Introduction
As an inflammatory immune disease, ankylosing
spondylitis (AS) is slow in taking effect that progresses over
time, with the global prevalence rate ranging from 0.1-1.4%
(1). AS mainly afflicts the
sacroiliac joints and lumbar vertebrae, leading to fibrosis, spinal
fusion, calcification and hence loss of flexibility (2). Spinal fusion due to chronic
inflammation gradually restricts spinal activity to result in the
partial loss of function, which causes the loss of normal working
ability and substantially debases the life quality of sufferers
with AS (3,4). Previous studies have reported the
alteration of cytokines in AS. Proinflammatory cytokines and tumor
necrosis factor-α (TNF-α) are involved in osteoclast stimulation.
However, the exact mechanism underlying AS remains enigmatic. It is
generally accepted that AS is related to heredity, immunity and
infection. It has been documented that the abnormal change of cell
factors is a significant characteristic of the development of AS
(5,6). Among all interleukins (ILs) secreted
by T helper type 17 (Th17) cells, IL-23 and IL-17 have the
strongest response to Th17 cells (7,8) and
IL-10 are an important anti-inflammatory factor. The Janus kinase
2/signal transducers and activators of transcription 3 (JAK2/STAT3)
signaling pathway is one of the critical pathways in inflammation
and is implicated in the etiology and pathogenesis of AS. The
JAK2/STAT3 signaling pathway induces the expression of TNF-α and
other proinflammatory cytokines (9,10).
JAK1/2/3, tyrosine kinase 2 (TYK2), and STAT are the main factors
for the signal transduction of proinflammatory and
anti-inflammatory cytokines. The JAK2/TYK2-STAT3 signaling pathway
has been identified to participate in IL-23-mediated Th17 cell
regulation (11,12). Therefore, the JAK2/STAT3 signaling
pathway can be regarded as the major promising target of AS
treatment (13).
The synovium is an important structure connecting
bone and joint capsule, which is prone to inflammatory erosion and
hyperplasia in inflammatory diseases (14). Fibroblast-like synoviocytes (FLSs)
decisively participate in the pathophysiology of synovitis in
numerous joint diseases including AS. FLSs can secrete a variety of
inflammatory cytokines, including tumor necrosis factor-α (TNF-α)
(15) and interleukin-6 (IL-6)
(16), to trigger inflammatory
signaling cascades and exacerbate inflammatory responses (16,17),
thus resulting in persistent local inflammation and joint
destruction (18). Peripheral
blood mononuclear cells (PBMCs) mainly include lymphocytes,
monocytes, phagocytes, dendritic cells and a small number of other
cell types. PBMCs are commonly used to simulate the blood immune
environment in vitro. Numerous studies have indicated the
critical role of monocytes and their secreted cytokines in the
pathogenesis of AS (19). In the
present study, AS-PBMCs were co-cultured with AS-FLSs. As a result,
inflammatory factors in AS-PBMCs can promote the inflammation and
proliferation of FLSs and respond in an improved way to the
internal environment of AS patients (20).
Long non-coding RNAs (lncRNAs) act as novel
regulators of gene expression to orchestrate biological processes
including chondrocyte proliferation and death, as well as
inflammation in PBMCs (12).
LncRNAs have attracted increasing attention in the field of AS.
Previously (21), lncRNA
NONHSAT227927.1 was identified by the authors as a key lncRNA
involved in the immune-inflammation of AS through high-throughput
sequencing and bioinformatics analysis. Despite intensive studies
on lncRNAs, their functional and pathological significance remains
largely unknown. In the present study, NONHSAT227927.1 was selected
as the object of the study to detect its expression in PBMCs of AS
patients and its relationship with immune-inflammation and
self-rating depression and anxiety levels of patients. In order to
explore the possible mechanism, AS-FLSs were further utilized to
assess the impact of altered NONHSAT227927.1 on the inflammatory
response.
Materials and methods
Subjects
AS patients and sex- and age-matched healthy
controls (HCs) were recruited from the First Affiliated Hospital of
Anhui University of Traditional Chinese Medicine between March 2021
and May 2021. Patients who did not meet the 1984 American College
of Rheumatology diagnostic criteria (22) with severe mental illness,
significantly impaired liver or renal function, pregnancy, or
immunosuppressants, were excluded. All participants provided
written informed consents. The present study was approved (approval
no. 2015-AH20) by the Medical Ethics Committee of the First
Affiliated Hospital of Anhui University of Traditional Chinese
Medicine (Hefei, China).
Culture of FLSs
Human primary FLSs isolated from sacroiliac joints
(cat. no. RAB-iCell-s004) and AS-FLSs (cat. no. JDBG200752) were
purchased from Subikang iCell Bioscience Inc. (Shanghai, China) and
cultured in RPMI-1640 medium [Saibaikang (Shanghai) Biotechnology
Co., Ltd.] containing 1% penicillin-streptomycin and 10% fetal
bovine serum (Gibco; Thermo Fisher Scientific, Inc.) in a 37˚C
incubator with 5% CO2 and 100% humidity (11). The medium was renewed once every
2-3 days. When the cells were cultured to 80-90% confluence, they
were washed twice with phosphate buffer saline, digested with 0.25%
trypsin, followed by the observation of the digestion under a x200
light microscope. The trypsin was discarded when the cells adhered
to the wall and became loose. Following the addition of the
complete medium, the cell layer was blown.
Culture of PBMCs
The venous blood (4 ml) of AS patients was collected
using an anticoagulant tube and mixed well with 4 ml of PBS. Then,
4 ml Ficoll solution (cat. no. 17-1440-02; GE Healthcare) was added
into a 15-ml centrifuge tube. The diluted blood was added gently
and slowly to the upper layer of Ficoll solution to avoid mixing
the two solutions. The tube was placed in a horizontal centrifuge
for centrifugation at 1,150 x g at 37˚C for 20 min. After
successful centrifugation, the PBMCs were located in the second
white layer from top to bottom. All the cells were moved to a new
centrifuge tube, added with 10-15 ml of PBS, and placed in a
centrifuge for centrifugation at 640 x g at 37˚C for 10 min. After
removing the supernatant, 5-10 ml of PBS was added to repeat the
same procedures. The cells were resuspended by adding 1 ml of PBS,
transferred to a 1.5-ml EP tube, and set aside.
AS-PBMC induction and AS-FLS
transfection
AS-PBMCs and AS-FLSs were seeded and cultured in a
Transwell chamber at a ratio of 3:1. PBMCs were added into the
apical chamber and FLSs were placed in the basolateral chamber. The
chamber was placed in the incubator for 24 h. Upon the cell
confluence reaching 70-90%, cells in each Transwell well were taken
out. Following the operating instructions, According to the
manufacturer's instructions, pcDNA3.1-negative control
(NC)(C11022), pcDNA3.1-NONHSAT227927.1 (C05008), small interfering
RNA (siRNA)-NC (A06001), and siRNA-NONHSAT227927.1 (A01001) (both
Shanghai Jima Pharmaceutical Technology Co., Ltd.) were transfected
into AS-FLSs using Lipofectamine® 2000 (cat. no.
11668-019; Thermo Fisher Scientific, Inc.) and cultured at 37˚C for
24 h. The transfection concentration of siRNA was 50 pmol/ml.
According to the manufacturer's instructions, >5 µg nucleic acid
was used, and then the cells were collected after incubation at
37˚C for 48 h. RT-qPCR was then used to observe cell transfection
efficiency. The oligonucleotide sequences were as follows: siRNA-NC
forward, 5'-UUCUCCGAACGUGUCACGUTT-3' and reverse,
5'-ACGUGACACGUUCGGAGAATT-3'; and siRNA-NONHSAT227927.1 forward,
5'-CGACUGACUCGAUCUUUGAAG-3' and reverse,
5'-UCAAAGAUCGAGUCAGUCGGG-3'.
Reverse transcription quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from AS-FLSs with
TRIzol® (cat. no. 15596026; Thermo Fisher Scientific,
Inc.), Reverse transcription of lncRNA was performed using
Novostart SYBR qPCR SuperMix Plus (cat. no. E096-01B; Novoprotein
Scientific, Inc.) according to the manufacturer's instructions.
PrimeScript™ RT Reagent Kit with gDNA Eraser kit (cat.
no. RR047A; Takara Biotechnology Co., Ltd.) was used according to
the manufacturer's instructions for qPCR (reaction conditions: 95˚C
pre-denaturation for 1 min, 95˚C denaturation for 20 sec and 60˚C
annealing for 1 min, for 40 cycles). Relative quantitative analysis
was performed using the 2-ΔΔCq method (23) with β-actin as an internal
reference. The sequences of primers used were as follows:
NONHSAT227927.1 forward, 5'-TGGGAACTCCTGAGCATACC-3' and reverse,
5'-ATGCTCCAGCAAGTCAGGAT-3'; and β-actin forward,
5'-CCCTGGAGAAGAGCTACGAG-3' and reverse,
5'-GGAAGGAAGGCTGGAAGAGT-3'.
Enzyme-linked immunosorbent assay
(ELISA)
The levels of IL-17 (JYM0082Hu), IL-23 (JYM0077Hu)
and IL-10 (JYM0155Hu) in the serum of patients or the supernatant
of FLSs were evaluated through ELISA according to the
manufacturer's instructions (Wuhan Genomics Technology Co., Ltd.).
Each sample was examined three times.
Cell counting Kit-8 (CCK-8) assay
Cell viability was measured using a CCK-8 assay kit
(BIOSS) following the manufacturer's protocols. At first,
3x104 AS-FLSs were seeded into 96-well plates and
cultured until 70 and 90% confluence. Logarithmically growing cells
were transfected with the aforementioned method with three wells in
each group. Cells were cultured for 0, 12, 24, 48 and 72 h,
respectively. Then, 10 µl CCK-8 solution was supplemented into each
well at each experimental site, followed by 1-4 h of cell culture
at 37˚C. Cell viability was assessed by measuring the optical
density at 450 nm.
Western blot analysis
A total of 600 µl RIPA lysate (cat. no. P0013B;
Beyotime Institute of Biotechnology) was used to extract the total
protein in the cell. SDS-PAGE gel preparation kit (cat. no. S8010;
Beijing Solarbio Science & Technology Co., Ltd.) was used to
prepare the gel (5% stacking gel, 10% separating gel
concentration), and then 30 µg protein per lane was added for
electrophoresis on PVDF membranes. The membranes were blocked with
5% skimmed dry milk for 2 h at room temperature, and then incubated
in anti-phosphorylated (p)-STAT3 (1:500; cat. no. ab76315; Abcam),
p-JAK2 (1:1,000; cat. no. ab32101; Abcam), JAK2 (1:500; cat. no.
ab39636; Abcam) and STAT3 (1:1,000; cat. no. ab68153; Abcam)
overnight with slow shaking at 4˚C. After washing, goat anti-mouse
(cat. no. ZB-2305; IgG; OriGene Technologies, Inc.) and goat
anti-rabbit (cat. no. ZB-2301; IgG; OriGene Technologies, Inc.)
secondary antibodies (labeled with horseradish peroxidase) were
added at a dilution of 1:1x104, and the membrane was
re-probed at room temperature for 2 h. After washing, proteins were
visualized using an ECL luminescence kit (cat. no. 34094; Thermo
Fisher Scientific, Inc.). Image capture was then performed. The
relative expression of target proteins was calculated as the ratio
of absorbance of GAPDH. ImageJ software (ImageJ 180; National
Institutes of Health) was used for band density analysis.
Transwell migration assays
All cells were incubated overnight in serum-free
DMEM, the cell concentration was adjusted to 2x105/ml,
100 µl of cell suspension was added to the Transwell chamber, The
lower cavity of the Transwell contained complete fibroblast medium
(containing 10% FBS and 1% double antibody). The upper Transwell
chamber was 6.5 mm in diameter, with an 8.0-µm pore size PC
membrane, while the lower Transwell chamber was 6.5 mm in diameter,
with a 0.4-µm pore size PC membrane. The culture was continued for
24 h. Cells were fixed with 4% paraformaldehyde for 30 min at room
temperature followed by staining with 0.5% crystal violet for 15-30
min. Cells were rinsed several times with clean water, the chamber
was dried at room temperature, images were captured from randomly
selected fields of view and counted under a x200 light
microscope.
Statistical analysis
GraphPad Prism (version 8.2; Dotmatics) was utilized
for statistical analysis and image rendering. The differences
between groups were analyzed using the paired Student's t-test or
the Kruskal-Wallis non-parametric test, followed by a Steel-Dwass
post-hoc test. Explanatory data were compared using the
χ2 test. Correlation analysis was conducted using the
Pearson's correlation coefficient method. Data are presented as the
mean ± standard deviation. P<0.05 was considered to indicate a
statistically significant difference.
Results
NONHSAT227927.1 expression is
upregulated in the PBMCs of AS patients
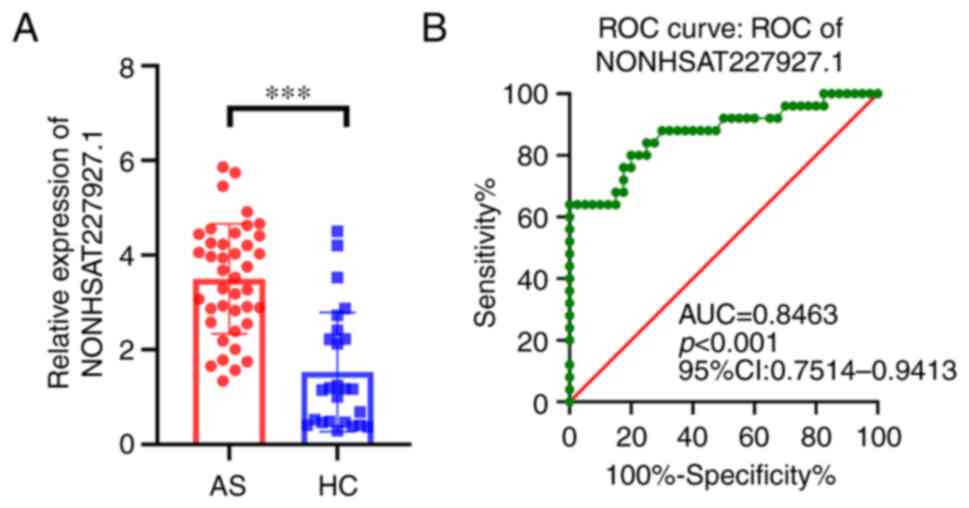
To investigate NONHSAT227927.1 expression in AS,
RT-qPCR was performed in the PBMCs from 50 AS patients and 30 HCs.
The results demonstrated that under different environments,
NONHSAT227927.1 was upregulated in AS (Fig. 1A). Subsequently, the receiver
operating characteristic (ROC) curve analysis was implemented to
evaluate NONHSAT227927.1 diagnostic utility, and the area under the
ROC curve (AUC) was 0.8463 [95% confidence interval (CI):
0.7514-0.9413; P<0.001] (Fig.
1B), suggesting that NONHSAT227927.1 had significant diagnostic
utility.
Changes of clinical
immune-inflammatory index and perception scale of patients with
AS
Compared with normal subjects, there were
substantial increases of C-reactive protein (CRP), erythrocyte
sedimentation rate (ESR), immunoglobin A (IgA), immunoglobin G,
immunoglobin M, self-rating anxiety scale (SAS) scores, self-rating
depression scale scores in AS patients, and visual analog scale
(VAS). (P<0.05; Table I).
 | Table IClinical immune-inflammatory markers
and perception score of patients with AS and NC. Data were
expressed as the n or mean ± standard deviation. |
Table I
Clinical immune-inflammatory markers
and perception score of patients with AS and NC. Data were
expressed as the n or mean ± standard deviation.
|
Characteristics | NC (n=30) | AS (n=50) | P-value |
|---|
| Sex | | | 0.952 |
|
Male | 19 | 32 | |
|
Female | 11 | 18 | |
| Age, years | | 56.50
(47.00-65.50) | <0.01 |
| Course of disease,
years | | 9.30 (4.54) | - |
| CRP, mg/dl | 7.13 (5.90) | 24.28
(12.34-55.56) | <0.01 |
| ESR, mm/h | 6.5
(2.00-30.00) | 39.90 (23.37) | <0.01 |
| IgA, mmol/l | 1.33
(0.63-4.41) | 1.65
(1.32-4.54) | <0.01 |
| IgG, mmol/l | 8.99
(7.97-10.78) | 10.63
(9.00-12.08) | <0.01 |
| IgM, mmol/l | 1.05
(0.70-1.58) | 1.31
(0.93-1.84) | 0.02 |
| C3, g/l | 1.01
(1.00-1.37) | 1.27
(1.13-1.40) | 0.245 |
| C4, g/l | 0.33
(0.25-0.37) | 0.35
(0.25-0.40) | 0.636 |
| SAS | 23.54
(15.32-51.56) | 48.75
(47.50-52.50) | <0.01 |
| SDS | 23.54
(15.04-46.44) | 56.55 (23.63) | <0.01 |
| VAS | 5.81
(4.49-6.84) | 6.80
(5.82-7.28) | <0.01 |
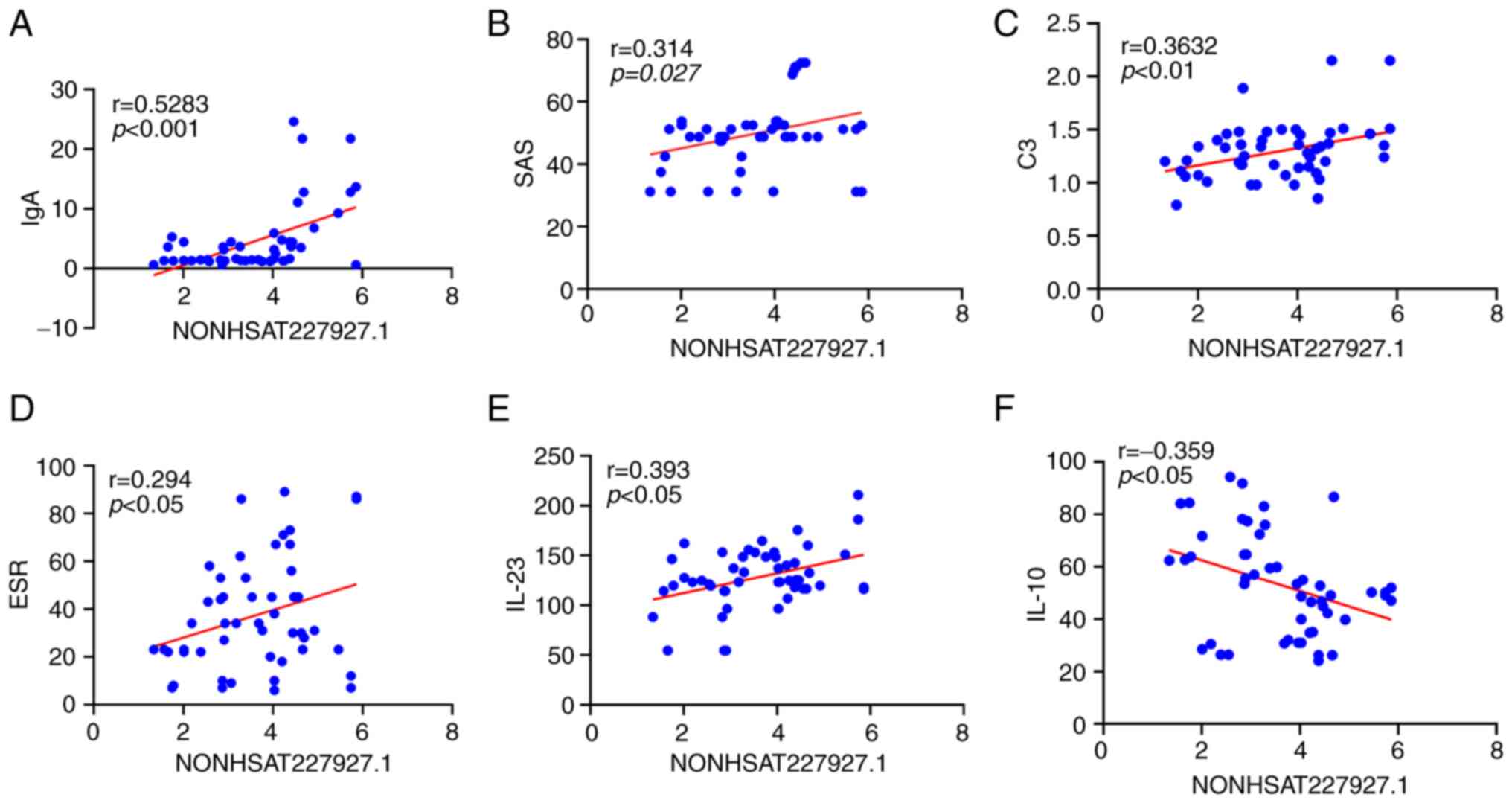
Correlations between NONHSAT227927.1
and clinical indicators in AS patients
The results of correlation analysis manifested the
positive correlation of NONHSAT227927.1 expression with the levels
of IgA, C3, ESR, SAS and IL-23 (P<0.05) and its negative
correlation with IL-10 expression (P<0.05) in AS patients
(Fig. 2A-F).
Association rules analysis between
laboratory indexes and NONHSAT227927.1 in AS patients
Association rules analysis displayed that the
upregulation of NONHSAT227927.1 in patients with AS was strongly
correlated with the elevation in the levels of CRP, VAS, ESR, C3
and IL-17, and the support was >55% and the confidence level was
>23% (Table II).
 | Table IIAssociation rules analysis of
laboratory indexes of NONHSAT227927.1 in patients with ankylosing
spondylitis. |
Table II
Association rules analysis of
laboratory indexes of NONHSAT227927.1 in patients with ankylosing
spondylitis.
| Items (LHS ⇒
RHS) | Confidence (%) | Support (%) |
|---|
| ↑ (NONHSAT227927.1
↑) ⇒ (CRP ↑) | 81.08 | 60 |
| ↑ (NONHSAT227927.1
↑) ⇒ (VAS ↑) | 78.38 | 58 |
| ↑ (NONHSAT227927.1
↑) ⇒ (ESR ↑) | 70.27 | 52 |
| ↑ (NONHSAT227927.1
↑) ⇒ (C3 ↑) | 62.16 | 46 |
| ↑ (NONHSAT227927.1
↑) ⇒ (IL-17 ↑) | 56.76 | 46 |
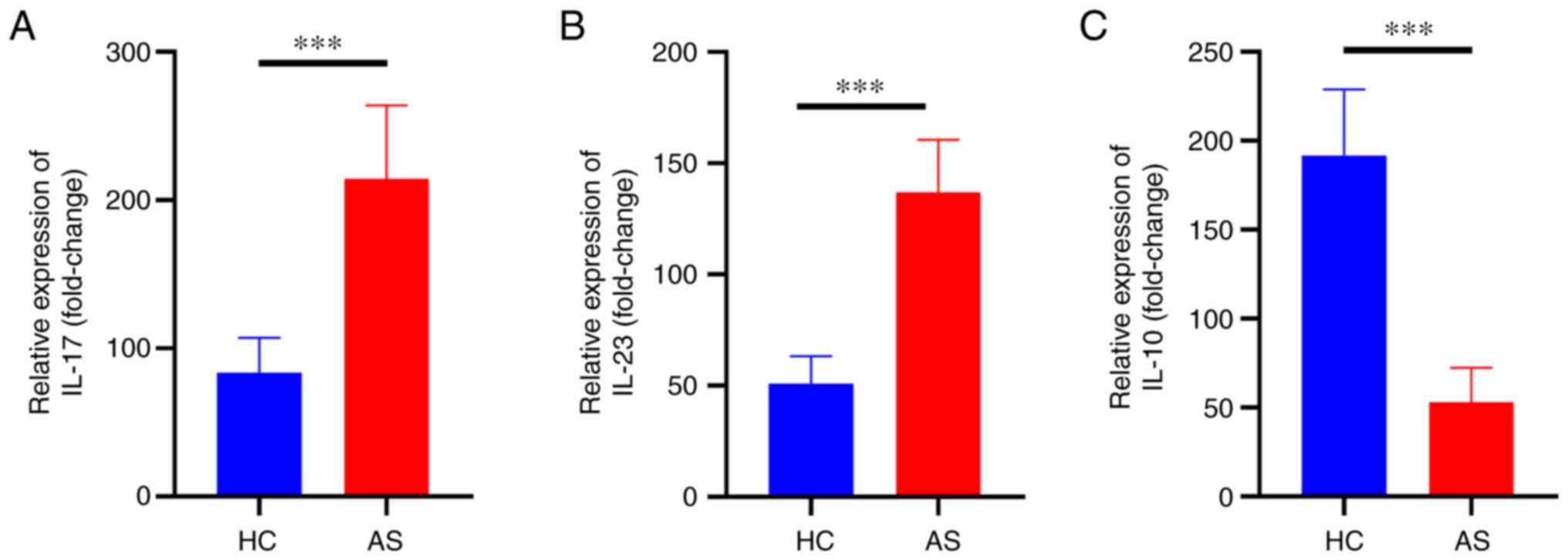
Expression of inflammatory cytokines
in patients with AS
ELISA analysis was performed to detect the level of
inflammation in patients with AS. The levels of IL-23 and IL-17 in
AS patients were significantly increased (P<0.01) compared with
those in the HC, while IL-10 level was significantly reduced
(P<0.01) (Fig. 3A-C).
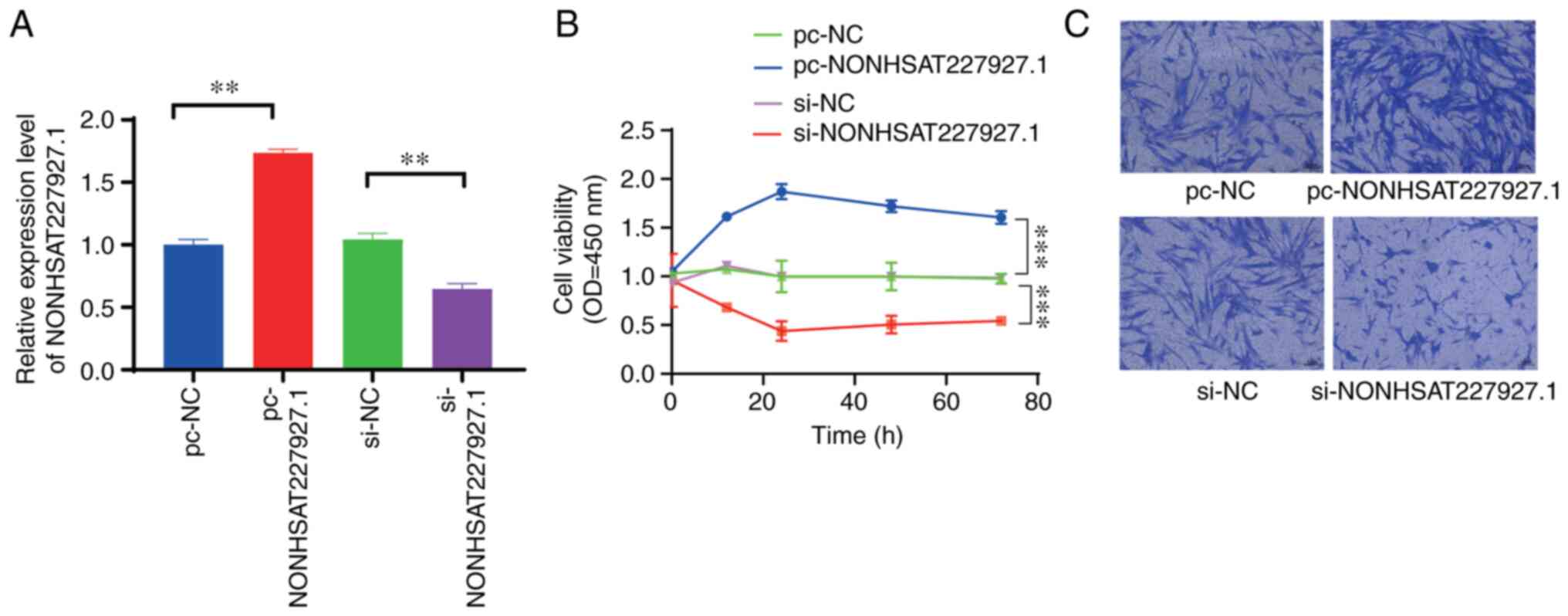
Expression of NONHSAT227927.1 in
AS-FLSs and its effect on cell viability
In order to ascertain whether NONHSAT227927.1 was
successfully transfected and expressed in AS-FLSs, NONHSAT227927.1
mRNA expression was examined by RT-qPCR (Fig. 4A). CCK-8 (Fig. 4B) and migration experiments
(Fig. 4C) were performed to
evaluate the effect of NONHSAT227927.1 on the viability and
migratory abilities of AS-FLS cells, respectively. NONHSAT227927.1
silencing significantly reduced AS-FLS migration and cell viability
(Fig. 4).
The effects of NONHSAT227927.1 on
inflammatory cytokines in AS-FLS
The expression of inflammatory cytokines was
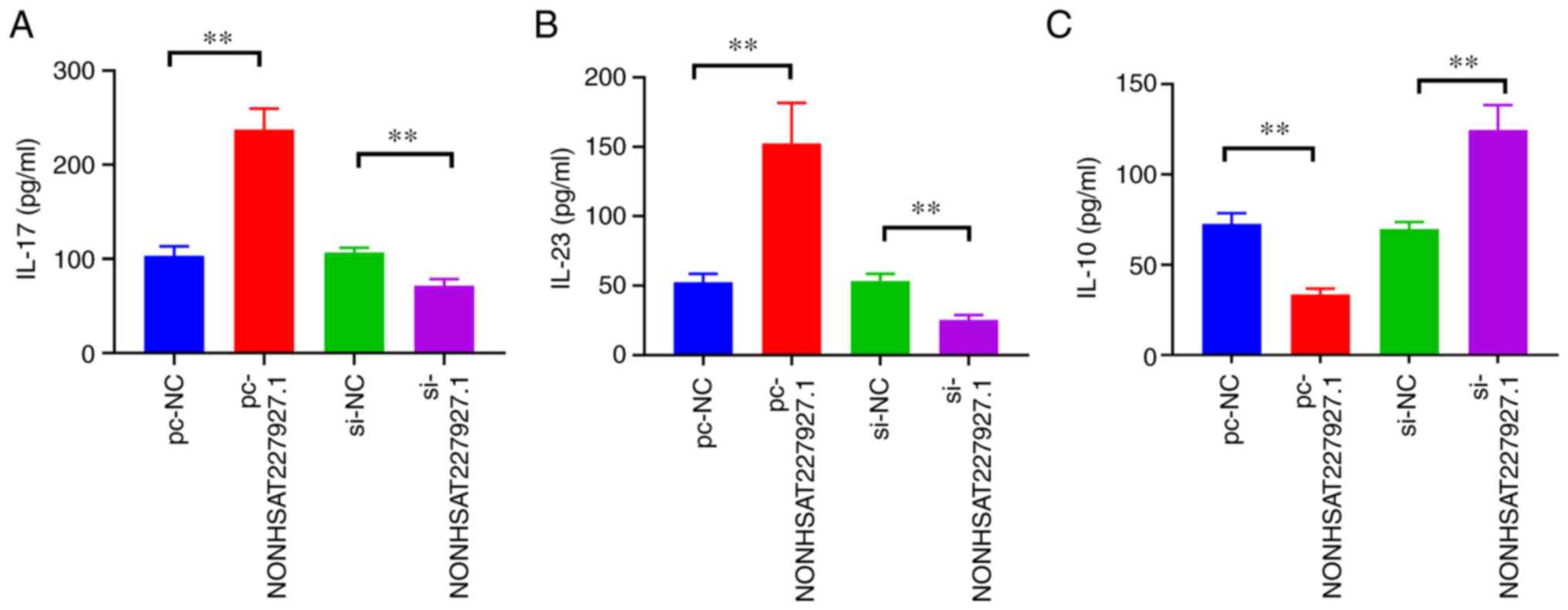
measured by ELISA, which exhibited that NONHSAT227927.1
overexpression augmented the expression of IL-17 and IL-23 but
diminished IL-10 expression, which was contrary after silencing
NONHSAT227927.1 (Fig. 5).
The impacts of NONHSAT227927.1 on the
expression of JAK2, STAT3, P-JAK2, and P-STAT3 in AS-PBMC-induced
AS-FLSs
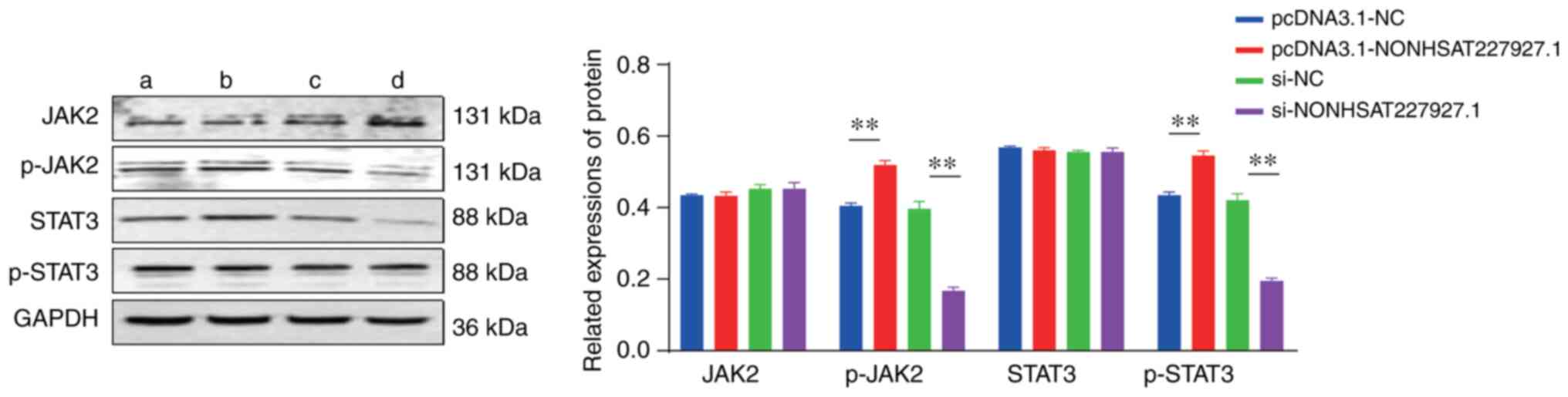
In order to identify whether the JAK2/STAT3
signaling pathway is involved in NONHSAT227927.1-manipulated cell
inflammation in AS, western blot analysis was performed. The
results demonstrated that the overexpression of NONHSAT227927.1
elevated the phosphorylation of JAK2 and STAT proteins in AS-FLSs
(Fig. 6).
Discussion
AS facilitates bone deterioration and aberrant bone
density due to inflammation and the pathogenesis of the disease
(24). Mechanical, inflammatory
and metabolic factors are non-negligible (25). Inflammatory cells including
macrophages and lymphocytes penetrate the joints, which contributes
to fibrosis and synovial thickening, thus the joints and bones
stiffening (26,27). RANKL signaling leads to disease
progression by promoting osteoclast formation (28,29).
Genetic factors have an obvious causal relationship, and it is
crucial to dissect out the underlying mechanism and the way leading
to disease from a genetic perspective (30). To expand the number of the research
records, lncRNAs participate in the orchestration of inflammatory
reactions and cell processes and are aberrantly expressed in a
range of immune-mediated diseases (31). However, the meaningful
understanding of the function of lncRNAs can only be realized
through relevant in-depth researches (32).
Initially, the current research provided evidence of
obvious high expression of NONHSAT227927.1 in PBMCs, PBMC-induced
AS-FLSs, and AS patients, and ROC curve analysis elucidated that
the AUC value was 0.8463, illustrating that NONHSAT227927.1 had
great diagnostic utility. There is an imbalance between
inflammatory cytokines in the serum of patients with AS. IL-17 and
IL-23 increased prominently, and IL-10 decreased noticeably. In
addition, clinical trials unraveled that NONHSAT227927.1 expression
shared a positive correlation with IL-23 expression and had an
inverse correlation with IL-10 expression. In order to further
ascertain the influences of NONHSAT227927.1 on AS-FLS viability and
the JAK2/STAT3 signaling pathway, AS-PBMCs were utilized to
stimulate AS-FLSs as a model to implement in vitro cell
experiments. CCK-8 was applied to assess the impacts of
NONHSAT227927.1 on the function of AS-FLSs. The results showed that
after transfection of NONHSAT227927.1 overexpression plasmid, the
activity and migration ability of AS-FLS were enhanced. By
contrast, AS-FLS viability and migration ability were reduced after
NONHSAT227927.1 silencing. Under the condition of the
overexpression or silencing of NONHSAT227927.1, the inflammatory
cytokines were further measured using ELISA. The results manifested
that the overexpression of NONHSAT227927.1 conspicuously increased
the levels of IL-17 and IL-23 and decreased IL-10 level. On the
contrary, the silencing of NONHSAT227927.1 exhibited the opposite
trends. These results suggested that NONHSAT227927.1 may be the key
lncRNA in the progress of AS.
However, there are also a large number of studies
elaborating the potential relevance and mechanism of lncRNAs at the
level of AS-FLSs. LncRNAs are mainly involved in the inflammatory
reaction and osteogenic differentiation of AS through the sponge
microRNA (33). They primarily
participate in the manipulation of gene expression after gene
transcription and post-transcription, and also are implicated in
the progression of AS. Li et al (34) found that MEG3 was downregulated in
patients with AS, and Wang et al (21) observed that lncRNA H19 was also
related to the pathogenesis of inflammatory diseases. The data
obtained by Gong et al (35) documented that lncRNAs (326C3.7 and
122K13.12) elevated considerably in AS patients with osseous
bridges. Inflammatory factors afflict the severity and potential
course of AS (36). A previous
study has unveiled that in patients with AS at the active stage,
the high level of inflammation (ESR and CRP) is positively
correlated with the percentage of regulatory T cells (Tregs) in
PMBCs (37). Another research
demonstrated that young AS patients had a reduced Vd Tregs ratio in
their PBMCs, which suppressed naive CD4 T cell proliferation and
the secretion of produced interferon-g through lowering IL-10 level
(38). The pathophysiology of AS
has been proven to involve an aberrant inflammatory cytokine
pathway (39). Stem cell-based
bone tissue regeneration has shown promising therapeutic potential
in AS (40). Besides, it has been
reported that JAK is the major sensor in Th17 cells (41,42).
Th17 cells assume critical roles in AS, which secrete a variety of
inflammatory mediators, including IL-17 to 22, IL-6, IL-26,
interferon, Gamma and TNF-α (43).
Additionally, the shaft of activating IL-23 and IL-17 cytokines has
been pointed out (44). Melis
et al (45) demonstrated
that AS patients possessed increased serum IL-23 level. The
combination of the JAK2/STAT3 signaling pathway and IL-23 has been
documented to stimulate IL-23R (13). The inhibition of the JAK2/STAT3
signaling pathway is hypothesized to be facilitated by IL-23. The
JAK/STAT signaling pathway plays a crucial role in growth factor
and cytokine-activated signal transduction (46).
In summary, the present data provided the notion
that NONHSAT227927.1 expression was high in AS, and that its
abnormal expression could modulate the viability of AS-FLSs and
mediate inflammatory response through the JAK2/STAT3 signaling
pathway, thus influencing the occurrence and development of AS.
Therefore, NONHSAT227927.1 may function as a biomarker for AS and
provide new insights into the pathogenesis of AS.
Acknowledgements
Not applicable.
Funding
Funding: The present study was approved by the 2021 Key Project
of Natural Science Research in Anhui Universities (grant no.
KJ2021A0558) and the 2021 Open Fund of Anhui Provincial Key
Laboratory of Applied Basic and Development Research in Modern
Traditional Chinese Medicine (grant no. 2021AKLMCM002).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XD and JL were both involved in the research design.
XD participated in the data analysis, authored the first draft and
made revisions to the paper. YQS participated in the collection and
analysis of experimental data. JL and YQS supervised the project
and contributed to the manuscript revision. JL and YQS confirm the
authenticity of all the raw data.
Ethics approval and consent to
participate
The present study was approved (approval no.
2015-AH20) by the Medical Ethics Committee of the First Affiliated
Hospital of Anhui University of Traditional Chinese Medicine
(Hefei, China). All procedures in the present study were conducted
in accordance to the approved protocols of the Medical Ethics
Committee of the First Affiliated Hospital of Anhui University of
Traditional Chinese Medicine.
Patient consent for publication
Written informed consent was obtained from the
patients for their anonymized information to be published in this
article.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Braun J and Sieper J: Ankylosing
spondylitis. Lancet. 369:1379–1390. 2007.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Zhu W, He X, Cheng K, Zhang L, Chen D,
Wang X, Qiu G, Cao X and Weng X: Ankylosing spondylitis: Etiology,
pathogenesis, and treatments. Bone Res. 7(22)2019.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Smith JA: Update on ankylosing
spondylitis: Current concepts in pathogenesis. Curr Allergy Asthma
Rep. 15(489)2015.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Vanaki N, Aslani S, Jamshidi A and
Mahmoudi M: Role of innate immune system in the pathogenesis of
ankylosing spondylitis. Biomed Pharmacother. 105:130–143.
2018.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Gracey E, Qaiyum Z, Almaghlouth I, Lawson
D, Karki S, Avvaru N, Zhang Z, Yao Y, Ranganathan V, Baglaenko Y
and Inman RD: IL-7 primes IL-17 in mucosal-associated invariant T
(MAIT) cells, which contribute to the Th17-axis in ankylosing
spondylitis. Ann Rheum Dis. 75:2124–2132. 2016.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Ranganathan V, Gracey E, Brown MA, Inman
RD and Haroon N: Pathogenesis of ankylosing spondylitis-recent
advances and future directions. Nat Rev Rheumatol. 13:359–367.
2017.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Moschen AR, Tilg H and Raine T: IL-12,
IL-23 and IL-17 in IBD: immunobiology and therapeutic targeting.
Nat Rev Gastroenterol Hepatol. 16:185–196. 2019.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Gaffen SL, Jain R, Garg AV and Cua DJ: The
IL-23-IL-17 immune axis: From mechanisms to therapeutic testing.
Nat Rev Immunol. 14:585–600. 2014.PubMed/NCBI View
Article : Google Scholar
|
|
9
|
Barakat W, Safwet N, El-Maraghy NN and
Zakaria MN: Candesartan and glycyrrhizin ameliorate ischemic brain
damage through downregulation of the TLR signaling cascade. Eur J
Pharmacol. 724:43–50. 2014.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Williams AJ, Dave JR and Tortella FC:
Neuroprotection with the proteasome inhibitor MLN519 in focal
ischemic brain injury: Relation to nuclear factor kappaB
(NF-kappaB), inflammatory gene expression, and leukocyte
infiltration. Neurochem Int. 49:106–112. 2006.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Ghoreschi K, Laurence A and O'Shea JJ:
Janus kinases in immune cell signaling. Immunol Rev. 228:273–287.
2009.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Feng X, Yang Q, Wang C, Tong W and Xu W:
Punicalagin exerts protective effects against ankylosing
spondylitis by regulating NF-κB-TH17/JAK2/STAT3 signaling and
oxidative stress. Biomed Res Int. 2020(4918239)2020.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Parham C, Chirica M, Timans J, Vaisberg E,
Travis M, Cheung J, Pflanz S, Zhang R, Singh KP, Vega F, et al: A
receptor for the heterodimeric cytokine IL-23 is composed of
IL-12Rbeta1 and a novel cytokine receptor subunit, IL-23R. J
Immunol. 168:5699–5708. 2002.PubMed/NCBI View Article : Google Scholar
|
|
14
|
MacFarlane LA, Arant KR, Kostic AM, Mass
H, Jones MH, Collins JE, Losina E and Katz JN: Identifying
inflammation in knee osteoarthritis: relationship of synovial fluid
white blood cell count to effusion-synovitis on magnetic resonance
imaging. Arthritis Care Res (Hoboken): Oct 17, 2022 (Epub ahead of
print).
|
|
15
|
Liu L, Chen H, Jiang T and He D:
MicroRNA-106b overexpression suppresses synovial inflammation and
alleviates synovial damage in patients with rheumatoid arthritis.
Mod Rheumatol. 32:1054–1063. 2022.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Garrido-Mesa J and Brown MA: T cell
repertoire profiling and the mechanism by which HLA-B27 causes
ankylosing spondylitis. Curr Rheumatol Rep. 24:398–410.
2022.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Shah NG, Keraliya A, Nunez DB, Schoenfeld
A, Harris MB, Bono CM and Khurana B: Injuries to the rigid spine:
What the spine surgeon wants to know. Radiographics. 39:449–466.
2019.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Gouveia EB, Elmann D and Morales MS:
Ankylosing spondylitis and uveitis: Overview. Rev Bras Reumatol.
52:742–756. 2012.PubMed/NCBI
|
|
19
|
Wang M, Wang L, Zhang X, Yang X, Li X, Xia
Q, Chen M, Han R, Liu R, Xu S and Pan F: Overexpression of miR-31
in peripheral blood mononuclear cells (PBMC) from patients with
ankylosing spondylitis. Med Sci Monit. 23:5488–5494.
2017.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Tang Y, Wang B, Sun X, Li H, Ouyang X, Wei
J, Dai B, Zhang Y and Li X: Rheumatoid arthritis fibroblast-like
synoviocytes co-cultured with PBMC increased peripheral
CD4+ CXCR5+ ICOS+ T cell numbers.
Clin Exp Immunol. 190:384–393. 2017.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Wang JX, Jing FY, Xu YC, Zong HX, Chu YR,
Wang C, Chen KM, Tong WQ, Wang XL and Xu SQ: The potential
regulatory mechanism of lncRNA 122K13.12 and lncRNA 326C3.7 in
ankylosing spondylitis. Front Mol Biosci. 8(745441)2021.PubMed/NCBI View Article : Google Scholar
|
|
22
|
van der Linden S, Valkenburg HA and Cats
A: Evaluation of diagnostic criteria for ankylosing spondylitis. A
proposal for modification of the New York criteria. Arthritis
Rheum. 27:361–368. 1984.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Shiau MY, Lo MK, Chang CP, Yang TP, Ho KT
and Chang YH: Association of tumour necrosis factor alpha promoter
polymorphisms with ankylosing spondylitis in Taiwan. Ann Rheum Dis.
66:562–563. 2007.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Hu Y, Chen X, Wang S, Jing Y and Su J:
Subchondral bone microenvironment in osteoarthritis and pain. Bone
Res. 9(20)2021.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Tam LS, Chan KY and Li EK: The influence
of illness and variables associated with functional limitations in
Chinese patients with ankylosing spondylitis. J Rheumatol.
34:1032–1039. 2007.PubMed/NCBI
|
|
27
|
Gu X, Wu H and Fu P: Allicin attenuates
inflammation and suppresses HLA-B27 protein expression in
ankylosing spondylitis mice. Biomed Res Int.
2013(171573)2013.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Hu Y, Li X, Zhi X, Cong W, Huang B, Chen
H, Wang Y, Li Y, Wang L, Fang C, et al: RANKL from bone marrow
adipose lineage cells promotes osteoclast formation and bone loss.
EMBO Rep. 22(e52481)2021.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Chen X, Zhi X, Wang J and Su J: RANKL
signaling in bone marrow mesenchymal stem cells negatively
regulates osteoblastic bone formation. Bone Res.
6(34)2018.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Chen XX, Baum W, Dwyer D, Stock M, Schwabe
K, Ke HZ, Stolina M, Schett G and Bozec A: Sclerostin inhibition
reverses systemic, periarticular and local bone loss in arthritis.
Ann Rheum Dis. 72:1732–1736. 2013.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Aune TM and Spurlock CF III: Long
non-coding RNAs in innate and adaptive immunity. Virus Res.
212:146–160. 2016.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Kopp F and Mendell JT: Functional
classification and experimental dissection of long noncoding RNAs.
Cell. 172:393–407. 2018.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Lan X, Ma H, Zhang Z, Ye D, Min J, Cai F
and Luo J: Downregulation of lncRNA TUG1 is involved in ankylosing
spondylitis and is related to disease activity and course of
treatment. Biosci Trends. 12:389–394. 2018.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Li Y, Zhang S, Zhang C and Wang M: LncRNA
MEG3 inhibits the inflammatory response of ankylosing spondylitis
by targeting miR-146a. Mol Cell Biochem. 466:17–24. 2020.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Gong ZM, Tang ZY and Sun XL: LncRNA PRNCR1
regulates CXCR4 expression to affect osteogenic differentiation and
contribute to osteolysis after hip replacement. Gene. 673:251–261.
2018.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Wielińska J, Świerkot J, Kolossa K, Bugaj
B, Chaszczewska-Markowska M, Jeka S and Bogunia-Kubik K:
Polymorphisms within genes coding for IL-17A and F and their
receptor as clinical hallmarks in ankylosing spondylitis. Mediators
Inflamm. 2021(3125922)2021.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Liao HT, Lin YF, Tsai CY and Chou CT:
Regulatory T cells in ankylosing spondylitis and the response after
adalimumab treatment. Joint Bone Spine. 82:423–427. 2015.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Wang H, Sun N, Li K, Tian J and Li J:
Assay of peripheral regulatory Vδ1 T cells in ankylosing
spondylitis and its significance. Med Sci Monit. 22:3163–3168.
2016.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Hreggvidsdottir HS, Noordenbos T and
Baeten DL: Inflammatory pathways in spondyloarthritis. Mol Immunol.
57:28–37. 2014.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Arora D and Robey PG: Recent updates on
the biological basis of heterogeneity in bone marrow stromal
cells/skeletal stem cells. Biomater Transl. 3:3–16. 2022.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Hammitzsch A, Chen L, de Wit J, Al-Mossawi
MH, Ridley A, Sekine T, Simone D, Doig K, Skapenko A and Bowness P:
Inhibiting ex-vivo Th17 responses in ankylosing spondylitis by
targeting Janus kinases. Sci Rep. 8(15645)2018.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Klasen C, Meyer A, Wittekind PS, Waqué I,
Nabhani S and Kofler DM: Prostaglandin receptor EP4 expression by
Th17 cells is associated with high disease activity in ankylosing
spondylitis. Arthritis Res Ther. 21(159)2019.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Dong C: TH17 cells in development: An
updated view of their molecular identity and genetic programming.
Nat Rev Immunol. 8:337–348. 2008.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Raychaudhuri SP and Raychaudhuri SK:
IL-23/IL-17 axis in spondyloarthritis-bench to bedside. Clin
Rheumatol. 35:1437–1441. 2016.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Melis L, Vandooren B, Kruithof E, Jacques
P, De Vos M, Mielants H, Verbruggen G, De Keyser F and Elewaut D:
Systemic levels of IL-23 are strongly associated with disease
activity in rheumatoid arthritis but not spondyloarthritis. Ann
Rheum Dis. 69:618–623. 2010.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Raychaudhuri SK and Raychaudhuri SP: Janus
kinase/signal transducer and activator of transcription pathways in
spondyloarthritis. Curr Opin Rheumatol. 29:311–316. 2017.PubMed/NCBI View Article : Google Scholar
|