Introduction
Among urological malignancies, bladder urothelial
carcinoma (BLCA) is the most common type of all urological
malignancy, including those not caused by transitional epithelial
tissue (1). Currently, surgery is
the initial treatment option for BLCA, followed by chemotherapy
with gemcitabine and other drugs (2). However, despite systemic therapy,
BLCA still has high recurrence and progression rates and poor
prognosis in China as at 2019, especially for those with muscular
infiltration (3). Although
numerous studies (4) have shown
that tumor suppressor genes and proto-oncogenes serve key roles in
the development of BLCA, to the best of our knowledge, their
underlying mechanisms have not been clarified. Therefore, it is key
to explore novel molecular markers and mechanisms of BLCA to
improve its early diagnosis and prognosis.
Cyclin B1 (CCNB1), a member of the cyclin family
located at locus 12 on the long arm of chromosome 5, is a key
initiator of mitosis that can promote cell cycle progression
(5,6) and exhibits changes in expression
throughout the cell cycle. Several studies have confirmed that
CCNB1 is highly expressed in numerous types of human tumor tissue
and is related to tumor cell proliferation, metastasis and poor
prognosis (7). Zhang et al
(8) demonstrated that the CCNB1
expression levels and apoptosis in pancreatic cancer tumor tissue
are increased and the cell proliferation is decreased following
silencing of CCNB1 via short hairpin (sh)CCNB1. Another report
indicated that sh-CCNB1 arrests colorectal cancer cells in the G2/M
phase and thereby inhibits proliferation (9). Zou et al (10) noted that the CCNB1 expression is
positively correlated with infiltration levels of various
inflammatory cells, including CD4+ and CD8 +
T cells and negatively associated with prognosis of hepatocellular
carcinoma and may serve as a potential prognostic biomarker for
this liver malignancy. In another study, the mitosis-promoting
factor CCNB1/CDK1 complex was found to regulate mitochondrial
metabolism and ATP production; this might be an effective
antineoplastic resistance-associated therapeutic target (11). Together, the aforementioned studies
indicated that CCNB1 is highly expressed in multiple types of
malignancy, including pancreatic, colorectal and liver cancer, and
affects cell cycle regulation, proliferation and ATP synthesis of
cancer cells and thus the development and prognosis of tumors.
Several research groups have investigated the putative effects of
CCNB1 in BLCA, but they mainly focused on the roles of
CCNB1-related genes (12,13) and the roles and underlying
mechanisms of CCNB1 in the development of BLCA remain
unexplored.
The present study aimed to investigate the effects
and possible mechanisms of CCNB1 in BLCA using public databases,
followed by primary confirmative experiments in vitro and
in vivo, such as cell cycle and proliferation analysis, to
determine its biological functions in BLCA cell models and the
effect of CCNB1 on prognosis of patients with BLCA to find
practical biomarkers for early diagnosis and intervention in
BLCA.
Materials and methods
Materials
Normal bladder cells (SV-HUC-1) and human BLCA cells
(T24, 5637 and UM-UC-3) were obtained from Shanghai Cell Bank
(Chinese Academy of Sciences). RPMI-1640 and fetal bovine serum
were obtained from Gibco (Thermo Fisher Scientific, Inc.).
Penicillin-streptomycin double antibody was obtained from Beijing
Solarbio Science & Technology Co., Ltd. Trypsin and CCNB1 and
GAPDH primers were obtained from Sangon Biotech Co., Ltd. RNA
extraction (Axygen) and Transwell chambers were obtained from
Corning, Inc. HiScript® Ⅲ RT SuperMix for qualitive
(q)PCR and Cell Counting Kit (CCK)-8 were obtained from Vazyme
Biotech Co., Ltd. RIPA lysis solution and BCA quantification kit
were obtained from Beyotime Institute of Biotechnology. Chickens
embryos (age, 7 days) were obtained from Guangxi Fufeng
Agricultural and Livestock Company.
Bioinformatics analysis
CCNB1 mRNA expression in pan-cancer was obtained
from The Cancer Genome Atlas (TCGA; https://www.cancer.gov/ccg/research/genome-sequencing/tcga)
database. CCNB1 mRNA expression data were obtained for both BLCA
and non-cancerous tissue, including 144 BLCA cases and 19
non-cancerous controls. BLCA-associated high-throughput sequencing
data were obtained from the Gene Expression Omnibus (GEO;
https://www.ncbi.nlm.nih.gov/geo)
database. A total of 20 datasets from 10 platforms were utilized
(accession nos. GPL96, GPL570, GPL6102, GPL14951, GSE19915,
GSE24152, GSE40355, GSE52519 and GSE76211), including 742 BLCA and
173 non-cancerous samples (Fig.
S1; Table SI). The expression
matrix was transformed by uniform Log2 calculation and the batch
effect was removed using the removeBatchEffect function in R
version 4.2.1 Limma package to normalize data. The summary
standardised mean differences (SMD) were estimated using
meta-analysis with random effects. Kaplan-Meier univariate survival
analysis (KM survival curve) of the ENCORI (http://starbase.sysu.edu.cn/index.php) online
database. The protein expression of CCNB1 in BLCA and normal
bladder tissue was investigated in randomly chosen clinical samples
(Patient ID: 2053, 2311, 2031 and 2699, n=4) from the Human Protein
Atlas (HPA) database (proteinatlas.org/).
Cell culture and transfection
T24 and 5637 BLCA cells were cultured at 37˚C and 5%
CO2 in RPMI-1640 medium with 10% fetal bovine serum and
1% penicillin-streptomycin double antibody. The cultured T24 and
5637 cells were randomly divided into experimental group and the
negative control (NC) group. The viral solution obtained following
lentiviral packaging of sh-CCNB1 (5'-gcCAAATACCTGATGGAACTA-3') and
sh-NC (5'-TTCTCCGAACGTGTCACGT-3') plasmid with the HIV
backbone-based lentiviral particle packaging kit and empty viral
backbone (Lenti-Pac™ HIV Expression Packaging Kit; GeneCopoeia,
Inc.) were used to transfect T24 and 5637 cells. The lentiviral
backbone was pCMV-hU6-MCS-Ubiquitin-EGFP-IRES-puromycin (Shanghai
GeneChem Co., Ltd.). Opti-MEM serum-free medium (200 µl) was added
with 2.5 µg of plasmid and 5 µl of Lenti-Pac and marked as tube A.
An additional 200 µl opti-MEM was used in the absence of serum
medium, a total of 15 µl of EndoFectin Lenti was added, and marked
as tube B. Tubes A and B were mixed to transfect 293T cells until 4
days in order to collect lentivirus. Temperature of transfection
for T24 and 5637 cells was 37˚C and lasted for 48 h. After
screening with 2 µg/ml puromycin for at least 3 days before
additional experiments were performed, the proportion of cells with
green fluorescent protein was observed under a fluorescent
microscope (magnification, x100). Subsequent experiments were
performed when the percentage of cells with green fluorescent
protein was >70% of the total number of cells.
Detection of CCNB1 mRNA expression in
BLCA cells by reverse transcription-quantitative (RT-q)PCR
Total RNA was harvested from cells (Axygen
Scientific Inc) and reverse transcriptase was used to synthesize
cDNA (HiScript Ⅲ RT SuperMix for qPCR). The system of comprised 4
µl 4X gDNA wiper mix, 1 µl RNA and 11 µl RNase-free water to 42˚C
for 2 min. 4 µl 5X HiScript Ⅲ q-RT SuperMix was added at 37˚C for
15 min, 85˚C for 5 sec and stored at 4˚C. The cDNA was diluted
three times for RT-qPCR reaction. The reaction system comprised 10
µl 2X FS Universal SYBR Green Master Mix (FS Universal SYBR Green
Master; Roche Diagnostics), 0.6 µl each of the upstream and
downstream primers, 2 µl cDNA and RNase-free water to a total
volume of 20 µl. Thermocycling conditions were as follows: 95˚C for
3 min, followed by 40 cycles of 95˚C for 10 sec, 65˚C for 1 min and
97˚C for 1 sec and final extension at 37˚C for 30 sec. The primer
sequences of CCNB1 and the internal reference control GAPDH were
amplified separately. The primer sequences were as follows: CCNB1
forward, 5'-GCCTGAGCCTATTTTGGTTGATAC-3' and reverse,
5'-TCCATCTTCTGCATCCACATCA-3' and GAPDH (internal control) forward,
5'-ACCACAGTCCATGCCATCAC-3' and reverse, 5'-TTCCCGTTCAGCTCAGGGAT-3'.
The 2-ΔΔCq method was used to calculate the relative
expression (2-ΔΔCq Livak and Schmittgen 2001).
CCK-8 assay
When the density of the stable cell lines at the
logarithmic growth stage reached 70-80%, the cells were digested
with trypsin and counted by counting board. Cell suspension (100 µl
containing 2,500 cells/well) was added to a 96-well plate with
triplicate wells. After 1 h incubation with 10 µl CCK-8 solution,
the cells were assessed at 490 nm using an enzyme marker to assess
the cells at 490 nm and the measured OD value indirectly reflects
the number of viable cells. A cell proliferation curve was
drawn.
Transwell assay for cell migration and
invasion
Transwell chambers were sterilized by radiation
under UV light for 2 h at room temperature. The RPMI-1640 medium
containing 5% fetal bovine serum was added to each well of the
24-well plate and Transwell chambers were gently placed into the
wells to avoid air bubbles. Then, the upper chamber was filled with
a cell suspension containing 6x104 cells in a volume of
100 µl. It was incubated for 24 h at the incubation temperature of
37˚C. The cells were gently wiped from the chamber with a cotton
swab. Following washing with PBS solution three times, 500 µl 70%
methanol solution was added, fixed at 37˚C for 30 min, and put into
a 3% crystal violet solution for overnight staining at room
temperature. Chambers were dried with a clean cotton swab and
images captured under a microscope. The quantity of cells that had
penetrated the membrane was recorded. For cell invasion assay, the
ratio of the Matrigel matrix gel at 4˚C to the serum-free RPMI-1640
medium was 1:8, and 60 µl solution was spread evenly on the upper
chamber for 30 min. Medium plated in upper chamber (usually without
serum) and medium and serum (type and concentration) plated in
lower chamber. The remaining procedures were the same as the
migration assay. Observation was with a light microscope
(magnification, x100). A total of three independent repeats was
performed.
Cell cycle and apoptosis analysis
When the cell density in the culture dish was ~70%,
the RPMI-1640 medium containing 10% fetal bovine serum was replaced
by serum-free RPMI-1640 medium. After 12 h at 37˚C, the 10% fetal
bovine serum of RPMI-1640 medium was substituted by the serum-free
RPMI-1640 medium to synchronize the cell cycle. After another 12 h
at 37˚C, ≥1x106 cells were extracted following
transfection, washed twice with pre-cooled PBS solution and
centrifuged at 300 x g for 5 min at 37˚C. A total of 1 ml PBS
solution was added and the sample was resuspended after supernatant
was discarded. The cells were homogenized by adding 1 ml PBS
solution and shaken on a vortex shaker while adding 3 ml 75%
isopropyl alcohol dropwise. The solution was fixed by 1 ml PBS
solution and 3 ml 75% isopropyl alcohol overnight at -20˚C in the
dark. It was centrifuged at 300 x g for 5 min at 37˚C. The
supernatant was discarded and cell precipitation were cleaned with
3 ml PBS. Then, 1 ml PI/RNase dye was added for cell cycle analysis
by flow cytometry (BD FACSCalibur; BD Biosciences). The cells were
shielded from light for 30 min before being tested. For apoptosis
detection, the cells were centrifuged at 300 x g at 37˚C for 5 min
and resuspended in 500 µl 1X binding buffer. A total of 5 µl
Annexin V-FITC and 10 µl PI dye were added to each sample,
incubated for 15 min with gentle shaking and then incubated at room
temperature for 1 h before apoptotic analysis by flow cytometry (BD
FACSCalibur; BD Biosciences). FlowJo 7.6 (FlowJo LLC) was used for
analysis.
Tumorigenic experiment on chick
chorioallantoic membrane (CAM)
The air chambers of CAM from 7-day-old chicken
embryos at 42˚C were outlined by a marker after checking for signs
of life: In the dark room, blood vessels and embryonic movements
were clearly visible under a flashlight. The tumorigenic experiment
was performed as follows. The eggshell was knocked on at the air
chamber of the circle and the air chamber membrane soaked with 0.9%
normal saline until blood vessels could be clearly observed.
Tweezers were used to tear the air chamber membrane where the blood
vessels were rare and exposed the CAM. Finally, a silicon ring
(diameter 7 mm) was placed between two large blood vessels.
BLCA cells (1.2x107) at the logarithmic
growth stage were collected and added into the silicon ring, which
was closed with sterile and breathable adhesive tape. The signs of
life, tumor size and the surrounding blood vessels of embryos were
observed with a torch every 24 h. At 6 days after cell
implantation, intact tumors were removed and measured, then stored
in 4% paraformaldehyde at room temperature for three days. The
sections were cut at 4 µm and stained with hematoxylin solution for
3-5 min, washed with 85% ethanol for 5 min, 95% ethanol for 5 min
and stained with eosin for 5 min. The sections were observed with a
light microscope (magnification, x400).
Statistical analysis
Data were analyzed using SPSS 25.0 (IBM Corp.) and
images were drawn using GraphPad Prism 8. Data (Dotmatics) were
tested for normality. Data adhering to a normal distribution are
presented as the mean ± SD. Independent unpaired t test was used
for comparisons between two groups; the Analysis of Variance
(ANOVA) was performed for comparisons between >2 groups. For
measures that did not have a normal distribution, descriptive
statistics are reported as the median and interquartile range.
Non-parametric tests were used to compare groups and correlation
was analyzed using Spearman's correlation coefficient. The real
standardized mean difference (SMD) was calculated using Stata 15.0
(StataCorp LLC) and forest plots and the summary receiver operating
characteristic (sROC) curves were plotted. P<0.05 was considered
to indicate a statistically significant difference.
Results
CCNB1 expression is increased in
BLCA
The expression of CCNB1 mRNA in numerous types of
cancers and non-cancer tissues of TCGA database were integrated for
analysis. Numerous types of tumor tissues had notably greater
levels of CCNB1 mRNA than normal tissue, except for certain tumors
such as thymic carcinoma (Fig.
1A). In the online database TCGA, 411 BLCA samples and 19
normal bladder tissue samples were analyzed. CCNB1 mRNA expression
was notably higher in BLCA than in normal bladder tissue (Fig. 1B).
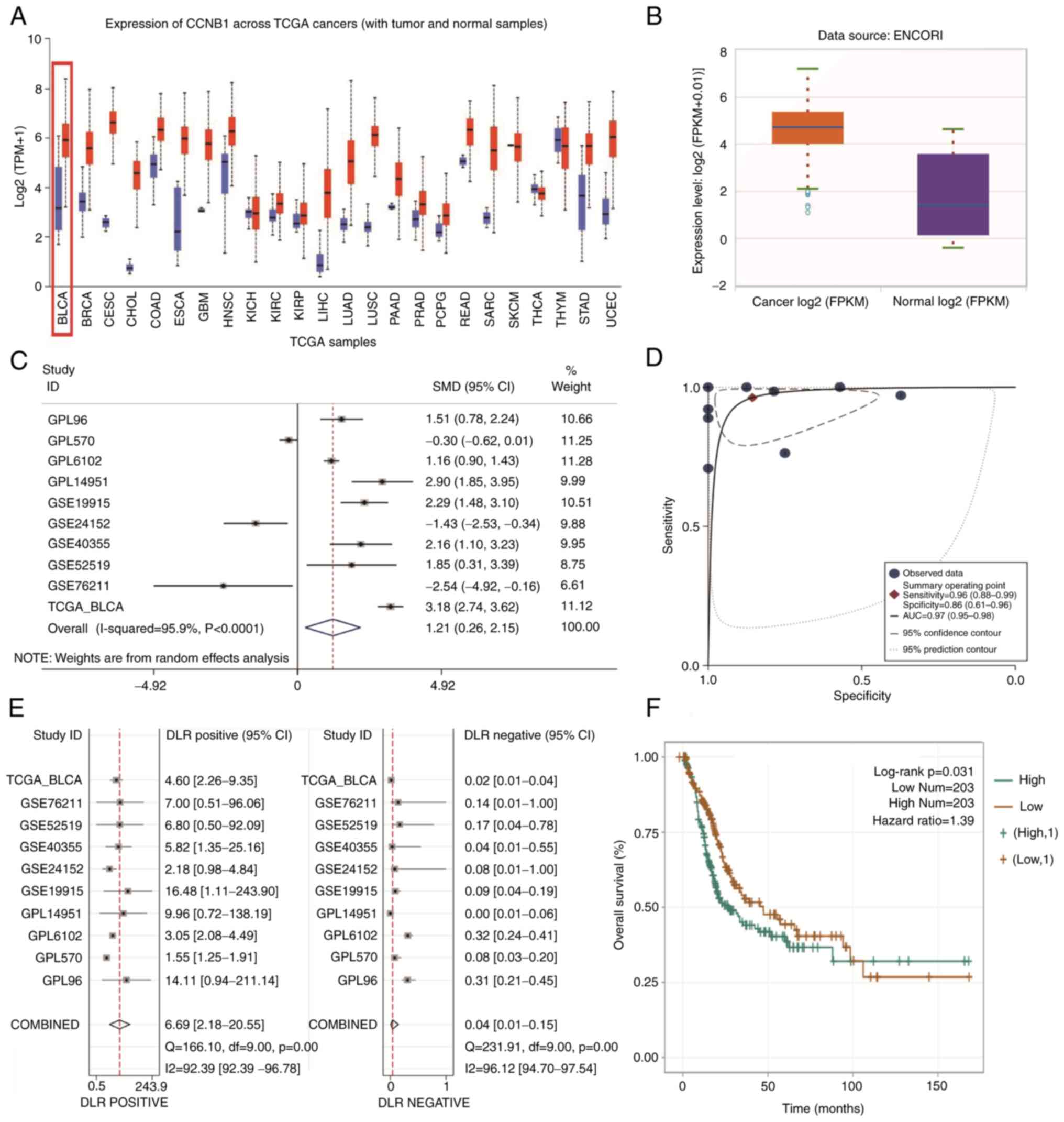 | Figure 1CCNB1 mRNA expression in pan-cancer
and BLCA. (A) CCNB1 was expressed at notably higher levels in
pan-cancer than in normal tissue. (B) Expression of CCNB1 was
notably higher in BLCA than in normal bladder tissue in the online
database ENCORI (P<0.05). (C) Forest plots of eligible datasets
in TCGA and GEO databases showed high expression of CCNB1 in BLCA
tissue. (D) sROC AUC suggested good diagnostic value of CCNB1. (E)
Combined positive likelihood ratio was 6.69 (2.18-20.55) and the
combined negative likelihood ratio was 0.04 (0.01-0.15)
(P<0.05). (F) Kaplan-Meier survival curve suggested that
patients with BLCA and high CCNB1 expression had worse prognosis
than those with low expression (P<0.05). CCNB1, ; BLCA, ; TCGA,
; GEO, ; sROC, ; AUC, ; KM, ; FPKM, ; TPM, ; DLR,. |
Meta-analysis of the GEO microarray data and TCGA
database data showed that CCNB1 mRNA expression levels were
markedly higher in BLCA than in non-cancerous bladder tissue
(SMD=1.21; 95% CI, 0.26-2.15; I²=95.9%; Fig. 1C); the area under the sROC curve
was 0.97 (95% CI: 0.95-0.98; Fig.
1D). Stata 15.0 yielded a combined positive likelihood ratio of
6.69 and a combined negative likelihood ratio of 0.04 for CCNB1
(Fig. 1E). These results suggested
that the high expression of CCNB1 mRNA distinguished BLCA from
normal bladder tissue samples. Kaplan-Meier univariate survival
analysis of the ENCORI online database demonstrated that patients
with BLCA and high CCNB1 expression had a significantly decreased
overall survival time and poorer prognosis than those with low
expression (Fig. 1F).
CCNB1 expression differed between BLCA and non-BLCA
bladder tissues in 11 datasets and showed significantly higher
expression in six datasets, including GPL6102, GPL14951, GSE19915,
GSE40355, GSE52519 and TCGA-GTEx (Fig.
2).
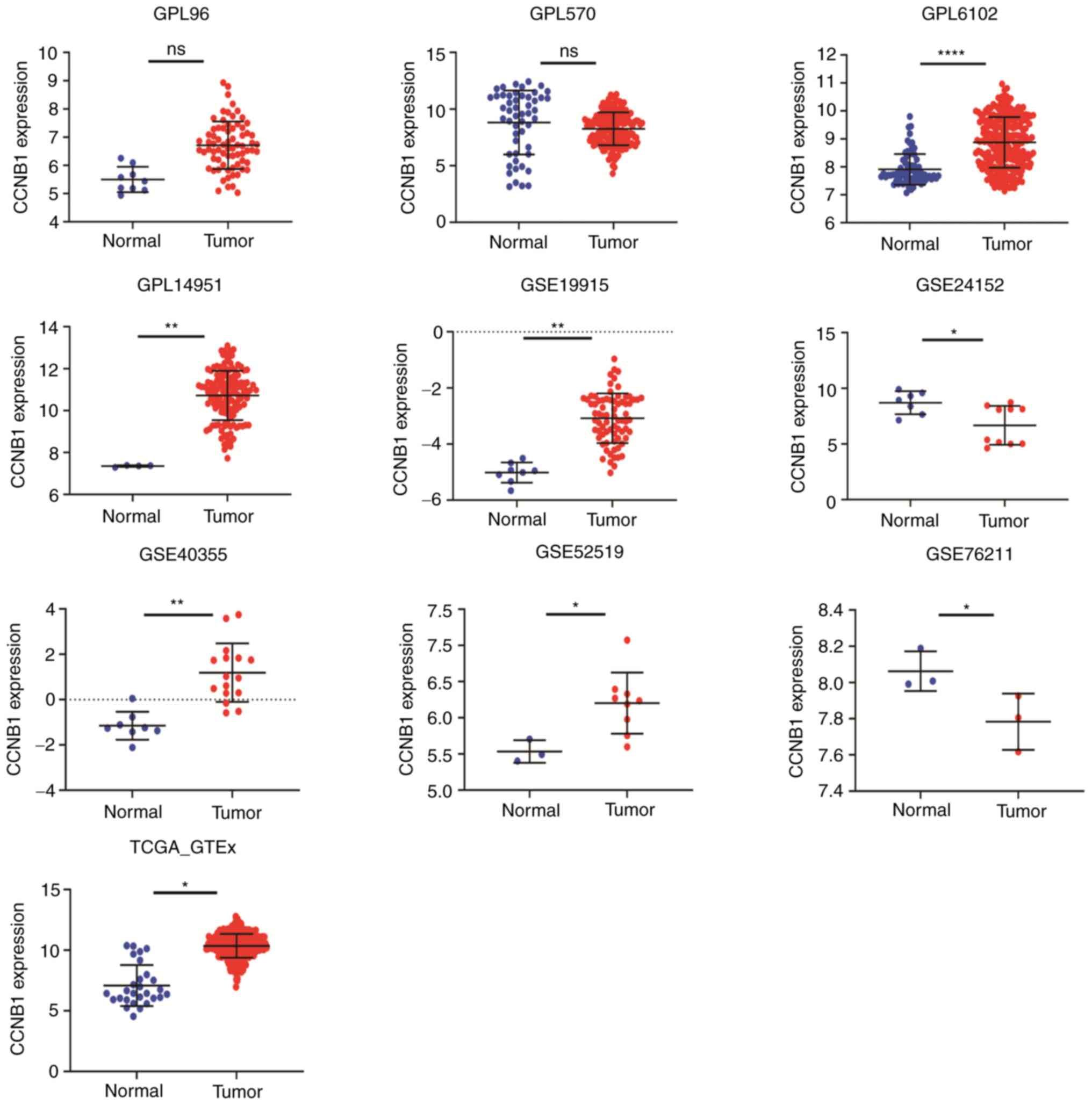 | Figure 2Expression of CCNB1 in datasets. A
total of six datasets showed significantly high expression,
including GPL6102, GPL14951, GSE19915, GSE40355, GSE52519 and
TCGA-GTEx. *P<0.05, **P<0.01,
****P<0.0001. CCNB1, cyclin B1; TCGA, The Cancer
Genome Atlas; ns, not significant. |
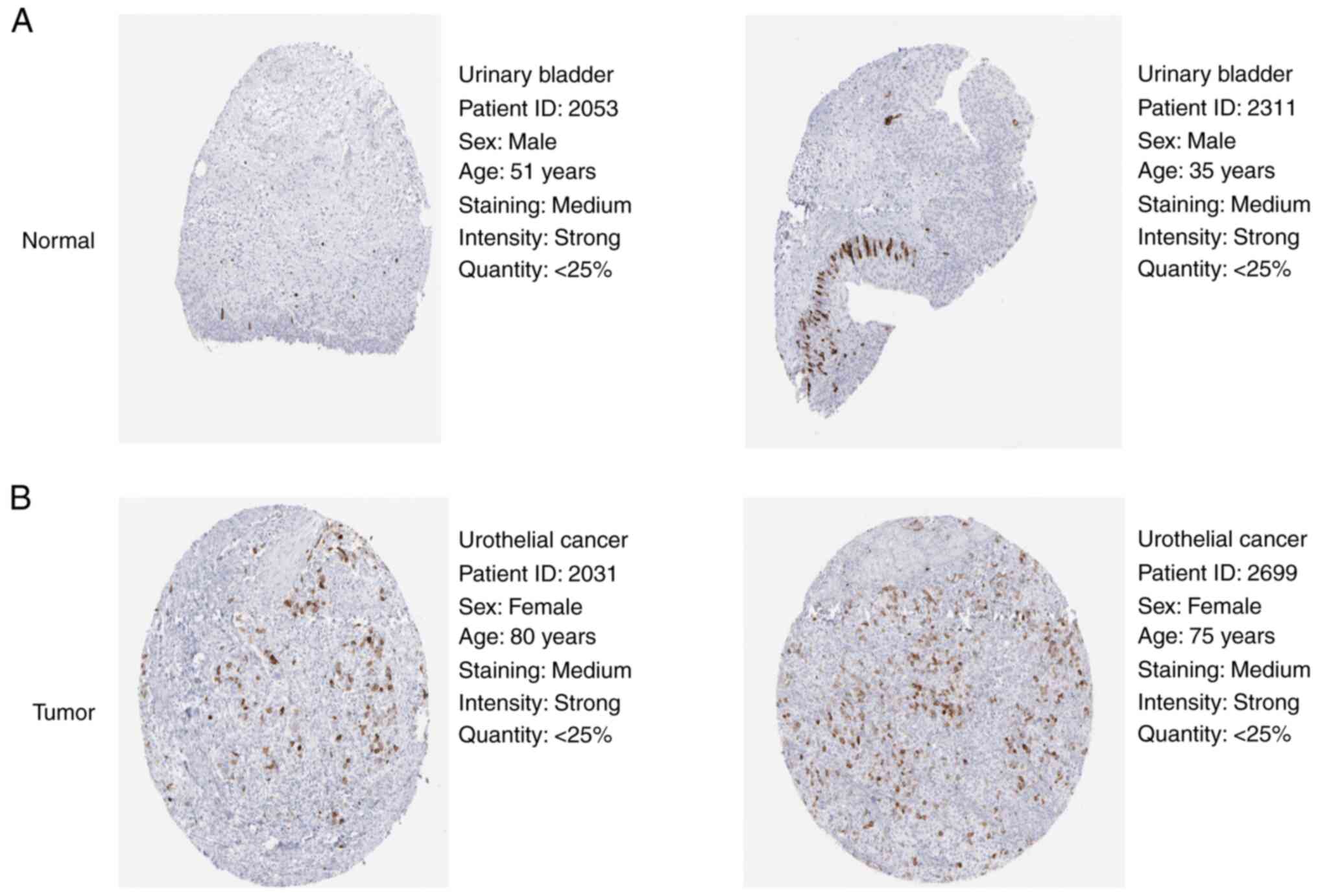
To validate the protein expression levels of CCNB1
in BLCA and normal bladder tissue, two groups of samples from the
HPA database were randomly selected for analysis. Protein levels
were substantially increased in BLCA compared with normal bladder
tissue (Fig. 3).
Expression of CCNB1 mRNA in normal and
bladder cancer cells
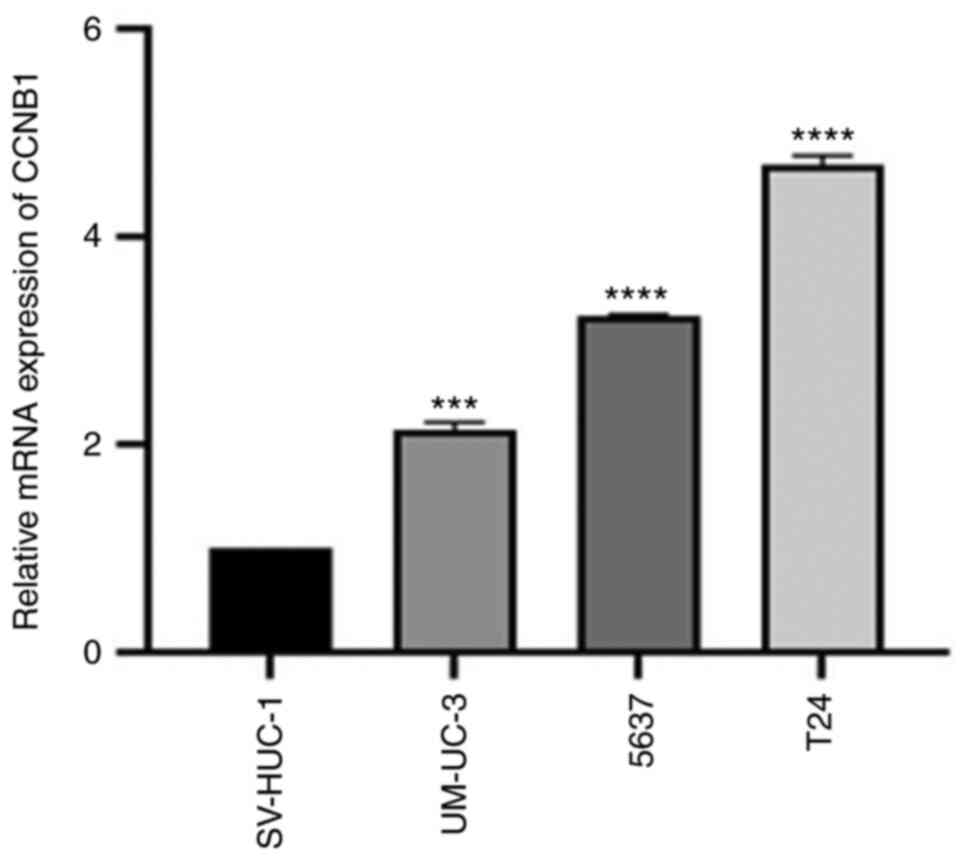
Expression of CCNB1 mRNA in normal bladder cells
(SV-HUC-1) and human BLCA cells (T24, 5637 and UM-UC-3) was
detected by RT-qPCR. The relative expression of CCNB1 mRNA in
bladder cancer cells was higher than in normal bladder cells and
the relative expression of CCNB1 mRNA in T24 and 5637 cell lines
was compared with UM-UC-3 cells (Fig.
4). Therefore, T24 and 5637 cell lines were selected for
silencing CCNB1.
Establishment of cell lines with
stable expression of sh-CCNB1
T24 and 5637 cell lines were infected with the virus
solution obtained after lentiviral packaging of sh-CCNB1 plasmid
and sh-CCNB1 stably expressed cell lines were established after
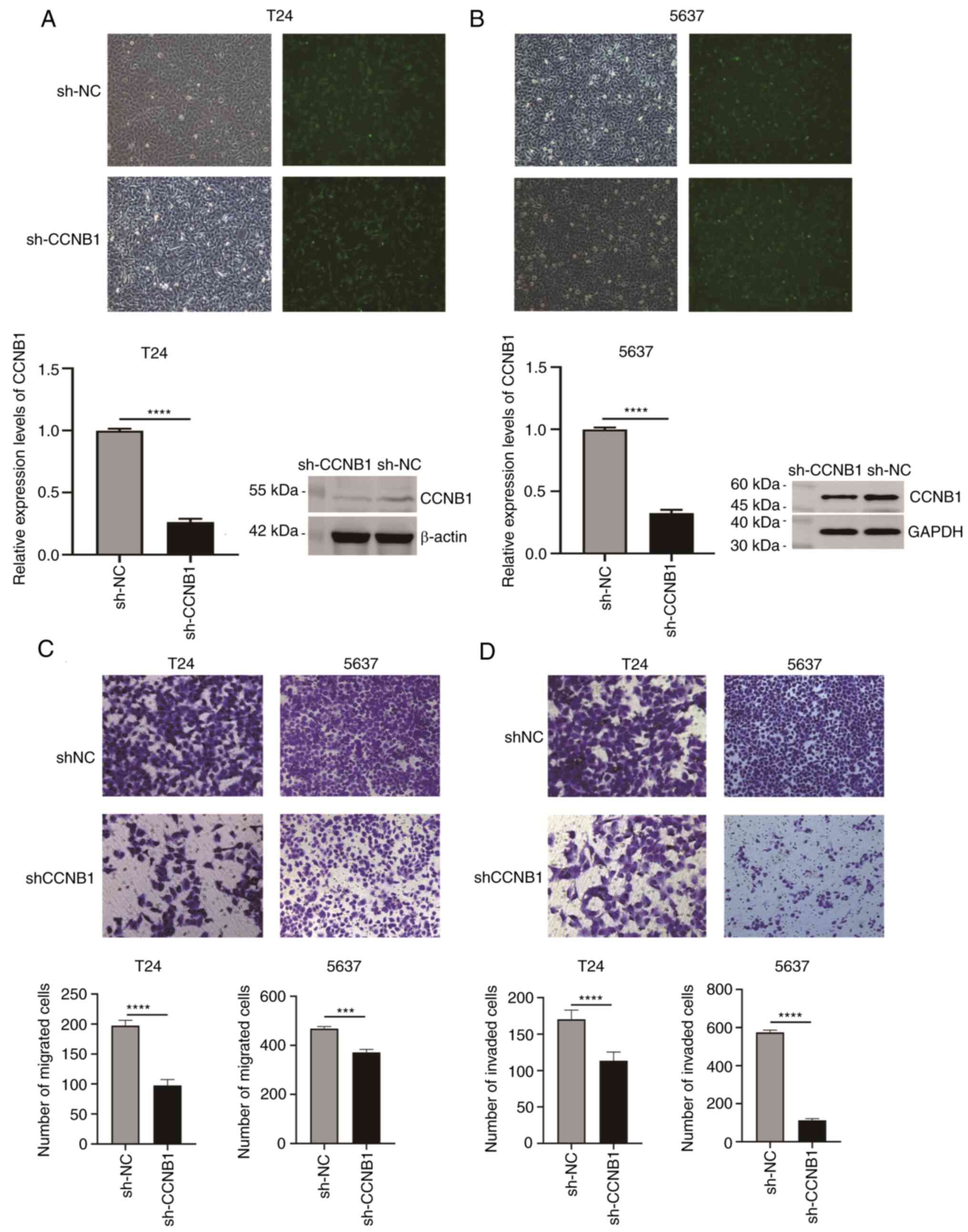
puromycin selection. Silencing efficiency of CCNB1 was measured by
RT-qPCR, yielding 75 and 71%, respectively (Fig. 5A and B). Western blotting and RT-qPCR showed
that the expression levels of CCNB1 protein and mRNA were
significantly decreased following sh-CCNB1 transfection compared
with sh-NC, indicating that stable expression of sh-CCNB1 was in
T24 and 5637 cells.
Inhibition of CCNB1 expression
decreases the migration and invasion of BLCA cells
The ability of cells to migrate and invade was
examined by Transwell assay. Compared with sh-NC group, sh-CCNB1
group in both cell lines showed a significant reduction in cell
migration and invasion (P<0.0001 and P<0.001, respectively;
Fig. 5C and D).
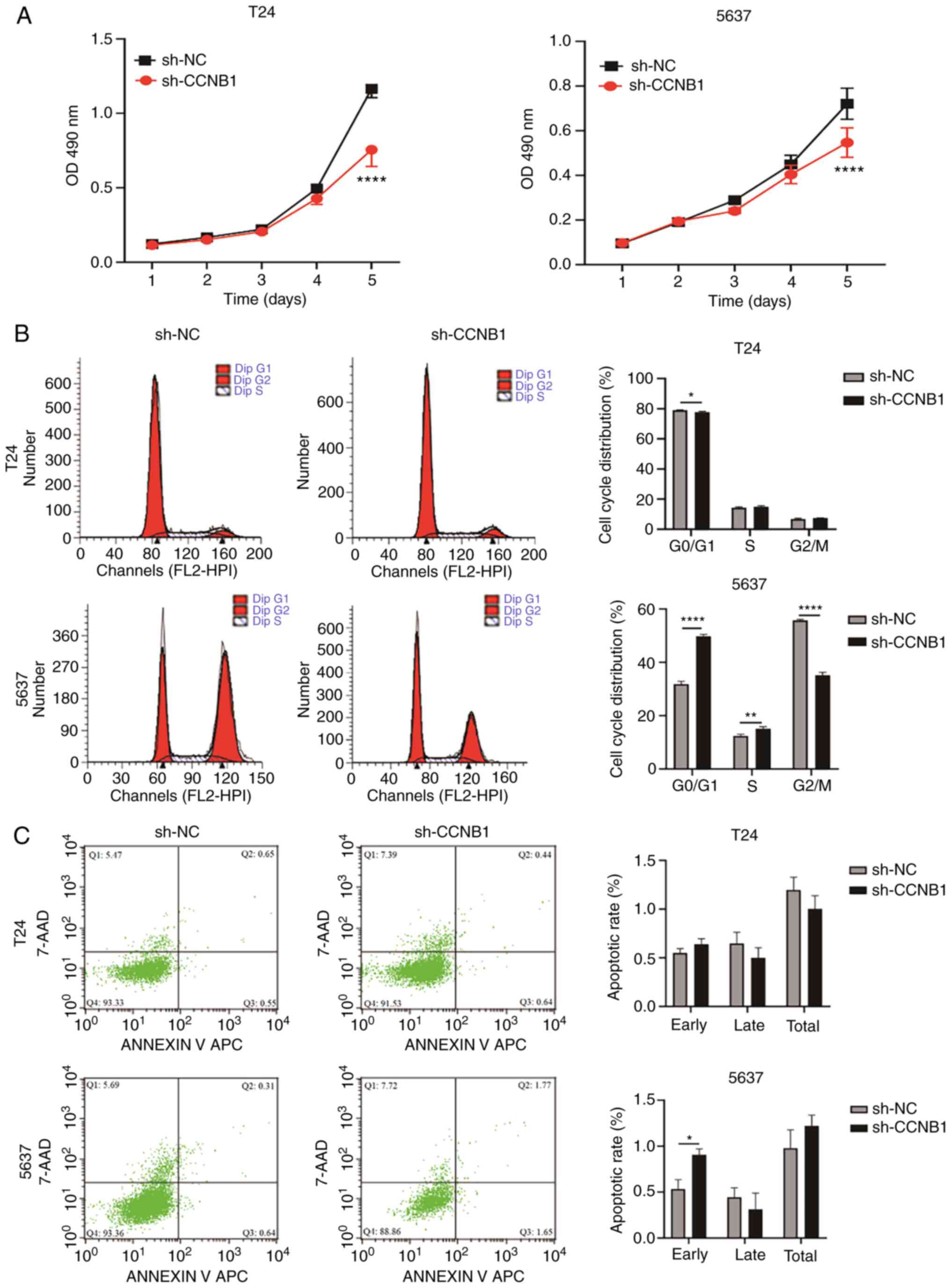
sh-CCNB1 inhibits the proliferation of
BLCA cells, affects the cell cycle distribution and promotes
apoptosis of 5637 cells
CCK-8 showed that compared with sh-NC group,
sh-CCNB1 group of T24 and 5637 cells exhibited significantly lower
proliferative capacity at day 6 (P<0.0001; Fig. 6A). The effect of sh-CCNB1 on BLCA
cell cycle and apoptosis was observed by flow cytometry, with
sh-CCNB1 decreasing the percentage of T24 cells in G0/G1
(P<0.05) 5637 cells in the G0/G1 phase (P<0.0001) and S phase
(P<0.01) and increasing percentage of 5637 cells in the G2/M
phase (P<0.0001; Fig. 6B).
sh-CCNB1 induced early apoptosis in 5637 cells (P<0.05; Fig. 6C), but had no significant effect on
the apoptosis of T24 cells.
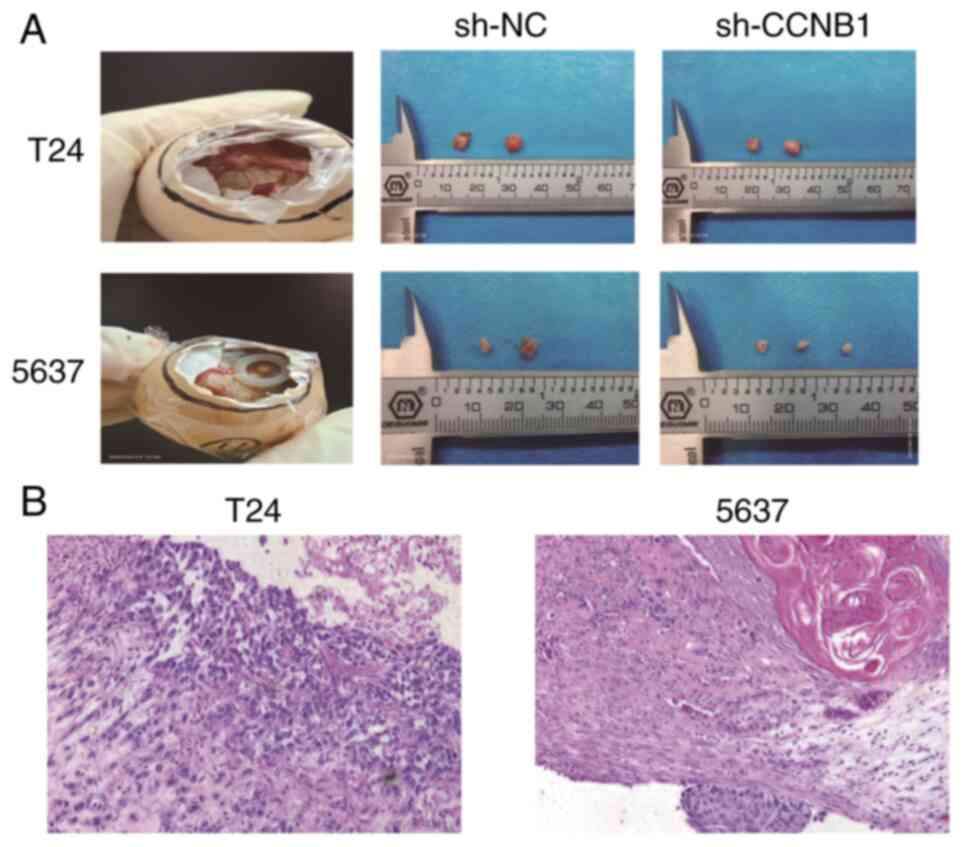
sh-CCNB1 inhibits tumorigenesis in
CAM
T24 and 5637 cells were inoculated into the CAM and
the survival of the chicken embryo was observed every 24 h. The
tumor cell suspension floated on the surface of the CAM in a thin
film within 24 h of inoculation, with no notable change in the
blood vessels. At day 6, the tumor protruded and small blood
vessels were observed in surrounding area. By day 6, the tumor
appeared in the AM, protruding from the CAM, which presented clear
borders. When small blood vessels were seen observed in a radial
pattern around the tumor center, the growth of chicken embryos were
terminated and tumors were removed at sixth day after inoculation.
Volumes of T24-sh-CCNB1 and 5637-sh-CCNB1 derived tumors were
smaller compared with sh-NC group (Fig. 7A). The diameter of the implanted
tumor was measured and its volume was calculated after it was
separated from CAM (Table I). This
experiment was repeated three times and it was concluded that the
volumes of T24-sh-CCNB1 and 5637-sh-CCNB1 were smaller than the
sh-NC group. Under light microscope in the group of T24-sh-CCNB1
and 5637-sh-CCNB1, HE-stained invasive cancer cells were
morphologically changed, exhibiting enlarged, angular or irregular,
dark-stained nuclei, some of which presented small or double
nucleoli. Fibrous connective tissue reaction was observed in the
interstitial tissue, with different degree of plasma cell and
lymphocyte infiltration. Squamous and glandular differentiation was
observed in focal cancer cells (Fig.
7B).
 | Table IBladder urothelial carcinoma cell
transplant tumor volume after 6 days. |
Table I
Bladder urothelial carcinoma cell
transplant tumor volume after 6 days.
| Group | Mean diameter,
mm | Mean volume,
mm³ |
|---|
| T24-sh-NC | 5.00 | 22.50 |
| T24-sh-CCNB1 | 2.75 | 12.38a |
| 5637-sh-NC | 3.25 | 14.63 |
| 5637-sh-CCNB1 | 1.33 | 5.985a |
Discussion
To date, curing cancer remains a challenge for
humans (14). The application of
cell cycle protein family in tumor therapy provides a novel
direction for cancer treatment (15). Many studies have demonstrated that
CCNB1 serves critical roles in tumor progression (16), but, to the best of our knowledge,
its effects and mechanisms in BLCA have not yet been reported. To
the best of our knowledge, the present study is the first to
demonstrate that CCNB1 was highly expressed in BLCA. sh-CCNB1 was
found to inhibit the proliferation, invasion, migration and tumor
growth of BLCA cells, indicating key roles in cell functions and
tumorigenesis of BLCA. Additionally, there was an association
between high CCNB1 expression and poor prognosis for patients with
BLCA. Therefore, exploring the precise roles of CCNB1 in BLCA may
provide a theoretical foundation for future clinical treatment of
BLCA.
As a member of the cell cycle protein family, CCNB1
is highly expressed in various types of tumor tissues and is
involved in tumorigenesis by regulating cell proliferation and
cycle distribution. For example, CCNB1 mRNA is highly expressed in
gastric cancer and serves as a downstream target gene of
heterogeneous nuclear ribonucleoprotein R (hnRNPR) protein, to
which it binds to form the hnRNPR/CCNB1/Centromere Protein F
(CENPF) mRNA axis, thus promoting proliferation of gastric cancer
cells (17); another study found
that CCNB1 partially reverses increased proliferation of gastric
cancer cells (18). In an ovarian
cancer study, high expression of CCNB1 is negatively regulated by
microRNA-559, thus promoting proliferation of ovarian cancer cells
in vitro and increasing the lung metastasis of ovarian
cancer cells in vivo (19).
By contrast, silencing of CCNB1 inhibits tumor cell proliferation
(20). Taken together, increasing
evidence suggests that a high expression of CCNB1 promotes cancer
cell proliferation and tumorigenesis (21). It was hypothesized that CCNB1 may
similarly impact the pathogenesis of BLCA. Therefore, the present
study constructed a BLCA and CAM model with sh-CCNB1. As expected,
the proliferation of BLCA cells and tumor growth in CAM was
significantly inhibited by sh-CCNB1.
As an allosteric modulator for cyclin-dependent
kinases CDK1 and Cdc2, CCNB1 had been documented to inhibit tumor
growth by influencing cell cycle events (22). For example, in renal cell
carcinoma, the downregulation of CCNB1 and CDK1 expression causes
G2/M arrest and apoptosis in tumor cells, indicating that upstream
eukaryotic translation initiation factor 3 subunit D (EIF3D) is
involved in development of renal cell carcinoma as a potential
proto-oncogene (23). Another
study demonstrated that diacetin, which is a symptomatic slow
acting drug in osteoarthritis, inhibits proliferation of
chondrosarcoma cells by inducing G2/M cell cycle arrest via
downregulation of CDK1/CCNB1(24).
Similarly, CCNB1, as a key regulator of cell cycle and promoter of
G2/M phase, improves the prognosis of platinum-based chemotherapy
in advanced non-small cell lung cancer (25). To determine the mechanism
underlying the effects of CCNB1 on BLCA, the present study analyzed
the cell cycle distribution and apoptotic rate in vitro.
Flow cytometry demonstrated that 5637 cells were significantly
arrested in the G2/M phase, the proportion of G0/G1 phase was
relatively decreased and the proportion of T24 cells in G0/G1 phase
decreased. These findings demonstrated that sh-CCNB1 arrested BLCA
cells at G2/M phase and inhibited BLCA cell proliferation.
Studies have investigated the potential roles of
CCNB1 in BLCA (26), demonstrating
that overexpressing G2 and S phase-expressed 1 (GTSE1)
promotes BLCA cell proliferation, invasion and migration through
the P53/FoxM1/CCNB1 pathway and is positively associated with
disease recurrence history, lymph node invasion and progression
(27). Differential gene
enrichment analysis by analyzing sequencing data from BLCA clinical
samples shows that CCNB1 is a hub gene expressed at 4.795-fold
higher levels in BLCA than in normal tissue, correlating with
overall survival (28). However,
the primary aim of the aforementioned studies was to explore the
role of the genes associated with CCNB1, not CCNB1. To the best of
our knowledge, the present study was the first to construct in
vivo and in vitro models to determine the role of CCNB1
in BLCA. CCNB1 mRNA expression and its clinical significance in
patients with BLCA was assessed. Analysis of 1149 BLCA and 201
non-cancerous bladder tissue samples in database sequencing data
confirmed high expression of CCNB1 in BLCA. Furthermore, the KM
survival curve suggested that patients with BLCA with high levels
of CCNB1 had a poorer prognosis than those with low levels of
CCNB1, which indicated that CCNB1 may serve as a prognostic
indicator for patients with BLCA. RT-qPCR, western blotting and
in vitro cell function and in vivo CAM xenograft
assay were performed to determine the effects of sh-CCNB1 on
proliferation, invasion, migration, cell cycle progression and
apoptosis of BLCA cells. The present results confirmed that
sh-CCNB1 inhibited proliferation, invasion and migration of BLCA
T24 and 5637 cells in vitro, significantly increased
proportion of cells in G2/M phase and inhibited growth of xenograft
tumors in vivo. Furthermore, the high expression of CCNB1
mRNA could distinguish BLCA from normal bladder tissue. CCNB1 in
tumor cells as a cell cycle progression modulator may be an
effective indicator to assess the malignancy of tumors (29). Therefore, targeting CCNB1 may be a
promising option for BLCA therapy.
The present study had limitations. First, bigger
clinical samples are required to validate the prognostic
significance of CCNB1 in BLCA. Second, although experiments on
chicken CAM transplantation tumors were performed, experimental
sample size should be increased. In vivo experiments on
other animals should also be conducted for validation, such as
tumor formation in nude mice. CCNB1 protein levels were
significantly higher in BLCA compared with normal bladder tissue
from immunohistochemical results of clinical samples in the HPA
database. This was not confirmed in clinical samples because it is
difficult to obtain normal bladder tissue as controls. Finally, the
present study only investigated the putative mechanism of action of
CCNB1 in BLCA. Information on the molecular mechanism is lacking
and the specific signaling pathways involved need further
investigation.
In summary, the present study demonstrated that
CCNB1 expression was upregulated in BLCA tumor tissue and sh-CCNB1
inhibited proliferation of BLCA cells, arresting most cells in the
G2/M phase and thereby inducing apoptosis. The present study
established a foundation for a understanding of CCNB1 function in
BLCA. CCNB1 may have a key role in the progression of BLCA.
Silencing of CCNB1 may have inhibited growth of BLCA tumors. The
present study may provide theoretical foundation for future
clinical treatment of BLCA.
Supplementary Material
Screening criteria for BLCA-associated
high-throughput sequencing data from GEO and TCGA. BLCA, bladder
urothelial carcinoma; GEO, Gene Expression Omnibus; TCGA, The
Cancer Genome Atlas; CCNB1, cyclin B1.
Datasets meeting the inclusion
criteria.
Acknowledgements
The authors would like to thank Dr Fang-Cheng Jiang
(Guangxi Medical University, Nanning, China) for assistance with
bioinformatics analysis.
Funding
Funding: The present study was supported by Guangxi Natural
Science Foundation (grant no. 2020GXNSFAA238027) and National
Natural Science Foundation of China (grant nos. 81860205 and
32060188).
Availability of data and materials
The datasets presented in this study (including
GSE2361, GSE3167, GSE5287, GSE2109, GSE7476, GSE17906, GSE31189,
GSE31684, GSE13507, GSE19423, GSE37815, GSE37817, GSE65635,
GSE86411, GSE19915, GSE24152, GSE40355, GSE52519, GSE76211,
TCGA-GTEx) can be found in online databases: Gene Expression
Omnibus (ncbi.nlm.nih.gov/geo/) and The Cancer
Genome Atlas (genome.gov/).
Authors' contributions
JP and JT confirm the authenticity of all the raw
data. XW performed all experiments, analyzed relevant data and
drafted the manuscript. HW participated in the design of most
experiments and proposed modifications. YY participated in the
design of some of the experiments. MM participated in the analysis
of the experimental data and revised the manuscript. YZ and HH
participated in the collection and analysis of bioinformatics data.
SL participated in the analysis of some of the experimental data
and proposed various suggestions. SP, JT and JP conceived and
designed the present study, provided financial support and revised
the manuscript. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Xu Y, Wu G, Li J, Li J, Ruan N, Ma L, Han
X, Wei Y, Li L, Zhang H, et al: Screening and identification of key
biomarkers for bladder cancer: A study based on TCGA and GEO Data.
Biomed Res Int. 82(83401)2020.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Schlack K, Boegemann M, Steinestel J,
Schrader A and Krabbe L: The safety and efficacy of gemcitabine for
the treatment of bladder cancer. Expert Rev Anticancer Ther.
16:255–271. 2016.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Shi S and Tian B: Identification of
biomarkers associated with progression and prognosis in bladder
cancer via co-expression analysis. Cancer Biomark. 24:183–193.
2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Cao R, Yuan L, Ma B, Wang G, Qiu W and
Tian Y: An EMT-related gene signature for the prognosis of human
bladder cancer. J Cell Mol Med. 24:605–617. 2020.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Xie B, Wang S, Jiang N and Li JJ: Cyclin
B1/CDK1-regulated mitochondrial bioenergetics in cell cycle
progression and tumor resistance. Cancer Lett. 443:56–66.
2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Petrachkova T, Wortinger LA, Bard AJ,
Singh J, Warga R and Kane D: Lack of Cyclin B1 in zebrafish causes
lengthening of G2 and M phases. Dev Biol. 451:167–179.
2019.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Marques HP, da Silva SG, De Martin E,
Agopian VG and Martins PN: Emerging biomarkers in HCC patients:
Current status. Int J Surg. 82s:70–76. 2020.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Zhang H, Zhang X, Li X, Meng WB, Bai ZT,
Rui SZ, Wang ZF, Zhou WC and Jin XD: Effect of CCNB1 silencing on
cell cycle, senescence, and apoptosis through the p53 signaling
pathway in pancreatic cancer. J Cell Physiol. 234:619–631.
2018.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Fang Y, Yu H, Liang X, Xu J and Cai X:
Chk1-induced CCNB1 overexpression promotes cell proliferation and
tumor growth in human colorectal cancer. Cancer Biol Ther.
15:1268–1279. 2014.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Zou Y, Ruan S, Jin L, Chen Z, Han H, Zhang
Y, Jian Z, Lin Y, Shi N and Jin H: CDK1, CCNB1, and CCNB2 are
prognostic biomarkers and correlated with immune infiltration in
hepatocellular carcinoma. Med Sci Monit. 26(e925289)2020.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Hu L, Pan X, Hu J, Zeng H, Liu X, Jiang M
and Jiang B: Proteasome inhibitors decrease paclitaxel-induced cell
death in nasopharyngeal carcinoma with the accumulation of
CDK1/cyclin B1. Int J Mol Med. 48(193)2021.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Zeng S, Yu X, Ma C, Song R, Zhang Z, Zi X,
Chen X, Wang Y, Yu Y, Zhao J, et al: Transcriptome sequencing
identifies ANLN as a promising prognostic biomarker in bladder
urothelial carcinoma. Sci Rep. 7(3151)2017.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Wu F, Sun Y, Chen J, Li H, Yao K, Liu Y,
Liu Q and Liu J: The oncogenic role of APC/C activator protein
Cdc20 by an integrated pan-cancer analysis in human tumors. Front
Oncol. 11(721797)2021.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Dobruch J and Oszczudłowski M: Bladder
cancer: Current challenges and future directions. Medicina
(Kaunas). 57(749)2021.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Jamasbi E, Hamelian M, Hossain MA and
Varmira K: The cell cycle, cancer development and therapy. Mol Biol
Rep. 49:10875–10883. 2022.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Fang L, Du WW, Awan FM, Dong J and Yang
BB: The circular RNA circ-Ccnb1 dissociates Ccnb1/Cdk1 complex
suppressing cell invasion and tumorigenesis. Cancer Lett.
459:216–226. 2019.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Chen EB, Qin X, Peng K, Li Q, Tang C, Wei
YC, Yu S, Gan L and Liu T: HnRNPR-CCNB1/CENPF axis contributes to
gastric cancer proliferation and metastasis. Aging (Albany NY).
11:7473–7491. 2019.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Li Y, Ji S, Fu LY, Jiang T, Wu D and Meng
FD: Knockdown of cyclin-dependent kinase inhibitor 3 inhibits
proliferation and invasion in human gastric cancer cells. Oncol
Res. 25:721–731. 2017.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Yang X, Zhou S, Yang C, Cao C, He M and Zi
S: CCNB1, negatively regulated by miR-559, promotes the
proliferation, migration, and invasion of ovarian carcinoma cells.
Mol Biotechnol. 64:958–969. 2022.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Gu J, Liu X, Li J and He Y: MicroRNA-144
inhibits cell proliferation, migration and invasion in human
hepatocellular carcinoma by targeting CCNB1. Cancer Cell Int.
19(15)2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Song Y, Zhao C, Dong L, Fu M, Xue L, Huang
Z, Tong T, Zhou Z, Chen A, Yang Z, et al: Overexpression of cyclin
B1 in human esophageal squamous cell carcinoma cells induces tumor
cell invasive growth and metastasis. Carcinogenesis. 29:307–315.
2008.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Sun J, Du Y, Song Q, Nan J, Guan PZ, Guo
JH, Wang X, Yang JB and Zhao CY: E2F is required for STAT3-mediated
upregulation of cyclin B1 and Cdc2 expressions and contributes to
G2-M phase transition. Acta Biochim Biophys Sin (Shanghai).
51:313–322. 2019.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Pan XW, Chen L, Hong Y, Xu DF, Liu X, Li
L, Huang Y, Cui LM, Gan SS, Yang QW, et al: Cui: EIF3D silencing
suppresses renal cell carcinoma tumorigenesis via inducing G2/M
arrest through downregulation of Cyclin B1/CDK1 signaling. Int J
Oncol. 48:2580–2590. 2016.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Lohberger B, Leithner A, Stuendl N,
Kaltenegger H, Kullich W and Steinecker-Frohnwieser B: Diacerein
retards cell growth of chondrosarcoma cells at the G2/M cell cycle
checkpoint via cyclin B1/CDK1 and CDK2 downregulation. BMC Cancer.
15(891)2015.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Ying Y, Wang Z, Tan Y, Cao H, Gao H, Zhang
Z, Zeng S and Xu C: Identification and validation of
immunohistochemical marker panels to predict the prognosis of
muscle invasive bladder cancer. Transl Androl Urol. 12:176–186.
2023.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Liu D, Xu W, Ding X, Yang Y, Su B and Fei
K: Polymorphisms of CCNB1 associated with the clinical outcomes of
platinum-based chemotherapy in Chinese NSCLC Patients. J Cancer.
8:3785–3794. 2017.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Liu A, Zeng S, Lu X, Xiong Q, Xue Y, Tong
L, Xu W, Sun Y, Zhang Z and Xu C: Overexpression of G2 and S
phase-expressed-1 contributes to cell proliferation, migration, and
invasion via regulating p53/FoxM1/CCNB1 pathway and predicts poor
prognosis in bladder cancer. Int J Biol Macromol. 123:322–334.
2019.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Chen Q, Hu J, Deng J, Fu B and Guo J:
Bioinformatics analysis identified key molecular changes in bladder
cancer development and recurrence. Biomed Res Int.
16(3917982)2019.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Brcic L, Heidinger M, Sever AZ, Zacharias
M, Jakopovic M, Fediuk M, Maier A, Quehenberger F, Seiwerth S and
Popper H: Prognostic value of cyclin A2 and B1 expression in lung
carcinoids. Pathology. 51:481–486. 2019.PubMed/NCBI View Article : Google Scholar
|