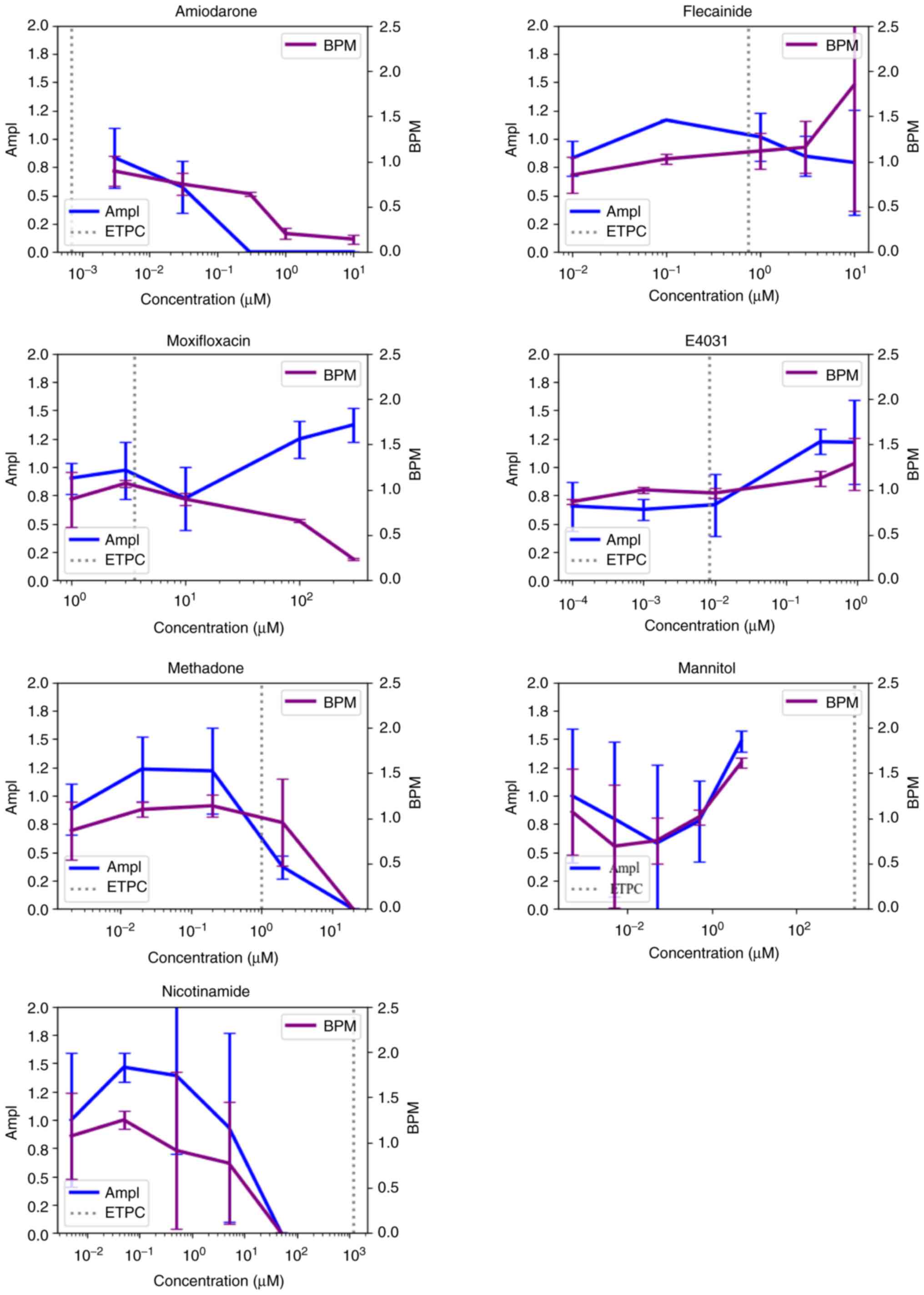
SII). In addition, three (Amiodarone, Methadone and
Nicotinamide) out of seven of the non-CiPA compounds decreased the
cell activity of cardiomyocytes at concentrations greater than ETPC
(Fig. 5).
BPM was used to evaluate the effect of compounds on
the contractile function of the cardiomyocytes. Of the high TdP
risk compounds, six out of eight lead to a decrease in BPM
(Fig. 2). In addition, ibutilide,
quinidine, sotalol and vandetanib all led to BPM decrease at
concentrations less than ETPC. In the intermediate TdP risk
compounds set, nine (Astemizole, Chlorpromazine, Cisapride,
Clarithromycin, Droperidol, Ondansetron, Pimozide, Risperidone and
Terfenadine) out of eleven were found to decrease BPM (Fig. 3, Table SII), with only domperidone
concentration having been lower than ETPC. BPM was decreased by
four (Diltiazem, Metoprolol, Mexiletine and Tamoxifen) out of nine
low TdP risk compounds at concentrations greater than ETPC
(Fig. 4, Table SII). Furthermore, four
(Amiodarone, Moxifloxacin, Methadone and Nicotinamide) out of seven
of the non-CiPA compounds decreased BPM at a concentration greater
than ETPC, excluding nicotinamide (Fig. 5).
By contrast, two out of eight, two out of eleven and
five out of nine compounds with high, intermediate and low TdP
risk, respectively, led to increases in BPM (Fig. 2, Fig.
3 and Fig. 4). In addition,
the incidence of arrhythmia and FPDc prolongation was 57.89 and
84.21%, respectively, when BPM was decreased (Table II). Spearman correlation analysis
showed that reducing BPM was significantly correlated with the
occurrence of FPDc prolongation (ρ=0.438, P<0.01). However, the
incidence of arrhythmia and FPDc prolongation were 66.67 and
44.44%, respectively (Table II),
when BPM was increased. However, no relationship was noted between
BPM increase, arrhythmia and FPDc prolongation.
 | Table IICorrelation of BPM with
Arrhythmia-like Events and FPD prolongation |
Table II
Correlation of BPM with
Arrhythmia-like Events and FPD prolongation
| TdP risk | BPM changes | Number of
Arrhythmia-like Events | Number of FPD
prolongation |
|---|
| High | Reduced (n=6) | 5 | 6 |
| | Elevated (n=2) | 2 | 1 |
| Intermediate | Reduced (n=9) | 5 | 8 |
| | Elevated (n=2) | 2 | 2 |
| Low | Reduced (n=4) | 1 | 2 |
| | Elevated (n=5) | 2 | 1 |
| Proportion of
Arrhythmia-like events due to reduced BPM | | | 57.89% |
| Proportion of
Arrhythmia-like events due to elevated BPM | | | 66.67% |
| Proportion of FPD
prolongation due to the reduced BPM | | | 84.21% |
| Proportion of FPD
prolongation due to the elevated BPM | | | 44.44% |
Effect of compounds on cardiomyocyte
rhythm
Of the high TdP risk compounds, seven (Azimilide,
Bepridil, Disopyramide, Ibutilide, Quinidine, Sotalol and
Vandetanib) out of eight were observed to induce arrhythmia-like
waveforms (Table I). Similarly,
ibutilide was observed to cause arrhythmia-like waveforms at 10% of
the ETPC, which was the minimum multiple of the ETPC. Bepridil
induced arrhythmia-like waveforms at 312.5 times of the ETPC, which
was the highest multiple of the ETPC. No arrhythmia-like waveforms
were observed in dofetilide up to 150 times of the ETPC (Table I).
In the intermediate TdP risk compound group, seven
(Astemizole, Clozapine, Domperidone, Droperidol, Ondansetron,
Pimozide and Risperidone) out of eleven caused arrhythmia-like
waveforms (Table I). Ondansetron
induced arrhythmia-like waveforms at 81% of the ETPC, which was the
minimum multiple of the ETPC. In addition, arrhythmia-like
waveforms were observed with risperidone at 555.6 times of the
ETPC, which was the highest multiple times of the ETPC. For the
other four compounds, no arrhythmia-like waveforms were observed at
the tested concentrations (Table
I).
Among the low TdP risk compounds, ranolazine,
nitrendipine and tamoxifen were observed with arrhythmia-like
waveforms at 15.4, 331.1 and 142.9 times of the ETPC, respectively.
Only flecainide was observed with arrhythmia-like waveforms at 1.33
times of the ETPC of the seven non-CiPA compounds, while no
arrhythmia-like waveforms were observed in other compounds
(Table I).
Effect of compounds on FPDc of
cardiomyocytes
Only bepridil was observed to not induce FPDc
prolongation among the eight high TdP risk compounds, with the
other seven compounds showing FPDc prolongation in hiPSC-CMs after
administration (Fig. 6, Table I), among which, dofetilide and
vandetanib caused FPDc prolongation at concentrations higher than
ETPC. Moreover, azimilide, disopyramide, ibutilide, quinidine and
sotalol were observed with FPDc prolongation at concentrations
lower than ETPC (Fig. S2).
Only clarithromycin had no FPDc prolongation among
the eleven intermediate TdP risk compounds, while the remaining ten
compounds were observed with FPDc prolongation (Fig. 6, Table
I), of which, clozapine's concentration was 42% of the ETPC.
The other nine compounds' concentrations were higher compared with
the ETPC (Fig. S3).
Among the nine compounds with low TdP risk,
ranolazine (51% of the ETPC), metoprolol (6% of the ETPC) and
mexiletine (1.2 times of the ETPC) could induce FPDc prolongation,
while no FPDc prolongation was observed in the remaining six
compounds (Figs. 6 and S4, Table
I). The seven non-CiPA compounds, except for E4031, were all
observed to cause FPDc prolongation (Figs. 6 and S5; Table
I).
Model establishment and
validation
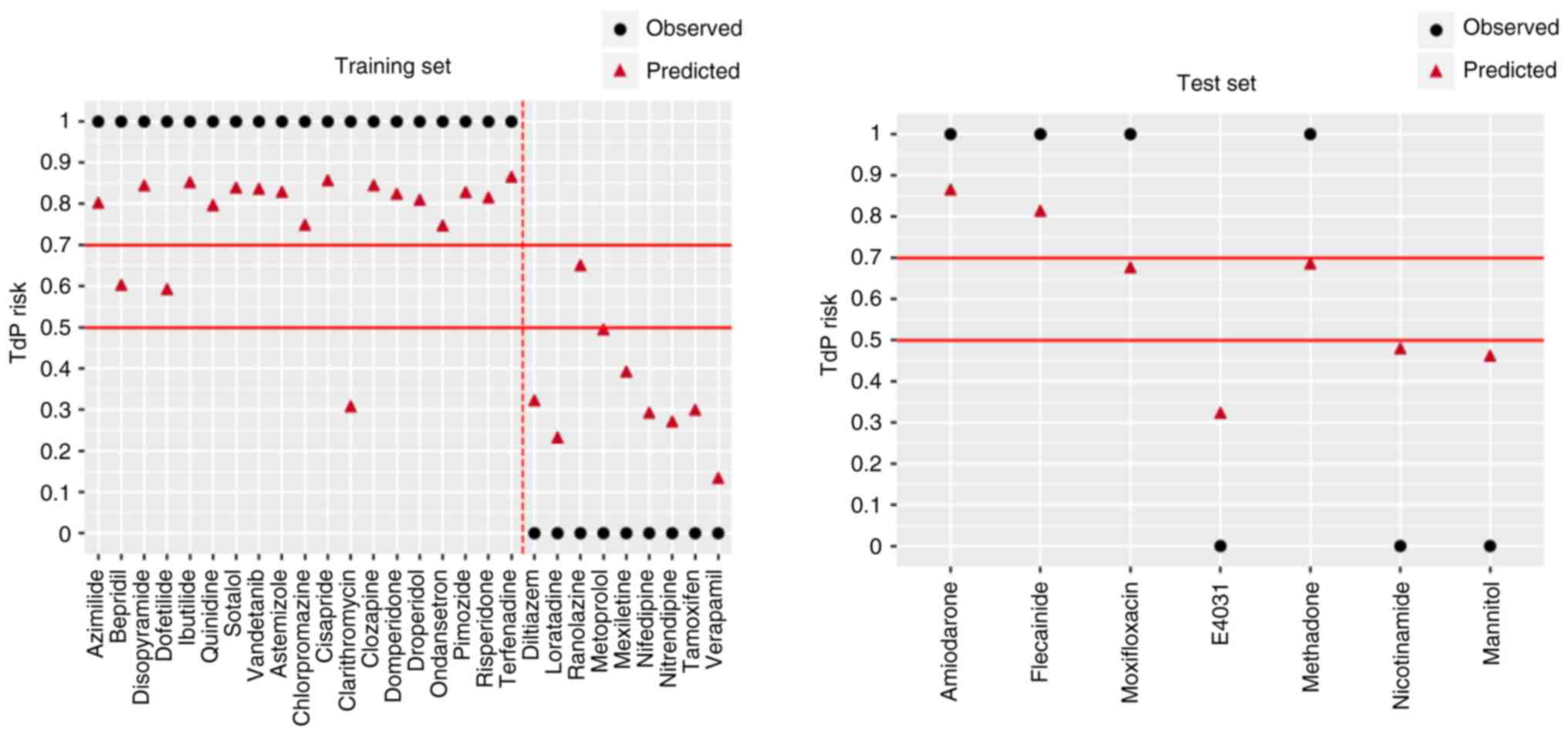
Model selection. The classification accuracy
of LR, SVM and DT was 0.86, 0.79 and 0.93, respectively, when the
training set was used for modeling; the recall rates of these three
models for intermediate- and high-risk compounds were 0.89, 0.74
and 0.89, respectively; and the AUC were 0.84, 0.81 and 0.95,
respectively (Table SIII). The
classification accuracy of KNN, AdaBoost, CatBoost and RF were all
1.00 and the recall rates in the training set were all 1.00, and
all the AUC were 1.00 (Table
SIII). According to the performance of each model in the
training and test sets (Table
III), a high AUC value indicates a strong distinguishing
ability of the model, while a small difference in AUC, observed
between the training and the test sets, indicates good model
stability. According to the discriminative ability and stability,
LR and AdaBoost were the two models with best performance in both
the training and test sets and were selected as sub-models going
forward. Each sub-model is assigned a similar weight and the new
prediction model (VC model) is established through soft voting.
 | Table IIIAUC results of seven models. |
Table III
AUC results of seven models.
| Sample set | LR | SVM | KNN | DT | AdaBoost | CatBoost | RF |
|---|
| Training set | 0.84 | 0.81 | 1.00 | 0.95 | 1.00 | 1.00 | 1.00 |
| Test set | 0.83 | 0.75 | 0.71 | 0.50 | 0.83 | 0.71 | 0.83 |
Model predictors evaluation. Arrhythmia-like
waveforms and FPDc prolongation the two most important modelling
indicators. Spearman correlation analysis showed that
arrhythmia-like waveforms were correlated with the predictive
values (r=0.40, P<0.05, data not shown). Pearson
correlation analysis showed that FPDc prolongation was also
significantly correlated with the predictive values
(r=0.744, P<0.001, data not shown)
Model evaluation. The newly established VC
model was compared with the LR and AdaBoost models. The LR and
AdaBoost models both performed poorly on the training and test sets
(Fig. 7, Table IV). However, the VC model
performed well in both the training and test sets. Only a small
difference was noted in all the parameters between the two sets,
indicating that the VC model had good stability.
 | Table IVModel evaluation results. |
Table IV
Model evaluation results.
| | Training set | Test set |
|---|
| Model | Accuracy | Recall | AUC | Accuracy | Recall | AUC |
|---|
| LR | 0.86 | 0.89 | 0.84 | 0.86 | 1.00 | 0.83 |
| AdaBoost | 1.00 | 1.00 | 1.00 | 0.86 | 1.00 | 0.83 |
| VC | 0.93 | 0.95 | 0.92 | 1.00 | 1.00 | 1.00 |
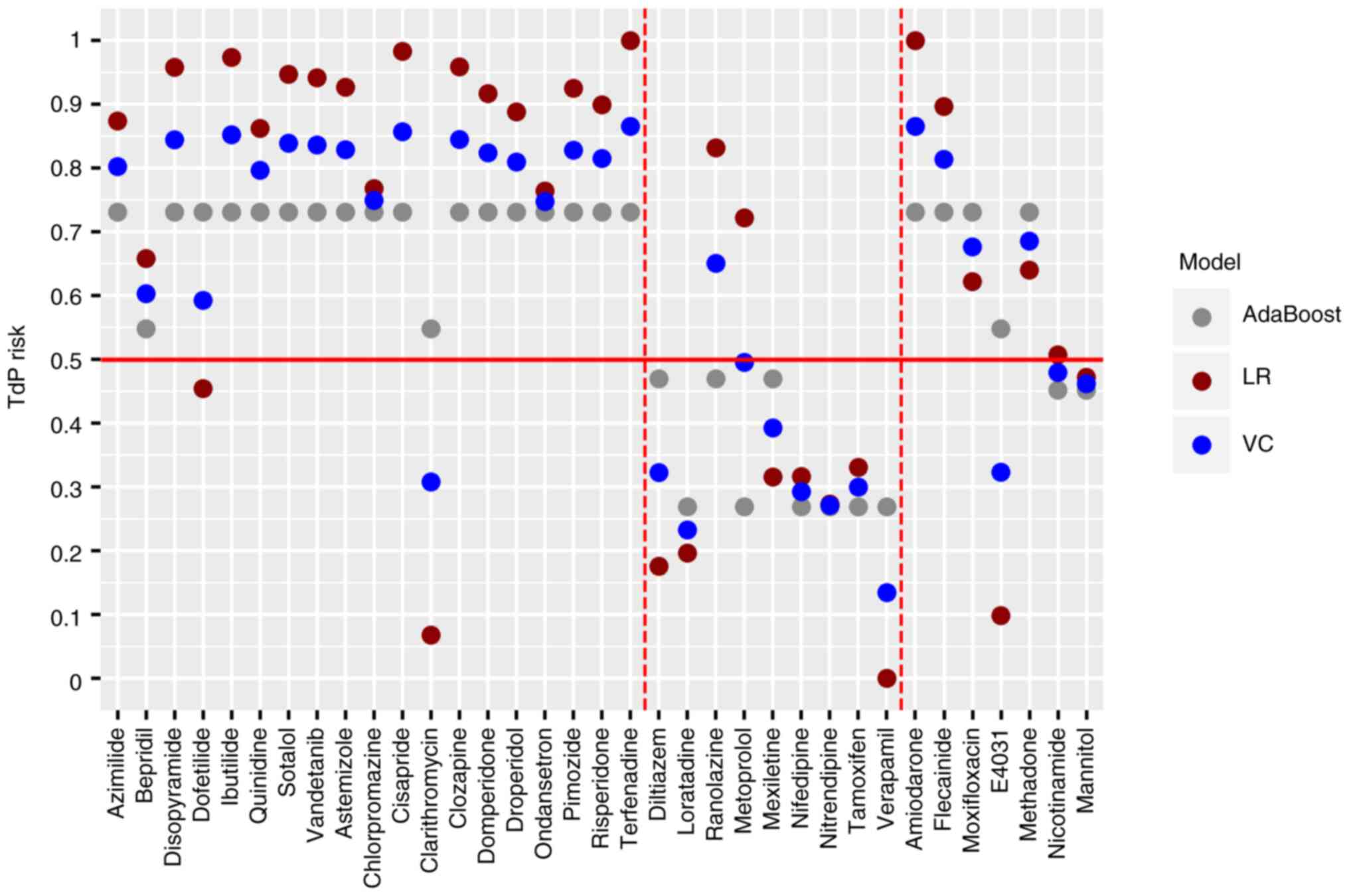
Threshold setting. The default threshold of
intermediate and high to low risk was 0.5 in the process of model
establishment. When the threshold was set as 0.5, clarithromycin at
intermediate-risk was predicted to be low-risk and ranolazine
(low-risk) was predicted to be an intermediate- or high-risk
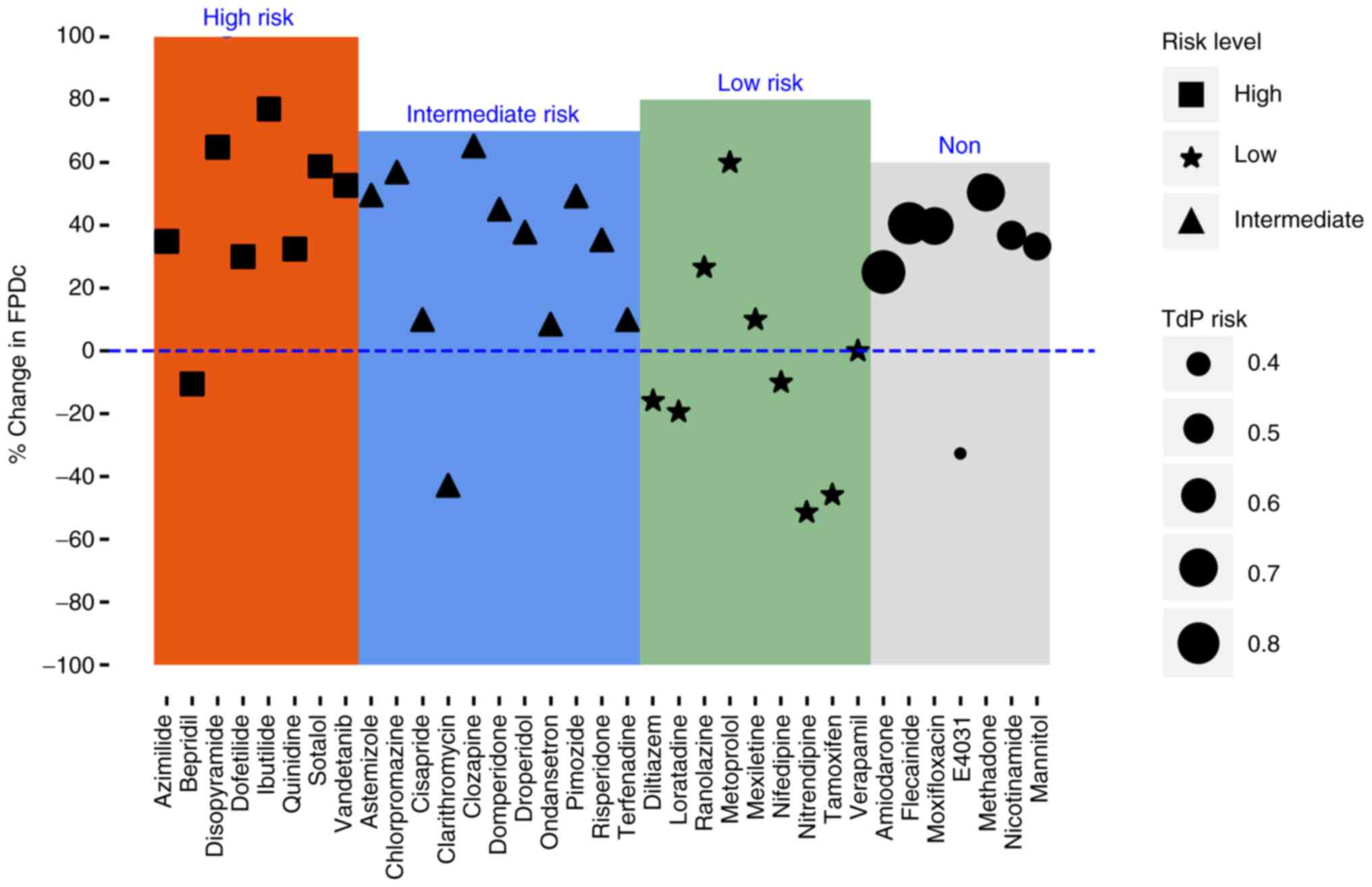
compound with a consistency of 92.8% (Fig. 8). In the test set, amiodarone,
flecainide, moxifloxacin and methadone were all predicted to be of
intermediate or high-risk, while E4031, nicotinamide and mannitol
were low risk with a consistency of 100% (Fig. 8).
Discussion
Primary rat cardiomyocytes are currently the main
cellular model for cardiac toxicity evaluation (30). However, pronounced differences in
species have been noted between mice and humans, resulting in 37%
tolerance of compounds in mice, which was 10 times higher compared
with that of humans (31).
Increasing attention has highlighted the advantages of hiPSC-CMs in
previous studies. Moreover, hiPSC-CMs express various human cardiac
ion channels and genes encoding key components of the
Ca2+ cycle, which greatly reduces the impact of
differences between species and reduces the discrepancy between rat
and adult primary human cardiomyocytes (32-34).
hiPSC-CMs display the complex physiological functions associated
with human myocardial cells in vitro and possess the
biochemical and molecular biological characteristics of human
cardiac myocytes. Moreover, hiPSC-CMs are easy to purify and can be
cultured in vitro for extended periods of time compared to
rat primary cardiomyocytes (data not shown). Hence, it is an
excellent model for in vitro prediction of arrhythmia in
vivo. In the present study, hiPSC-CMs has a clonal morphology
and gene expression that highly resembles human embryonic stem
cells. It is probable that different hiPSC-CM lines can behave
differently in response to electrophysiological changes, so three
hiPSC-CM lines were evaluated in previous studies. The prolongation
of FPDc was used as a single predictor for the TdP risk, showing
the sensitivity and specificity of the hiPSC-CM line from Cellapy
were optimal, as such this cell line was selected for further
investigations.
The effects of different compounds on cell activity,
which aimed to observe cytotoxicity and the rationality of dose
design, were evaluated in the present study. Ampl also reflects the
beating pattern of the hiPSC-CMs and the change in beating patterns
reflects which ion channels in the cell membrane are affected
(35). The results showed that no
cytotoxicity was noted at all concentrations of the compounds
(Table SI), but the correlation
between the change of Ampl and TdP risk needs further evaluation.
Simultaneously, the effects of compounds on the BPM of
cardiomyocytes were tested. Abnormal heart rate correlates with
drug-induced cardiotoxicity (36).
According to the present data, compounds that inhibited the
contractile function of cardiomyocytes resulted in reduced BPM,
which is more likely to cause arrhythmia and more likely to induce
prolonged FPDc. This suggested that inhibition of cell beating may
be a risk factor for TdP.
Drug effects and adverse reactions are closely
related to dose, for some compounds, the risk of side effects can
increase with increasing dose, ETPC is usually proportional to the
dose, and is a common indicator of the dose administered (37). Thus, ETPC was also accepted as a
model predictor. The arrhythmia-like waveforms are another
important model predictor. Arrhythmia-like waveforms were observed
in all three TdP risk compound groups and high-risk compounds were
more likely to induce arrhythmia-like waveforms. A notable event,
which advises compound risk prediction, is the possibility of
increased TdP if arrhythmia-like waveforms appeared at a dose lower
than the ETPC.
AP detection is based on individual cardiomyocytes
and in this context there is a lack intercellular communication, so
instead small populations cardiomyocytes were chosen (38). Instead of recording individual APs
and early depolarization, the CardioExcyte 96 recorded field
potential. FPDc prolongation was considered to be closely related
to TdP occurrence. In the multi-electrode array (MEA) test,
adjacent recording electrodes can record spontaneous APs of the
cell population as external field potential. The recording
electrode can monitor the change in field potential when an AP
spreads across monolayer cells (39). Halbach et al (40) revealed a direct relationship
between the rise in time of APs and field potentials, showing that
APs and field potentials are linearly correlated. In the present
study, FPDc change was observed to be consistent with instances of
arrhythmia, with the higher the FPDc prolongation probability, the
higher the risk of TdP. Arrhythmia-like events and the type of FPDc
change were the most important among the six predictors. Though no
FPDc prolongation was observed for bepridil and clarithromycin in
the present study, thus, they were classified as low-risk
compounds; no arrhythmia-like events were observed during
administration of dofetilide, so dofetilide was also classified as
a low-risk compound.
Sharifi et al (41) developed a TdP prediction model
based on the chemical structure of the compounds, though this model
works only if compounds with similar structures possess the same
electrophysiological responses. Additionally, the drugs' effects on
different ion channels have also been used as predictors for TdP
risk (42). However, previous
studies of TdP prediction models have mainly been based on the
electrophysiological effects of compounds on hiPSC-CMs. The
classical models for predicting TdP risk based on hiPSC-CMs mainly
include the LR model (43).
However, in the current study, its classification accuracy was only
0.86 and the recognition rate of intermediate- and high-risk
compounds was only 0.89, which showed that the model classification
ability was not sufficient. Blinova et al (21) used the LR model to assess the risk
of dysrhythmia, one high-risk compound (bepridil) and four
intermediate-risk compounds (risperidone, terfenadine,
chlorpromazine and clozapine) were incorrectly predicted to be
low-risk; this indicated that the logical regression classifier may
not be able to identify some significant nonlinear relationships
and detect the correlation between predictive variables, making it
prone to false negatives in practical application (22).
In the current study, the classification accuracy,
the recall rate and the AUC of KNN, CatBoost and RF were all 1.00
in the training set, but these models performed poorly in the test
set. Thus, these models may be overfitted. The main influencing
factors of overfitting may include small sample size in the
training set and the inconsistency of data distribution in the
training and test sets (44). The
difference between data distribution may have introduced bias into
the models. Considering that a precise classification is difficult
to achieve with only one model, the combination of multiple models
was used in the current study. Thus, LR and AdaBoost (a black box
model widely used in computer-aided diagnosis), were chosen as the
sub-models according to the classification performance of each
model (45). Finally, a new model
named VC was established through a soft voting strategy. The AUC of
the VC model had little difference between training set and test
set and the consistency of the VC model between the predicted and
observed results was optimal. This suggested that this model could
tolerate a certain degree of differential data distribution through
the soft voting strategy.
Moreover, 28 CiPA compounds were considered to be a
small sample size (46). The data
showed that AdaBoost had improved performance compared with VC in
the training set, but VC outperformed AdaBoost in the test set.
Whether data distribution of an unknown compound is consistent with
the training set is unknown, thus the soft voting strategy was used
and the VC model was chosen to avoid overfitting and enhance the
generalization of the model.
The prediction results of VC for the training set of
28 known compounds showed that bepridil, dofetilide and
clarithromycin were classified as a low-risk at a threshold of 0.7,
but bepridil and dofetilide are high TdP risk compounds, which can
cause QT interval prolongation and induce TdP (47,48).
In the present study, bepridil was predicted as a low-risk compound
owing to no FPDc prolongation was observed. Bepridil is a calcium
antagonist and also blocks sodium and potassium channels, so the
effect of potassium channel blocking may be counterbalanced by
calcium channel blocking (49).
This may be one of the reasons for the shortening of its FPDc. No
arrhythmia-like waveforms were observed in dofetilide, which may be
the main reason why this compound was predicted to be low-risk. The
minimum and maximum concentrations of dofetilide were 0.03 (15
times of the ETPC) and 0.3 µm (150 times of the ETPC),
respectively. The cells stopped beating at the maximum
concentration, thus, the concentrations used may have been
inappropriate and further experimental confirmation is
required.
Clarithromycin is an intermediate-risk compound and
an hERG channel blocker (50). No
QT prolongation was found in healthy individuals after
clarithromycin administration (51). However, women, the elderly and
patients with underlying heart disease may exhibit increased QTc
prolongation and increased risk of TdP induced by clarithromycin
(52). In the present study, FPDc
showed shortening after clarithromycin administration and was
predicted as low risk. Ando et al (17) reported that clarithromycin at 13
times (42.9 µM) of the free plasma concentration can lead to 10%
FPDc prolongation. The experiment for clarithromycin was repeated
in triplicate and no FPDc prolongation was observed at the highest
concentration of 100 µM, therefore, experimentation should be
continued for verification.
According to the prediction results with 0.7 as the
threshold, amiodarone and flecainide, both known compounds with
associated TdP risk, were predicted to be high- or
intermediate-risk compounds. Amiodarone is a class III
antiarrhythmic drug, which can inhibit the hERG channel and cause
QT prolongation (53). The TdP
incidence of amiodarone ~15% after intravenous administration
(54). Flecainide is a class IC
antiarrhythmic drug, which can inhibit K+ channels,
causing prolonged QT interval and TdP (55). According to the established model,
the prediction results of amiodarone and flecainide are consistent
with literature reports.
The risk threshold was adjusted to 0.5 because
numerous false negatives were noted in the training set at 0.7
threshold value. Clarithromycin should be predicted as a high- or
intermediate-risk compound, and ranolazine should be predicted as a
low-risk compound, so clarithromycin was not identified and
ranolazine was misidentified in the training set. However,
amiodarone, flecainide, moxifloxacin and methadone in the test set
were all correctly predicted to be high or intermediate risk
compounds, while E4031, nicotinamide and mannitol were all
predicted to have low-risk compounds. Tisdale (53)
previously reported moxifloxacin to have known TdP risk, which
supports the findings of the present study. Moxifloxacin is more
prone to TdP in patients with multiple risk factors for prolonged
QT interval (56). Methadone is
also high risk at low doses and has the potential to prolong QT
interval and develop TdP (57).
E4031 is an hERG blocker that can prolong QT interval (58). However, to the best of our
knowledge, no clinical report has been on the risk of E4031-induced
TdP. Niacinamide and mannitol had not been reported to induce TdP
risk. Therefore, the consistency between the predicted and the
observed results was 92.8 and 100% in the training and test sets,
respectively, when 0.5 was used as the threshold value. In toxicity
evaluation, false positives could be supported by other
experiments, while false negatives could cause serious clinical
risks. Thus, the low false-negative value of 0.5 was chosen as the
threshold value.
A number of uncertainties have been noted in
extrapolating animal data to humans in the nonclinical safety
evaluations of new drugs and remains a troubling issue in toxicity
research (59). Thus, hiPSC-CMs
are a viable way to reduce uncertainty caused by species
differences. As such, the false positive and false negative rates
in toxicity evaluation caused by a single predictor was avoided in
the present study. The prediction model was able to reveal the
relationship between prediction indicators and results based on the
specific algorithms. Each model had its advantages in a particular
sample set, though the two models with improved performance were
selected to establish a new model. Thus, avoiding the defects of a
single model methodology and improving the prediction accuracy and
stability of the model.
Though a functional TdP prediction model was
established based on hiPSC-CMs in the present study, data was
generated from only a single cell batch. Hence, assessing the
differentiation variability and reproducibility of this model is
still necessary. Although hiPSC-CMs are considered an improvement
on rodent cells, this type of CMs bear more resemblance to neonatal
CMs or those in early development rather than adult CMs, and their
functional and electrophysiological properties are not fully
mature, so there are still some limitations in their application
(60). In the present study, small
sample size was used for both training and test sets and no risk
labels were assigned to the test set, which is inconsistent with
the training set. These two factors may have led to over-fitting of
the models. Thus, expanding the sample size for further
verification is needed in future investigations. Additionally,
consistent dataset risk labels are needed and ‘Transfer learning’
may also be useful to overcome differences in data distribution,
for small data sets, transfer learning is a useful strategy to
increase model power for specific tasks (61).
In the present study, a normal cell model was used,
therefore, it is mainly applicable to the prediction of TdP in
healthy patients after drug treatment or during treatment. For
specific clinical contexts, such as patients with long QT syndrome,
a disease model cell line should be used as the research subject to
establish a risk prediction model for TdP. In total, 28 compounds
were used to establish the TdP risk prediction model based on
hiPSC-CMs. LR and AdaBoost were taken as sub-models, with the same
assigned weight, to create a new TdP risk prediction model using a
soft voting strategy. This improved upon the accuracy of the
individual models and enhanced the generalization ability. In the
established TdP risk prediction model, 0.5 was set as the threshold
of TdP risk. All the seven compounds in the test sets were
accurately classified, achieving good consistency. The current
study also found that the multiple compounds inhibited the beating
of cardiomyocytes and those with reduced beating frequency were
more likely to have arrhythmic waveforms and more likely to induce
prolonged FPDc.
Supplementary Material
Representative impedance traces of
cardiomyocyte cultures during treatment with different compounds.
(A) Azimilide. (B) Chlorpromazine; (C) Metoprolol; (D)
Amiodarone.
Effects of high torsades de pointes
compounds on field potential duration. Pre, pre-administration;
Post, post-administration; FPDc, field potential duration
correction.
Effects of intermediate torsades de
pointes compounds on FPD. Pre, pre-administration; Post,
post-administration; FPDc, corrected field potential duration.
Effects of low torsades de pointes
compounds on FPD. Pre, pre-administration; Post,
post-administration; FPDc, corrected field potential duration.
Effects of non-Comprehensive in
vitro Proarrhythmia Assay compounds on FPD. Pre,
pre-administration; Post, post-administration; FPDc, corrected
field potential duration.
Specifications and experimental
conditions and concentrations of tested compounds
Changes in AMPL and BPM
Model output based on the training
set
Acknowledgements
The authors would like to thank Dr Gaojian Cheng
(National Institutes for Food and Drug Control, Beijing, China) for
technical guidance and Dr Hong Zhi Zhang (National Institutes for
Food and Drug Control) for statistical analysis. The authors would
also like to thank Dr Yu Chang and Professor Feng Lan (Chinese
Academy of Medical Sciences & Peking Union Medical College,
Beijing, China) for language editing.
Funding
Funding: This work was supported by the National Major
Scientific and Technological Special Project for ‘Significant New
Drugs Development’ (grant no. 2018ZX09201017-001) from the Ministry
of Science and Technology of the People's Republic of China.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request
Authors' contributions
DP, SW and BL contributed to the conception of the
study. DP and SW conducted the experiments. DP analyzed data and
wrote the manuscript. DP and SW confirm the authenticity of all the
raw data. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Frommeyer G and Eckardt L: Drug-induced
proarrhythmia: Risk factors and electrophysiological mechanisms.
Nat Rev Cardiol. 13:36–47. 2016.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Vicente J, Zusterzeel R, Johannesen L,
Mason J, Sager P, Patel V, Matta MK, Li ZH, Liu J, Garnett C, et
al: Mechanistic model-informed proarrhythmic risk assessment of
drugs: Review of the ‘CiPA’ initiative and design of a prospective
clinical validation study. Clin Pharmacol Ther. 103:54–66.
2018.PubMed/NCBI View
Article : Google Scholar
|
|
3
|
Niemeijer MN, van den Berg ME, Eijgelsheim
M, Rijnbeek PR and Stricker BH: Pharmacogenetics of drug-induced QT
interval prolongation: An update. Drug Saf. 38:855–867.
2015.PubMed/NCBI View Article : Google Scholar
|
|
4
|
El-Sherif N and Turitto G: Electrolyte
disorders and arrhythmogenesis. Cardiol J. 18:233–245.
2011.PubMed/NCBI
|
|
5
|
Shah RR: Drugs, QT interval prolongation
and ICH E14: The need to get it right. Drug Saf. 28:115–125.
2005.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Darpo B and Ferber G: The new S7B/E14
question and answer draft guidance for industry: Contents and
commentary. J Clin Pharmacol. 61:1261–1273. 2021.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Vargas HM, Rolf MG, Wisialowski TA,
Achanzar W, Bahinski A, Bass A, Benson CT, Chaudhary KW, Couvreur
N, Dota C, et al: Time for a fully integrated nonclinical-clinical
risk assessment to streamline QT prolongation liability
determinations: A pharma industry perspective. Clin Pharmacol Ther.
109:310–318. 2021.PubMed/NCBI View
Article : Google Scholar
|
|
8
|
Sanguinetti MC and Mitcheson JS:
Predicting drug-hERG channel interactions that cause acquired long
QT syndrome. Trends Pharmacol Sci. 26:119–124. 2005.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Gintant G, Sager PT and Stockbridge N:
Evolution of strategies to improve preclinical cardiac safety
testing. Nat Rev Drug Discov. 15:457–471. 2016.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Zhang S, Zhou Z, Gong Q, Makielski JC and
January CT: Mechanism of block and identification of the verapamil
binding domain to HERG potassium channels. Circ Res. 84:989–998.
1999.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Fenichel RR, Malik M, Antzelevitch C,
Sanguinetti M, Roden DM, Priori SG, Ruskin JN, Lipicky RJ and
Cantilena LR: Independent Academic Task Force. Drug-induced
torsades de pointes and implications for drug development. J
Cardiovasc Electrophysiol. 15:475–495. 2004.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Yim DS: Five years of the CiPA project
(2013-2018): What did we learn? Transl Clin Pharmacol. 26:145–149.
2018.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Park JS, Jeon JY, Yang JH and Kim MG:
Introduction to in silico model for proarrhythmic risk assessment
under the CiPA initiative. Transl Clin Pharmacol. 27:12–18.
2019.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Wallis R, Benson C, Darpo B, Gintant G,
Kanda Y, Prasad K, Strauss DG and Valentin JP: CiPA challenges and
opportunities from a non-clinical, clinical and regulatory
perspectives. An overview of the safety pharmacology scientific
discussion. J Pharmacol Toxicol Methods. 93:15–25. 2018.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Roden DM: Predicting drug-induced QT
prolongation and torsades de pointes. J Physiol. 594:2459–2468.
2016.PubMed/NCBI View
Article : Google Scholar
|
|
16
|
Crestani T, Steichen C, Neri E, Rodrigues
M, Fonseca-Alaniz MH, Ormrod B, Holt MR, Pandey P, Harding S, Ehler
E and Krieger JE: Electrical stimulation applied during
differentiation drives the hiPSC-CMs towards a mature cardiac
conduction-like cells. Biochem Biophys Res Commun. 533:376–382.
2020.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Ando H, Yoshinaga T, Yamamoto W, Asakura
K, Uda T, Taniguchi T, Ojima A, Shinkyo R, Kikuchi K, Osada T, et
al: A new paradigm for drug-induced torsadogenic risk assessment
using human iPS cell-derived cardiomyocytes. J Pharmacol Toxicol
Methods. 84:111–127. 2017.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Huo J, Wei F, Cai C, Lyn-Cook B and Pang
L: Sex-related differences in drug-induced QT prolongation and
torsades de Pointes: A new model system with human iPSC-CMs.
Toxicol Sci. 167:360–374. 2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
da Rocha AM, Creech J, Thonn E, Mironov S
and Herron TJ: Detection of drug-induced torsades de Pointes
arrhythmia mechanisms using hiPSC-CM syncytial monolayers in a
high-throughput screening voltage sensitive dye assay. Toxicol Sci.
173:402–415. 2020.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Raphel F, De Korte T, Lombardi D, Braam S
and Gerbeau JF: A greedy classifier optimization strategy to assess
ion channel blocking activity and pro-arrhythmia in
hiPSC-cardiomyocytes. PLoS Compute Biol.
16(e1008203)2020.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Blinova K, Dang Q, Millard D, Smith G,
Pierson J, Guo L, Brock M, Lu HR, Kraushaar U, Zeng H, et al:
International multisite study of human-induced pluripotent stem
cell-derived cardiomyocytes for drug proarrhythmic potential
assessment. Cell Rep. 24:3582–3592. 2018.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Tu JV: Advantages and disadvantages of
using artificial neural networks versus logistic regression for
predicting medical outcomes. J Clin Epidemiol. 49:1225–1231.
1996.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Heo J, Yoon JG, Park H, Kim YD, Nam HS and
Heo JH: Machine learning-based model for prediction of outcomes in
acute stroke. Stroke. 50:1263–1265. 2019.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Sherazi SW, Bae JW and Lee JY: A soft
voting ensemble classifier for early prediction and diagnosis of
occurrences of major adverse cardiovascular events for STEMI and
NSTEMI during 2-year follow-up in patients with acute coronary
syndrome. PLoS One. 16(e0249338)2021.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Asakura K, Hayashi S, Ojima A, Taniguchi
T, Miyamoto N, Nakamori C, Nagasawa C, Kitamura T, Osada T, Honda
Y, et al: Improvement of acquisition and analysis methods in
multi-electrode array experiments with iPS cell-de rived
cardiomyocytes. J Pharmacol Toxicol Methods. 75:17–26.
2015.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Gerds TA, Cai T and Schumacher M: The
performance of risk prediction models. Biom J. 50:457–479.
2008.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Pencina MJ and D'Agostino RB Sr:
Evaluating discrimination of risk prediction models: The C
statistic. JAMA. 314:1063–1064. 2015.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Alba AC, Agoritsas T, Walsh M, Hanna S,
Iorio A, Devereaux PJ, McGinn T and Guyatt G: Discrimination and
calibration of clinical prediction models: Users' guides to the
medical literature. JAMA. 318:1377–1384. 2017.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Ziegler R, Häusermann F, Kirchner S and
Polonchuk L: Cardiac safety of kinase inhibitors-improving
understanding and prediction of liabilities in drug discovery using
human stem cell-derived models. Front Cardiovasc Med.
8(639824)2021.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Harary I and Farley B: In vitro studies of
single isolated beating heart cells. Science. 131:1674–1675.
1960.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Price PS, Keenan RE and Swartout JC:
Characterizing interspecies uncertainty using data from studies of
anti-neoplastic agents in animals and humans. Toxicol Appl
Pharmacol. 233:64–70. 2008.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Sheng CC, Amiri-Kordestani L, Palmby T,
Force T, Hong CC, Wu JC, Croce K, Kim G and Moslehi J: 21st century
cardio-oncology: Identifying cardiac safety signals in the era of
personalized medicine. JACC Basic Transl Sci. 1:386–398.
2016.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Haraguchi Y, Ohtsuki A, Oka T and Shimizu
T: Electrophysiological analysis of mammalian cells expressing hERG
using automated 384-well-patch-clamp. BMC Pharmacol Toxicol.
16(39)2015.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Higa A, Hoshi H, Yanagisawa Y, Ito E,
Morisawa G, Imai JI, Watanabe S and Takagi M: Evaluation system for
arrhythmogenic potential of drugs using human-induced pluripotent
stem cell-derived cardiomyocytes and gene expression analysis. J
Toxicol Sci. 42:755–761. 2017.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Abassi YA, Xi B, Li N, Ouyang W, Seiler A,
Watzele M, Kettenhofen R, Bohlen H, Ehlich A, Kolossov E, et al:
Dynamic monitoring of beating periodicity of stem cell-derived
cardiomyocytes as a predictive tool for preclinical safety
assessment. Br J Pharmacol. 165:1424–1441. 2012.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Wang SY, Wang XJ and Ma J: Progress in
real time xCELLigence analysis system on drug cardiotoxicity
screening. Chin J Pharmacol Toxicol,. 27:908–912. 2013.
|
|
37
|
Yue Peng, Ying BB, He Min and Yang
ChaoWen: Study on hepatotoxicity of different doses of cisplatin in
rats. J Toxico l,. 33:302–306. 2019.
|
|
38
|
Marcu IC, Illaste A, Heuking P, Jaconi ME
and Ullrich ND: Functional characterization and comparison of
intercellular communication in stem cell-derived cardiomyocytes.
Stem Cells. 33:2208–2218. 2015.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Clements M: Multielectrode array (MEA)
assay for profiling electrophysiological drug effects in human stem
cell-derived cardiomyocytes. Curr Protoc Toxicol.
68:22.24.21–22.24.32. 2016.PubMed/NCBI View
Article : Google Scholar
|
|
40
|
Halbach M, Egert U, Hescheler J and Banach
K: Estimation of action potential changes from field potential
recordings in multicellular mouse cardiac myocyte cultures. Cell
Physiol Biochem. 13:271–284. 2003.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Sharifi M, Buzatu D, Harris S and Wilkes
J: Development of models for predicting Torsade de Pointes cardiac
arrhythmias using perceptron neural networks. BMC Bioinformatics.
18 (Suppl 4)(S497)2017.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Liu H, Ji M, Luo X, Shen J, Huang X, Hua
W, Jiang H and Chen K: New p-methylsulfonamido phenylethylamine
analogues as class III antiarrhythmic agents: Design, synthesis,
biological assay, and 3D-QSAR analysis. J Med Chem. 45:2953–2969.
2002.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Chamberlain AM, Boyd CM, Manemann SM,
Dunlay SM, Gerber Y, Killian JM, Weston SA and Roger VL: Risk
factors for heart failure in the community: Differences by age and
ejection fraction. Am J Med. 133:e237–e248. 2020.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Belkin M, Hsu D, Ma S and Mandal S:
Reconciling modern machine-learning practice and the classical
bias-variance trade-off. Proc Natl Acad Sci USA. 116:15849–15854.
2019.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Hatwell J, Gaber MM and Atif Azad RM:
Ada-WHIPS: Explaining AdaBoost classification with applications in
the health sciences. BMC Med Inform Decis Mak.
20(250)2020.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Kanda Y, Yamazaki D, Osada T, Yoshinaga T
and Sawada K: Development of torsadogenic risk assessment using
human induced pluripotent stem cell-derived cardiomyocytes: Japan
iPS cardiac safety assessment (JiCSA) update. J Pharmacol Sci.
138:233–239. 2018.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Aktas MK, Shah AH and Akiyama T:
Dofetilide-induced long QT and torsades de pointes. Ann Noninvasive
Electrocardiol. 12:197–202. 2007.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Jaiswal A and Goldbarg S: Dofetilide
induced torsade de pointes: Mechanism, risk factors and management
strategies. Indian Heart J. 66:640–648. 2014.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Thomine S, Zimmerman S, Duijn BV,
Barbier-Brygoo H and Guern J: Calcium channel antagonists induce
direct inhibition of the outward rectifying potassium channel in
tobacco protoplasts. FEBS Lett. 340:45–50. 1994.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Duncan RS, Ridley JM, Dempsey CE, Leishman
DJ, Leaney JL, Hancox JC and Witchel HJ: Erythromycin block of the
HERG K+ channel: Accessibility to F656 and Y652. Biochem Biophys
Res Commun. 341:500–506. 2006.PubMed/NCBI View Article : Google Scholar
|
|
51
|
van Haarst AD, van 't Klooster GA, van
Gerven JM, Schoemaker RC, van Oene JC, Burggraaf J, Coene MC and
Cohen AF: The influence of cisapride and clarithromycin on QT
intervals in healthy volunteers. Clin Pharmacol Ther. 64:542–546.
1998.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Vieweg WV, Hancox JC, Hasnain M, Koneru
JN, Gysel M and Baranchuk A: Clarithromycin, QTc interval
prolongation and torsades de pointes: The need to study case
reports. Ther Adv Infect Dis. 1:121–138. 2013.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Tisdale JE: Drug-induced QT interval
prolongation and torsades de pointes: Role of the pharmacist in
risk assessment, prevention and management. Can Pharm J (Ott).
149:139–152. 2016.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Lo YC and Kuo CC: Temperature dependence
of the biophysical mechanisms underlying the inhibition and
enhancement effect of amiodarone on hERG channels. Mol Pharmacol.
96:330–344. 2019.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Shenthar J, Rachaiah JM, Pillai V, Chakali
SS, Balasubramanian V and Chollenhalli Nanjappa M: Incidence of
drug-induced torsades de pointes with intravenous amiodarone.
Indian Heart J. 69:707–713. 2017.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Khan F, Ismail M, Khan Q and Ali Z:
Moxifloxacin-induced QT interval prolongation and torsades de
pointes: A narrative review. Expert Opin Drug Saf. 17:1029–1039.
2018.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Behzadi M, Joukar S and Beik A: Opioids
and cardiac arrhythmia: A literature review. Med Princ Pract.
27:401–414. 2018.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Kasama M, Furukawa Y, Oguchi T, Hoyano Y
and Chiba S: Effects of low temperature on the chronotropic and
inotropic responses to zatebradine, E-4031 and ver apamil in
isolated perfused dog atria. Jpn J Pharmacol. 78:493–499.
1998.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Saeidnia S, Manayi A and Abdollahi M: From
in vitro Experiments to in vivo and clinical studies; Pros and
cons. Curr Drug Discov Technol. 12:218–224. 2015.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Zuppinger C, Gibbons G, Dutta-Passecker P,
Segiser A, Most H and Suter TM: Characterization of cytoskeleton
features and maturation status of cultured human iPSC-derived
cardiomyocytes. Eur J Histochem. 61(2763)2017.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Cai C, Wang S, Xu YJ, Zhang WL, Tang K,
Ouyang Q, Lai LH and Pei JF: Transfer learning for drug discovery.
J Med Chem. 63:8683–8694. 2020.PubMed/NCBI View Article : Google Scholar
|