Introduction
The COVID-19 pandemic continues to challenge current
medical practice and scientific research. Bacterial infections with
antimicrobial-resistant pathogens had caused therapeutic challenges
long before the current pandemic broke out. Empirical
overprescription of antibiotics worldwide at the beginning of the
COVID-19 pandemic has increased the burden of this issue,
increasing the risk of bacterial superinfection during
hospitalization and the risk of antimicrobial resistance (1,2).
The future waves of COVID 19 infections will
probably not overload health systems, but will continue to raise
serious problems in patients with severe forms of the disease. The
previous experience in managing superinfections with MDR bacteria
in COVID-19 patients will be valuable for outcomes in patients.
Patients vulnerable to viral lung infections are at
significant risk of developing multidrug-resistant (MDR) bacterial
infections (3). Bacterial
superinfections in COVID 19 patients have not been well studied yet
and are considered a significant risk factor for COVID 19 mortality
(4,5).
A recent Italian study suggests that pulmonary
superinfections with MDR bacteria might negatively impact the
outcome in COVID 19 patients (6).
Bacterial superinfections occur at least 48 h after
hospital admission and are most often caused by hospital-acquired
MDR bacteria (7). Patients with
severe COVID-19 require extended hospitalization in intensive care
units and are frequently treated with empiric, broad-spectrum
antibiotics, which increases the risk of MDR bacteria selection
(8). COVID-19 patients have a
broad spectrum of clinical manifestations, which may change with
emerging strains. Consequently, biological parameters are critical
predictors of adverse outcomes, having a significant role in
selecting the cases to be closely monitored. A real challenge for
the clinician is to discern between COVID-19 severe inflammatory
reaction and bacterial secondary infection (9).
Procalcitonin (PCT) is a glycoprotein synthesized in
the C cells of the thyroid gland. In case of a microbial invasion,
PCT is released from all parenchymal tissues (10). Serum PCT has normal values in
patients with non-complicated COVID-19. A significant increase in
the PCT serum levels suggests a non-viral coinfection or a
development of a severe form of COVID-19 disease (11). It is unclear whether PCT can be
used as a marker of bacterial coinfection for COVID-19 patients in
the same way it has been used in other respiratory diseases
(12).
The present study aimed to evaluate the etiological
spectrum of bacterial superinfection in adult patients with
COVID-19 and to investigate the correlation between superinfection
with MDR bacteria and serum PCT.
Materials and methods
Data source
This retrospective observational study of bacterial
superinfection in adult patients with confirmed COVID-19 was
conducted in the Clinical Analysis Laboratory of the Arad County
Hospital in Romania.
Definitions
A COVID-19 confirmed case was defined by a positive
result for SARS-CoV-2 virus at admission, based on a reverse
transcription-polymerase-chain-reaction (RT-PCR) test on a
nasopharyngeal swab.
The cases with superinfection were carefully
selected according to the following definition: The identification
of a likely pathogen 48 h after admission (≥48 h) in a clinical
sample taken for a diagnostic purpose. Coinfection was defined as
identifying a pathogen within the first 24 h of hospital admission
(7).
MDR bacteria are non-susceptible to at least one
antimicrobial agent in three or more different antibiotic classes,
except for intrinsic resistance.
The definition of a severe case was meeting one of
the two criteria: The need for intensive care admission or assisted
respiration (invasive ventilation or non-invasive oxygen support)
in an intensive care unit (ICU). The non-severe cases were the
patients admitted in non-ICUs, with oxygen saturation ≥94% on room
air and no need for oxygen support.
Study design and population
The present single-center retrospective study
included adult patients aged >18 years, diagnosed with COVID-19
and association bacterial infection hospitalized in Arad County
Hospital between 01.04.2020 and 01.06.2021. Arad County Hospital
serves ~470,000 individuals, which is a representative sample of
the Romanian population. The period from which the cases were
studied included the entire second pandemic wave in Romania and an
optimal number of COVID-19 patients was provided in a short
time.
Eligible patients presented COVID-19 infection at
admission, an associated bacterial infection and available serum
PCT within 24 h before and after the patients suspected of
bacterial infection were taken samples for cultures.
Exclusion criteria were: Serum PCT determined
outside the pre-set range of time, Clostridium difficile
infection during hospitalization, surgical patients and presence of
coinfections. Patients diagnosed with Clostridium difficile
infection were excluded due to their particular progression and
need for specific antibiotic treatment.
The present study evaluated the etiological spectrum
of superinfection and observed the relation between superinfection
with MDR bacteria and serum PCT. Also, an analysis was performed to
identify the time from hospital admission to the diagnosis of
superinfection. Based on these, the present study classified
infections into early (3-7 days) and late (>7 days)
infections.
Data collection
Electronic medical records of all SARS-CoV-2
positive patients (including both sexes and all age groups) were
searched between 01.04.2020 and 01.06.2021. Personal data was
removed. Patients who met the criteria of bacterial coinfection or
superinfection were carefully selected. Fields collected were:
Demographics (age, sex); hospital admission details (date of
admission, number of days to the bacterial superinfection
diagnosis); microbiological results; inflammatory biomarkers; the
presence of underlying medical conditions; and COVID 19 vaccination
status.
The present study reviewed the microbiological
results (bacteria identified in cultures and antibiogram), time
(days) from admission to superinfection and serum PCT as an
inflammatory biomarker.
Microbiological results were: Standard positive
culture from sputum, urine, tracheal aspirate, bronchoalveolar
lavage and surgical wounds. Culture results were excluded if they
were considered to represent colonization or contamination. The
cultures were performed on VITEK 2C (bioMérieux), an automatic
analyzer for bacterial identification and antibiogram. The present
study used Vitek 2 GP cards (cat. no. 21342 (bioMérieux) and Vitek
2 AST P592 cards cat. no. 22287; bioMérieux) to identify
Gram-positive cocci, respectively, for their antibiograms.
Gram-negative bacilli were identified with Vitek 2 GN cards (cat.
no. 21341; bioMérieux). The present study tested the susceptibility
of the bacilli to antibiotics using Vitek 2 AST N233 cards (cat.
no. 413117; bioMérieux) and Vitek 2 AST XN05 (cat. no. 413230;
bioMérieux).
At admission, the RT-PCR specimen was collected for
all patients on a nasopharyngeal swab using SARS-CoV-2 automated
analysis on the DLITE System (cat. no CTSC820001PCn; Certest
Biotec) according to the manufacturer's protocol.
Serum PCT was measured by Electro-Chemiluminescence
immunoassay kits (cat. no. 09318712190; Roche Diagnostics GmbH) on
the Cobas e 601 analyzer (Roche Diagnostics GmbH) using the
manufacturer's recommendations. The results were expressed as
nanogram of PCT per microliter of serum (ng/ml). The normal ranges
were considered to be between 0-0.5 ng/ml. Serum PCT values were
analyzed in all studied patients within 24 h before and after
taking samples for cultures. Patients with serum PCT determined
outside this time range were excluded from the study.
Statistical analysis
Statistical analyses were performed using MedCalc
v20.015 (MedCalc Software Ltd). The proportion of bacterial
pathogens was recorded. The association between categorical data
was performed by Chi-square with Fisher's exact test. The
distribution of numerical results was calculated using the
Shapiro-Wilk normality test. Normally distributed data were
presented as mean ± standard deviation. Data following a
nonparametric distribution were presented as the median and
interquartile range (IQR) and the comparisons between two
independent groups were performed with the Mann-Whitney U test.
P<0.05 was considered to indicate a statistically significant
difference.
Ethics
The study was carried out in accordance with the
declaration of Helsinki. It was approved by the Ethics Commission
for Clinical Trials of Arad County Hospital (approval no.
37/25.02.2021) and of Scientific Research Ethics Commission of
Vasile Goldis, University of Arad (approval no. 10/29.03.2021).
Results
Superinfection profile
Overall, during the present study research period,
165 eligible patients hospitalized for COVID-19 with bacterial
infection association were documented. Of these patients, 52
infections were identified in surgical patients, which were
excluded due to the specificity of the germs related to the
surgical wounds.
Another 113 infections were found in non-surgical
patients, with 31 (27.4%) cases of coinfections and 82 cases of
superinfection. Former patients met inclusion criteria and
represented the study population (n=82 cases).
In these included patients, 66 (80.49%)
superinfections occurred in severe cases, patients admitted in ICUs
and 16 (19.51%) superinfections occurred in non-severe cases,
patients admitted in non-ICUs-COVID-19 medical departments
(Fig. 1).
The mean age of the study population was 65.51±11.23
years. A total of 49 (59.75%) patients were male. Of the 82
patients included, only five were COVID-19 vaccinated, three with
Pfizer BioNTech COVID-19 vaccine and two with AstraZeneca COVID-19
vaccine.
The main underlying medical conditions found were
hypertension (63.4% of patients), obesity (48.8% of patients),
diabetes mellitus (39% of patients, chronic lung disease (36.6% of
patients) and cardiovascular disease (32.9% of patients).
In the period after 48 h from hospital admission to
discharge, 93 bacterial infections were identified in the study
population (n=82 patients), 32 (34.40%) potential pathogens from
days 3-7 and 61 (65.59%) potential pathogens from day eight
onwards. Polymicrobial infections were found in 11 patients
(13.41%).
The median time from hospital admission to the
diagnosis of superinfection was 11 days (IQR 6-17).
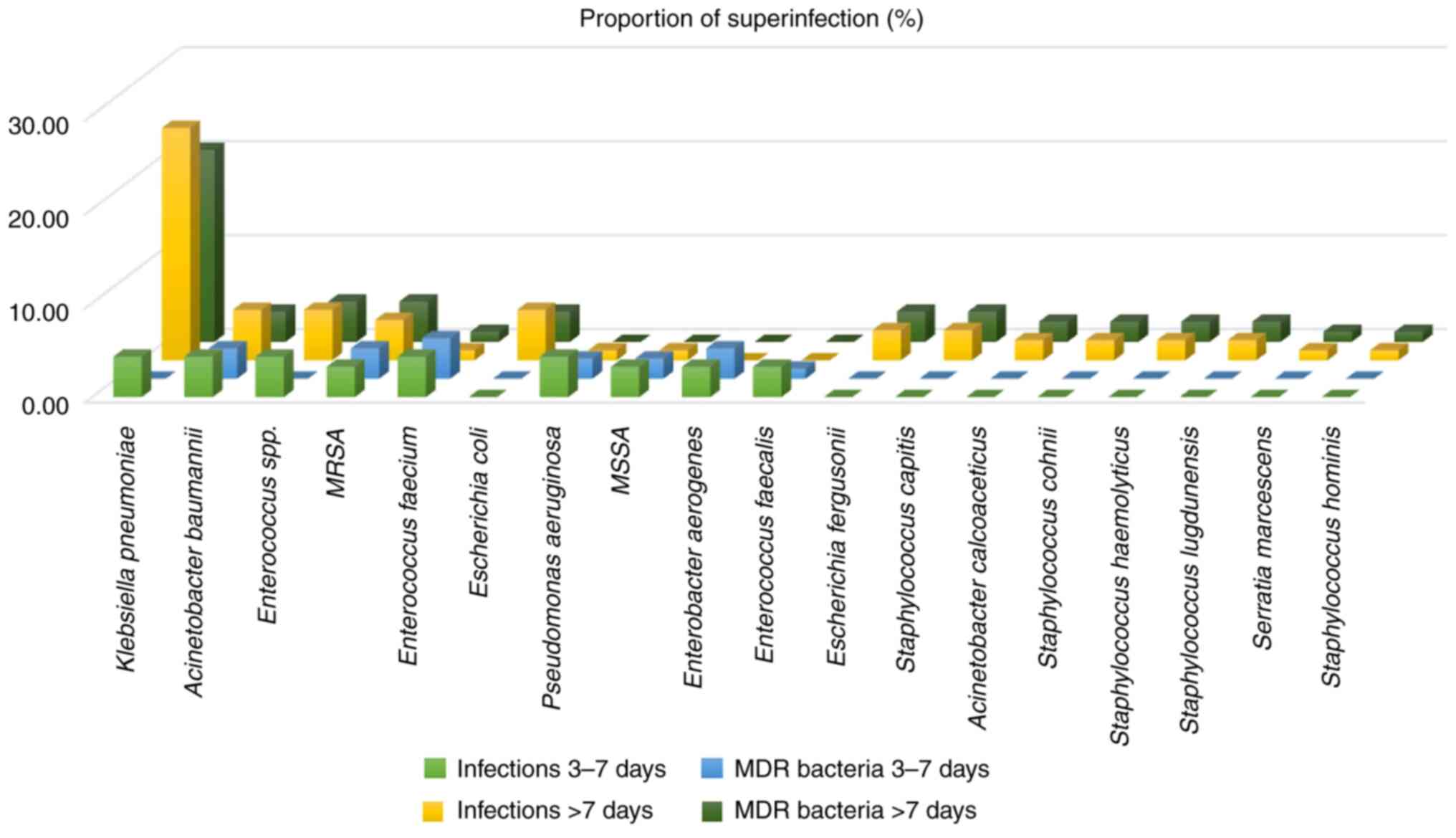
A total of 18 types of bacterial pathogens were
identified. The most frequently isolated microorganisms were
Klebsiella pneumoniae (29.03% of superinfections),
Acinetobacter baumannii (9.68% of superinfections) and
Enterococcus spp. (9.68% of superinfections). Klebsiella
pneumoniae was detected early (days 3-7) in four (4.30%)
superinfections and late (>7 days) in 23 (24.73%)
superinfections. Enterobacter aerogenes and Enterococcus
faecalis were detected only on days 3-7 (early detection).
Escherichia coli, Escherichia fergusonii,
Acinetobacter calcoaceticus, Staphylococcus capitis,
Staphylococcus cohnii, Staphylococcus haemolyticus,
Staphylococcus lugdunensis, Staphylococcus hominis
and Serratia marcescens were detected only after 7 days
(late detection). Methicillin-resistant Staphylococcus
aureus (MRSA), Methicillin-susceptible Staphylococcus
aureus (MSSA), Pseudomonas aeruginosa and
Enterococcus faecium were identified both in early and late
detection period.
MDR bacteria profile. Documented MDR bacteria
superinfections were found in 60 (73.17%) patients (Fig. 2). Of MDR bacteria superinfections,
73.52% occurred in the late infection period. The most frequently
identified MDR bacteria on days 3-7 were Enterococcus
faecium, MRSA, Acinetobacter baumannii and
Enterobacter aerogenes in a proportion between 3.22 and
4.30%. Klebsiella pneumoniae, Enterococcus spp. and
MRSA were the most common MDR bacteria identified in late
infections after hospitalization in 20.43, 4.30 and 4.30% of all
infections, respectively. Klebsiella pneumoniae and
Enterococcus spp. were associated with antibiotic resistance
only in late infections.
No antibiotic resistance was observed in late
infections with MSSA, Pseudomonas aeruginosa,
Enterobacter aerogenes and Enterococcus faecalis.
There were no significant differences between sex
and the presence of MDR bacteria (P=0.448). No association was
found between MDR bacteria superinfections and age (P=0.565).
Site of infections. The site of infections
according to the etiology is reported in Table I; there were 41 (44.09%)
hospital-acquired pneumonia, 29 (31.18%) ventilator-associated
pneumonia, nine bacteremia (9.68%), eight (8.60%) urinary tract
infection and six (6.45%) acute bacterial skin and skin structure
infection.
 | Table IEtiology by the site of
superinfections. |
Table I
Etiology by the site of
superinfections.
| Site of
superinfections | Bacteria | Total |
|---|
| HAP | | 41 (44.09%) |
| | Klebsiella
pneumoniae | 13 |
| | Acinetobacter
baumannii | 7 |
| | Enterococcus
spp. | 6 |
| | Escherichia
coli | 5 |
| | Enterobacter
aerogenes | 2 |
| | MRSA | 2 |
| | Serratia
marcescens | 2 |
| | Staphylococcus
capitis | 2 |
| | Staphylococcus
lugdunensis | 2 |
| | | |
| VAP | | 29 (31.18) |
| | Klebsiella
pneumoniae | 9 |
| | Pseudomonas
aeruginosa | 5 |
| | Acinetobacter
calcoaceticus | 2 |
| | Enterococcus
faecalis | 2 |
| | Enterococcus
faecium | 2 |
| | Enterococcus
spp. | 2 |
| | MRSA | 2 |
| | Staphylococcus
cohnii | 2 |
| | Staphylococcus
haemolyticus | 2 |
| | Escherichia
fergusonii | 1 |
| | | |
| Bacteremia | | 9 (9.68%) |
| | Enterococcus
faecium | 3 |
| | Enterococcus
faecalis | 2 |
| | Klebsiella
pneumoniae | 2 |
| | MSSA | 1 |
| | Staphylococcus
hominis | 1 |
| | | |
| UTI | | 8 (8.60%) |
| | Klebsiella
pneumoniae | 3 |
| | Escherichia
fergusonii | 2 |
| | MSSA | 2 |
| | Enterococcus
spp. | 1 |
| | | |
| ABSSSI | | 6 (6.45%) |
| | MRSA | 3 |
| | Acinetobacter
baumannii | 2 |
| | MSSA | 1 |
PCT-MDR bacteria association
The values of serum PCT (median, IQR) were
significantly higher in patients with MDR bacteria superinfection
than in patients with sensitive bacteria (SB) superinfections
(5.665, 3.08-8.12 ng/ml vs. 3.150 ng/ml, 2.48-4.6 ng/ml,
P=0.009).
Discussion
Superinfection in COVID-19
patients
There are differences in microbial resistance to
antibiotics in urban compared with rural areas and from one
geographical area to another. Microbial resistance to antibiotics
differs even more from one country to another, depending on
therapeutic protocols, specific indications and occasionally on the
clinical experience of the prescribing physician.
On the other hand, it is difficult to estimate how
the COVID-19 pandemic will change the MDR bacteria spectrum.
Therefore, a comparative synthesis of several studies from
different regions and countries would help to fight against
microbial resistance to antibiotics.
Bacterial coinfection within 48 h of hospital
admission for COVID-19 infection in adults was uncommon: 1.6% at
admission and 5.5% within 48 h (13). In a study that evaluated
Streptococcus pneumoniae coinfection in hospitalized
patients with COVID-19, the most frequent pathogens identified
within the first 48 h of hospital admission were Staphylococcus
aureus and Streptococcus pneumoniae (14). In the present study, 27.4% of the
total number of bacterial associations were coinfections, being
diagnosed within the first 48 h from admission. This significantly
higher proportion of already superinfected patients was probably
due to a particularity, well known by the medical professionals
working in the studied population, the late presentation of
patients in the emergency room, often with clinically manifest
respiratory failure and oxygen desaturation.
Most studies performed on COVID-19 patients show no
clear distinction between early and late superinfections (15,16).
By contrast, the high case fatality of COVID-19 in a relatively
short time may exclude the development of a possible late
superinfection, which may lead to an underestimation of the risk of
superinfection (17).
In the present study, the number of superinfections
was higher in severe compared with non-severe cases (80.49% vs.
19.1% of all diagnosed superinfections).
The results of the present study are superposable
with the available literature data. In a study published by
Baskaran et al (9) in 2021,
30.3% of bacterial infections in ICU patients represented
hospital-acquired infections. The most frequently identified
bacteria were Klebsiella pneumoniae (commonly associated
with hospital and ventilator-acquired pneumonia) and Escherichia
coli. The median time to superinfection was 9 days in the same
study compared with 11 days in the present study.
In the vast majority of studies, the proportion of
pathogens detected increased with the duration of ICU stay. It
mainly consisted of Gram-negative bacteria, especially
Klebsiella pneumoniae and Escherichia coli.
The present study found mostly Klebsiella
pneumoniae, Acinetobacter baumannii and
Enterococcus spp. in the early infection period (3 to
7 days from admission). In the late infection period (after 7 days
from admission), the same bacteria occurred, to which
Escherichia coli was added in the same proportion as
Acinetobacter baumannii and Enterococcus spp.
Klebsiella pneumoniae was predominant after 7 days from
admission.
According to a recent meta-analysis, the prevalence
of bacterial superinfection was estimated to be ~14% in ICU
patients (15). An observational
cohort study from England (2021) reported a higher prevalence of
superinfection (27.2%) in critically ill patients with COVID-19,
with the majority of superinfection involving Gram-negative
bacteria (Klebsiella pneumoniae and Escherichia coli)
(10). A high prevalence of
superinfection reported in a study on ICU patients was 45%
(18). This significant
variability is probably due to the vast diversity of patients,
diagnostic and therapeutic procedures and environment-specific
factors (19).
Superinfections are frequently caused by
hospital-acquired MDR bacteria (7). A prolonged stay in an ICU associated
with broad-spectrum antibiotic therapy significantly increases the
risk of MDR bacteria selection (8).
In a study of 315 patients, conducted in 2020 by
Falcone, which evaluated superinfection in hospitalized COVID-19
patients, MDR bacteria caused 61.5% of all superinfections
(20). The present study
identified MDR bacteria in 73.17% of all patients with bacterial
superinfection. This high proportion of MDR bacteria raises
significant concern about the future efficiency of antibiotics in
treating bacterial infections.
Serum PCT levels
Serum PCT is widely used in the management of
COVID-19 patients and its role in disease diagnosis, prognosis and
clinical decision-making was reassessed in the current pandemic.
According to a meta-analysis by Stegeman et al in
2020(21), PCT has low sensitivity
ranging (0-48%) and specificity from 26-95% in diagnosing COVID-19.
By contrast, a number of studies show that PCT levels are
significantly associated with the severity and potential
complications of the disease (22-24).
Serum PCT levels increase markedly between the first
6 to 12 h after the onset of sepsis, systemic infection and severe
inflammation (25). Serum PCT
kinetics in infections with SB is well-known (26). However, there are insufficient data
about serum PCT kinetics and its practical utility in critically
ill patients with infections caused by MDR bacteria. In this
regard, a previous study showed that catheter-related bloodstream
infections caused by MDR bacteria are associated with higher PCT
levels and slower decline of its levels compared with those caused
by SB, probably due to a more robust and prolonged inflammatory
response (26).
To the best of the authors' knowledge, there is a
lack of data in the literature regarding PCT levels in COVID-19
critically ill patients suffering from superinfection with MDR
bacteria.
In critically ill COVID-19 patients, the release of
PCT from extrathyroidal parenchymal tissues is increased by
infectious insults and accelerated by the inflammatory cytokines
storm (27). It is complicated to
differentiate between the increase in PCT levels caused by the
cytokine storm and that caused by a possible superinfection.
Therefore, it appears crucial to corroborate serum PCT with other
biomarkers, cultures and clinical data if superinfection is
suspected.
The results of the present study demonstrate that
serum PCT obtained within 24 h before and after the patients were
suspected of superinfection and were taken samples for cultures are
significantly higher (P=0.009) in patients with infection caused by
MDR bacteria compared with those with infection caused by SB,
suggesting that the increase in serum PCT levels is caused by
superinfection rather than by de progression of COVID-19.
Strengths and limitations of the
present study
The present study provided novel data on bacterial
superinfection, including MDR bacteria superinfections and the
association between MDR bacteria superinfections and serum PCT
levels in hospitalized COVID-19 adult patients.
The principal findings were that the high prevalence
of superinfection with MDR bacteria among the COVID-19 patients
with bacterial superinfections and the presence of a statistically
significant correlation between serum PCT levels and the presence
of superinfection with MDR bacteria.
A notable limitation of the present study was its
retrospective observational design, which mainly reported on the
selection of cases. The fact that severe and non-severe patients
were studied together resulted in a heterogeneity of the study
population regarding the severity of the viral disease. The number
of included patients was relatively small. Serial measurements of
PCT were not performed to report its kinetics in dynamics. There is
a possibility of empirical antibiotic treatment before admission,
which could affect the studied laboratory parameters;
unfortunately, no such details were found in the data records of
the included patients.
Although COVID 19 has become a significantly less
severe disease, patients with severe forms are still exposed to
bacterial superinfections, especially during prolonged ICU
hospitalizations. The MDR bacteria superinfection spectrum found in
the present study and its correlation with PCT values may be
helpful in future clinical judgment.
The principal findings of the present study were the
high prevalence of superinfection with MDR bacteria among the
COVID-19 patients with bacterial superinfections and the presence
of a statistically significant correlation between serum
procalcitonin levels and the presence of superinfection with MDR
bacteria.
MDR bacteria were involved in almost three-quarters
of COVID 19 patients with bacterial superinfections. Most MDR
bacteria superinfections appeared in the late infection period.
Serum PCT values were significantly higher in
patients with MDR bacteria superinfection compared with patients
with SB superinfection. Serum PCT is a possible biomarker for
individualizing antibiotic treatment in COVID-19 patients with MDR
bacteria superinfection. More prospective studies are needed to
establish the serum PCT role in monitoring these patients.
Corroboration of laboratory biomarkers in dynamics
with clinical data is essential and COVID-19 patients with
bacterial superinfection need to be monitored not only by
hematological, biochemical and immunological markers, but also from
the microbiological point of view.
An improved understanding of superinfection
mechanisms, its favoring and precipitating factors and the variety
of pathogens involved in COVID-19 patients with bacterial
superinfection reinforces the need for more reviews and prospective
clinical studies with standardized protocols.
The most effective way to fight against microbial
resistance to antibiotics and subsequently preserve the
antibiotics' effectiveness in the treatment of infectious diseases
is to pursue a national policy for the rational use of
antibiotics.
As pandemics have no borders, several countries must
find common paths to fight against MDR bacterial infections,
whether they occur independently or overlap with viral infections,
as in the case of COVID 19.
Acknowledgements
Not applicable
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
MS and DM together designed the study and were
responsible for data analysis and writing the paper. RS, RB, AlI
and AnI searched the literature and were responsible for data
analysis and interpretation. CD and DL interpreted the data and
critically revised the manuscript. VL, IB and AM were responsible
for data collection, obtaining ethics approval and confirm the
authenticity of all the raw data. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
The present study was performed in accordance with
the declaration of Helsinki. It was approved by the Ethics
Commission for Clinical Trials of Arad County Hospital (approval
no. 37/25.02.2021) and of Scientific Research Ethics Commission of
‘Vasile Goldis’ Western University of Arad (approval no.
10/29.03.2021).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Rawson TM, Moore LS, Castro-Sanchez E,
Charani E, Davies F, Satta G, Ellington MJ and Holmes AH: COVID-19
and the potential long-term impact on antimicrobial resistance. J
Antimicrob Chemother. 75:1681–1684. 2020.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Huttner BD, Catho G, Pano-Pardo JR,
Pulcini C and Schouten J: COVID-19: don't neglect antimicrobial
stewardship principles! Clin Microbiol. Infect. 26:808–810.
2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
McCullers JA: The co-pathogenesis of
influenza viruses with bacteria in the lung. Nat Rev Microbiol.
12:252–262. 2014.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Mirzaei R, Goodarzi P, Asadi M, Soltani A,
Aljanabi HAA, Jeda AS, Dashtbin S, Jalalifar S, Mohammadzadeh R,
Teimoori A, et al: Bacterial co-infections with SARS-CoV-2. IUBMB
Life. 72:2097–2111. 2020.PubMed/NCBI View
Article : Google Scholar
|
|
5
|
Zhang G, Hu C, Luo L, Fang F, Chen Y, Li
J, Peng Z and Pan H: Clinical features and short-term outcomes of
221 patients with COVID-19 in Wuhan, China. J Clin Virol.
127(104364)2020.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Mazzariol A, Benini A, Unali I, Nocini R,
Smania M, Bertoncelli A, De Sanctis F, Ugel S, Donadello K, Polati
E and Gibellini D: Dynamics of SARS-CoV2 infection and multi-drug
resistant bacteria superinfection in patients with assisted
mechanical ventilation. Front Cell Infect Microbiol.
11(683409)2021.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Garcia-Vidal C, Sanjuan G, Moreno-García
E, Puerta-Alcalde P, Garcia-Pouton N, Chumbita M, Fernandez-Pittol
M, Pitart C, Inciarte A, Bodro M, et al: Incidence of co-infections
and superinfections in hospitalized patients with COVID-19: A
retrospective cohort study. Clin Microbiol Infect. 27:83–88.
2021.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Bengoechea JA and Bamford CG: SARS-CoV-2,
bacterial co-infections, and AMR: the deadly trio in COVID-19? EMBO
Mol Med. 12(e12560)2020.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Baskaran V, Lawrence H, Lansbury LE, Webb
K, Safavi S, Zainuddin NI, Huq T, Eggleston C, Ellis J, Thakker C,
et al: Co-infection in critically ill patients with COVID-19: An
observational cohort study from England. J Med Microbiol.
70(001350)2021.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Kim JH, Seo JW, Mok JH, Kim MH, Cho WH,
Lee K, Kim KU, Jeon D, Park HK, Kim YS, et al: Usefulness of plasma
procalcitonin to predict severity in elderly patients with
community-acquired pneumonia. Tuberc Respir Dis (Seoul).
74:207–214. 2013.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Lippi G and Plebani M: Procalcitonin in
patients with severe coronavirus disease 2019 (COVID-19): A
meta-analysis. Clin Chim Acta. 505:190–191. 2020.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Wolfisberg S, Gregoriano C and Schuetz P:
Procalcitonin for individualizing antibiotic treatment: An update
with a focus on COVID-19. Crit Rev Clin Lab Sci. 59:54–65.
2022.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Metersky ML, Masterton RG, Lode H, File TM
Jr and Babinchak T: Epidemiology, microbiology, and treatment
considerations for bacterial pneumonia complicating influenza. Int
J Infect Dis. 16:e321–e331. 2012.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Anton-Vazquez V and Clivillé R:
Streptococcus pneumoniae coinfection in hospitalised patients with
COVID-19. Eur J Clin Microbiol Infect Dis. 40:1353–1355.
2021.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Lansbury L, Lim B, Baskaran V and Lim WS:
Co-infections in people with COVID-19: A systematic review and
meta-analysis. J Infect. 81:266–275. 2020.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Rawson TM, Moore LS, Zhu N, Ranganathan N,
Skolimowska K, Gilchrist M, Satta G, Cooke G and Holmes A:
Bacterial and fungal coinfection in individuals with coronavirus: A
rapid review to support COVID-19 antimicrobial prescribing. Clin
Infect Dis. 71:2459–2468. 2020.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Bassetti M, Kollef MH and Timsit JF:
Bacterial and fungal superinfections in critically ill patients
with COVID-19. Intensive Care Med. 46:2071–2074. 2020.PubMed/NCBI View Article : Google Scholar
|
|
18
|
d'Humières C, Patrier J, Lortat-Jacob B,
Tran-Dinh A, Chemali L, Maataoui N, Rondinaud E, Ruppé E, Burdet C,
Ruckly S, et al: Two original observations concerning bacterial
infections in COVID-19 patients hospitalized in intensive care
units during the first wave of the epidemic in France. PLoS One.
16(e0250728)2021.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Adalbert JR, Varshney K, Tobin R and
Pajaro R: Clinical outcomes in patients co-infected with COVID-19
and Staphylococcus aureus: A scoping review. BMC Infect Dis.
21:1–7. 2021.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Falcone M, Tiseo G, Giordano C, Leonildi
A, Menichini M, Vecchione A, Pistello M, Guarracino F, Ghiadoni L,
Forfori F, et al: Predictors of hospital-acquired bacterial and
fungal superinfections in COVID-19: A prospective observational
study. J Antimicrob Chemother. 76:1078–1084. 2021.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Stegeman I, Ochodo EA, Guleid F, Holtman
GA, Yang B, Davenport C, Deeks JJ, Dinnes J, Dittrich S, Emperador
D, et al: Routine laboratory testing to determine if a patient has
COVID-19. Cochrane Database Syst Rev. 11(CD013787)2020.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Garrido P, Cueto P, Rovira C, Garcia E,
Parra A, Enriquez R, Pinos A, Sosa M, Hernández-Aguilera A and
Vallverdú I: Clinical value of procalcitonin in critically ill
patients infected by SARS-CoV-2. Am J Emerg Med. 46:525–531.
2021.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Hariyanto TI, Japar KV, Kwenandar F, Damay
V, Siregar JI, Lugito NP, Tjiang MM and Kurniawan A: Inflammatory
and hematologic markers as predictors of severe outcomes in
COVID-19 infection: A systematic review and meta-analysis. Am J
Emerg Med. 41:110–119. 2021.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Del Sole F, Farcomeni A, Loffredo L,
Carnevale R, Menichelli D, Vicario T, Pignatelli P and Pastori D:
Features of severe COVID-19: A systematic review and meta-analysis.
Eur J Clin Invest. 50(e13378)2020.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Müller B and Becker KL: Procalcitonin: How
a hormone became a marker and mediator of sepsis. Swiss Med Wkly.
131:595–602. 2001.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Huespe I, Prado E, Staneloni I, Contrera
N, Denaday L, San Roman E and Sinner J: Kinetics of procalcitonin
in infections caused by multidrug-resistant bacteria. Medicina (B
Aires). 80:599–605. 2020.PubMed/NCBI
|
|
27
|
Ponti G, Maccaferri M, Ruini C, Tomasi A
and Ozben T: Biomarkers associated with COVID-19 disease
progression. Crit Rev Clin Lab Sci. 57:389–399. 2020.PubMed/NCBI View Article : Google Scholar
|