Introduction
Globally, colorectal cancer (CRC) is the third most
common malignant cancer in terms of incidence and the second in
terms of mortality (1). In 2018,
~10% of all new cases of cancer and cancer-associated mortalities
were recorded as being due to CRC (2). The Dukes' staging system classifies
CRC into four groups known as Dukes' A, B, C and D, which represent
stages I, II, III and IV, respectively (3). CRC develops through a multistep
process, including morphological, histological and genetic
alterations that accumulate over time (4). Furthermore, metabolic reprogramming
allows cancer cells to maintain redox homeostasis while increasing
ATP production to meet their high energy demand (5).
Recently, purine metabolism has become a topic of
interest in CRC research, and alteration of the purine metabolism
pathway, including the modification of enzyme activities, has been
shown to be associated with disease progression (6). Purine metabolites supply cells with
vital energy and cofactors to enhance cell survival and
proliferation (7). Purine
nucleosides (purine metabolites), such as adenosine, guanine and
inosine, function as signaling molecules (8) and precursors to ATP and GTP
nucleotides during DNA and RNA synthesis (7). An insufficient supply of nucleotides
during DNA replication may reduce the rate of cell replication,
eventually triggering DNA damage and genomic instability (9).
Purine levels are maintained by the de novo
and salvage biosynthetic pathways, whereby under normal
physiological conditions, degraded bases are recycled via a salvage
pathway to sustain the cellular purine pool (10). Moreover, existing bases in the
extracellular matrix can be transported into the cells to generate
nucleotides (7). Conversely, high
purine levels are required in the environment of cancer cells,
which necessitates upregulation of the de novo biosynthetic
pathway (7). Previous research has
indicated that the treatment of cancer cells with purine
anti-metabolites blocks DNA synthesis in the cells, thereby
inhibiting their growth (11,12).
Hydrophilic purine nucleosides require specific
membrane transport proteins known as nucleoside transporters (NTs)
to transport them across plasma membranes. NTs are classified
according to their transport mechanisms as Na+-dependent
concentrative transporters, which are encoded by the solute carrier
family 28 (SLC28) gene, and equilibrative NTs (ENTs), which
are encoded by the SLC29 gene (13). The ENTs are bidirectional
transporter proteins that facilitate the movement of nucleosides
across the cell membrane driven solely via the concentration
gradient between nucleoside permeants inside and outside the cell
(14). The ENT2 protein is able to
transport nucleosides and a wide range of purine and pyrimidine
nucleobases. Although the exact function of ENT2 is unclear, its
capacity and affinity for the translocation of hypoxanthine has
been reported to be higher than that of ENT1 (15,16).
Hypoxanthine is a purine substrate in the salvage
pathway for nucleotide synthesis, as well as the first metabolite
in the purine catabolism process (17). Consequently, any interruption in
the expression or activity of ENT2 could be deleterious to
the cells, as the intracellular accumulation of reactive oxygen
species (ROS), a by-product of the purine catabolism pathway, might
disrupt the redox balance and ultimately kill the cancer cells
(18). Furthermore, ENT2 and other
NTs play crucial roles in the clearance from the body of certain
anticancer nucleoside analogs, such as 5-fluorouracil, which is
widely used as a chemotherapeutic drug for the treatment of
gastrointestinal cancer, particularly CRC (19), and antiviral drugs, such as
remdesivir, which is used as a treatment for COVID-19(20). The current study therefore aimed to
compare the levels of hypoxanthine phosphoribosyl transferase
(HPRT), hypoxanthine/xanthine and uric acid (UA), as well as the
activity of xanthine oxidase (XO) in CRC cell lines and a normal
colorectal cell line. It may be noted that enzyme activity
measurements are typically used to estimate the concentration of
enzyme present in a sample under certain conditions (21). In addition, the levels of
ENT2 mRNA expression were evaluated and compared in cells
derived from CRC of different stages.
Materials and methods
Cell culture
All cell lines were purchased from the American Type
Culture Collection (ATCC). A panel of human CRC cell lines was
used, where each cell line represented a different CRC stage, as
well as a normal colon epithelial cell line. CCD-841CoN (cat. no.
CRL-1790) was used as the normal colon epithelial cell line, and
SW480 (cat. no. CCL-228; Dukes' B), HCT15 (cat. no. CCL-225; Dukes'
C) and HCT116 (cat. no. CCL-247; Dukes' D) were used as the CRC
cell lines. These CRC stages were classified according to the tumor
stage following Dukes' classification criteria by the ATCC and
previous studies (22,23). All cells were cultured in
Dulbecco's modified Eagle's medium (high glucose; Nacalai Tesque,
Inc.) supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo
Fisher Scientific, Inc.) and 1% penicillin-streptomycin
(MilliporeSigma). The cells were incubated at 37˚C in a humidified
incubator (BINDER GmbH) with 5% CO2.
Cell lysate preparation
Following cell quantification, 1x106
cells were resuspended in 100 µl appropriate buffer as provided
with the kit used for each assay. The cells were hen homogenized
using an Ultrasonic Vibra-Cell™ homogenizer (Sonics &
Materials, Inc.) with pulses of 5 sec on and 25 sec off for 10
cycles. The tube containing the cell lysate was homogenized in ice
to reduce the heat produced by the ultrasonic treatment. Tubes
containing cell lysate and debris were then centrifuged at 24,104 x
g for 10 min at 4˚C. The supernatants were transferred to new tubes
and kept at -20˚C until required for further use.
HPRT enzyme-linked immunosorbent assay
(ELISA) assay
The HPRT level in the cells was measured using a
sandwich ELISA kit for HPRT (cat. no. SEA717Hu; Cloud-Clone Corp.)
according to the manufacturer's instructions. The cells were lysed
as aforementioned. The detection method is based on the
colorimetric reaction of horseradish peroxidase following the
addition of a tetramethylbenzidine substrate. The absorbance was
measured immediately at 450 nm using a microplate reader (BMG
Labtech GmbH).
XO assay
The activity of XO was quantified
spectrophotometrically using a Xanthine Oxidase Activity
Colorimetric/Fluorimetric Assay Kit (cat. no. K710; BioVision,
Inc.; Abcam). In this assay, XO in the sample oxidizes xanthine to
hydrogen peroxide (H2O2), which reacts
stoichiometrically with OxiRed Probe to generate a color that can
be detected at 570 nm. Briefly, 44 µl assay buffer, 2 µl enzyme
mix, 2 µl substrate mix and 2 µl OxiRed probe were mixed and added
to the designated wells. The reactions were initiated by the
addition of 50 µl prepared standard or cell lysate into each well.
Absorbance (A) measurements A1 and A2 were taken after 20 min
incubation in the dark at room temperature using a microplate
reader (BMG Labtech GmbH) at a wavelength of 570 nm. A standard
curve was plotted for several dilutions of
H2O2 standard and the volume was adjusted to
50 µl/well using distilled water. The XO activity of each sample
was calculated using the following equation: XO activity (mU/ml)=(B
x dilution factor)/(T2-T1) x V, in which B is the amount of
H2O2 generated by XO according to the
standard curve (nmol), T1 is the time of the first reading (A1,
min), T2 is the time of the second reading (A2, min) and V is the
pre-treated sample volume added to the reaction well (ml).
Hypoxanthine/xanthine assay
The hypoxanthine/xanthine level in all cell lines
was detected using a Xanthine/Hypoxanthine
Colorimetric/Fluorimetric Assay Kit (cat. no. K685; BioVision,
Inc.; Abcam) according to the manufacturer's instructions. In
brief, 1x106 cells were incubated on ice with 100 µl
ice-cold xanthine assay buffer for 10 min. After centrifugation at
24,104 x g for 5 min, the supernatant was collected and incubated
with the reaction mix for 30 min at room temperature. A standard
curve was plotted using several dilutions of xanthine standard and
the volume was adjusted to 50 µl/well with xanthine assay buffer.
The absorbance was measured at a wavelength of 570 nm.
UA assay
UA quantification was performed using a Uric Acid
Colorimetric/Fluorometric Assay Kit (cat. no. K608; BioVision, Inc.
Abcam) according to the manufacturer's instructions. Briefly, 46 µl
UA assay buffer, 2 µl UA enzyme mix and 2 µl UA probe were mixed
and added to the designated wells on the plate. The reactions were
initiated by the addition of 50 µl prepared UA standard or cell
lysate to each well. The plates were then incubated for 30 min at
37˚C and protected from light. A standard curve was plotted using
several dilutions of UA standard and the volume was adjusted to 50
µl/well with UA assay buffer. The absorbance was measured at a
wavelength of 570 nm.
RNA isolation and cDNA synthesis
Total RNA from all cell lines was extracted using a
NucleoSpin RNA mini kit for RNA purification (cat. no. 740955;
Macherey-Nagel, GmbH & Co. KG) according to the manufacturer's
instructions. The total RNA concentration and the A230/208 and
A260/280 nm ratios of the initial RNA were measured to assess the
RNA quality using a SpectraMax® QuickDrop™ UV-Vis
Spectrophotometer (Molecular Devices, LLC). RNA integrity was
assessed via a gel electrophoresis assay. Next, 200 ng total RNA
was used for reverse transcription (RT) using a SensiFAST™ cDNA
Synthesis kit (cat. no. BIO-65053l Bioline; Meridian Bioscience,
Inc.) according to the manufacturer's instructions.
Quantitative PCR (qPCR) of ENT2
Following RT, qPCR was performed using a CFX96™
Touch Real-Time PCR detection system (Bio-Rad Laboratories, Inc.).
The reactions were carried out in triplicate using a SensiFAST™
SYBR® & Fluorescein Kit (cat. no. BIO96005; Bioline;
Meridian Bioscience, Inc.) following the manufacturer's
instructions. The sequences of the oligonucleotide primers that
were used for qPCR amplification are listed in Table I. Each reaction was performed in a
final volume of 10 µl and the primer concentration was 400 nM.
Thermocycling was conducted with an initial start cycle at 95˚C for
2 min, followed by 39 cycles at 95˚C for 5 sec and 60˚C for 30 sec.
To confirm product specificity, melting curve analysis was
performed after each amplification. As normalization against a
single reference gene may not be considered adequate (24), the mRNA level of ENT2 was
normalized by the geometric averaging of two reference genes,
GAPDH and HPRT, to obtain the most reliable results
in the gene transcription analysis. The stability and reliability
of GAPDH and HPRT1 as reference genes in the CRC and
normal colon cell lines were determined via measurement of their
quantification cycle (Cq) value and calculating the coefficient of
variation using CFX96 software (Bio-Rad Laboratories, Inc.). The
fold-change in ENT2 expression of the human CRC cell lines
relative to the normal colon cell line was determined using the
2-ΔΔCq method (25). The geometric average (GeoMean) of
the relative quantities [(EREF)∆Cq REF] of
the two reference genes was utilized to determine the expression of
the gene of interest (GOI) as shown in the following equation:
Relative gene expression=[(EGOI)ΔCq
GOI]/GeoMean[(EREF)ΔCq
REF].
 | Table IPrimer sequences used for
quantitative PCR. |
Table I
Primer sequences used for
quantitative PCR.
| Gene | Forward
(5'-3') | Reverse
(5'-3') |
|---|
| ENT2 |
CCACTCTCTCACCGAAGCCTAA |
GCAGGAAGAACAGCACCAACA |
| GAPDH |
GCATCCTGGGCTACACTGAG |
TCCTCTTGTGCTCTTGCTGG |
| HPRT1 |
GAGTCCTATTGACATCGCCAGT |
TCCGCCCAAAGGGAACTGAT |
Statistical analysis
All data were statistically analyzed using IBM SPSS
Statistical Software version 27 (IBM Corp.). The results are
expressed as the mean ± SD obtained from three independent
experiments where each assay was performed in triplicate. The
significant differences between multiple groups were assessed by
one-way analysis of variance (ANOVA) followed by Tukey's post hoc
multiple comparisons tests. Statistical evaluation of ENT2
gene expression was performed using CFX96 software followed by
one-way ANOVA with Tukey's multiple comparisons tests. The
correlations between ENT2 expression and the different
stages of CRC were determined by Spearman's correlation test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
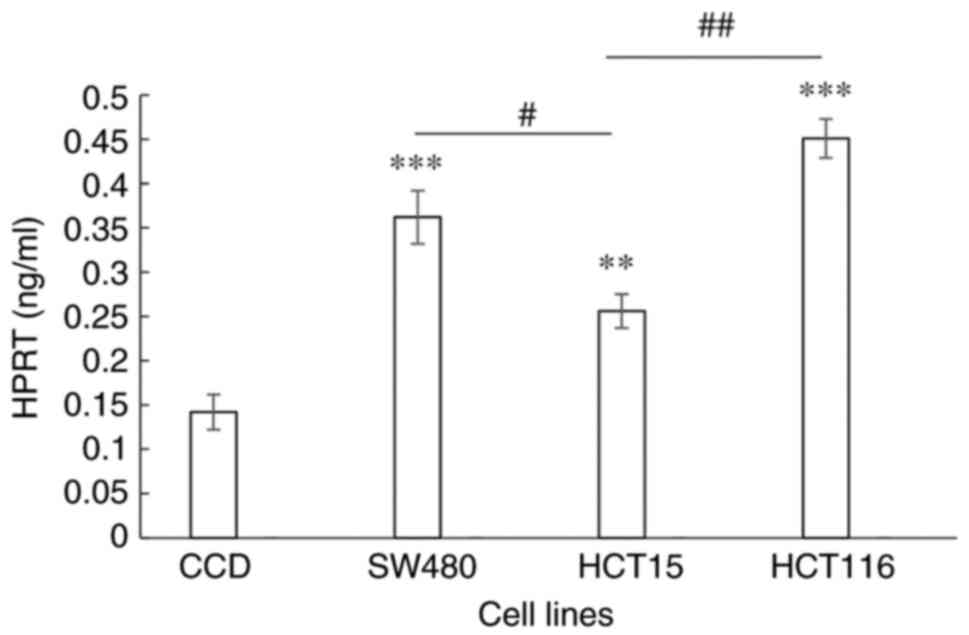
Determination of HPRT levels
HPRT levels in the normal colon and CRC cell lines
were determined by the binding of HPRT to an antibody in an ELISA
experiment. When compared with normal colon cells, the levels of
HPRT in the SW480 (P=0.0001), HCT15 (P=0.013) and HCT116
(P<0.0001) cell lines were significantly higher. HPRT levels in
the cell lines representing Dukes' stages B and D CRC were
significantly higher compared with that in the cell line
representing Dukes' stage C (P=0.0079 and P=0.0003, respectively;
Fig. 1).
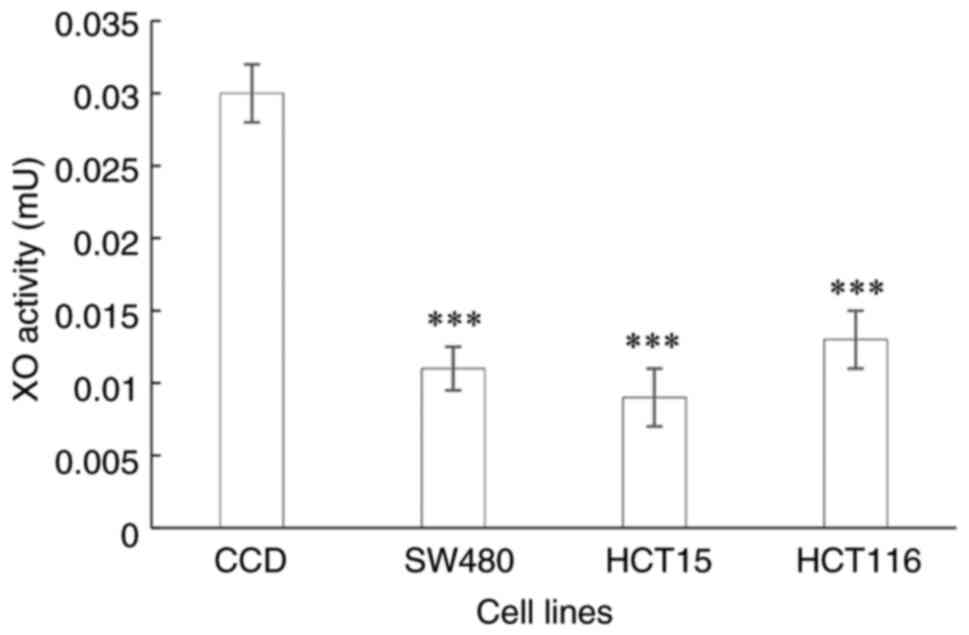
Determination of XO activity
The XO enzymatic activity in the CRC cell lines
representing the different stages of CRC were all significantly
lower (all P<0.0001) than those in the normal colon cells
(Fig. 2).
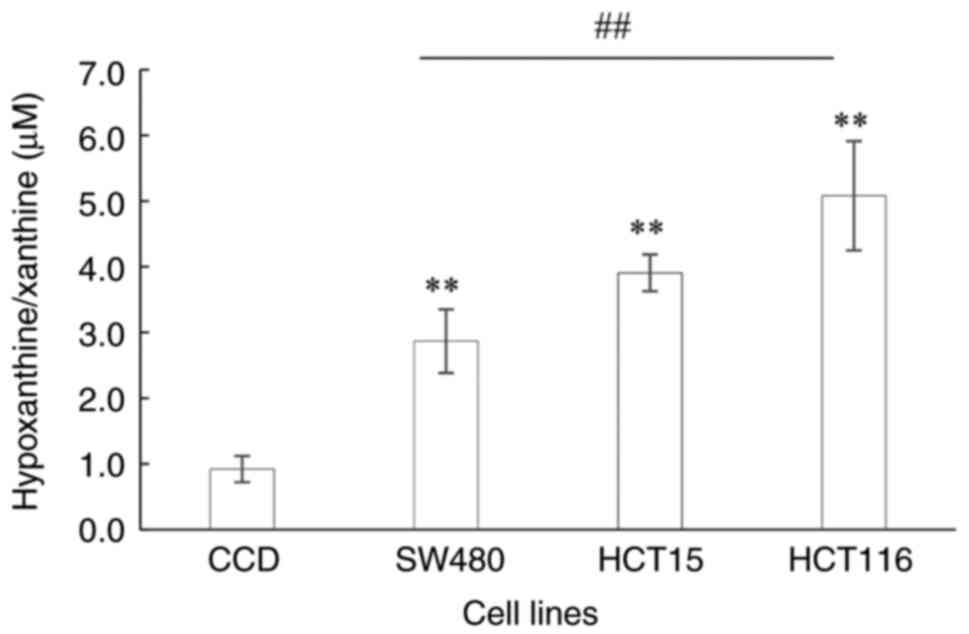
Determination of hypoxanthine/xanthine
level
The hypoxanthine/xanthine levels of the Dukes' B, C
and D stage CRC cell lines were significantly higher compared with
those in the normal colon cells (P=0.007, P=0.004 and P<0.0001,
respectively; Fig. 3). The
hypoxanthine/xanthine levels ranged from 2.86 to 5.08 µM in the CRC
cell lines compared with 0.92 µM in the normal colon cells. The
hypoxanthine/xanthine levels were observed to increase as the CRC
stage advanced, with 3.1-, 4.2- and 5.5-fold increases in Dukes' B,
C and D stages, respectively, compared with the normal colon
control. The hypoxanthine/xanthine level was significantly higher
in the Dukes' D CRC cell line than that in the Dukes' B cell line
(P=0.003).
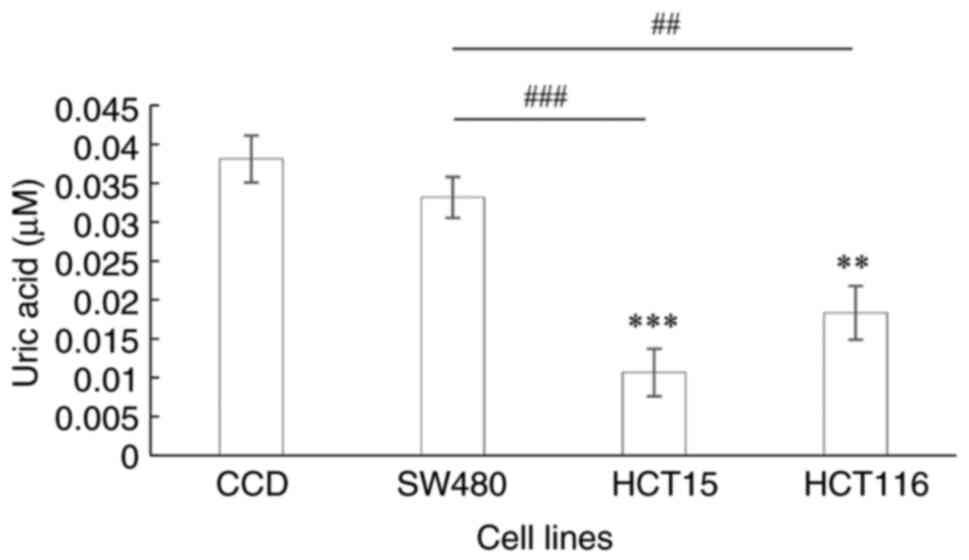
Determination of UA level
The end product of purine catabolism is UA. When
compared with that in the normal colon cells, the UA level in the
HCT15 and HT116 cell lines was significantly downregulated
(P<0.0001 and P=0.0002, respectively; Fig. 4). Furthermore, UA levels were
significantly decreased in the CRC cell lines representing Dukes' C
and D stages compared with those in the Dukes' B CRC cell line
(P<0.0001 and P=0.0008, respectively).
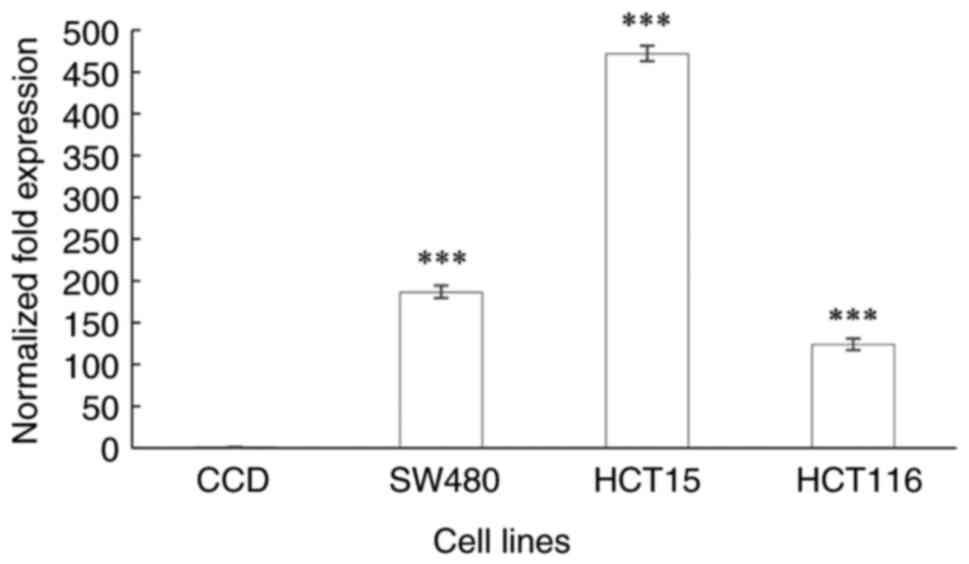
ENT2 gene expression
ENT2 was significantly upregulated in all CRC
cell lines when compared with that in the normal colon cells
(P<0.0001; Fig. 5). The
ENT2 expression level was found to be 186-, 471- and
123-fold higher in cells from the Dukes' B, C and D stages of CRC,
respectively, compared with that in the normal colon cell line. The
ENT2 expression level was the highest in the Dukes' C stage
CRC cells. Notably, ENT2 expression was lower in the cells
of Dukes' stage D compared with those of Dukes' stage B, however
this was insignificant. However, Spearman's correlation analysis
for ENT2 expression and CRC stage did not reveal a
significant correlation (ρ=0.475; P=0.128; data not shown).
Discussion
Purine metabolism has been observed to be one of the
most affected pathways in CRC (26,27)
and other cancer types, including breast cancer (28) and urothelial carcinoma (29). Moreover, alterations in its
enzymatic pattern (30) and purine
metabolism dysfunction have been found to be associated with cancer
progression. For example, several cancer types have been
demonstrated to express a high level of HPRT, the key enzyme in the
salvage pathway, thus promoting metabolic flux to produce purine
mononucleotides, which are precursors in nucleic acid synthesis
(31). While the hypoxanthine
concentration gradient alone may not provide a compelling argument
for the involvement of ENT2 in hypoxanthine transport, as
hypoxanthine influx/efflux was not observed in the present study,
the molecules and substrates involved in the intracellular purine
metabolic pathway, including HPRT level, XO activity and UA level,
were examined. The present study recorded a significant
upregulation of HPRT and hypoxanthine/xanthine levels in different
stages of CRC, notably in advanced stages, whereas the XO
activities and UA levels in the CRC cell lines were markedly
decreased compared with those in the normal colon cells.
The upregulation of HPRT in several solid cancer
types, including breast, lung, colon and prostate cancer (32), the significantly higher HPRT
activities in colorectal (33) and
breast (34) cancer, and a notable
increase in HPRT activity with the clinical progression of human
colon carcinoma (35) have been
documented, which support the findings of the current study. A
metabolomic study reported elevated hypoxanthine levels in
lymphoblastic leukemia and lung and prostate cancer (36), whereas under normal physiological
conditions, hypoxanthine levels are very low (37). Similarly, another study recorded
elevated hypoxanthine and decreased UA levels in CRC tissues
(38). The data in the present
study suggest that increased hypoxanthine levels are associated
with CRC progression. However, further investigation into the
association between hypoxanthine elevation and cancer progression
in vivo is required to verify this.
Consistent with the findings of the present study,
several investigations have reported diminished XO activity in
cancer types such as colon (39),
kidney and bladder (40) cancer.
Low XO expression has been shown to be associated with more
aggressive ovarian cancer (41),
indicating that it plays a vital role in cancer progression and
could be a risk factor (42).
Furthermore, the reduction in XO activity may benefit cancer cells
by lowering the generation of ROS, which can trigger apoptosis and
cell death. It has been demonstrated that reduced XO activity is
advantageous to cancer cells, as it attenuates the detrimental
effect of ROS generation on hypoxanthine degradation and catabolic
pathway activation (43). Another
parameter examined in the present study is UA levels. Previous
studies have recorded elevated UA levels in advanced stages of
rectal cancer (44) and head and
neck carcinoma (45). Conversely,
the current study recorded significantly decreased UA levels in the
CRC cells of different stages compared with those in normal colon
cells. The decrease in UA levels may be due to the low XO activity
in all CRC stages evaluated.
The role of UA in tumorigenesis has not yet been
determined. Several investigations have suggested that an elevated
serum level of UA (hyperuricemia) is associated with an increased
cancer risk due to its pro-inflammatory properties (46). It has also been suggested that UA
plays an essential role as a potent antioxidant (31). Moreover, elevated UA levels might
serve as a tumor defense, by protecting the tumor cells against
free-radical oxidative damage by acting as antioxidant and by
stimulating the immune system (47). Elevated UA levels have been shown
to be associated with an increased survival time in patients with
CRC (48). It is notable that the
investigated metabolites and enzyme levels in the present study did
not exhibit a strict association with Dukes' stage. This may be due
to different mutational statuses in the different stages (49). Therefore, the rate of purine
metabolism may not increase with higher staging. Advanced stages of
cancer may have alternative cancer-protective/resistance mechanisms
that are not limited to the purine metabolism pathway. Cells are
particularly adaptable to intrinsic oxidative stress in the
advanced stages of cancer, as they develop a strong ability to
maintain the homeostatic balance between the generation and
elimination of ROS (50-52).
The current study found that ENT2 expression
was significantly higher in cells from all stages of CRC than that
in normal colon cells. Similarly, in previous studies, the level of
ENT2 was observed to be 2- to 5.5-fold higher in breast,
kidney and prostate cancer cells than in corresponding normal cells
(18,53), with the highest expression of
ENT2 detected in cancer derived from digestive organ tissues
(54). ENT2 has been
reported to be highly expressed in colorectal biopsy samples
(55), and to be highly expressed
in four metastatic cell lines, namely LoVo, Colo205, SK-CO-1 and
T84, and four primary CRC cell lines, namely HT29, Caco2, Colo320
and HCT116(56). Furthermore, high
levels of ENT2 gene expression have been associated with the
advanced stages of a variety of malignancies, including
hepatocellular carcinoma, mantle cell lymphoma and ovarian
carcinoma (57).
In the present study, ENT2 expression was
significantly higher in CRC cell lines that representing Dukes' B,
C, and D compared with that in normal colon cells, but was not
significantly lower in Dukes' D stage cell line (stage IV) than in
the Dukes' B and C stage cell lines (stage II and III)This suggests
that the expression of ENT2 is not associated with CRC
stage. This may imply that cancer cells, particularly in advanced
stages, regulate ENT2 expression for protection and
survival. However, the mechanism of action of ENT2, specifically on
nucleoside permeant influx/efflux, requires further exploration via
in vivo studies. ENT2 is a bidirectional transporter that
has the ability to translocate a wide range of nucleosides into and
out of cells; it has been documented that ENT2 is the main
transporter responsible for cellular hypoxanthine efflux (58). In the current study, the low
expression of ENT2 in Dukes' D stage cells corresponded to
the highest level of hypoxanthine, an endogenous substrate of
ENT2 (31). Increased
ENT2 expression may allow cancer cells to escape from the
damaging effects of ROS on hypoxanthine degradation, while lowered
ENT2 expression may limit hypoxanthine efflux, in a process
whereby hypoxanthine recycling supplies nucleotides to cancer cells
to meet the demand for rapid cell proliferation.
During salvage purine metabolism, hypoxanthine is
recycled to inosine monophosphate (IMP) by HPRT or synthesized
through the deamination of adenosine by adenosine deaminase to
produce inosine, which is converted to hypoxanthine by purine
nucleoside phosphorylase (33).
Subsequently, hypoxanthine is oxidized to produce xanthine via the
purine catabolism pathway and then converted to UA via XO. During
the catabolism pathway, ROS are produced as a by-product (42).
Studies have shown that intrinsic oxidative stress,
such as ROS-induced stress, can effectively kill tumor cells via
the activation of apoptotic processes (59-62).
It has been observed that when ROS accumulation reaches a specific
threshold that is incompatible with cell survival, cytotoxic
effects are exerted, which induce apoptosis and slow cancer growth
(63). ROS generation mediated by
XO or NADH dehydrogenase has been linked with mitochondrial
malfunction, which is considered to be the first indication of
apoptotic intrinsic pathway activation (64). Excess hypoxanthine in cancer cells
is then transported out of the cancer cells via the ENT2
transporter (58) to maintain low
to average levels of ROS, or recycled to generate new nucleotides
for DNA synthesis in order to meet the demand for rapid cell
proliferation. Cancer cells may alter their metabolic regulation
processes, in order to avoid the harmful effects of ROS and allow
the cells to adapt and thrive in a hostile environment (38). These findings may explain the
contradictory data observed for hypoxanthine levels in the serum of
patients with CRC (6) and in CRC
cells (65), particularly in
advanced stages (27).
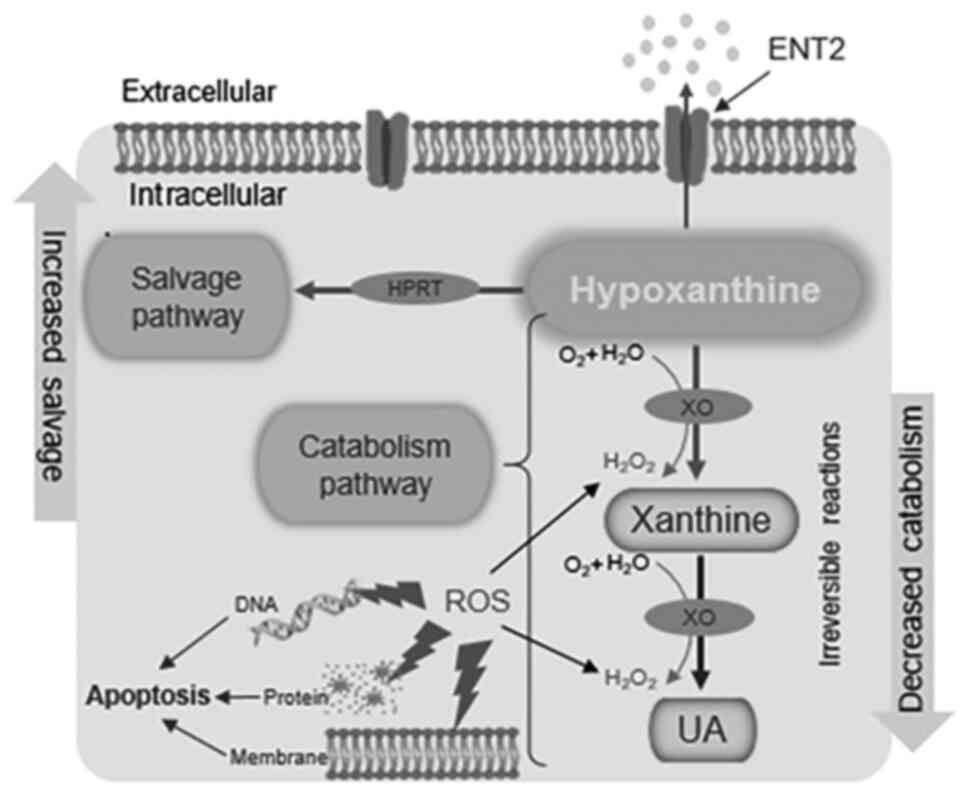
The upregulated HPRT and hypoxanthine levels
reported in the CRC cells in the present study may imply that the
activity of the salvage pathway is increased to meet the demand for
rapid cell proliferation via the generation of nucleotides for DNA
synthesis by hypoxanthine recycling, as previously proposed
(31). Furthermore, the activity
of the catabolism pathway may be reduced, as suggested by the
decreased XO and UA levels, in order to preserve the antioxidant
balance and avoid ROS-induced damage, which would lead to apoptosis
(Fig. 6). Changes in the levels of
enzymes and metabolites associated with the purine salvage and
catabolism pathways may have diagnostic and prognostic importance
for CRC progression. Furthermore, ENT2 may play a crucial role in
the regulation of metabolite transport to enhance cancer cell
survival and proliferation.
Since hypoxanthine influx/efflux was not observed in
the present study, the role of ENT2 in hypoxanthine transport and
the hypoxanthine concentration gradient has not been clearly
demonstrated. However, various molecules and substrates involved in
the purine metabolic pathway were examined, including XO activity,
and HPRT and UA levels, which act intracellularly.
Increased ENT2 expression in cells of higher
Dukes' stages suggests that hypoxanthine may be being transported
out of the cells as a form of defense, resulting in fewer
substrates being available for the purine metabolism pathway.
Evidence for this is provided by the inverse correlation of
hypoxanthine levels with XO activity and UA levels. In this case,
the degradation pathway is unfavorable to cells, and extracellular
transport avoids the generation of high levels of ROS, which are
toxic to cancer cells, and the antioxidant UA.
Notably, the level of HPRT was found to be
upregulated in the CRC cell lines of the present study, which could
imply that the hypoxanthine available inside the cells is converted
to IMP via the salvage pathway to produce the nucleosides GMP and
AMP for conversion to the nucleotides required for DNA synthesis
and cell growth. However, further investigation is required for a
definitive conclusion on the latter process. An in vivo
experiment, which would give tangible support for the findings, is
lacking and is a limitation of the present study; therefore, it
could be the subject of future research.
In conclusion, CRC cells may utilize ENT2 in dual
roles for protection and survival, that is, for avoiding the
detrimental effects of ROS accumulation via purine catabolic
metabolism, as demonstrated by decreased XO enzyme and UA
production, and for utilizing hypoxanthine to meet the energy
demands of rapid cell proliferation via the purine salvage pathway,
as indicated by increased HPRT levels. Accordingly, the purine
metabolism pathway appears to be an important metabolic pathway in
the progression of CRC. However, the mechanism of action, in
particular, whether additional pathways are affected by aberrant
ENT2 expression, requires further investigation.
In the current study, HPRT, XO, hypoxanthine and UA
levels, and ENT2 expression, were detected in a panel of CRC
cell lines. However, further investigations such as measuring
hypoxanthine influx/efflux and its association with low and high
staging, assessment of protein expression levels by western
blotting, performing ENT2 gene silencing and detecting the
ROS level in different stages of colorectal carcinogenesis are
recommended to determine the relevance and role of ENT2, and
its association with purine metabolism in colorectal
carcinogenesis. However, although the metabolomic analysis of cell
lines has been extensively used in disease research due to certain
advantages, including lower costs, ease of control and simpler data
interpretation than the analysis of in vivo models using
human tissues and bio-fluids (66), in vivo experiments are
necessary to provide concrete support to the findings and a solid
conclusion. Despite these limitations, the findings from the
present study suggest that targeting ENT2 could potentially
be a therapeutic strategy for the enhancement of CRC treatment
efficacy.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the Ministry of
Higher Education Government of Malaysia under a Long-Term Research
Grant (grant no. LRGS/2014/UKM-UiTM/K/03) and the Fundamental
Research Grant Scheme (no. FRGS/1/2019/SKK08/UITM/02/11). It was
funded by the National Cancer Council [Majlis Kanser Nasional; no.
100-IRMI/16/6/2 (011/2019)].
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
SMN drafted the manuscript. SMN and NAAH conducted
the experiments, and analyzed and interpreted the data. SAR, MM and
AAR designed the study, discussed experimental approaches and
outcomes, and critically revised the manuscript. MM and AAR
obtained funding. AAR, SAR and MM confirm the authenticity of all
the raw data. All authors read and approved the final version of
the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Wong MC, Huang J, Lok V, Wang J, Fung F,
Ding H and Zheng ZJ: Differences in incidence and mortality trends
of colorectal cancer worldwide based on sex, age, and anatomic
location. Clin Gastroenterol Hepatol. 19:955–966.e61.
2021.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Xie YH, Chen YX and Fang JY: Comprehensive
review of targeted therapy for colorectal cancer. Signal Transduct
Target Ther. 5:1–30. 2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Peravali R and Hall N: Colorectal cancer:
Features and investigation. Medicine. 43:299–302. 2015.
|
|
4
|
Simon K: Colorectal cancer development and
advances in screening. Clin Interv Aging. 11:967–976.
2016.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Martinez-Outschoorn UE, Peiris-Pagés M,
Pestell RG, Sotgia F and Lisanti MP: Cancer metabolism: A
therapeutic perspective. Nat Rev Clin Oncol. 14:11–31.
2017.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Hashim NAA, Ab-Rahim S, Ngah WZW, Nathan
S, Ab Mutalib NS, Sagap I, Jamal ARA and Mazlan M: Global
metabolomics profiling of colorectal cancer in Malaysian patients.
Bioimpacts. 11:33–43. 2021.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Pedley AM and Benkovic SJ: A new view into
the regulation of purine metabolism: The purinosome. Trends Biochem
Sci. 42:141–154. 2017.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Di Virgilio F and Adinolfi E:
Extracellular purines, purinergic receptors and tumor growth.
Oncogene. 36:293–303. 2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Bester AC, Roniger M, Oren YS, Im MM,
Sarni D, Chaoat M, Bensimon A, Zamir G, Shewach DS and Kerem B:
Nucleotide deficiency promotes genomic instability in early stages
of cancer development. Cell. 145:435–446. 2011.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Yamaoka T, Kondo M, Honda S, Iwahana H,
Moritani M, Ii S, Yoshimoto K and Itakura M:
Amidophosphoribosyltransferase limits the rate of cell
growth-linked de novo purine biosynthesis in the presence of
constant capacity of salvage purine biosynthesis. J Biol Chem.
272:17719–17725. 1997.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Yin J, Ren W, Huang X, Deng J, Li T and
Yin Y: Potential mechanisms connecting purine metabolism and cancer
therapy. Front Immunol. 9(1697)2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Rosenthal EL, Chung TK, Parker WB, Allan
PW, Clemons LL, Lowman D, Hong J, Hunt FR, Richman J, Conry RM, et
al: Phase I dose-escalating trial of Escherichia coli purine
nucleoside phosphorylase and fludarabine gene therapy for advanced
solid tumors. Ann Oncol. 26:1481–1487. 2015.PubMed/NCBI View Article : Google Scholar
|
|
13
|
dos Santos-Rodrigues A, Grañé-Boladeras N,
Bicket A and Coe IR: Nucleoside transporters in the purinome.
Neurochem Int. 73:229–237. 2014.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Cass CE, Young JD, Baldwin SA, Cabrita MA,
Graham KA, Griffiths M, Jennings LL, Mackey JR, Ng AM, Ritzel MW,
et al: Nucleoside transporters of mammalian cells. Pharm
Biotechnol. 12:313–352. 2002.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Altaweraqi RA, Yao SY, Smith KM, Cass CE
and Young JD: HPLC reveals novel features of nucleoside and
nucleobase homeostasis, nucleoside metabolism and nucleoside
transport. Biochim Biophys Acta Biomembr.
1862(183247)2020.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Naes SM, Ab-Rahim S, Mazlan M and Rahman
AA: Equilibrative nucleoside transporter 2: Properties and
physiological roles. Biomed Res Int. 3(5197626)2020.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Young JD, Yao SY, Baldwin JM, Cass CE and
Baldwin SA: The human concentrative and equilibrative nucleoside
transporter families, SLC28 and SLC29. Mol Aspects Med. 34:529–547.
2013.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Tang PC, Yang C, Li RWS, Lee SMY, Hoi MPM,
Chan SW, Kwan YW, Tse CM and Leung GPH: Inhibition of human
equilibrative nucleoside transporters by 4-((4-(2-fluorophenyl)
piperazin-1-yl)
methyl)-6-imino-N-(naphthalen-2-yl)-1,3,5-triazin-2-amine. Eur J
Pharmacol. 791:544–551. 2016.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Phua LC, Mal M, Koh PK, Cheah PY, Chan ECY
and Ho HK: Investigating the role of nucleoside transporters in the
resistance of colorectal cancer to 5-fluorouracil therapy. Cancer
Chemother Pharmacol. 71:817–823. 2013.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Al-Tawfiq JA, Al-Homoud AH and Memish ZA:
Remdesivir as a possible therapeutic option for the COVID-19.
Travel Med Infect Dis. 34(101615)2020.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Scopes RK: Enzyme activity and assays. e
LS. 2001.
|
|
22
|
Gatzidou E, Mantzourani M, Giaginis C,
Giagini A, Patsouris E, Kouraklis G and Theocharis S: Augmenter of
liver regeneration gene expression in human colon cancer cell lines
and clinical tissue samples. J BUON. 20:84–91. 2015.PubMed/NCBI
|
|
23
|
Yusof HM, Ab-Rahim S, Ngah WZW, Nathan S,
Jamal ARA and Mazlan M: Metabolomic characterization of colorectal
cancer cell lines highlighting stage-specific alterations during
cancer progression. Bioimpacts. 11:147–156. 2021.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Bustin SA, Benes V, Garson JA, Hellemans
J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL,
et al: The MIQE guidelines: Minimum information for publication of
quantitative real-time PCR experiments. Clin Chem. 55:611–622.
2009.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative CT method. Nat Protoc.
3:1101–1108. 2008.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Brown DG, Rao S, Weir TL, O'Malia J, Bazan
M, Brown RJ and Ryan EP: Metabolomics and metabolic pathway
networks from human colorectal cancers, adjacent mucosa, and stool.
Cancer Metab. 4(11)2016.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Long Y, Sanchez-Espiridion B, Lin M, White
L, Mishra L, Raju GS, Kopetz S, Eng C, Hildebrandt MAT, Chang DW,
et al: Global and targeted serum metabolic profiling of colorectal
cancer progression. Cancer. 123:4066–4074. 2017.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Luo X, Yu H, Song Y and Sun T: Integration
of metabolomic and transcriptomic data reveals metabolic pathway
alteration in breast cancer and impact of related signature on
survival. J Cell Physiol. 234:13021–13031. 2019.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Sahu D, Lotan Y, Wittmann B, Neri B and
Hansel DE: Metabolomics analysis reveals distinct profiles of
nonmuscle-invasive and muscle-invasive bladder cancer. Cancer Med.
6:2106–2120. 2017.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Zhu J, Djukovic D, Deng L, Gu H, Himmati
F, Chiorean EG and Raftery D: Colorectal cancer detection using
targeted serum metabolic profiling. J Proteome Res. 13:4120–4130.
2014.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Garcia-Gil M, Camici M, Allegrini S, Pesi
R, Petrotto E and Tozzi M: Emerging role of purine metabolizing
enzymes in brain function and tumors. Int J Mol Sci.
19(3598)2018.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Townsend MH, Felsted AM, Ence ZE, Piccolo
SR, Robison RA and O'Neill K: Elevated expression of hypoxanthine
guanine phosphoribosyltransferase within malignant tissue. Cancer
Clin Oncol. 6:19–34. 2017.
|
|
33
|
Sedano MJ, Ramos EI, Choudhari R, Harrison
AL, Subramani R, Lakshmanaswamy R, Zilaie M and Gadad SS:
Hypoxanthine phosphoribosyl transferase 1 is upregulated, predicts
clinical outcome and controls gene expression in breast cancer.
Cancers (Basel). 12(1522)2020.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Camici M, Tozzi MG, Allegrini S, Del Corso
A, Sanfilippo O, Daidone MG, De Marco C and Ipata PL: Purine
salvage enzyme activities in normal and neoplastic human tissues.
Cancer Biochem Biophys. 11:201–209. 1990.PubMed/NCBI
|
|
35
|
Sanfilippo O, Camici M, Tozzi MG, Turriani
M, Faranda A, Ipata P and Silvestrini R: Relationship between the
levels of purine salvage pathway enzymes and clinical/biological
aggressiveness of human colon carcinoma. Cancer Biochem Biophys.
14:57–66. 1994.PubMed/NCBI
|
|
36
|
Kami K, Fujimori T, Sato H, Sato M,
Yamamoto H, Ohashi Y, Sugiyama N, Ishihama Y, Onozuka H, Ochiai A,
et al: Metabolomic profiling of lung and prostate tumor tissues by
capillary electrophoresis time-of-flight mass spectrometry.
Metabolomics. 9:444–453. 2013.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Nemkov T, Sun K, Reisz JA, Song A, Yoshida
T, Dunham A, Wither MJ, Francis RO, Roach RC, Dzieciatkowska M, et
al: Hypoxia modulates the purine salvage pathway and decreases red
blood cell and supernatant levels of hypoxanthine during
refrigerated storage. Haematologica. 103:361–372. 2018.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Ong ES, Zou L, Li S, Cheah PY, Eu KW and
Ong CN: Metabolic profiling in colorectal cancer reveals signature
metabolic shifts during tumorigenesis. Mol Cell Proteomics 10: doi:
10.1074, 2010.
|
|
39
|
Durak İ, Cetin R, Devrim E and Ergüder İB:
Effects of black grape extract on activities of DNA turn-over
enzymes in cancerous and non cancerous human colon tissues. Life
Sci. 76:2995–3000. 2005.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Battelli MG, Polito L, Bortolotti M and
Bolognesi A: Xanthine oxidoreductase in cancer: More than a
differentiation marker. Cancer Med. 5:546–557. 2016.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Linder N, Bützow R, Lassus H, Lundin M and
Lundin J: Decreased xanthine oxidoreductase (XOR) is associated
with a worse prognosis in patients with serous ovarian carcinoma.
Gynecol Oncol. 124:311–318. 2012.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Wang Y, Liu S, Tian S, Du R, Lin T, Xiao
X, Wang R, Chen R, Geng H, Subramanian S, et al: C1QBP regulates
apoptosis of renal cell carcinoma via modulating xanthine
dehydrogenase (XDH) mediated ROS generation. Int J Med Sci.
19:842–857. 2020.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Linder N, Martelin E, Lundin M, Louhimo J,
Nordling S, Haglund C and Lundin J: Xanthine
oxidoreductase-Clinical significance in colorectal cancer and in
vitro expression of the protein in human colon cancer cells. Eur J
Cancer. 45:648–655. 2009.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Yuan C, Xu XH, Wang XL, Xu L, Chen Z and
Li YQ: Relationship between serum uric acid and metastatic and
nonmetastatic rectal cancer patients with undergoing no
chemotherapy. Medicine (Baltimore). 95(e5463)2016.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Dhankhar R, Dahiya K, Sharma TK, Ghalaut
VS, Atri R and Kaushal V: Diagnostic significance of adenosine
deaminase, uric acid and C-reactive protein levels in patients of
head and neck carcinoma. Clin Lab. 57:795–798. 2011.PubMed/NCBI
|
|
46
|
Fini MA, Elias A, Johnson RJ and Wright
RM: Contribution of uric acid to cancer risk, recurrence, and
mortality. Clin Transl Med. 1:1–15. 2012.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Shi L, Chen S, Yang L and Li Y: The role
of PD-1 and PD-L1 in T-cell immune suppression in patients with
hematological malignancies. J Hematol Oncol. 6:1–6. 2013.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Dziaman T, Banaszkiewicz Z, Roszkowski K,
Gackowski D, Wisniewska E, Rozalski R, Foksinski M, Siomek A,
Speina E, Winczura A, et al: 8-Oxo-7, 8-dihydroguanine and uric
acid as efficient predictors of survival in colon cancer patients.
Int J Cancer. 134:376–383. 2014.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Gamage CD, Park SY, Yang Y, Zhou R, Taş İ,
Bae WK, Kim KK, Shim JH, Kim E, Yoon G and Kim H:
Deoxypodophyllotoxin exerts anti-cancer effects on colorectal
cancer cells through induction of apoptosis and suppression of
tumorigenesis. Int J Mol Sci. 20(2612)2019.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Trachootham D, Alexandre J and Huang P:
Targeting cancer cells by ROS-mediated mechanisms: A radical
therapeutic approach? Nat Rev Drug Dis. 8:579–591. 2009.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Lu H, Li X, Lu Y, Qiu S and Fan Z: ASCT2
(SLC1A5) is an EGFR-associated protein that can be co-targeted by
cetuximab to sensitize cancer cells to ROS-induced apoptosis.
Cancer Lett. 381:23–30. 2016.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Ray PD, Huang BW and Tsuji Y: Reactive
oxygen species (ROS) homeostasis and redox regulation in cellular
signaling. Cell Signal. 24:981–990. 2012.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Dueregger A, Guggenberger F, Barthelmes J,
Stecher G, Schuh M, Intelmann D, Abel G, Haunschild J, Klocker H,
Ramoner R and Sampson N: Attenuation of nucleoside and anti-cancer
nucleoside analog drug uptake in prostate cancer cells by
Cimicifuga racemosa extract BNO-1055. Phytomedicine. 20:1306–1314.
2013.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Al-Abdulla R, Perez-Silva L, Abete L,
Romero MR, Briz O and Marin JJ: Unraveling ‘The Cancer Genome
Atlas’ information on the role of SLC transporters in anticancer
drug uptake. Expert Rev Clin Pharmacol. 12:329–341. 2019.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Mukhopadhya I, Murray GI, Berry S, Thomson
J, Frank B, Gwozdz G, Ekeruche-Makinde J, Shattock R, Kelly C,
Iannelli F, et al: Drug transporter gene expression in human
colorectal tissue and cell lines: Modulation with antiretrovirals
for microbicide optimization. J Antimicrob Chemother. 71:372–386.
2015.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Liu Y, Zuo T, Zhu X, Ahuja N and Fu T:
Differential expression of hENT1 and hENT2 in colon cancer cell
lines. Genet Mol Res 16: doi: 10.4238, 2017.
|
|
57
|
Pastor-Anglada M and Pérez-Torras S:
Emerging roles of nucleoside transporters. Front Pharmacol.
9(606)2018.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Senyavina N and Tonevitskaya S: Effect of
hypoxanthine on functional activity of nucleoside transporters ENT1
and ENT2 in caco-2 polar epithelial intestinal cells. Bull Exp Biol
Med. 160:160–164. 2015.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Cho HD, Lee JH, Moon KD, Park KH, Lee MK
and Seo KI: Auriculasin-induced ROS causes prostate cancer cell
death via induction of apoptosis. Food Chem Toxicol. 111:660–669.
2018.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Pan H, Wang BH, Lv W, Jiang Y and He L:
Esculetin induces apoptosis in human gastric cancer cells through a
cyclophilin D-mediated mitochondrial permeability transition pore
associated with ROS. Chem Biol Interact. 242:51–60. 2015.PubMed/NCBI View Article : Google Scholar
|
|
61
|
García V, Lara-Chica M, Cantarero I,
Sterner O, Calzado MA and Muñoz E: Galiellalactone induces cell
cycle arrest and apoptosis through the ATM/ATR pathway in prostate
cancer cells. Oncotarget. 7:4490–4506. 2016.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Jung SN, Shin DS, Kim HN, Jeon YJ, Yun J,
Lee YJ, Kang JS, Han DC and Kwon BM: Sugiol inhibits STAT3 activity
via regulation of transketolase and ROS-mediated ERK activation in
DU145 prostate carcinoma cells. Biochem Pharmacol. 97:38–50.
2015.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Fruehauf JP and Meyskens FL Jr: Reactive
oxygen species: A breath of life or death? Clin Cancer Res.
13:789–794. 2007.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Pelin M, Fusco L, Martín C, Sosa S,
Frontiñán-Rubio J, González-Domínguez JM, Vázquez E, Prato M and
Tubaro A: Graphene and graphene oxide induce ROS production in
human HaCaT skin keratinocytes: The role of xanthine oxidase and
NADH dehydrogenase. Nanoscale. 10:11820–11830. 2018.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Kim K, Yeo SG and Yoo BC: Identification
of hypoxanthine and phosphoenolpyruvic Acid as serum markers of
chemoradiotherapy response in locally advanced rectal cancer.
Cancer Res Treat. 47:78–89. 2015.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Zhang A, Sun H, Xu H, Qiu S and Wang X:
Cell metabolomics. OMICS. 17:495–501. 2013.PubMed/NCBI View Article : Google Scholar
|