Introduction
Lung cancer ranks among the top malignancies in
terms of morbidity and mortality globally, accounting for 11.4% of
all cancer cases and 18% of all cancer-associated deaths (1). Lung cancer includes non-small cell
lung cancer (NSCLC) and SCLC. Among the different types of NSCLC,
including adenocarcinoma and squamous and large cell carcinoma,
adenocarcinoma is the most common type (2). Lung adenocarcinoma (LUAD) is prone to
bone metastasis and patients with bone metastasis have been shown
to have poor prognosis. Additionally, the pain and bone breakage
caused by bone metastasis decreases quality of life of patients
(3). Molecular-targeted therapy
and immunotherapy have been widely used in clinical practice as a
supplement to traditional surgery and chemoradiotherapy; however,
the 5-year survival rate of patients is still not ideal and more
effective therapies need to be found (4,5).
Therefore, it is of clinical significance to study the pathogenesis
of LUAD metastasis and find effective treatments.
Deoxyribonuclease 1-like 3 (DNASE1L3) is a member of
DNASE1 gene family and serves a key role in DNA degradation during
cell death and apoptosis (6,7).
DNASE1L3 has been shown to be involved in signal transduction in
breast cancer, exhibiting intracellular signal cascade receptor
activity and serving as a GTPase regulatory factor (8). DNASE1L3 has also been shown to
suppress apoptosis and reprogram glucose metabolism, thus
inhibiting hepatocellular carcinoma (HCC) progression (9). The senescence-associated secretory
phenotypes are damaged by DNASE1L3 interacted with H2BE to suppress
tumor angiogenesis in HCC (10).
Underexpression of DNASE1L3 in colon cancer samples is a potential
prognostic biomarker associated with immune invasion of colon
cancer and overexpression of DNASE1L3 inhibits cell proliferation
and motility (11). A previous
study reported that DNASE1L3 is downregulated in LUAD (12). An additional study found that mRNA
and protein expression levels of DNASE1L3 in patients with LUAD are
significantly lower than those in normal tissue and low expression
of DNASE1L3 is significantly correlated with higher pathological
stage, T stage and poor prognosis, making it an independent factor
for predicting the overall survival rate (13). The specific role of DNASE1L3 in
LUAD, to the best of our knowledge, has not yet been reported.
Previous studies have shown that transcription
factor forkhead-box P2 (FOXP2) is involved in the regulation of
tumor invasion and metastasis (14-17).
FOXP2 protein has been shown to promote cell proliferation,
invasion and metastasis in breast cancer (14). FOXP2 has also been shown to
activate TGF-β to promote migration and invasion of prostate cancer
cells (15). Upregulated FOXP2
expression inhibits proliferation, migration, invasion and
epithelial-mesenchymal transformation (EMT) and promotes cell
apoptosis in NSCLC (16,17). All these findings imply that FOXP2
may function in opposing ways in different tissues.
Therefore, it was hypothesized that as DNASE1L3 is
underexpressed in LUAD, overexpression of DNASE1L3 may inhibit
malignant progression of LUAD. In addition, DNASE1L3 transcription
may be positively regulated by FOXP2.
Materials and methods
Bioinformatic analysis
The expression of DNASE1L3 in LUAD (n=483) and
adjacent tissue (n=347) as well as overall survival were analyzed
using Gene Expression Profiling Interactive Analysis (GEPIA;
gepia.cancer-pku.cn/) (18). The JASPAR database (jaspar.genereg.net/) predicted the potential binding
between FOXP2 and DNASE1L3 promoter.
Cell culture
Human bronchial epithelial cell line (16HBE cells)
and three LUAD cell lines (PC-9, NCI-H1975 and A549 cells) were
provided by Ningbo Mingzhou Biotechnology Co., Ltd. Human umbilical
vein endothelial cells (HUVECs) were obtained from PromoCell GmbH.
The cells were cultured at 37˚C in 5% CO2 in RPMI-1640
medium (Sigma-Aldrich, Merck KGaA) with 10% FBS (Sigma-Aldrich,
Merck KGaA). When cells reached the logarithmic growth phase, they
were digested with 0.25% trypsin and collected for experiments.
Cell transfection
PcDNA3.1(+) Overexpression-negative control (empty
vector plasmid; Oe-NC), Oe-DNASE1L3 (accession no. NM_001256560.2)
or Oe-FOXP2 (accession no. NM_001172766.3), small interfering RNAs
targeting FOXP2 (si-FOXP2; #1, 5'-GACATTCAGACAAATACAACATT-3'; #2,
5'-GACAATAAGCAACAGTTCAATGA-3') and negative control sequence (NC,
5'-AAGACAUUGUGUGUCCGCCTT-3'; both 50 nM) were designed and provided
by Guangzhou RiboBio Co., Ltd. A549 cells in logarithmic growth
phase were seeded in 6-well plates. When cells grew to a density of
80%, plasmids carrying the target gene and siRNAs were transfected
into the cells using Lipofectamine® 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.) for 48 h at 37˚C in 5%
CO2 as previously described (19). Subsequent experiments were
performed 48 h post-transfection.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
The expression of DNASE1L3 and FOXP2 in LUAD cells
or transfected LUAD cells was detected by RT-qPCR. A549 cells were
collected and total RNA was extracted using TRIzol®
reagent (Invitrogen; Thermo Fisher Scientific, Inc.). Next, total
RNA was reverse-transcribed into cDNA using a
PrimeScript™ RT reagent kit (Takara Bio, Inc.) according
to the manufacturer's protocol and qPCR was conducted with
SYBR-Green PCR Master Mix (MedChemExpress). The following
thermocycling conditions were used: Initial denaturation at 94˚C
for 5 min, 36 cycles at 94˚C for 20 sec, 54˚C for 20 sec and 72˚C
for 20 sec. The primer sequences were as follows: DNASE1L3,
forward, 5'-AGCCCTTTGTGGTCTGGTTC-3' and reverse,
5'-TCCTTAACGGATGTCTCTGGG-3'; FOXP2 forward,
5'-AATCTGCGACAGAGACAATAAGC-3' and reverse,
5'-TCCACTTGTTTGCTGCTGTAAA-3' and GAPDH forward,
5'-GGAGCGAGATCCCTCCAAAAT-3' and reverse,
5'-GGCTGTTGTCATACTTCTCATGG-3'. Relative expression of DNASE1L3 and
FOXP2 was quantified using the 2-ΔΔCq method (20) and GAPDH served as the internal
control.
Western blotting
The expression of DNASE1L3 and FOXP2 in LUAD cells
or transfected LUAD cells and expression of E-cadherin, N-cadherin,
Snail and vascular endothelial growth factor (VEGF) in transfected
LUAD cells was determined by western blotting. Following
transfection, A549 cells were homogenized in
radioimmunoprecipitation assay (RIPA) lysis buffer (Beyotime
Institute of Biotechnology) to extract total protein. The protein
concentration was detected by BCA Protein Assay kit (Pierce; Thermo
Fisher Scientific, Inc.). Protein samples (40 µg/lane) were loaded
into a 10% SDS-PAGE gel, which was then electrophorized and
transferred to a PVDF membrane. The membrane was blocked with 5%
BSA (Beyotime Institute of Biotechnology) at room temperature for 1
h before incubation with primary antibodies against DNASE1L3 (cat.
no. ab152118; 1/1,000; Abcam), E-cadherin (cat. no. ab40772;
1/10,000; Abcam), N-cadherin (cat. no. ab76011; 1/5,000; Abcam),
Snail (cat. no. ab216347; 1/1,000; Abcam), VEGF (cat. no. ab32152;
1/1,000; Abcam), FOXP2 (cat. no. ab16046; 1/1,000; Abcam) and GAPDH
(cat. no. ab9485; 1/2,500; Abcam) at 4˚C overnight. Afterwards, the
membranes were incubated with HRP-conjugated Goat Anti-Rabbit IgG
H&L secondary antibody (cat. no. ab97051; 1/2,000; Abcam) at
room temperature for 1 h. A BeyoECL Plus kit (cat. no. P0018S;
Beyotime Institute of Biotechnology) was used to visualize protein
bands, which were quantified by ImageJ 1.8.0 software (National
Institutes of Health).
MTT assay
The viability of transfected LUAD cells was analyzed
by MTT assay. Transfected A549 cells (5x103/well) were
seeded into 96-well plates and incubated at 37˚C with 5%
CO2 for 24, 48 and 72 h. Following incubation, 10 µl MTT
(5 mg/ml; Sigma-Aldrich; Merck KGaA) was added to cells in each
well, then incubated for 4 h at 37˚C. The supernatant was discarded
and 100 µl DMSO was added to each well before plates were shaken at
low speed for 10 min. The absorbance values at 490 nm were measured
using a microplate reader.
EdU staining
EdU staining was used to determine proliferation of
the transfected LUAD cells. The proliferation of A549 cells was
determined using the Cell-Light EdU Apollo488 In Vitro Kit
(cat. no. C10310-3; Guangzhou Ribobio Co., Ltd.). Briefly, A549
cells were incubated with EdU solution for 2 h at 37˚C and fixed
with PBS containing 4% paraformaldehyde at room temperature for 15
min. Subsequently, the nucleus was stained with 0.01 mg/ml DAPI at
37˚C for 30 min. Finally, five areas were randomly selected at x200
magnification to count the number of green fluorescence-positive
cells under a fluorescence microscope (Olympus Corporation) using
ImageJ (version no. 1.52; National Institutes of Health).
Wound healing assay
The migration of transfected LUAD cells was detected
by wound healing assay. Following transfection, A549 cells were
seeded into a six-well plate (3x105 cells/well) in
RPMI-1640 medium and grown to 90% confluence at 37˚C. A 100-µl
pipette tip was used to scratch a straight wound across the well
and incubated at 37˚C in serum-free medium. After 24 h, images of
the migrated cells were captured using a light microscope (Olympus
Corporation) at x100 magnification and scratch widths were recorded
by ImageJ 1.8.0 software (National Institutes of Health). Multiple
measurements were made along the scratch (n=3).
Transwell assay
The invasion of transfected LUAD cells was detected
by Transwell assay. A549 cells were inoculated into 24-well
invasion chambers (1x105 cells/well). The upper chamber
was coated with Matrigel (8 µm pores) at 37˚C for 30 min. The lower
chamber contained RPMI-1640 medium supplemented with 10% FBS as the
chemoattractant. After incubation for 24 h at 37˚C, the
non-invasive cells inside the upper chamber were removed and the
remaining cells were fixed with 4% paraformaldehyde for 15 min at
room temperature and stained with 1% Giemsa for 5 min at room
temperature. The invasive cells were observed by a light microscope
(Olympus Corporation) at x100 magnification and the number of
invasive cells was counted by a cell counter.
Tube formation assay
The tube formation of HUVECs in culture medium from
transfected LUAD cells was determined by tube formation assay.
Matrigel-coated 24-well Transwell plates (8 µm pores) at 37˚C for
30 min were used to conduct the tube formation assay. HUVECs were
suspended in medium from control, si-NC, si-FOXP2, si-FOXP2+Oe-NC
and si-FOXP2+Oe-DNASE1L3 groups at a density of 1x105
cells/ml. In total, ~100 µl cell suspension was added to the
surface of the Matrigel, which was incubated at 37˚C for 6 h. The
tube formation was visualized using a light microscope (Olympus
Corporation) at x40 magnification.
Dual-luciferase reporter assay
Dual-luciferase reporter assay was performed to
verify whether transcription factor FOXP2 interacted with the
DNASE1L3 promoter. The wild-type (DNASE1L3-WT) or mutant
(DNASE1L3-MUT) fragments of DNASE1L3 (accession no. NM_001256560.2)
with the binding site of FOXP2 were constructed by Shanghai
GenePharma Co., Ltd. WT or MUT sequence were cloned into the pGL3
luciferase reporter vector (Promega Corporation). A549 cells
(5x105) were seeded in 24-well plates for 24 h at 37˚C
and co-transfected with WT or MT plasmid and Oe-NC or Oe-FOXP2
using Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). After 48 h at 37˚C, the luciferase activity was
detected using dual luciferase reporter assay kit (cat. no. E1910;
Promega Corporation). Renilla luciferase activity was used
for normalization.
Chromatin immunoprecipitation
(ChIP)
ChIP analysis was performed to confirm whether
transcription factor FOXP2 bound to the DNASE1L3 promoter. ChIP
analysis was carried out using ChIP Assay kit (Beyotime Institute
of Biotechnology) according to manufacturer's protocol. Briefly, 1%
formaldehyde was added to transfected A549 cells for 10 min at room
temperature. The fixed cells were washed twice with
phosphate-buffered saline and were lysed using a lysis buffer (0.1%
SDS, 0.5% Triton X-100, 20 mM Tris-HCl, pH 8.1) that contained 150
mM NaCl and a protease inhibitor, after which chromatin fragments
were obtained using sonication using a 10 sec on and 10 sec off
mode for 12 cycles at 4˚C. Samples were centrifuged at 13,000 x g
for 10 min at 4˚C, and 100 µl of supernatant was pre-absorbed by 2
µg of anti-FOXP2 antibody (cat. no. #5337; 1/200; Cell Signaling
Technology) or IgG control (cat. no. ab172730; 1:50; Abcam)
overnight at 4˚C. Samples were supplemented with protein
agarose/sepharose (cat. no. #9863; Cell Signaling Technology) to
precipitate the endogenous DNA-protein complex. The
immunoprecipitated complex was centrifuged (5,000 x g for 1 min at
4˚C) and washed with low salt, high salt, LiCl and TE buffers in
the kit according to the manufacturer's protocols. The complex was
eluted from the antibody using a solution of 1% SDS, 0.1 mol/l
NaHCO3 and 200 mmol/l NaCl. The complex was de-crosslinked at 65˚C
and the DNA fragment was recovered by phenol/chloroform extraction
and purification. The enrichment of specific fragments was
determined by RT-qPCR, as aforementioned.
Statistical analysis
Data from three independent replicates are presented
as the mean ± SD. GraphPad Prism 8.0.1 (GraphPad Software, Inc.)
was used for the statistical analysis. Overall survival was
calculated by Kaplan-Meier survival analysis. An unpaired Student's
t test was applied for the comparison between two groups.
Comparisons between >2 groups were made using one-way ANOVA
followed by Tukey's post hoc test. P<0.05 was considered to
indicate a statistically significant difference.
Results
DNASE1L3 underexpression in LUAD is
associated with poor prognosis
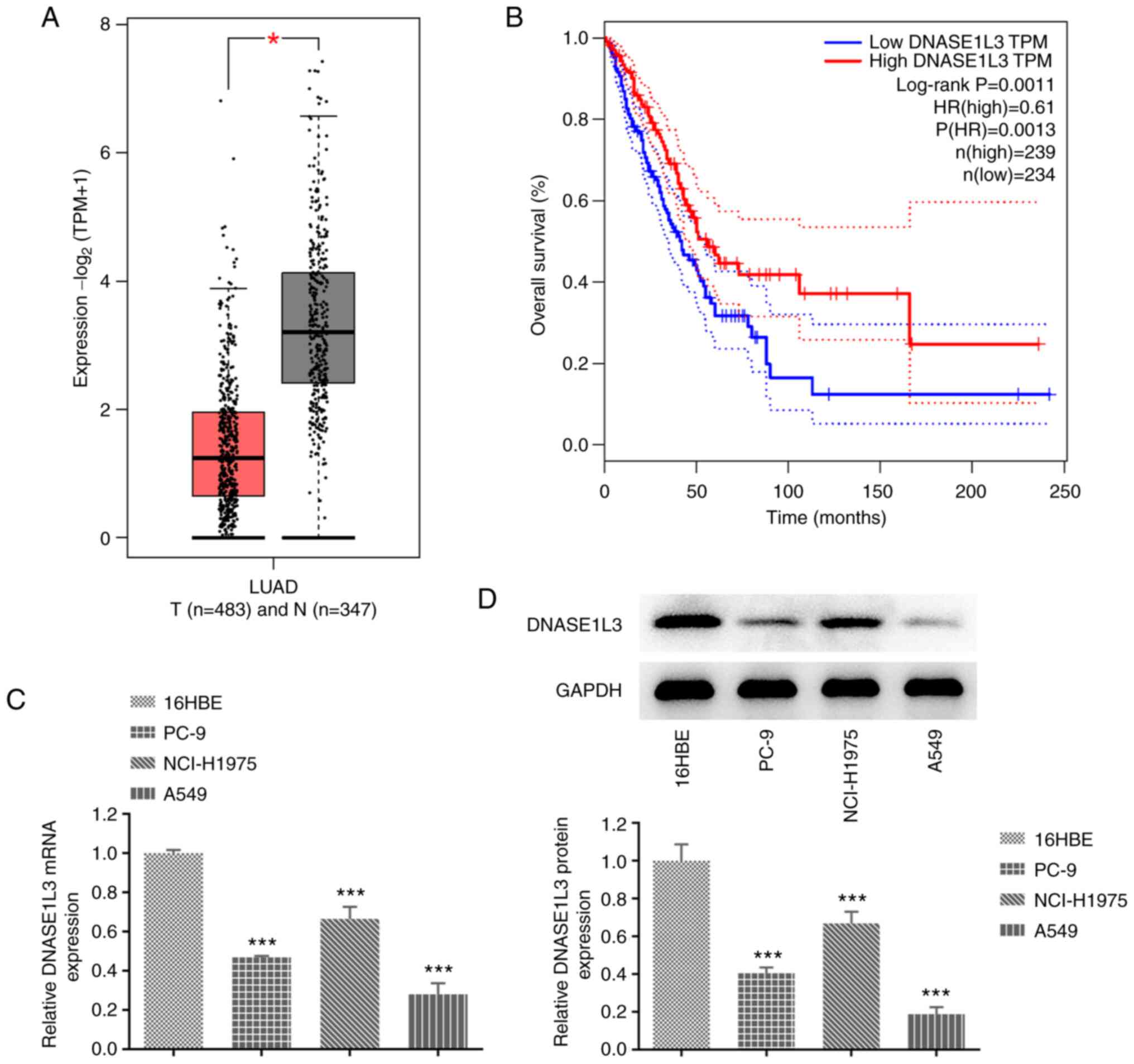
The GEPIA database indicated that the expression of
DNASE1L3 in LUAD tissue was decreased (Fig. 1A) and low expression of DNASE1L3
was significantly associated with poor prognosis in patients with
LUAD (Fig. 1B). RT-qPCR and
western blotting were used to detect the mRNA and protein
expression of DNASE1L3 in LUAD cells. DNASE1L3 was significantly
downregulated in PC-9, NCI-H1075 and A549 cells compared with 16HBE
cells and the lowest expression of DNASE1L3 was in the A549 cell
line (Fig. 1C and D). Thereafter, A549 cells were chosen for
the subsequent experiments.
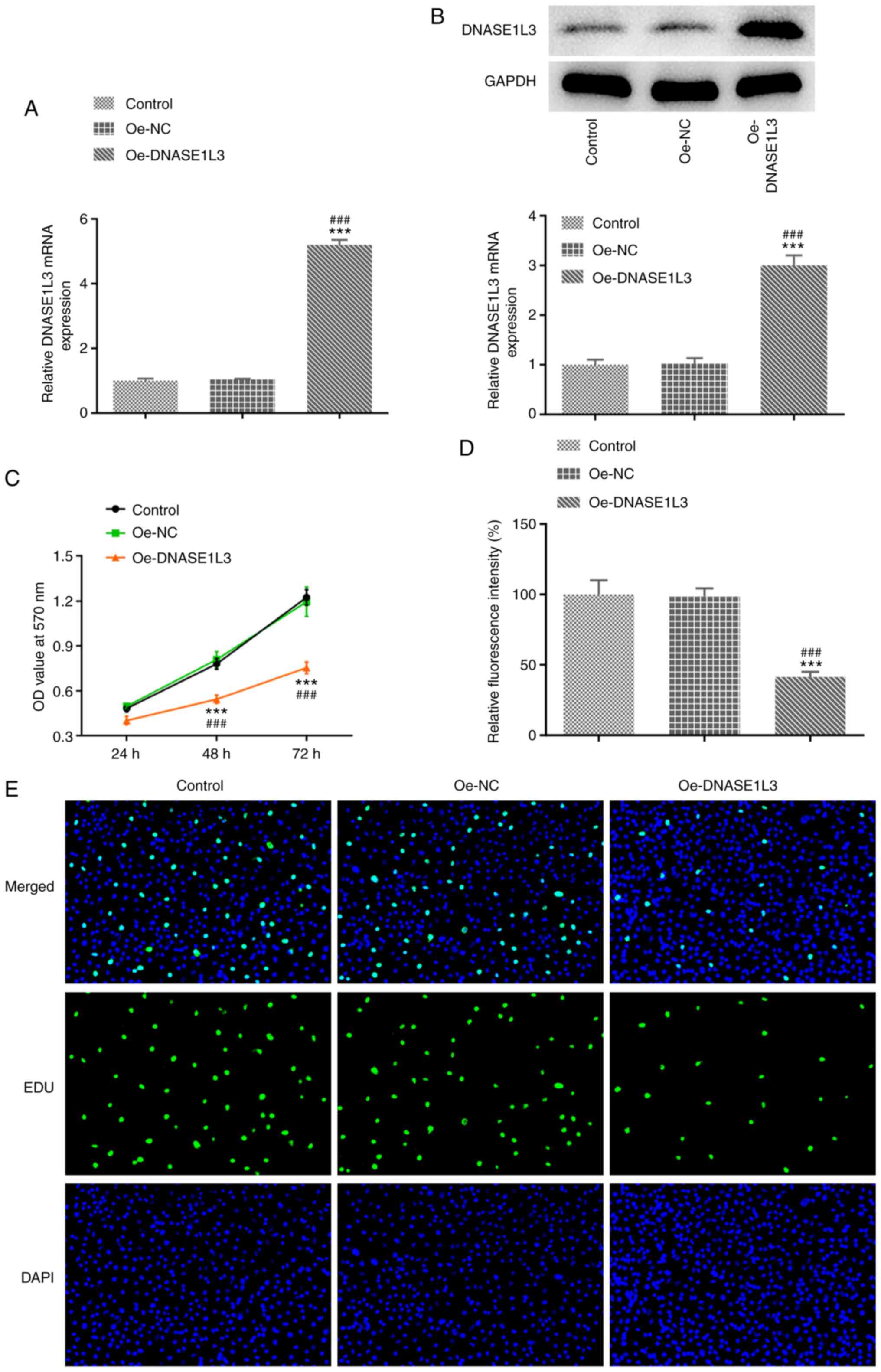
Oe-DNASE1L3 inhibits proliferation of
A549 cells
The transfection efficiency of Oe-DNASE1L3 in A549
cells was confirmed by RT-qPCR and western blotting. The viability
and proliferation of Oe-DNASE1L3-transfected A549 cells were
determined by MTT assay and EdU staining. When A549 cells were
transfected with Oe-DNASE1L3, expression of DNASE1L3 significantly
increased (Fig. 2A and B). The viability of A549 cells was
significantly decreased when cells were transfected with
Oe-DNASE1L3 (Fig. 2C). Oe-DNASE1L3
significantly suppressed the proliferation of A549 cells (Fig. 2D and E).
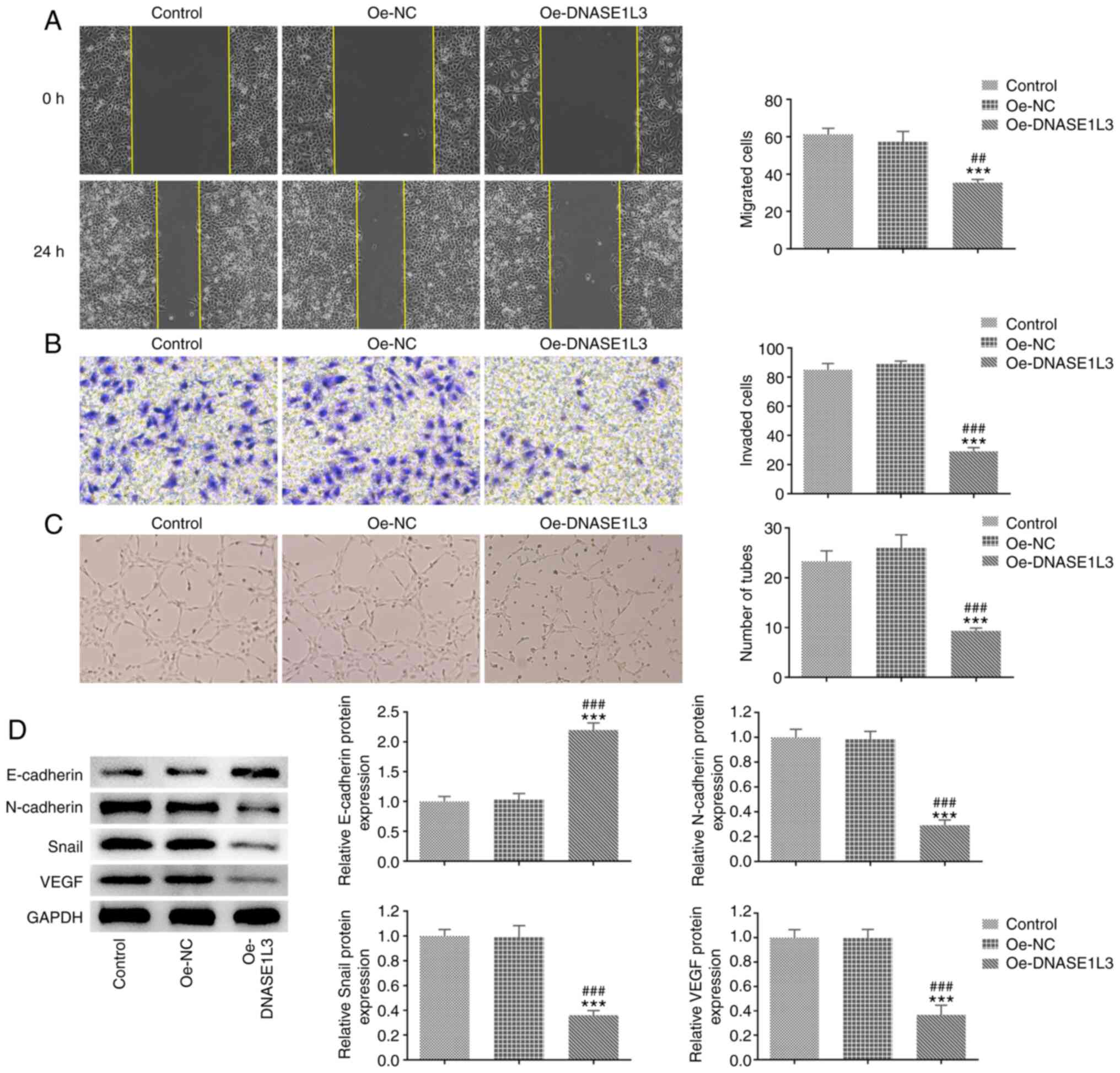
Oe-DNASE1L3 inhibits migration,
invasion and tube formation of A549 cells
The migration, invasion and tube formation of A549
cells transfected with Oe-DNASE1L3 were detected by wound healing,
Transwell and tube formation assays, respectively. The expression
of EMT- and tube formation-associated proteins in A549 cells
transfected with Oe-DNASE1L3 was analyzed by western blotting. The
migration rate and number of invaded cells were both significantly
decreased in the Oe-DNASE1L3 group by contrast with the Oe-NC group
(Fig. 3A and B). The number of tubes was also
significantly decreased in cells cultured in medium of A549 cells
transfected with Oe-DNASE1L3 relative to the Oe-NC group (Fig. 3C). The expression of E-cadherin was
significantly increased while the expression of N-cadherin, Snail
and VEGF was significantly decreased in A549 cells transfected with
Oe-DNASE1L3 relative to the Oe-NC group (Fig. 3D).
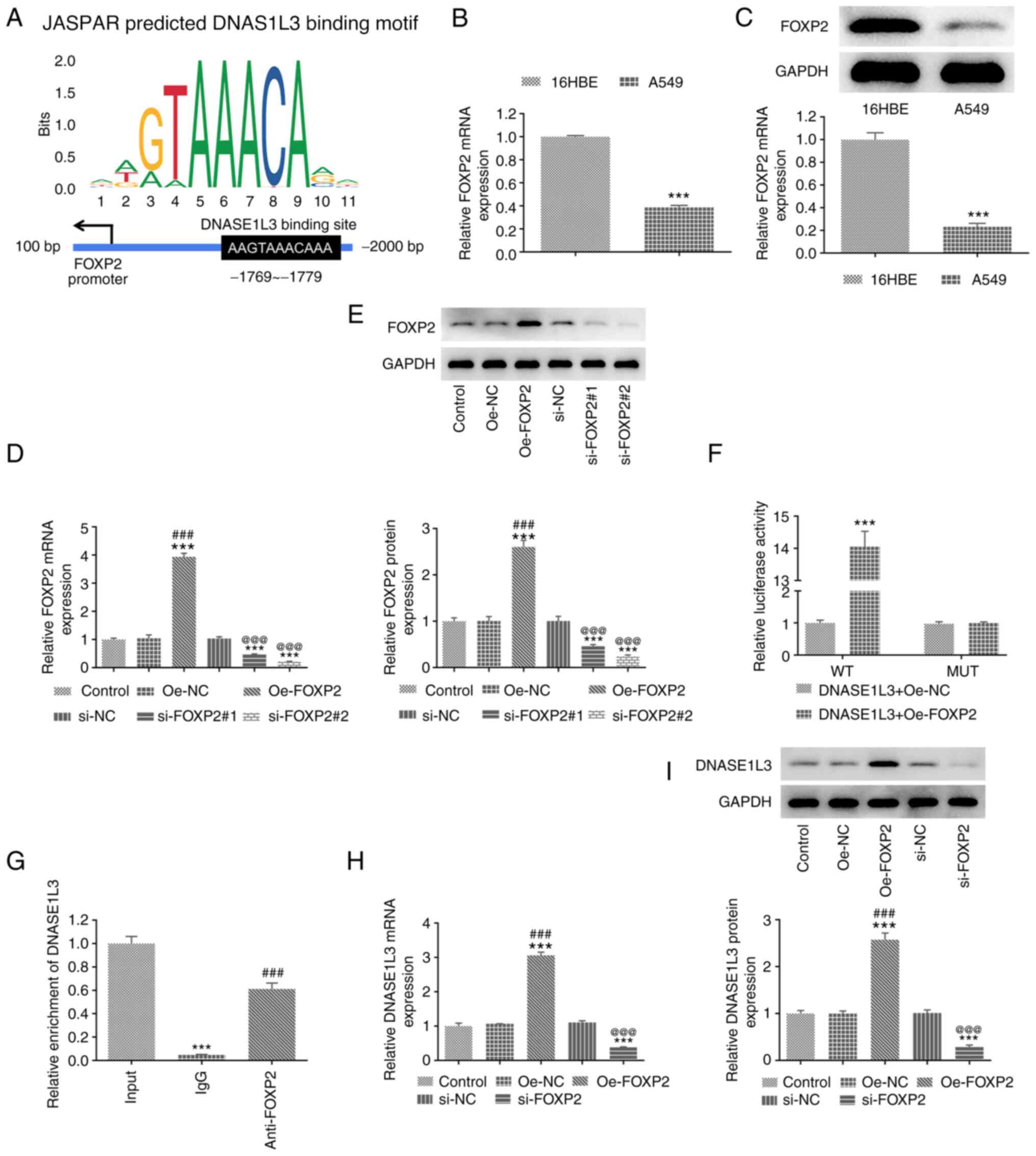
Transcription factor FOXP2 positively
regulates DNASE1L3 transcription in A549 cells
The binding sites between FOXP2 and DNASE1L3 were
predicted by the JASPAR database (Fig.
4A). FOXP2 expression was also significantly decreased in A549
compared with 16HBE cells (Fig. 4B
and C). The transfection
efficiency of Oe-FOXP2 and si-FOXP2 in A549 cells was confirmed by
RT-qPCR and western blotting. Dual-luciferase reporter assay and
ChIP analysis were performed to verify the binding between FOXP2
and DNASE1L3. The expression of FOXP2 in A549 cells transfected
with Oe-FOXP2 was significantly increased and significantly
decreased in A549 cells transfected with si-FOXP2#1 and si-FOXP2#2.
si-FOXP2#2 was selected for subsequent study as it displayed a more
excellent interference efficacy (Fig.
4D and E). The luciferase
activity was significantly increased in A549 cells co-transfected
with DNASE1L3-WT and Oe-FOXP2 (Fig.
4F). The binding of FOXP2 to DNASE1L3 was detected by the
addition of anti-FOXP2 antibody and the results elaborated that
DNASE1L3 promoter was abundant in anti-FOXP2 antibody (Fig. 4G). These results indicated that
FOXP2 may regulate DNASE1L3 expression. When A549 cells were
transfected with Oe-FOXP2 and si-FOXP2, expression of FOXP2 was
significantly increased and decreased, respectively (Fig. 4H and I).
Transcription factor FOXP2 regulates
transcription of DNASE1L3 and promotes proliferation of A549
cells
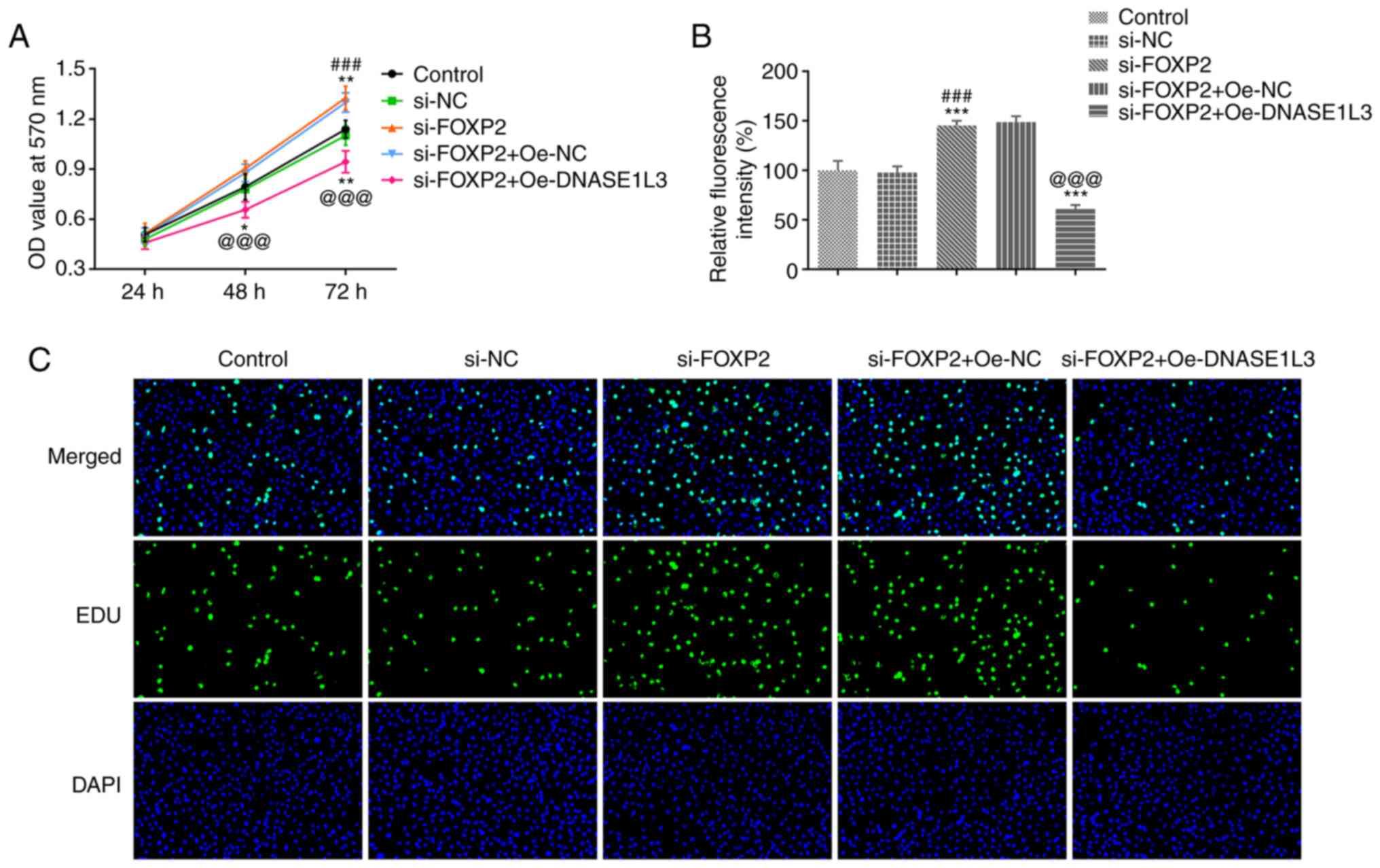
The viability and proliferation of A549 cells
transfected with si-FOXP2#2 and Oe-DNASE1L3 were detected by MTT
assay and EdU staining, respectively. When A549 cells were
transfected with si-FOXP2#2, viability significantly increased
after 72 h. However, the viability of A549 cells was significantly
decreased when si-FOXP2-transfected A549 cells were co-transfected
with Oe-DNASE1L3 both at 48 and 72 h (Fig. 5A). Proliferation of A549 cells was
significantly increased by suppressing FOXP2 expression and
significantly decreased in cells co-transfected with si-FOXP2#2 and
Oe-DNASE1L3 (Fig. 5B and C).
Transcription factor FOXP2 regulates
transcription of DNASE1L3 and promotes the migration, invasion and
tube formation of A549 cells
The migration, invasion and tube formation of A549
cells transfected with si-FOXP2#2 and Oe-DNASE1L3 were detected by
wound healing, Transwell and tube formation assays, respectively.
The expression of EMT- and tube formation-associated proteins in
A549 cells transfected with si-FOXP2 and Oe-DNASE1L3 was analyzed
by western blotting. Suppression of FOXP2 expression significantly
improved migration and invasion of A549 cells and tube formation of
HUVECs. These behaviors were all significantly decreased following
transfection with Oe-DNASE1L3 (Fig.
6A-C). The expression of E-cadherin was significantly decreased
while expression levels of N-cadherin, Snail and VEGF were
significantly increased by si-FOXP2#2 transfection; these effects
were significantly reversed by co-transfection with Oe-DNASE1L3
(Fig. 6D).
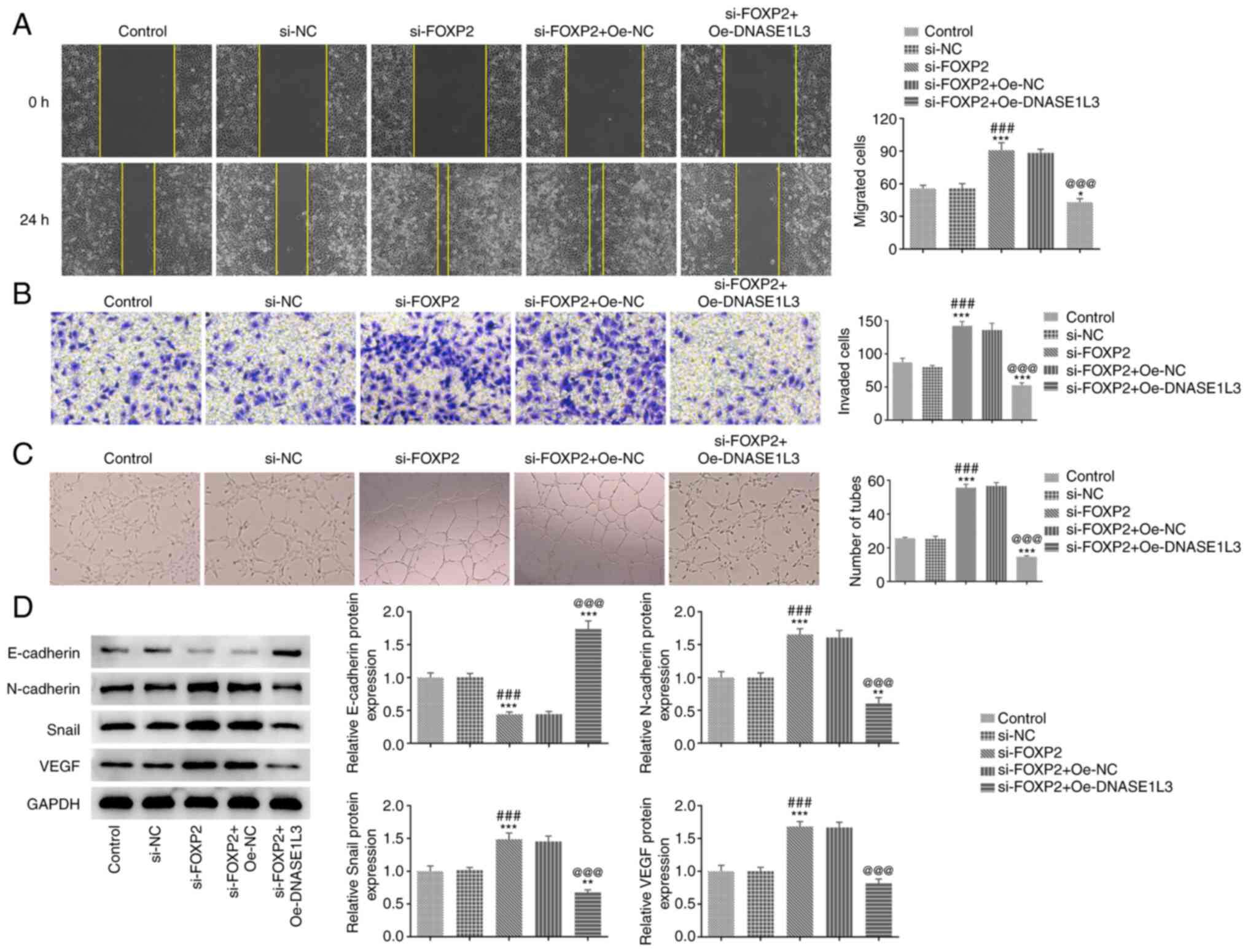 | Figure 6Transcription factor FOXP2 regulates
transcription of DNASE1L3 and promotes migration, invasion and tube
formation of A549 cells. (A) Migration of A549 cells transfected
with si-FOXP2 and Oe-DNASE1L3 was detected by wound healing assay.
Magnification, x100. (B) Invasion of A549 cells transfected with
si-FOXP2 and Oe-DNASE1L3 was determined by Transwell assay.
Magnification, x100. (C) Tube formation of human umbilical vein
endothelial cells cultured in the medium from A549 cells
transfected with si-FOXP2#2 and Oe-DNASE1L3 was observed by tube
formation assay. Magnification, x40. (D) Expression of
epithelial-mesenchymal transformation- and tube
formation-associated proteins in A549 cells transfected with
si-FOXP2#2 and Oe-FOXP2 was detected by western blotting.
*P<0.05, **P<0.01,
***P<0.001 vs. control. ###P<0.001 vs.
si-NC. @@@P<0.001 vs. si-FOXP2#2+Oe-NC. DNASE1L3,
deoxyribonuclease 1-like 3; FOXP2, forkhead-box P2; Oe,
overexpression; NC, negative control; si, small interfering; VEGF,
vascular endothelial growth factor. |
Discussion
LUAD, the most common type of lung cancer, grows and
migrates faster than other subtypes and most patients are already
in the middle and advanced stage when treated (21). The proliferation, invasion and
migration of lung cancer cells are often accompanied by EMT
progression (22,23). EMT describes epithelial cells that
have morphologically transformed into mesenchymal cells and
acquired migration ability, which serves a key role in the process
of embryonic growth, tissue remodeling and tumor metastasis
(24,25). EMT is a key process in lung cancer
cell migration, during which the cytoplasmic skeleton is rebuilt
and connectivity between cells is weakened, thus promoting cell
migration (26). VEGFα is a highly
specific regulatory factor that induces tumor angiogenesis and
promotes migration of vascular endothelial cells by increasing
mitosis of blood tubules. Remodeling of extracellular matrix and
increased vascular permeability are associated with occurrence,
development and metastasis of lung cancer (27,28).
DNASE1L3 suppresses tumor angiogenesis in HCC and overexpression of
DNASE1L3 inhibits proliferation and motility of colon cancer cells
(9,10). The present study showed that
Oe-DNASE1L3 inhibited proliferation, invasion, migration and tube
formation of A549 cells by suppressing EMT progression and VEGF
expression.
The loss or increase of expression of FOX protein
may change the fate of cells and promote the occurrence of tumors
and the progression of cancers through modulation of gene
expression or signaling. FOXP2 is expressed in a number of cell
types; however, it is underexpressed in biopsies of breast, liver
and gastric cancer (29-31).
In the present study, FOXP2 expression was also decreased in A549
cells. Underexpression of FOXP2 results in increased invasion of
tumor cells and vimentin expression and decreased E-cadherin
expression (30). In HCC, microRNA
(miR)-196b promotes the migration and invasion of tumor cells by
directly binding to the 3' untranslated region of FOXP2 mRNA to
inhibit epitaxy (32).
Additionally, a previous study on pancreatic ductal carcinoma
showed miR-23a significantly promotes proliferation and invasion of
tumor cells and subsequent restoration of FOXP2 expression limits
the effect of miR-23a on proliferation and invasion. This suggests
that FOXP2 serves a role in inhibiting tumor proliferation and
invasion (33). FOXP2 has been
found to regulate angiogenesis of U87 glioma-exposed endothelial
cells (34). The present study
found that underexpression of FOXP2 promoted the proliferation,
invasion, migration and tube formation in A549 cells by promoting
EMT progression and VEGF expression. Oe-DNASE1L3 reversed the
effect of downregulation of FOXP2 on malignant behaviors of A549
cells, indicating that FOXP2 positively regulates DNASE1L3
transcription.
In conclusion, the present study illustrated the
role of DNASE1L3 in LUAD and showed that DNASE1L3 was positively
regulated by transcription factor FOXP2, which in turn inhibited
proliferation, migration, invasion and tube formation of LUAD
cells. The present study is limited by the use of only the A549
cell line; other cell lines and in vivo models should be
assessed to confirm the results in future research.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by Tianjin Education
Bureau (grant no. 2020KJ161).
Availability of data and material
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
FM designed and conceived the study, performed
experiments and wrote the manuscript. FM, XY and PX analyzed the
data. XY and PX edited the manuscript. FM, XY and PX confirm the
authenticity of all the raw data. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249.
2021.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Reck M and Rabe KF: Precision diagnosis
and treatment for advanced non-small-cell lung cancer. N Engl J
Med. 377:849–861. 2017.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Qu BL, Cai BN, Yu W, Liu F, Huang YR, Ju
ZJ, Wang XS, Ou GM and Feng LC: Radiotherapy effects on brain/bone
metastatic adenocarcinoma lung cancer and the importance of EGFR
mutation test. Neoplasma. 63:158–162. 2016.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Wang Y, Yang N, Zhang Y, Li L, Han R, Zhu
M, Feng M, Chen H, Lizaso A, Qin T, et al: Effective treatment of
lung adenocarcinoma harboring EGFR-activating mutation, T790M, and
cis-C797S triple mutations by brigatinib and cetuximab combination
therapy. J Thorac Oncol. 15:1369–1375. 2020.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Marinelli D, Mazzotta M, Scalera S,
Terrenato I, Sperati F, D'Ambrosio L, Pallocca M, Corleone G,
Krasniqi E, Pizzuti L, et al: KEAP1-driven co-mutations in lung
adenocarcinoma unresponsive to immunotherapy despite high tumor
mutational burden. Ann Oncol. 31:1746–1754. 2020.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Napirei M, Wulf S, Eulitz D, Mannherz HG
and Kloeckl T: Comparative characterization of rat
deoxyribonuclease 1 (Dnase1) and murine deoxyribonuclease 1-like 3
(Dnase1l3). Biochem J. 389:355–364. 2005.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Sisirak V, Sally B, D'Agati V,
Martinez-Ortiz W, Özçakar ZB, David J, Rashidfarrokhi A, Yeste A,
Panea C, Chida AS, et al: Digestion of chromatin in apoptotic cell
microparticles prevents autoimmunity. Cell. 166:88–101.
2016.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Sjöblom T, Jones S, Wood LD, Parsons DW,
Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, et al:
The consensus coding sequences of human breast and colorectal
cancers. Science. 314:268–274. 2006.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Xiao Y, Yang K, Liu P, Ma D, Lei P and Liu
Q: Deoxyribonuclease 1-like 3 inhibits hepatocellular carcinoma
progression by inducing apoptosis and reprogramming glucose
metabolism. Int J Biol Sci. 18:82–95. 2022.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Guo D, Ma D, Liu P, Lan J, Liu Z and Liu
Q: DNASE1L3 arrests tumor angiogenesis by impairing the
senescence-associated secretory phenotype in response to stress.
Aging (Albany NY). 13:9874–9899. 2021.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Liu J, Yi J, Zhang Z, Cao D, Li L and Yao
Y: Deoxyribonuclease 1-like 3 may be a potential prognostic
biomarker associated with immune infiltration in colon cancer.
Aging (Albany NY). 13:16513–16526. 2021.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Deng Z, Xiao M, Du D, Luo N, Liu D, Liu T,
Lian D and Peng J: DNASE1L3 as a prognostic biomarker associated
with immune cell infiltration in cancer. Onco Targets Ther.
14:2003–2017. 2021.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Chen J, Ding J, Huang W, Sun L, Chen J,
Liu Y, Zhan Q, Gao G, He X, Qiu G, et al: DNASE1L3 as a novel
diagnostic and prognostic biomarker for lung adenocarcinoma based
on data mining. Front Genet. 12(699242)2021.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Wu J, Liu P, Tang H, Shuang Z, Qiu Q,
Zhang L, Song C, Liu L, Xie X and Xiao X: FOXP2 promotes tumor
proliferation and metastasis by targeting GRP78 in triple-negative
breast cancer. Curr Cancer Drug Targets. 18:382–389.
2018.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Song XL, Tang Y, Lei XH, Zhao SC and Wu
ZQ: miR-618 inhibits prostate cancer migration and invasion by
targeting FOXP2. J Cancer. 8:2501–2510. 2017.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Ren T, Liu C, Hou J and Shan F:
Hsa_circ_0043265 suppresses proliferation, metastasis, EMT and
promotes apoptosis in non-small cell lung cancer through
miR-25-3p/FOXP2 pathway. Onco Targets Ther. 13:3867–3880.
2020.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Li ZY, Zhang ZZ, Bi H, Zhang QD, Zhang SJ,
Zhou L, Zhu XQ and Zhou J: Upregulated microRNA-671-3p promotes
tumor progression by suppressing forkhead box P2 expression in
non-small-cell lung cancer. Mol Med Rep. 20:3149–3159.
2019.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Tang Z, Kang B, Li C, Chen T and Zhang Z:
GEPIA2: An enhanced web server for large-scale expression profiling
and interactive analysis. Nucleic Acids Res. 47:W556–W560.
2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Zhu Y, Li B, Xu G, Han C and Xing G:
lncRNA MIR4435-2HG promotes the progression of liver cancer by
upregulating B3GNT5 expression. Mol Med Rep. 25(38)2022.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Zagryazhskaya A, Gyuraszova K and
Zhivotovsky B: Cell death in cancer therapy of lung adenocarcinoma.
Int J Dev Biol. 59:119–129. 2015.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Nasim F, Sabath BF and Eapen GA: Lung
cancer. Med Clin North Am. 103:463–473. 2019.PubMed/NCBI View Article : Google Scholar
|
|
23
|
O'Leary K, Shia A and Schmid P: Epigenetic
regulation of EMT in non-small cell lung cancer. Curr Cancer Drug
Targets. 18:89–96. 2018.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Dongre A and Weinberg RA: New insights
into the mechanisms of epithelial-mesenchymal transition and
implications for cancer. Nat Rev Mol Cell Biol. 20:69–84.
2019.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Tan-Garcia A, Lai F, Sheng Yeong JP, Irac
SE, Ng PY, Msallam R, Tatt Lim JC, Wai LE, Tham CYL, Choo SP, et
al: Liver fibrosis and CD206+ macrophage accumulation
are suppressed by anti-GM-CSF therapy. JHEP Rep.
2(100062)2020.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Baek SH, Ko JH, Lee JH, Kim C, Lee H, Nam
D, Lee J, Lee SG, Yang WM, Um JY, et al: Ginkgolic acid inhibits
invasion and migration and TGF-β-induced EMT of lung cancer cells
through PI3K/Akt/mTOR inactivation. J Cell Physiol. 232:346–354.
2017.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Wu JB, Tang YL and Liang XH: Targeting
VEGF pathway to normalize the vasculature: An emerging insight in
cancer therapy. Onco Targets Ther. 11:6901–6909. 2018.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Frezzetti D, Gallo M, Maiello MR,
D'Alessio A, Esposito C, Chicchinelli N, Normanno N and De Luca A:
VEGF as a potential target in lung cancer. Expert Opin Ther
Targets. 21:959–966. 2017.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Cuiffo BG, Campagne A, Bell GW, Lembo A,
Orso F, Lien EC, Bhasin MK, Raimo M, Hanson SE, Marusyk A, et al:
MSC-regulated microRNAs converge on the transcription factor FOXP2
and promote breast cancer metastasis. Cell Stem Cell. 15:762–774.
2014.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Yan X, Zhou H and Zhang T, Xu P, Zhang S,
Huang W, Yang L, Gu X, Ni R and Zhang T: Downregulation of FOXP2
promoter human hepatocellular carcinoma cell invasion. Tumour Biol.
36:9611–9619. 2015.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Jia WZ, Yu T, An Q, Yang H, Zhang Z, Liu X
and Xiao G: MicroRNA-190 regulates FOXP2 genes in human gastric
cancer. Onco Targets Ther. 9:3643–3651. 2016.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Yu Z, Lin X, Tian M and Chang W:
microRNA-196b promotes cell migration and invasion by targeting
FOXP2 in hepatocellular carcinoma. Oncol Rep. 39:731–738.
2018.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Diao H, Ye Z and Qin R: miR-23a acts as an
oncogene in pancreatic carcinoma by targeting FOXP2. J Investig
Med. 66:676–683. 2018.PubMed/NCBI View Article : Google Scholar
|
|
34
|
He Q, Zhao L, Liu Y, Liu X, Zheng J, Yu H,
Cai H, Ma J, Liu L, Wang P, et al: circ-SHKBP1 regulates the
angiogenesis of U87 glioma-exposed endothelial cells through
miR-544a/FOXP1 and miR-379/FOXP2 pathways. Mol Ther Nucleic Acids.
10:331–348. 2018.PubMed/NCBI View Article : Google Scholar
|