Introduction
Glioma is the most common primary brain tumor, among
which World Health Organization (WHO) grade IV glioma, such as
glioblastoma (GBM), is a highly aggressive and fatal malignant
tumor (1). In recent years,
numerous efforts have been made to improve the treatment strategy
of glioma; however, the prognosis of patients with malignant glioma
has not significantly improved (2). Therefore, it is necessary to
investigate the genetic and epigenetic mechanisms of gliomas to
identify objective diagnostic, classification and prognostic
indicators and develop novel treatment strategies (3). Previous studies have preliminarily
defined several molecular pathological subtypes of GBM and numerous
molecular markers for prognosis (4). However, the pathogenesis and
prognosis of gliomas are not yet fully understood. Therefore,
malignant glioma has been the focus of tumor therapy research.
MicroRNAs (miRNAs/miRs) are a type of non-coding RNA
with a length of 21-24 nucleotides, which are evolutionarily
conserved (5). miRNAs mainly
regulate the expression of target genes at the post-transcriptional
level by binding to the 3'-untranslated region (UTR) complementary
site of target mRNAs (6).
Therefore, miRNAs play an indispensable regulatory role in a number
of important physiological processes, including embryonic
development, cell differentiation, proliferation, apoptosis and
metabolism (7). According to the
association between miRNAs and the occurrence and development of
malignant tumors, they can be divided into two categories:
Oncogenic miRNAs (onco-miRNAs) and tumor-suppressive miRNAs
(TS-miRNAs) (8). It is known that
the abnormal increase in onco-miRNA expression and/or the abnormal
decrease in TS-miRNA expression are important factors leading to
the occurrence and development of a variety of malignant tumors
(9). Previous studies have
demonstrated that miR-137 is expressed at low levels in primary
gliomas, and glioma cell proliferation and invasion are
significantly inhibited following the overexpression of miR-137
(10-14).
The tumor inhibitory effects of miR-137 have been confirmed in a
variety of tumors, including pancreatic cancer, osteosarcoma,
gastric, oral, ovarian, liver and lung cancer (15-21).
The authors have previously reported the association
between sphingosine kinase (SPHK) 2 expression and
glioma-associated macrophages and the Ki-67 proliferation index
(22). It was demonstrated that
SPHK2 was overexpressed in glioma and was positively associated
with the Ki-67 index and M2-type tumor-associated macrophages
(TAMs) (22). TAMs play an
important role in the development, progression and invasion of
gliomas. Activated macrophages include M1 and M2. M1-type
macrophages can protect the organism from viruses and bacterial
infection and eliminate tumor cells, while M2-type macrophages, in
contrast to M1-type macrophages, exert immunosuppressive effects,
which not only promote the growth and invasion of glioma, but also
stimulate the formation of tumor-related blood vessels (23). However, the relevance of miR-137 to
TAMs in gliomas remains unknown.
In the present study, it was confirmed that the
downregulation of miR-137 resulted in the overexpression of SPHK2
in glioma, and that the upregulation of miR-137 may promote M1 TAMs
by directly targeting SPHK2. The data presented herein suggested
that miR-137 may be a novel diagnostic biomarker and a potential
therapeutic target for human malignant gliomas.
Materials and methods
Patient samples
A retrospective study was designed and included 60
patients with glioma (21 female patients, 39 male patients; age,
40.78±11.61 years, 18-64 years), who were admitted to The Shenzhen
Second People's Hospital (Shenzhen, China) between January 2014 and
December 2017. The following patients were excluded: i) Aged <18
years or ≥65 years; ii) those with other tumors or neurological
diseases. After obtaining the written consent from all participants
at The Second People's Hospital of Shenzhen (Shenzhen, China),
formalin-fixed paraffin-embedded (FFPE) samples were collected from
the Department of Pathology after diagnosis and treatment. The
tissue was fixed in 10% formalin at room temperature for 12-24 h
before embedding with paraffin. FFPE samples sections with a
thickness of 5-µm were cut for hematoxylin and eosin (H&E)
staining and miR-137 in situ hybridization (ISH). The
pathological diagnosis was made independently by two
neuropathologists according to the WHO classification of central
nervous system tumors in 2016(24). In a previous study, the authors
summarized the WHO classification and histopathological subtypes of
these gliomas (22). The present
study followed the principles of The Declaration of Helsinki and
was approved by the Ethics Committee of Shenzhen Second People's
Hospital (approval no. XZ2019103101).
ISH
The deparaffinized tissue was partially hybridized
with 50 nm LNA-modified and digoxin-labeled miR-137 oligonucleotide
probes (Exiqon A/S) for 1 h at 55˚C, incubated with 5 µg/ml
anti-digoxin-Rhodamine antibody (cat. no. 11093274910; Roche
Applied Science) overnight at 4˚C, and stained with DAPI (cat. no.
ZLI-9557; OriGene Technologies, Ins.) in the dark at room
temperature for 15 min. The labeling index (LI) was expressed as
the percentage of positive cells to the total number of cells.
Cell lines and culture
In the present study, the human GBM cell lines U251,
U373, SK-MG3, U-343, A172, LNZ-308, U118 and U138, and normal human
astrocytes (HAs) were used. These cell lines were purchased from
The Cell Bank of Type Culture Collection of the Chinese Academy of
Sciences and cultured in DMEM (Gibco; Thermo Fisher Scientific,
Inc.) supplemented with 10% fetal bovine serum (Gibco; Thermo
Fisher Scientific, Inc.) (25,26).
Authentications of U118MG and U373MG cell lines were detected with
STR profiling. According to the authentication, U118MG and U373MG
cell lines are from American Type Culture Collection. All cell
lines were cultured in a humidified incubator at 37˚C in 5%
CO2/95% air.
Lentiviral vector transduction
The 2nd generation system was used for lentivirus
transduction. 293T cells (The Cell Bank of Type Culture Collection
of The Chinese Academy of Sciences) were used as the interim cell
line. Lentivirus packaging was performed by Hanbio Biotechnology
Co., Ltd. in 293T cells using pSPAX2 (10 µg), pMD2G (5 µg), shuttle
plasmids (10 µg) and Lipofiter (75 µl; cat. no. HB-LF-1000; Hanbio
Biotechnology Co., Ltd.) for 16 h. A total of 48 and 72 h after
transfection, lentiviruses were collected because more viruses are
collected at two time points to ensure enough for the later
experiments. Lentiviruses collected 48 and 72 h after transfection
were both used in the subsequent experiments.
Hblv-cmv-miR-137-GFP-puro, hblv-GFP-puro (control),
hblv-cmv-SPHK2-GFP-puro [short hairpin RNA (sh)-SPHK2] and
hblv-GFP-puro (NC) vectors (Hanbio Biotechnology Co., Ltd.) were
used to transduce U251 and U373 cell lines at a multiplicity of
infection of 3 at 37˚C for 24 h. A total of 72 h after
transduction, puromycin (3 µg/ml; Gibco; Thermo Fisher Scientific,
Inc.) was added to the culture medium of transduced cells to create
stable cell lines for 7 days. The overexpression of miR-137 and
knockdown of SPHK2 following transduction with lentiviral vector
was detected according to the manufacturer's protocol.
Measurement of cytokine secretion
The culture supernatant was collected and the
cytokines were analyzed. The quantity of cytokine secretion was
quantified using the Quantibody® Human Inflammation
Array 1 (RayBiotech, Inc.) to measure the levels of 10 types of
human cytokines, including IFN-γ, IL-1α, IL-1β, IL-10, IL-13, IL-4,
IL-6, IL-8, monocyte chemoattractant protein-1 (MCP-1) and TNFα.
IL-13 was examined using an ELISA kit (cat. no. CHE0004; Beijing 4A
Biotech Co., Ltd.). TNF-α and IFN-γ were detected with ELISA Kits
purchased from PeproTech, Inc. (cat. nos. 900-M25 and 900-M27,
respectively).
Luciferase plasmid construction
The candidate targets of miR-137 were predicted
using TargetScan (http://www.targetscan.org/). The wild-type (p-WT) and
mutant-type (p-MT) reporter vectors of the SPHK2 3'-UTR were
constructed using the pEZX-MT01 Luciferase miRNA Expression
Reporter Vector (GeneCopoeia, Inc.). The predicted target coding
sequence of miR-137 was deleted from SPHK2 3'-UTR cDNA for p-MT
construction by site-directed mutagenesis. The sequence and
direction of the two vectors were verified with Sanger DNA
sequencing by Genscript, Inc.
Transfection with miR-137 mimics and
plasmids
dsRNA oligonucleotides of miR-137 mimics (cat. no.
miR10000429) and scramble sequence (Scr; cat. no. miR1N0000001-1-5)
were purchased from Guangzhou RiboBio Co., Ltd. dsRNA
oligonucleotides (2,500 ng; 50 nM) were transfected either alone or
with 0.75 µg/ml luciferase plasmids (p-WT and p-MT; GeneCopoeia,
Inc.) using X-tremeGENE small interfering RNA (siRNA) Transfection
reagent (cat. no. 4476093001; MilliporeSigma) for 48 h at 37˚C. A
total of 48 h after transfection, subsequent experimentation was
performed.
Dual-luciferase reporter assay
U373 and U251 cells (5,000 cells/well) inoculated
into 96-well plates were transfected with 0.15 µg p-WT or p-MT and
0.08 µg miR-137 mimics or scrambled sequences using X-tremeGENE
siRNA Transfection reagent for 48 h. Subsequently, the activities
of Renilla and Firefly luciferase were detected on a Synergy
2 Microplate Reader Fluorometer (BioTek Instruments, Inc.) using
the Dual-Luciferase Reporter Assay system (Promega Corporation).
The results were normalized to the measured value of Firefly
luciferase.
Reverse transcription-quantitative PCR
(RT-qPCR)
TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) was used to extract total RNA from the two
groups of cell lines. miR-137 was quantified using the Bulge-loop
miRNA RT-qPCR Detection kit (Guangzhou RiboBio Co., Ltd.). The qPCR
thermocycling conditions were as follows: Initial denaturation at
95˚C for 10 min, followed by 40 cycles of denaturation at 95˚C for
2 sec, annealing at 60˚C for 20 sec and extension at 70˚C for 10
sec; final dissociation was performed at 95˚C for 15 sec, 60˚C for
1 min and 95˚C for 15 sec. The expression of SPHK2 mRNA was
detected using the Reverse Transcription System and GoTaq qPCR
Master Mix kit (Promega Corporation) according to the
manufacturer's instructions. U6 and GAPDH were used as internal
controls for miR-137 and SPHK2 mRNA, respectively. All reactions
were performed on the CFX Connect™ Real-Time PCR
Detection system (Bio-Rad Laboratories, Inc.). The expression of
miR-137 and SPHK2 were calculated using the 2-ΔΔCq
method (27). The miR-137-3P
Primer Set (cat. no. MQPS0000619-1-100; Guangzhou RiboBio Co.,
Ltd.) and the U6 qPCR Primer Set (cat. no. MQPS0000002-1-100;
Guangzhou RiboBio Co., Ltd.) were used in these experiments.
Western blot analysis
Western blot analysis was performed as previously
described (28). The primary
antibodies used were rabbit anti-SPHK2 (1:1,000; cat. no. ab264042;
Abcam) and mouse anti-β-actin (1:1,000; cat. no. sc-81178; Santa
Cruz Biotechnology, Inc.), used for incubation at 4˚C overnight.
The secondary HRP-conjugated antibodies (used for incubation at
room temperature for 1 h) were as follows: Anti-mouse IgG (1:2,000;
cat. no. 7076) and Anti-rabbit IgG (1:2,000; cat. no. 7074) both
purchased from Cell Signaling Technology, Inc.
Statistical analyses
All statistical analysis was performed using SPSS
24.0 software (IBM Corp.). The data are expressed as the mean ± SD.
The normality of the distribution was estimated using the
Kolmogorov-Smirnov test. The unpaired double-tailed Student's
t-test was used to analyze the statistical difference between the
two groups. The differences among the sample groups were analyzed
by one-way ANOVA followed by multiple comparison Tukey's honestly
significant difference post hoc test. In order to determine the
optimal cut-off level of miR-137 expression for the diagnosis of
gliomas, receiver operating characteristic (ROC) curve analysis was
performed. The Youden index (J=sensitivity + specificity-1) was
used to determine the optimal cutoff level. The optimal cutoff
value was the cutoff value used to obtain the highest Youden index.
P<0.05 was considered to indicate a statistically significant
difference. All cell lines were tested at least three times and the
samples were repeated three times.
Results
miR-137 is downregulated in human
glioma and is associated with tumor grade
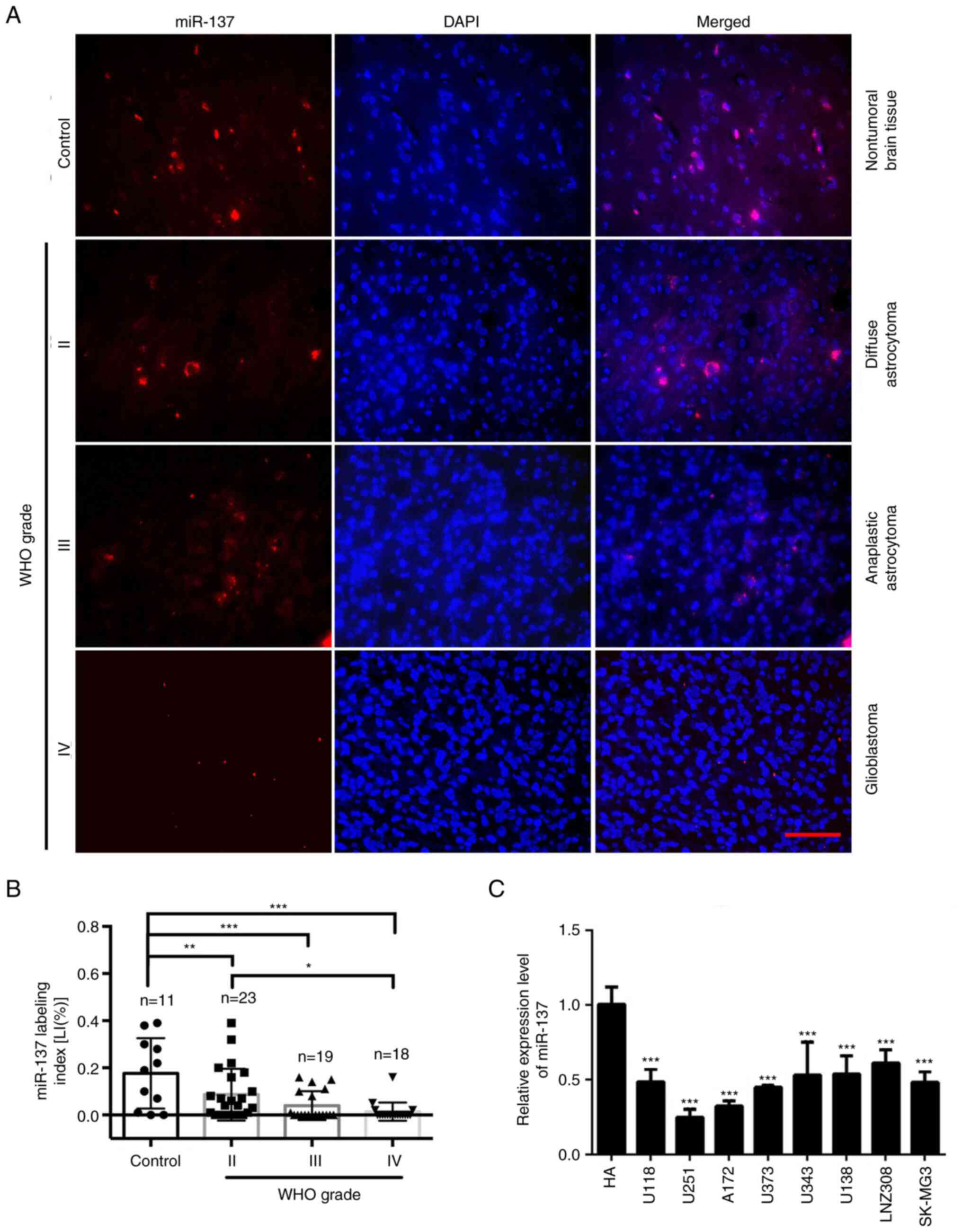
ISH with LNA-modified probes was applied to detect
endogenous miR-137 expression in FFPE specimens of 60 gliomas and
11 non-tumorous brain tissues. It was found that the expression of
miR-137 in gliomas was decreased compared with non-tumorous brain
tissues, and that its expression was significantly decreased as the
glioma grade increased; however, there was no significant
difference between two high-grade gliomas (WHO III grade and WHO IV
grade) (Fig. 1A and B). miR-137 expression was also detected
in 8 glioma cell lines and normal HAs. The results revealed that
the expression of miR-137 in all glioma cell lines was
significantly lower compared with normal HAs (Fig. 1C). The expression curve of miR-137
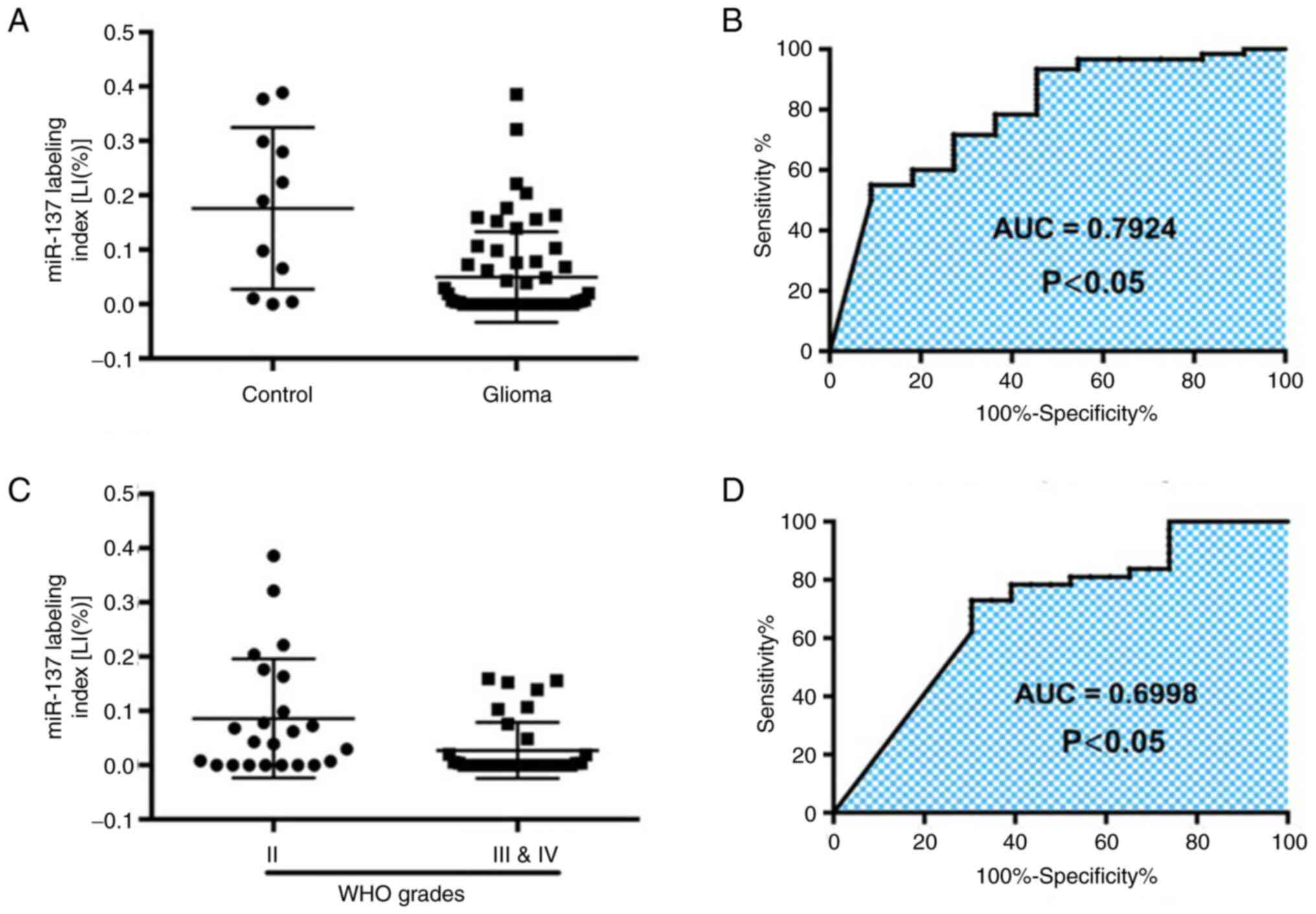
between non-tumorous brain tissues (control group) and glioma
tissues revealed that the area under the ROC curve was 0.7924 [95%
Confidence Interval (CI), 0.6425-0.9423; P<0.05]. As
demonstrated in Fig. 2A and
B, the expression of miR-137 may
prove helpful for the diagnosis of glioma. The optimal cutoff value
of the miR-137 LI was 0.37% (sensitivity, 55%; specificity,
90.91%). The ROC curve of miR-137 between low-grade (WHO II) and
high-grade (WHO III and V) gliomas revealed an area under the ROC
curve of 0.6998 (95% CI, 0.5584-0.8411; P<0.05; Fig. 2C and D), indicating that the expression of
miR-137 in gliomas contributes to the differentiation of glioma
grades. The optimal cutoff value of the miR-137 LI in the diagnosis
of glioma was 0.90% (sensitivity, 60%; specificity, 81.82%). These
data indicated the inverse association of miR-137 expression with
glioma and revealed that miR-137 may be a potential biomarker for
the diagnosis of patients with glioma.
miR-137 inhibits the release of IL-13,
and promotes the release of TNFα and IFNγ
To examine the effects of miR-137 on TAM-associated
release of cytokines, miR-137-overexpression cells were constructed
by transducing miR-137 overexpression lentiviral vector (empty
lentiviral vector transduced cells as control). The
Quantibody® Human Inflammation Array 1 was used to
detect the 10 human cytokines, including IFN-γ, IL-1α, IL-1β,
IL-10, IL-13, IL-4, IL-6, IL-8, MCP-1 and TNFα. The results
revealed that miR-137 overexpression reduced the IL-13 levels,
whereas it promoted TNFα and IFNγ production (Fig. 3A-C). As previously reported, IL-13
can promote M2 polarization, and TNFα and IFNγ can promote M1
polarization (29,30). Hence, it was hypothesized that
miR-137 may promote M1 TAM polarization by inhibiting M2-associated
cytokine and promoting M1-associated cytokine release. There were
no differences in the levels of IL-1α, IL-1β, IL-10, IL-4, IL-6,
IL-8 and MCP-1 between NC and miR-137 overexpression (data not
shown). To analyze the effect of SPHK2 on TAMs associated release
of cytokines, knockdown of SPHK2 was carried out in U251 and U373
cells at mRNA and protein levels (Fig.
3D-F). Two cell lines were transduced with lentivirus
containing knockdown shRNA sequence (HK as control group, and SH1,
SH2, SH3 as knockdown groups). After confirming SPHK2 knockdown,
IL-13, TNFα and IFNγ released by NC and sh-SPHK2 cells were
examined using ELISA. It was found that SPHK2 knockdown reduced
IL-13 concentration in U373 and U251 cells, promoted TNFα release
in U373 cells, and increased IFNγ release both in U373 and U251
cells (Fig. 3G-I).
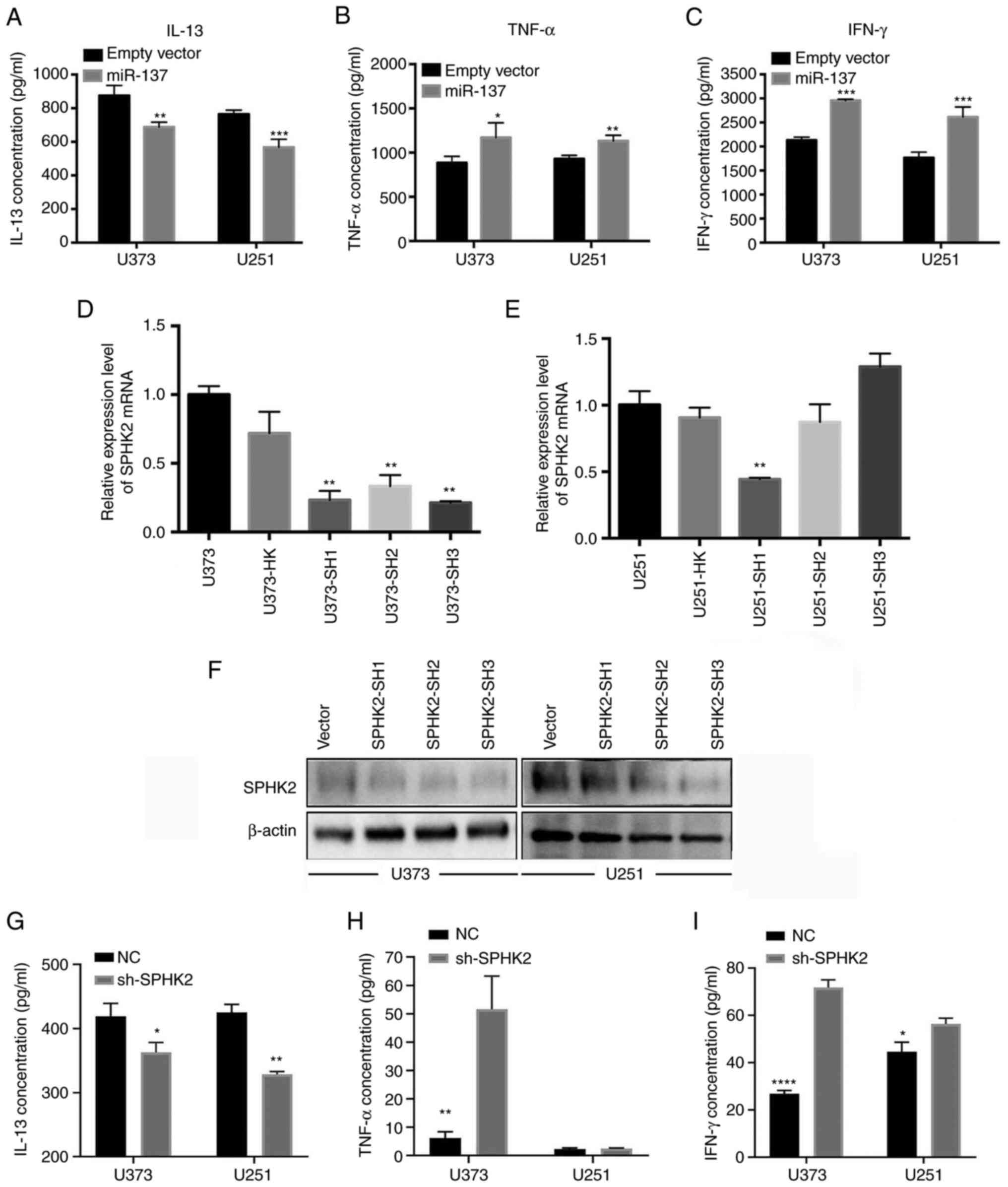 | Figure 3miR-137 inhibits the release of
IL-13, and promotes the release of TNFα and IFNγ. HK was used as
control group, and SH1, SH2, SH3 as knockdown groups. (A) miR-137
overexpression in glioma cells reduced IL-13 production. (B)
miR-137 overexpression in glioma cells promoted TNFα production.
(C) miR-137 overexpression in glioma cells promoted IFNγ
production.*P<0.05, **P<0.01 and
***P<0.001 vs. empty vector. (D and E) After
transduction with lentiviral vectors, SPHK2 mRNA was detected using
reverse transcription-quantitative PCR. **P<0.01 vs.
U373-HK. (F) SPHK2 protein level was assessed using western blot
analysis. (G-I) The supernatant from NC and sh-SPHK2 was collected,
and IL-13, TNF-α and IFN-γ were detected using ELISA.
*P<0.05, **P<0.01, and
****P<0.0001 vs. NC. miR, microRNA; SPHK2,
sphingosine kinase 2; sh-, short hairpin; NC, negative control. |
SPHK2 is a direct target of miR-137 in
human glioma cells
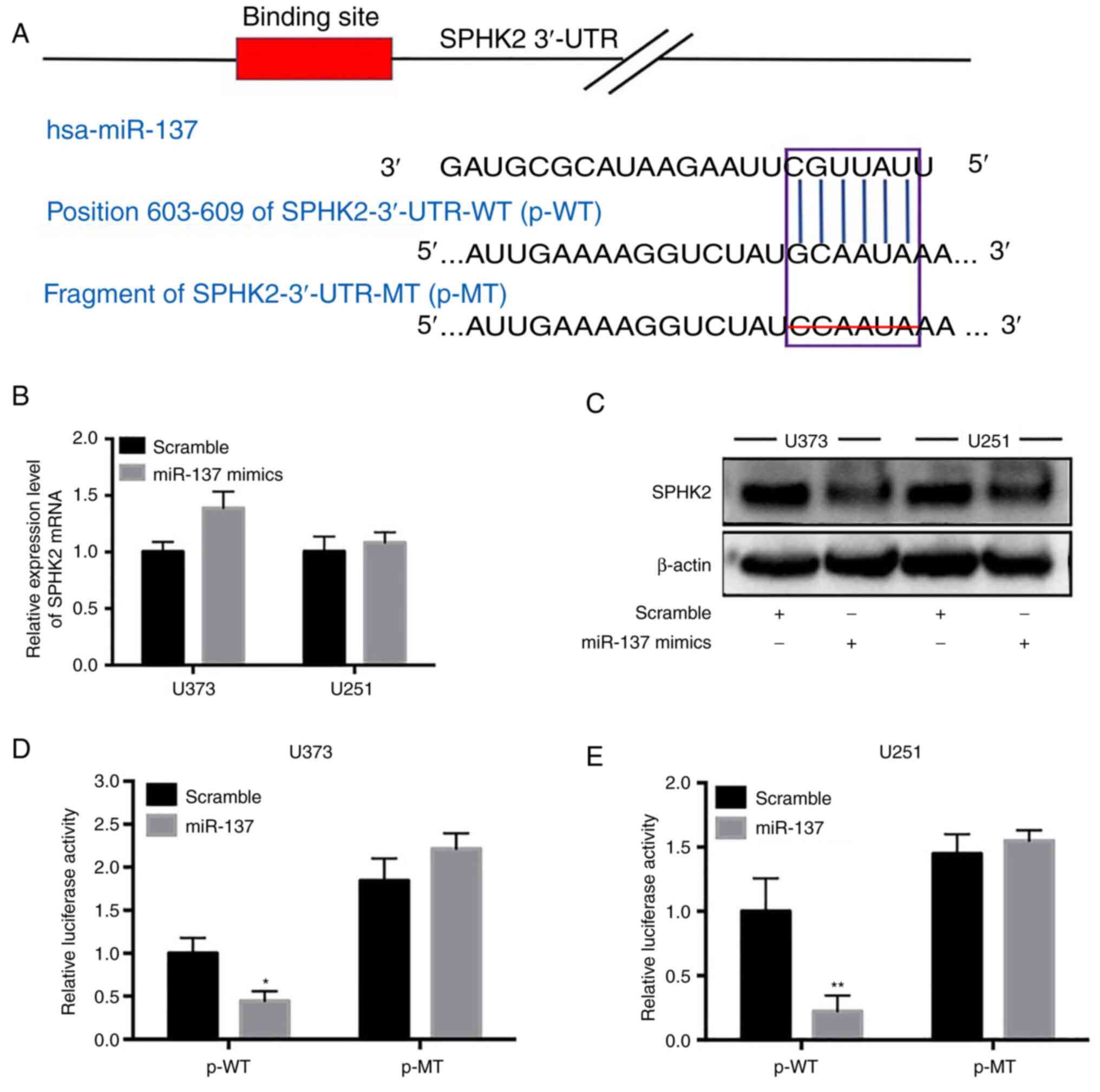
Using TargetScan, it was found that SPHK2 was one of
the potential targets of miR-137. miR-137 only had one putative
binding site in the SPHK2 3'-UTR (Fig.
4A). In order to obtain direct evidence of miR-137 targeting
SPHK2, two recombinant luciferase reporter gene vectors of SPHK2
3'-UTR were constructed: p-WT and p-MT. The recombinant luciferase
mRNA transcribed by p-WT carried the predicted miR-137 target
(SPHK2-3'-UTR-WT) in SPHK2-3'-UTR, while the mRNA transcribed by
p-MT lacked the predicted target (SPHK2-3'-UTR-MT) (Fig. 4A). The results of the
dual-luciferase reporter assay revealed that the relative
luciferase activity significantly decreased following p-WT
co-transfection with the miR-137 mimic (the scramble sequence was
used in the control group); however, no change was observed
following p-MT transfection (P<0.05; Fig. 4D and E). To further verify whether miR-137
directly targeted SPHK2, RT-qPCR and western blot analysis were
performed to detect the changes in SPHK2 expression in
miR-137-transfected cell lines. As revealed in Fig. 4B and C, SPHK2 protein expression in the
miR-137-transfected cell lines was significantly decreased compared
with the scramble control-transfected cell lines (Fig. 4C); however, the SPHK2 mRNA level
was not altered (Fig. 4B). These
results suggested that miR-137 may directly target SPHK2 in glioma
cells and downregulate SPHK2 protein expression by inhibiting its
mRNA translation.
Discussion
Over the years, research focusing on the
characteristics of glioma cells has made certain progress in
elucidating the molecular mechanisms responsible for the
occurrence, progress and prognosis of gliomas; however, the
treatment efficacy for gliomas has not improved. In recent years,
an increasing number of studies have demonstrated that tumor
resistance to cancer treatment is not necessarily an internal
problem, and the interaction between glioma and the tumor
microenvironment will also render the tumor resistant to drugs
(31-33).
It has been previously reported that during the process of tumor
development, a large number of immune cells accumulate around the
tumor, most of which are TAMs. TAMs are a ‘double-edged sword’,
which can play two different roles, either promoting inflammation
and exerting antitumor effects, or inhibiting inflammation and
exerting tumor-promoting effects (34). Therefore, it is of utmost
importance to guide the research and development of antitumor drugs
by inducing and amplifying the pro-inflammatory and antitumor
effects of TAMs.
Ceramide (Cer), sphingosine (Sph) and
sphingosine-1-phosphate (S1P), the metabolites of sphingomyelin,
play an important role in the development of a number of tumors
(35) by regulating cell
proliferation, survival and apoptosis. SPHK is the key enzyme which
maintains the metabolic balance between Cer, Sph and S1P (36). SPHK consists of two isomers, SPHK1
and SPHK2. SPHK1 has been proven to be a type of oncogenic kinase,
which is highly expressed in a number of types of tumors, and is
closely related to the occurrence and development of tumors
(35,37,38).
SPHK2 is highly expressed in colon, lung, liver and breast cancer,
and the inhibition of its expression can effectively suppress the
proliferation of tumor cells and promote cell apoptosis (39-42).
In papillary thyroid carcinoma (PTC), miR-613 can target SPHK2,
thus inhibiting the proliferation, migration and invasion of PTC
cells (43). In breast cancer, the
stimulation of EGF can promote the phosphorylation and activation
of SPHK2, thus increasing the production of S1P and promoting the
migration of breast cancer cells (44). In addition, as previously
demonstrated, in a mouse xenograft model of breast cancer following
SPHK2 knockdown, TAMs presented an antitumor phenotype and
expressed pro-inflammatory markers, indicating that SPHK2 was
closely related to the phenotype of TAMs (36). In a previous study, it was revealed
that SPHK2 protein was highly expressed in gliomas, and it was
positively associated with the infiltration of M2-type TAMs and the
proliferation index of Ki-67(23).
Therefore, the abnormal increase in SPHK2 expression may be an
important reason for TAM infiltration and M2 polarization in
glioma, and thus an important factor for the progression of
glioma.
An increasing number of studies have demonstrated
that miRNA plays an important role in determining the functional
state and differentiation performance of TAMs. Numerous miRNAs in
macrophages can increase pro-inflammatory signaling (including
miR-155 and miR-125a/b) or weaken the function of M2 macrophages
(including miR-511-3p, miR-378 and miR-155) to maintain the M1 TAM
phenotype, while others (such as miR-146a, miR-187, miR-21 and
miR-147) can inhibit the function of positive regulatory factors in
the macrophage pro-inflammatory signaling pathway, thus promoting
the M2 polarization of TAMs (45-47).
Previous studies have demonstrated that miR-137 expression is
downregulated in gliomas, and glioma cell proliferation and
invasion are significantly inhibited following the overexpression
of miR-137 (11,12). The tumor-suppressive effects of
miR-137 have been confirmed in a variety of tumors, including
pancreatic cancer, osteosarcoma, gastric, oral, ovarian, liver and
lung cancer (13-17).
The findings of the present study demonstrated that the expression
levels of miR-137 were downregulated in glioma cell lines and
tissues, which further suggests the antitumor role of miR-137 in
glioma. Bioinformatics analysis predicted that SPHK2 was a
potential target gene of miR-137 and the experimental results also
confirmed that prediction. Moreover, miR-137 inhibited the release
of IL-13, and promoted the release of TNFα and IFNγ, and
subsequently promoted M1 TAM polarization. Research has indicated
that the intra-nuclear SPHK2-S1P axis facilitates the M1-to-M2
shift of the microglia by suppressing histone deacetylase
1-mediated Kruppel-like factor 4 deacetylation (48). The aforementioned study also
demonstrated the positive association between SPHK2 and M2-type TAM
infiltration. Thus, it was hypothesized that miR-137 may promote M1
TAM polarization by inhibiting M2-associated cytokine release and
promoting M1-associated cytokine release by targeting SPHK2.
In conclusion, the present study demonstrated that
miR-137 expression was downregulated in glioma and that its
overexpression promoted M1-type TAM polarization by targeting
SPHK2. miR-137 functions as a tumor suppressor and may thus be used
as a diagnostic biomarker and therapeutic target for glioma.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the Shenzhen Science
and Technology Innovation Commission (grant nos.
JCYJ20170306090714854 and JCYJ20200109120205924), and the
University of South China Innovation Foundation for Postgraduate
(grant no. 203YXC038).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
JL, YX and HT performed the experiments and acquired
data. JL and YX confirm the authenticity of all the raw data. YZ
and GH contributed to conception, design and revision. XL, YS, TW,
MG, PC and HH were involved in acquisition and analysis of data and
drafting the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Shenzhen Second People's Hospital. Signed informed
consents were obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ahmed R, Oborski MJ, Hwang M, Lieberman FS
and Mountz JM: Malignant gliomas: Current perspectives in
diagnosis, treatment, and early response assessment using advanced
quantitative imaging methods. Cancer Manag Res. 6:149–170.
2014.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Wen PY and Kesari S: Malignant gliomas in
adults. N Engl J Med. 359:492–507. 2008.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Bastien JI, McNeill KA and Fine HA:
Molecular characterizations of glioblastoma, targeted therapy, and
clinical results to date. Cancer. 121:502–516. 2015.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Cohen AL and Colman H: Glioma biology and
molecular markers. Cancer Treat Res. 163:15–30. 2015.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Carthew RW and Sontheimer EJ: Origins and
mechanisms of miRNAs and siRNAs. Cell. 136:642–655. 2009.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Chipman LB and Pasquinelli AE: miRNA
targeting: Growing beyond the seed. Trends Genet. 35:215–222.
2019.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Esteller M: Non-coding RNAs in human
disease. Nat Rev Genet. 12:861–874. 2011.PubMed/NCBI View
Article : Google Scholar
|
|
8
|
Svoronos AA, Engelman DM and Slack FJ:
OncomiR or tumor suppressor? The duplicity of MicroRNAs in cancer.
Cancer Res. 76:3666–3670. 2016.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Nakamura K, Sawada K, Yoshimura A, Kinose
Y, Nakatsuka E and Kimura T: Clinical relevance of circulating
cell-free microRNAs in ovarian cancer. Mol Cancer.
15(48)2016.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Ji ZG, Jiang HT and Zhang PS: FOXK1
promotes cell growth through activating wnt/β-catenin pathway and
emerges as a novel target of miR-137 in glioma. Am J Transl Res.
10:1784–1792. 2018.PubMed/NCBI
|
|
11
|
Chen L, Wang X, Wang H, Li Y, Yan W, Han
L, Zhang K, Zhang J, Wang Y, Feng Y, et al: miR-137 is frequently
down-regulated in glioblastoma and is a negative regulator of
Cox-2. Eur J Cancer. 48:3104–3111. 2012.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Xiao J, Peng F, Yu C, Wang M, Li X, Li Z,
Jiang J and Sun C: microRNA-137 modulates pancreatic cancer cells
tumor growth, invasion and sensitivity to chemotherapy. Int J Clin
Exp Pathol. 7:7442–7450. 2014.PubMed/NCBI
|
|
13
|
Wang L, Liu J, Zhong Z, Gong X, Liu W, Shi
L and Li X: PTP4A3 is a target for inhibition of cell proliferatin,
migration and invasion through Akt/mTOR signaling pathway in
glioblastoma under the regulation of miR-137. Brain Res.
1646:441–450. 2016.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Li KK, Yang L, Pang JC, Chan AK, Zhou L,
Mao Y, Wang Y, Lau KM, Poon WS, Shi Z and Ng HK: MIR-137 suppresses
growth and invasion, is downregulated in oligodendroglial tumors
and targets CSE1L. Brain Pathol. 23:426–439. 2013.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Ding F, Zhang S, Gao S, Shang J, Li Y, Cui
N and Zhao Q: MiR-137 functions as a tumor suppressor in pancreatic
cancer by targeting MRGBP. J Cell Biochem. 119:4799–4807.
2018.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Li ZM, Zhang HY, Wang YX and Wang WB:
MicroRNA-137 is downregulated in human osteosarcoma and regulates
cell proliferation and migration through targeting FXYD6. J Drug
Target. 24:102–110. 2016.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Zheng X, Dong J, Gong T, Zhang Z, Wang Y,
Li Y, Shang Y, Li K, Ren G, Feng B, et al: MicroRNA library-based
functional screening identified miR-137 as a suppresser of gastric
cancer cell proliferation. J Cancer Res Clin Oncol. 141:785–795.
2015.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Kozaki K, Imoto I, Mogi S, Omura K and
Inazawa J: Exploration of tumor-suppressive microRNAs silenced by
DNA hypermethylation in oral cancer. Cancer Res. 68:2094–2105.
2008.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Chen W, Du J, Li X, Zhi Z and Jiang S:
microRNA-137 downregulates MCL1 in ovarian cancer cells and
mediates cisplatin-induced apoptosis. Pharmacogenomics. 21:195–207.
2020.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Wu DC, Zhang MF, Su SG, Fang HY, Wang XH,
He D, Xie YY and Liu XH: HEY2, a target of miR-137, indicates poor
outcomes and promotes cell proliferation and migration in
hepatocellular carcinoma. Oncotarget. 7:38052–38063.
2016.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Chang TH, Tsai MF, Gow CH, Wu SG, Liu YN,
Chang YL, Yu SL, Tsai HC, Lin SW, Chen YW, et al: Upregulation of
microRNA-137 expression by Slug promotes tumor invasion and
metastasis of non-small cell lung cancer cells through suppression
of TFAP2C. Cancer Lett. 402:190–202. 2017.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Liu J, Zhou Q, Wu CP, Xu YW, Liu WL, Zhao
HF and Li WP: SPHK2 protein expression, Ki-67 index and
infiltration of tumor-associated macrophages (TAMs) in human
glioma. Histol Histopathol. 33:987–994. 2018.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Domingues P, González-Tablas M, Otero Á,
Pascual D, Miranda D, Ruiz L, Sousa P, Ciudad J, Goncalves JM,
Lopes MC, et al: Tumor infiltrating immune cells in gliomas and
meningiomas. Brain Behav Immun. 53:1–15. 2016.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Brouland JP and Hottinger AF: Revised WHO
classification 2016 of gliomas: What's new? Rev Med Suisse.
13:1805–1809. 2017.PubMed/NCBI(In French).
|
|
25
|
Zhu J, Cai Y, Liu P and Zhao W.: Frequent
Nek1 overexpression in human gliomas. Biochem Biophys Res Commun.
476:522–527. 2016.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Barrett JW, Alston LR, Wang F, Stanford
MM, Gilbert PA, Gao X, Jimenez J, Villeneuve D, Forsyth P and
McFadden G: Identification of host range mutants of myxoma virus
with altered oncolytic potential in human glioma cells. J
Neurovirol. 13:549–560. 2007.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Liu J, Yang J, Yu L, Rao C, Wang Q, Sun C,
Shi C, Hua D, Zhou X, Luo W, et al: miR-361-5p inhibits glioma
migration and invasion by targeting SND1. Onco Targets Ther.
11:5239–5252. 2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Orihuela R, McPherson CA and Harry GJ:
Microglial M1/M2 polarization and metabolic states. Br J Pharmacol.
173:649–665. 2016.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Batra R, Suh MK, Carson JS, Dale MA,
Meisinger TM, Fitzgerald M, Opperman PJ, Luo J, Pipinos II, Xiong W
and Baxter BT: IL-1β (interleukin-1β) and TNF-α (tumor necrosis
factor-α) impact abdominal aortic aneurysm formation by
differential effects on macrophage polarization. Arterioscler
Thromb Vasc Biol. 38:457–463. 2018.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Berindan-Neagoe I and Calin GA: Molecular
pathways: microRNAs, cancer cells, and microenvironment. Clin
Cancer Res. 20:6247–6253. 2014.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Zheng Y, Bao J, Zhao Q, Zhou T and Sun X:
A spatio-temporal model of macrophage-mediated drug resistance in
glioma immunotherapy. Mol Cancer Ther. 17:814–824. 2018.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Khan S, Mittal S, McGee K, Alfaro-Munoz
KD, Majd N, Balasubramaniyan V and de Groot JF: Role of neutrophils
and myeloid-derived suppressor cells in glioma progression and
treatment resistance. Int J Mol Sci. 21(1954)2020.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Hambardzumyan D, Gutmann DH and Kettenmann
H: The role of microglia and macrophages in glioma maintenance and
progression. Nat Neurosci. 19:20–27. 2016.PubMed/NCBI View
Article : Google Scholar
|
|
35
|
Zheng X, Li W, Ren L, Liu J, Pang X, Chen
X, Kang D, Wang J and Du G: The sphingosine
kinase-1/sphingosine-1-phosphate axis in cancer: Potential target
for anticancer therapy. Pharmacol Ther. 195:85–99. 2019.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Weigert A, Schiffmann S, Sekar D, Ley S,
Menrad H, Werno C, Grosch S, Geisslinger G and Brüne B: Sphingosine
kinase 2 deficient tumor xenografts show impaired growth and fail
to polarize macrophages towards an anti-inflammatory phenotype. Int
J Cancer. 125:2114–2121. 2009.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Hatoum D, Haddadi N, Lin Y, Nassif NT and
McGowan EM: Mammalian sphingosine kinase (SphK) isoenzymes and
isoform expression: Challenges for SphK as an oncotarget.
Oncotarget. 8:36898–36929. 2017.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Liu H, Ma Y, He HW, Zhao WL and Shao RG:
SPHK1 (sphingosine kinase 1) induces epithelial-mesenchymal
transition by promoting the autophagy-linked lysosomal degradation
of CDH1/E-cadherin in hepatoma cells. Autophagy. 13:900–913.
2017.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Nemoto S, Nakamura M, Osawa Y, Kono S,
Itoh Y, Okano Y, Murate T, Hara A, Ueda H, Nozawa Y and Banno Y:
Sphingosine kinase isoforms regulate oxaliplatin sensitivity of
human colon cancer cells through ceramide accumulation and Akt
activation. J Biol Chem. 284:10422–10432. 2009.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Dai L, Smith CD, Foroozesh M, Miele L and
Qin Z: The sphingosine kinase 2 inhibitor ABC294640 displays
anti-non-small cell lung cancer activities in vitro and in vivo.
Int J Cancer. 142:2153–2162. 2018.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Xiao G, Wang Q, Li B, Wu X, Liao H, Ren Y
and Ai N: MicroRNA-338-3p suppresses proliferation of human liver
cancer cells by targeting SphK2. Oncol Res. 26:1183–1189.
2018.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Wang W, Hind T, Lam BWS and Herr DR:
Sphingosine 1-phosphate signaling induces SNAI2 expression to
promote cell invasion in breast cancer cells. FASEB J.
33:7180–7191. 2019.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Qiu W, Yang Z, Fan Y and Zheng Q:
MicroRNA-613 inhibits cell growth, migration and invasion of
papillary thyroid carcinoma by regulating SphK2. Oncotarget.
7:39907–39915. 2016.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Hait NC, Sarkar S, Le Stunff H, Mikami A,
Maceyka M, Milstien S and Spiegel S: Role of sphingosine kinase 2
in cell migration toward epidermal growth factor. J Biol Chem.
280:29462–29469. 2005.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Squadrito ML, Etzrodt M, De Palma M and
Pittet MJ: MicroRNA-mediated control of macrophages and its
implications for cancer. Trends Immunol. 34:350–359.
2013.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Yin R, Zhu X, Wang J, Yang S, Ma A, Xiao
Q, Song J and Pan X: MicroRNA-155 promotes the ox-LDL-induced
activation of NLRP3 inflammasomes via the ERK1/2 pathway in THP-1
macrophages and aggravates atherosclerosis in ApoE-/-mice. Ann
Palliat Med. 8:676–689. 2019.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Zhang L, Fu Y, Wang H, Guan Y, Zhu W, Guo
M, Zheng N and Wu Z: Severe fever with thrombocytopenia syndrome
virus-induced macrophage differentiation is regulated by miR-146.
Front Immunol. 10(1095)2019.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Ji J, Wang J, Yang J, Wang XP, Huang JJ,
Xue TF and Sun XL: The intra-nuclear SphK2-S1P axis facilitates
M1-to-M2 shift of microglia via suppressing HDAC1-mediated KLF4
deacetylation. Front Immunol. 10(1241)2019.PubMed/NCBI View Article : Google Scholar
|