Introduction
Gastric cancer is one of the commonest malignant
tumors in the world and one of the four major causes of
malignancy-related deaths (1).
According to GLOBOCAN 2020 statistics, the incidence of gastric
cancer in East Asia, including China, holds the first place. Nearly
1/3 of the patients with gastric cancer have advanced stage when
diagnosed and this is usually associated with poor prognosis
(2,3). The overall survival rate of patients
with gastric cancer is still very low, although various clinical
methods are available for the treatment of gastric cancer, such as
surgery, chemoradiotherapy and immunotherapy (4,5).
Therefore, the identification of new molecular targets of gastric
cancer and the investigation of the potential mechanism of action
of this disease have become the focus of the current research field
in gastric cancer.
Caspase recruitment domain-containing protein (CARD)
11 is an important member of the CARD family of proteins and is
mainly present in lymphocytes (6).
Phosphorylation of CARD11 by protein kinase C family isoenzymes
causes conformational change in the interior of the CARD11
molecule, which leads to recruitment of B-cell lymphoma/leukemia 10
(Bcl-10) to CARD11. Bcl-10 binds to muco-associated lymphoid tissue
lymphoma translocation protein 1 (MALT1) to form an oligomeric
CARMA/CARD-Bcl-10-MALT1 (CBM) complex (7). In activated B-cell-like diffuse large
B cell lymphoma, mutation in the CARD11 gene can activate CARD11
activity and selectively enhance its binding activity towards
Bcl-10, further leading to the activation of the NF-κB signaling
pathway and the development of tumorigenicity (8). Patients with chronic lymphocytic
leukemia and high CARD11 expression exhibit a poor survival rate
(9). Furthermore, CTD-2020K17.1
regulates CARD11 expression to promote the migration, invasion and
proliferation of ovarian cancer cells (10). In addition, CARD11 is mutated in
human skin squamous cell carcinoma, leading to abnormal NF-κB
regulation and increased CARD11 mRNA levels in skin squamous cell
carcinoma (11). However, the role
of CARD11 in gastric cancer has not been previously reported. The
encyclopedia of RNA interactomes (ENCORI) database (https://starbase.sysu.edu.cn/index.php,
version 3.0) indicated that CARD11 was highly expressed in gastric
cancer and that it was associated with poor disease prognosis.
HumanTFDB (http://bioinfo.life.hust.edu.cn/HumanTFDB#!/, version
3.0) predicted the binding of the transcription factor Krüppel-like
factor 5 (KLF5) to the CARD11 promoter. Previous studies have shown
that KLF5 activates long non-coding RNA DANCR to inhibit autophagy
of cancer cells, thereby accelerating the progression of gastric
cancer (12). KLF5 is activated by
gene amplification in gastric cancer and promotes gastric cancer
cell proliferation (13).
Therefore, the present study aimed to detect the expression levels
of KLF5 and CARD11 in gastric cancer cells and to investigate the
underlying mechanism of action.
Materials and methods
Cell culture and cell
transfection
Human normal gastric epithelial cells (GES-1) and
human gastric cancer cells (MKN-45, KE-39 and AGS) were provided by
BioVector NTCC Inc. The cells were cultured in RPMI-1640 medium
(Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine
serum (FBS; Gibco; Thermo Fisher Scientific, Inc.), 100 µg/ml
streptomycin and 100 units/ml penicillin at 37˚C in the presence of
5% CO2.
Small interfering (si) RNA of CARD11 (si-CARD11#1,
AGCTAAAGCACCGGTTGAATAAG; si-CARD11#2, CAGTCTCTAAAACTGAAGAATGA) or
siRNA negative control (NC, AAGACAUUGUGUGUCCGCCTT) were synthesized
by Guangzhou RiboBio Co., Ltd. The KLF5 overexpression vector
pcDNA3.0-KLF5 (oe-KLF5) and the empty plasmid pcDNA3.0 (oe-NC) were
obtained from MiaolingBio. AGS cells were seeded into 6-well plates
at a density of 2x105 cells/well and cultured until the
cell confluence reached 80%. Subsequently, a total of 100 nM
plasmids were transfected into AGS cells using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) at 37˚C for 48 h. Following 48 h of culture at
37˚C, the cells were harvested and used for further
experiments.
Reverse transcription-quantitative
(RT-q) PCR
Following transfection, 1x104 AGS cells
were collected and treated with TRIzol® (Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions to
obtain the total RNA. The first-strand cDNA was reverse transcribed
from RNA using the PrimeScript RT Reagent kit (Takara Bio, Inc.)
following the manufacturer's instructions and qPCR was carried out
by SYBR Real-time PCR kit (Takara Bio, Inc.) on an ABI 7500 system
(Applied Biosystems; Thermo Fisher Scientific, Inc.) according to
the manufacturer's instructions. The thermocycling conditions were:
Initial denaturation at 95˚C for 3 min; followed by 40 cycles of
denaturation at 95˚C for 30 sec, annealing at 60˚C for 30 sec and
extension at 72˚C for 30 sec. The following were the sequences of
primers: CARD11 forward primer: 5'-GCTCACAACCGCATCCCAA-3', reverse
primer: 5'-CTCCTCATGACCGCCATGTT-3'; KLF5 forward primer:
5'-AGCTACAATACGCTTGGCCT-3', reverse primer:
5'-ATGTGTGTTACGCACGGTCT-3'; GAPDH forward primer:
5'-GGAGCGAGATCCCTCCAAAAT-3', reverse primer:
5'-GGCTGTTGTCATACTTCTCATGG-3'. The mRNA expression levels of CARD11
and KLF5 were quantified using the 2-∆∆Cq method and the
internal control used was GAPDH (14). All experiments were replicated
three times.
Western blot analysis
Following transfection, total proteins were
extracted from AGS cells using RIPA lysis buffer (Beyotime
Institute of Biotechnology) and protein concentration was
determined using bicinchoninic protein quantification kit (Beyotime
Institute of Biotechnology). Equal amounts of protein samples (60
µg/lane) separated by 10% SDS-PAGE were transferred to
polyvinylidene fluoride (PVDF) membranes (Millipore). Subsequently,
the membranes were blocked with 5% non-fat dry milk at room
temperature for 2 h and incubated with primary antibodies,
including those against CARD11 (1:1,000; cat. no. ab124730; Abcam),
E-cadherin (1:1,000; cat. no. ab40772; Abcam), N-cadherin (1:5,000;
cat. no. ab76011; Abcam), vimentin (1:1,000; cat. no. ab92547;
Abcam), KLF5 (1:1,000; cat. no. ab137676; Abcam), phosphorylated
(p)-mTOR (1:1,000; cat. no. ab109268; Abcam), p-P70S6K (1:1,000;
cat. no. ab59208; Abcam), mTOR (1:10,000; cat. no. ab134903;
Abcam), P70S6K (1:5,000; cat. no. ab32529; Abcam) and GAPDH
(1:2,500; cat. no. ab9485; Abcam) at 4˚C overnight; the following
day, the membrane was incubated with horseradish peroxidase
(HRP)-conjugated secondary antibodies (1:2,000; cat. no. ab6721;
Abcam) at room temperature for 1 h. The protein bands were
presented using ECL Plus (Thermo Fisher Scientific Inc.) and
quantified by ImageJ 1.51 software (National Institutes of
Health).
Cell Counting Kit-8 (CCK-8) assay
The cell viability was detected using CCK-8 assay
(Beyotime Institute of Biotechnology). Following transfection, AGS
cells were cultured for 48 h, seeded into a 96-well plate
(3x103 cells/well) and further cultured at 37˚C, in the
presence of 5% CO2. Subsequently, 10 µl CCK-8 was added
to the cultured cells at 24, 48 and 72 h and further cultured for
an additional 1 h. The absorbance value (OD value) of the cells at
490 nm was detected by a microplate reader.
5-ethynyl-2'-deoxyuridine (EdU)
staining
An EdU kit (BeyoClick™ EdU Cell
Proliferation Kit with Alexa Fluor 488; Beyotime Institute of
Biotechnology) was used to detect cell proliferation. Following
culture for three days at 37˚C in 24-well plates, AGS cells were
incubated with EdU, fixed with 4% paraformaldehyde for 15 min at
room temperature and stained with Hoechst 33342 for 2, 0.5 h and 10
min. Finally, the proliferation of the cells was observed and
images captured by an inverted fluorescence microscope.
Flow cytometry analysis
Following transfection, AGS cells were incubated
into 24-well plates and cultured for 24 h. The cells were
centrifuged at 300 x g for 5 min and the medium was discarded.
Subsequently, they were washed with pre-chilled PBS twice and fixed
with 75% ethanol at 4˚C overnight. Following centrifugation, the
cells were fixed with 100 µg/ml RNase A at 37˚C for 30 min and
subsequently stained with 50 µg/ml PI (Thermo Fisher Scientific,
Inc.) for 5 min in the dark. Finally, the cell cycle was analyzed
by flow cytometry (NovoCyte 2060R; ACEA Biosciences, Inc.) with
Software NovoExpress 1.4.0 (ACEA Biosciences, Inc.).
Wound healing assay
Following transfection, 2x105 AGS cells
were added to the each well of the 6-well plate and was cultured to
form a cell monolayer. The cell monolayer was scratched by a
sterilized 10-µl pipette tip, followed by PBS washing to remove the
detached cells. Subsequently, the obtained cells were cultured in
RPMI-1640 medium containing 2% FBS for 24 h at 37˚C. Then, the
images of cells were observed under a light microscope (Thermo
Fisher Scientific, Inc.) and the migration distance was analyzed by
ImageJ software (version 1.8.0; National Institutes of Health).
Transwell assay
Following transfection, AGS cells were collected and
5x104 cells were added to the serum-free medium (200 µl)
in the upper chambers coated with Matrigel at 37˚C for 1 h.
RPMI-1640 medium containing 10% FBS was added to the lower
chambers. Following culture for 24 h at 37˚C, the cells in the
upper chamber were removed and the obtained cells were fixed with
95% ethanol, stained with 0.1% crystal violet for 10 min at room
temperature and washed by PBS. The observation of the invasive
cells was performed using an inverted microscope.
Dual luciferase reporter assay
Wild-type (WT) and mutant (mut) sequences of
pleckstrin homology-like domain family A member 3 were cloned and
inserted into the pGL4.13 reporter vector (Promega Corporation).
Subsequently, AGS cells were transfected with the luciferase
reporter, oe-KLF5 and oe-NC using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) for 48 h at 37˚C. At
48 h post-transfection, the Luc-Pair Duo-Luciferase HS Assay kit
(cat. no. LF004, GeneCopoeia Expressway to Discovery) was used to
calculate the luciferase activity referring to the standard curve
of luciferin activity.
Chromatin immunoprecipitation (ChIP)
assay
The binding ability of KLF5 to the CARD11 promoter
was confirmed by a ChIP assay kit (Beyotime Institute of
Biotechnology) according to the manufacturer's protocol. Briefly,
1x106 AGS cells were cultured in 1% formaldehyde at 37˚C
for 10 min to cross-link the target protein with the corresponding
genomic DNA. Cells were collected via centrifugation at 13,000 x g
for 10 min at 4˚C and washed twice with pre-chilled PBS. A high
intensity ultrasonic processor (Cole-Parmer; Antylia Scientific)
was used to shear the genomic DNA on ice so that its majority was
broken into 200-500 bp fragments. An equal amount of chromatin was
immunoprecipitated at 4˚C overnight. ChIP was performed using 2 µg
anti-KLF5 (1:5,000; cat. no. 21017-1-AP, Proteintech Group, Inc.).
Total chromatin was used as the input. Immunoprecipitated products
were collected after incubation with magnetic beads coupled with
anti-rabbit IgG (1:100; cat. no. 30000-0-AP, Proteintech Group,
Inc.). The precipitated chromatin DNA was recovered and analyzed by
qPCR. The sequences of primers were as follows: CARD11 forward
primer: 5'-CCCAGGAGGAGAGAGAATTTGAG-3', reverse primser:
5'-CGTTCATCAGGAAGTGCGTG-3'. The reaction conditions included an
initial pre-denaturation step at 94˚C for 10 min followed by 50
cycles at 94˚C for 20 sec and 60˚C for 1 min. Data were analyzed
using the 2-ΔΔCq method and normalized to input samples
(14).
Bioinformatics
HumanTFDB was used to predict the binding of the
transcription factor KLF5 to the CARD11 promoter.
Statistical analysis
The experimental data were expressed as mean ±
standard deviation of at least three independent experiments, which
were analyzed using GraphPad Prism 8.0.1 (Dotmatics). An unpaired
student's t-tests were used for comparisons between the two groups
and one-way ANOVA with Tukey's post hoc test was used for multiple
group comparisons. P<0.05 was considered to indicate a
statistically significant difference.
Results
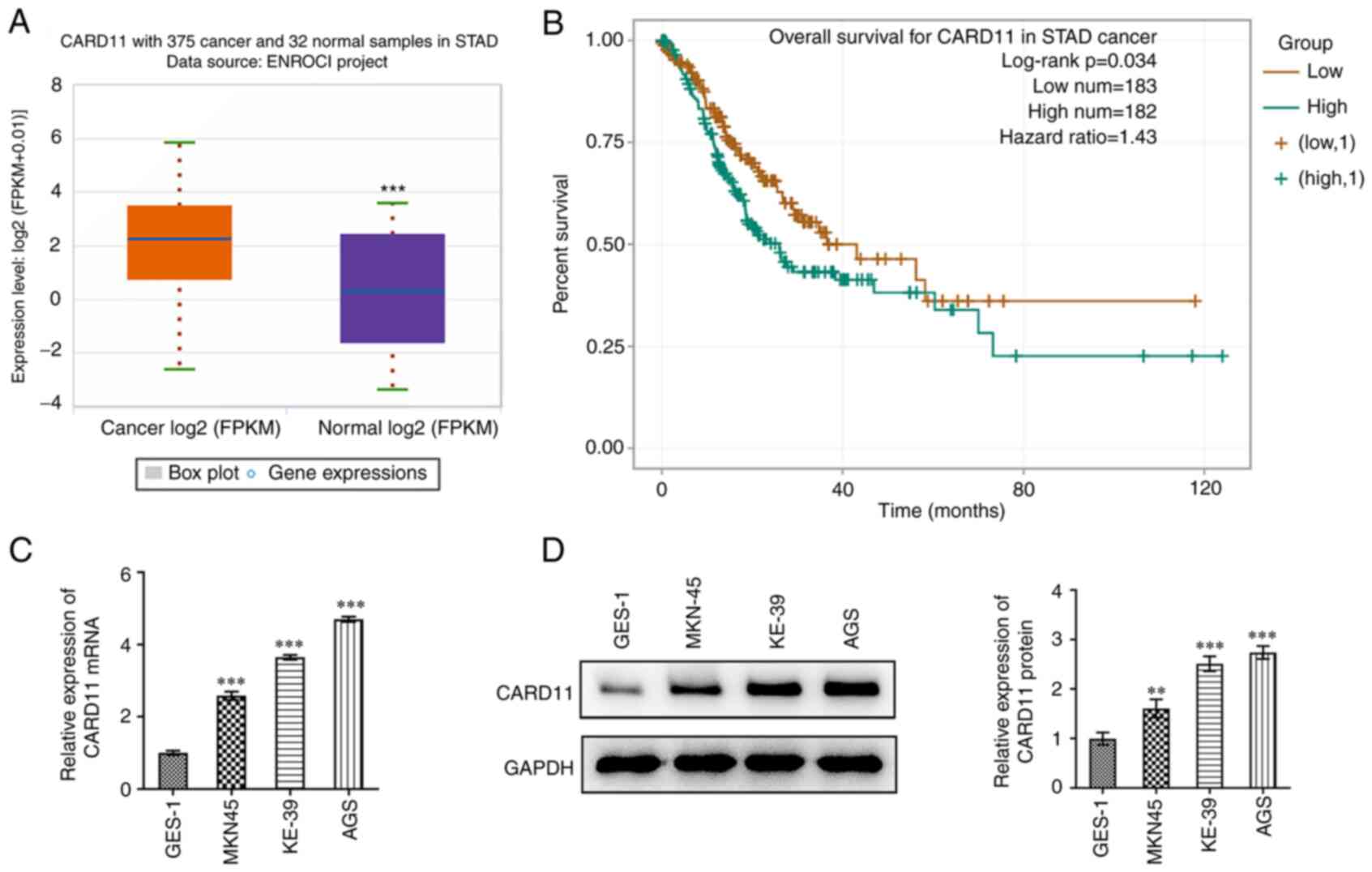
CARD11 is highly expressed in gastric
cancer and is associated with poor prognosis
First, CARD11expression in patients with gastric
cancer evaluated using ENCORI database. As shown in Fig. 1A, CARD11 was highly expressed in
tissues of patients with gastric cancer. Additionally, it was also
found from ENCORI database that high expression of CARD11 was
significantly associated with low overall survival in patients with
gastric cancer (Fig. 1B).
Additionally, the expression of CARD11 was significantly
upregulated in gastric cancer cells (MKN-45, KE-39 and AGS)
compared with that in human normal gastric epithelial cell line
GES-1 cells (Fig. 1C and D). As the highest CARD11 expression level
was observed in AGS cells, this cell line was selected to perform
the following experiments. These results indicated the abnormal
expression of CARD11 in gastric cancer.
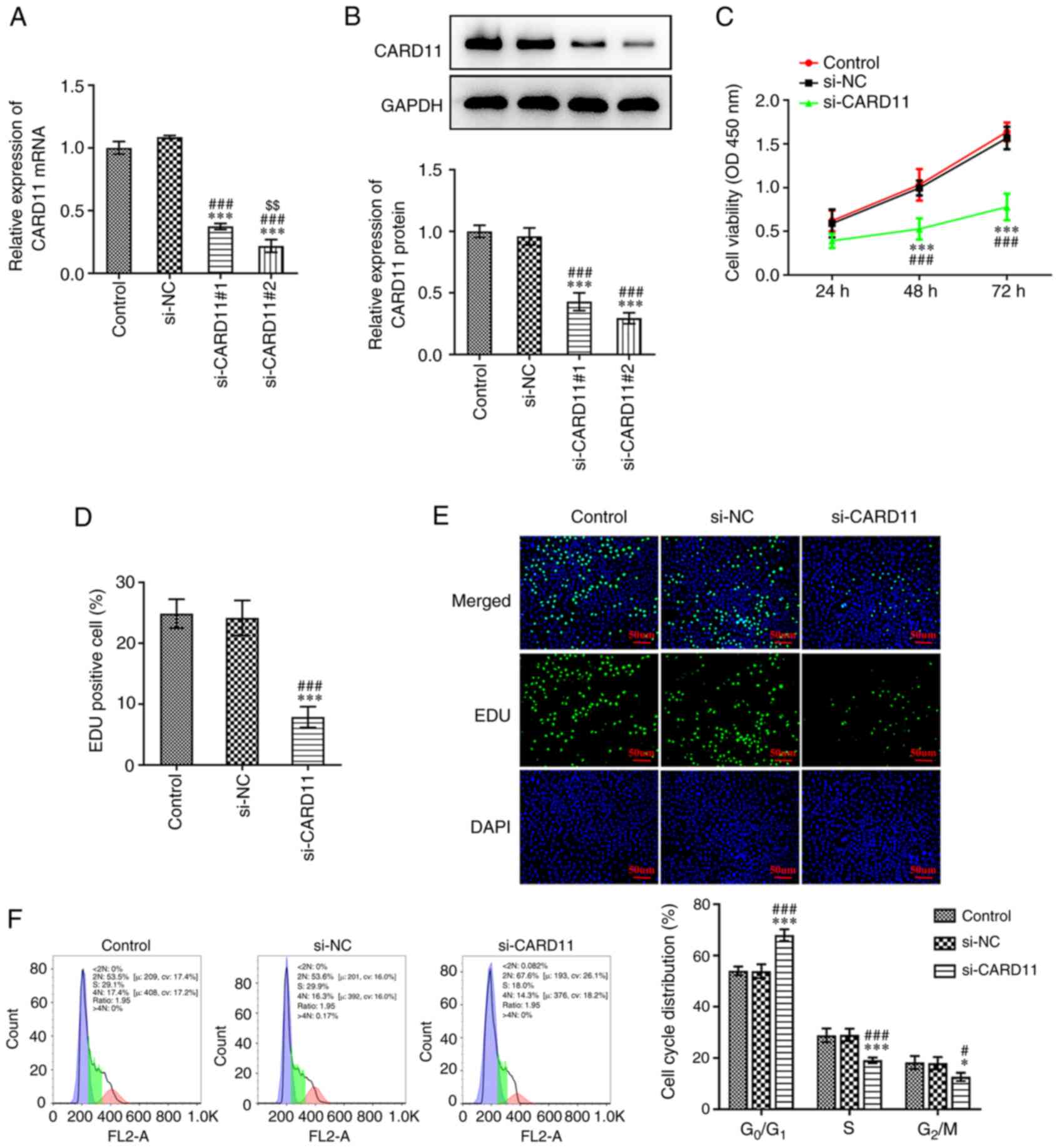
Interference of CARD11 expression
inhibits the proliferation of gastric cancer cells and induces cell
cycle arrest
To investigate the effects of CARD11 in the
progression of gastric cancer, AGS cells were transfected with
si-CARD11#1 and si-CARD11#2 to silence CARD11 expression. As shown
in Fig. 2A and B, when compared with the si-NC group, the
expression levels of CARD11 in AGS cells transfected with
si-CARD11#1 and si-CARD11#2 were significantly downregulated.
si-CARD11#2 demonstrated a stronger inhibitory effect on CARD11
expression and was therefore selected for subsequent experiments.
When AGS cells were transfected with si-CARD11, their viability and
proliferation were suppressed (Fig.
2C-E). Furthermore, interference of CARD11 expression increased
the proportion of cells at the G0/G1 phase and decreased
the proportion of the cells at the S and G2/M phases,
indicating that the cell cycle was arrested (Fig. 2F). These data suggested that CARD11
silencing inhibits the proliferation and induces cell cycle arrest
of gastric cancer cells.
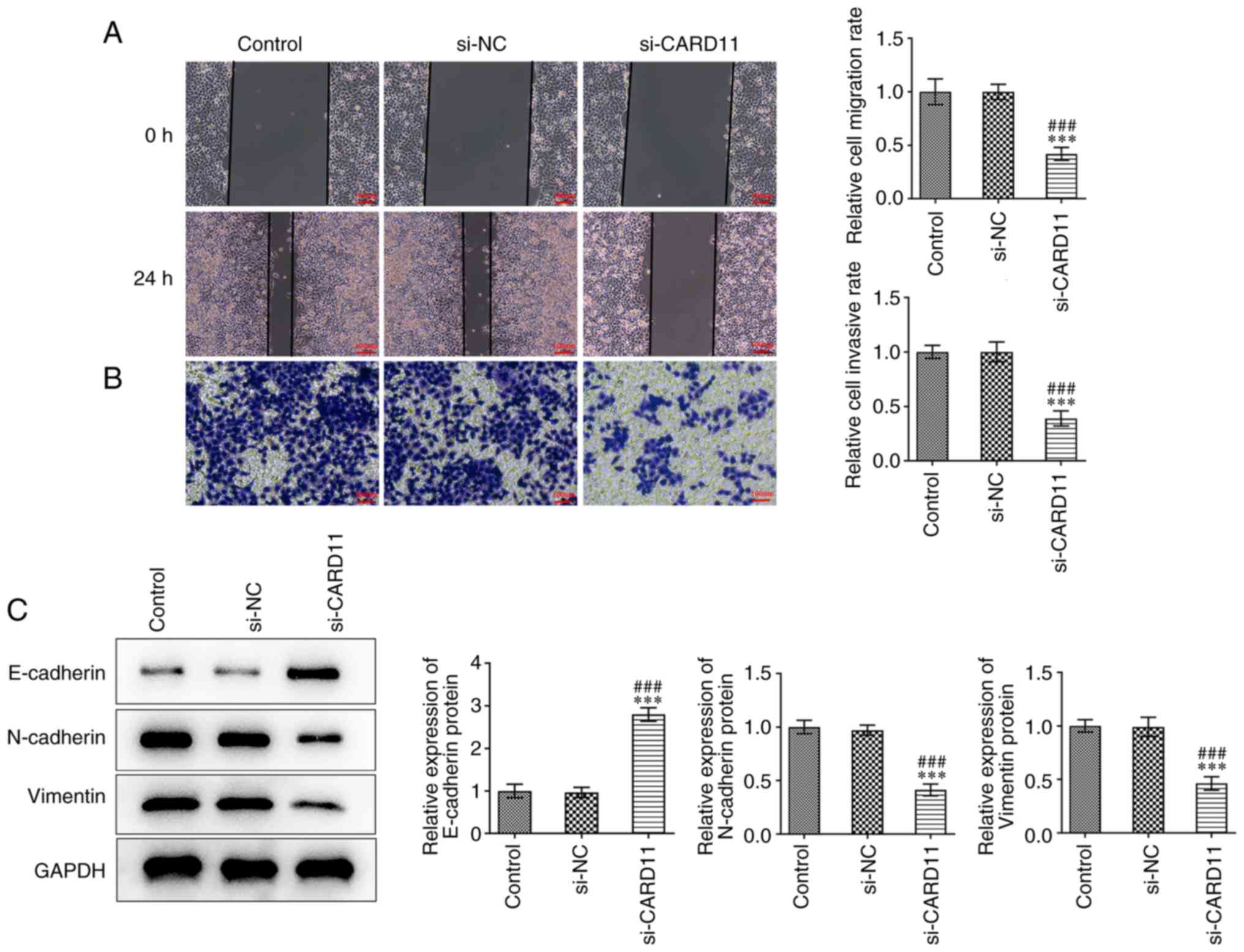
Interference of CARD11 expression
inhibits gastric cancer cell metastasis
To study the effects of CARD11 knockdown on the
metastasis of gastric cancer cells, the capacities of cell
migration, invasion and epithelial-to-mesenchymal transition (EMT)
were determined. The migratory and invasive abilities of AGS cells
were apparently suppressed by the downregulation of CARD11
expression compared with those noted in the control and si-NC
groups (Fig. 3A and B). Meanwhile, the expression levels of
E-cadherin were increased and those of N-cadherin and vimentin were
decreased in AGS cells following transfection with si-CARD11
(Fig. 3C). These data revealed
that CARD11 silencing suppressed the metastasis of gastric cancer
cells.
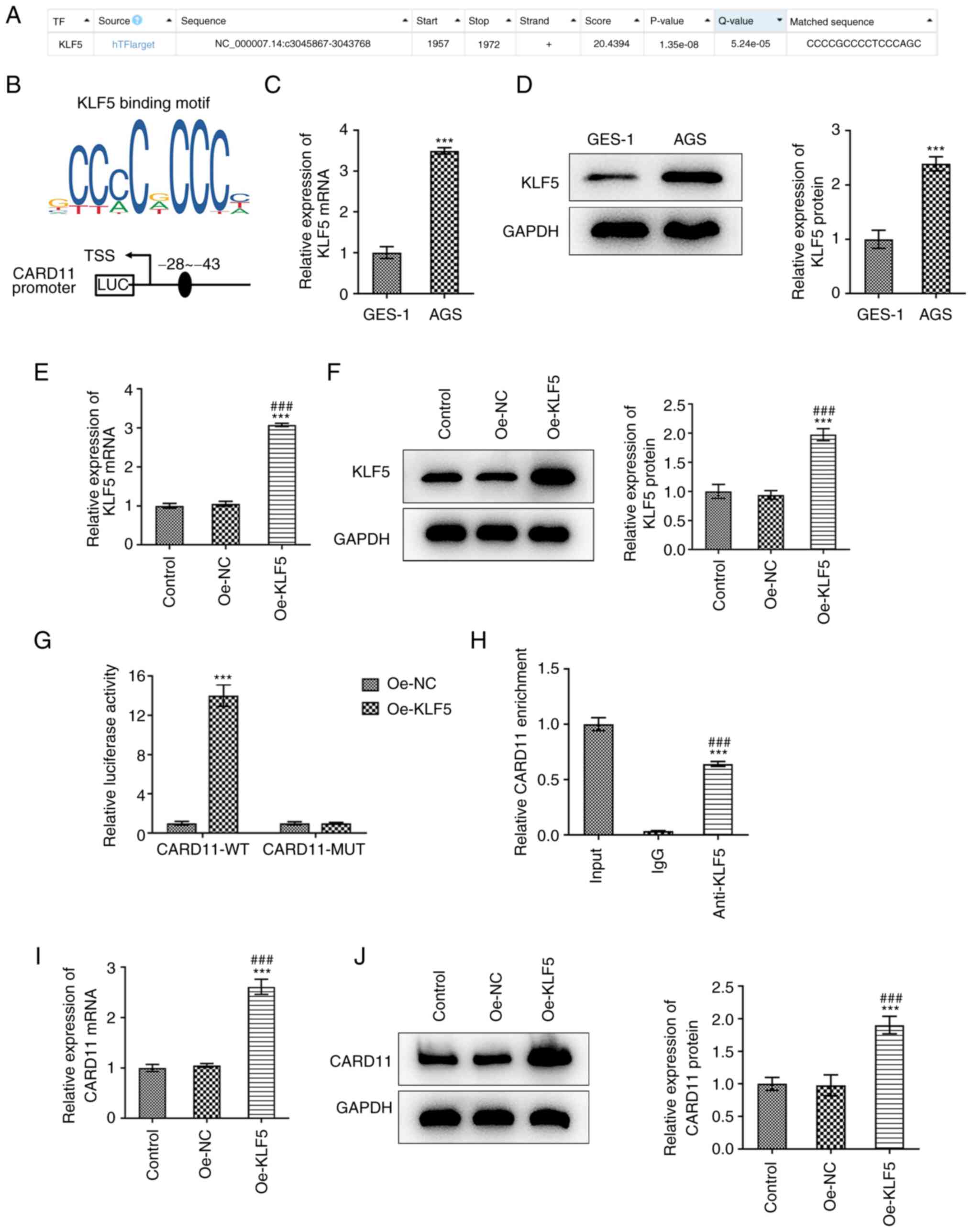
Transcription factor KLF5 positively
regulates the transcription of CARD11 in gastric cancer
To explore the potential mechanism of CARD11 on the
progression of gastric cancer, HumanTFDB predicted the binding of
the transcription factor KLF5 to the CARD11 promoter (Fig. 4A). The binding sites between the
transcription factor KLF5 and CARD11 are shown in Fig. 4B. The expression levels of KLF5 and
CARD11 in AGS cells were also upregulated (Fig. 4C and D). When AGS cells were transfected with
oe-KLF5, the expression levels of KLF5 and CARD11 were upregulated
(Fig. 4E and F). Additionally, the luciferase activity
of AGS cells co-transfected with CARD11-WT and oe-KLF5 were
increased compared with those in AGS cells co-transfected with
CARD11-mut and oe-KLF5 (Fig. 4G).
Moreover, ChIP assay indicated that the CARD11 promoter was
specifically pulled down by a KLF5-specific antibody; however, this
was not noted for the control antibody (Fig. 4H). Furthermore, the expression
levels of CARD11 in AGS cells were upregulated following
transfection of the cells with oe-KLF5 (Fig. 4I and J). These observations suggested that
transcription factor KLF5 positively regulated the transcription of
CARD11 in gastric cancer.
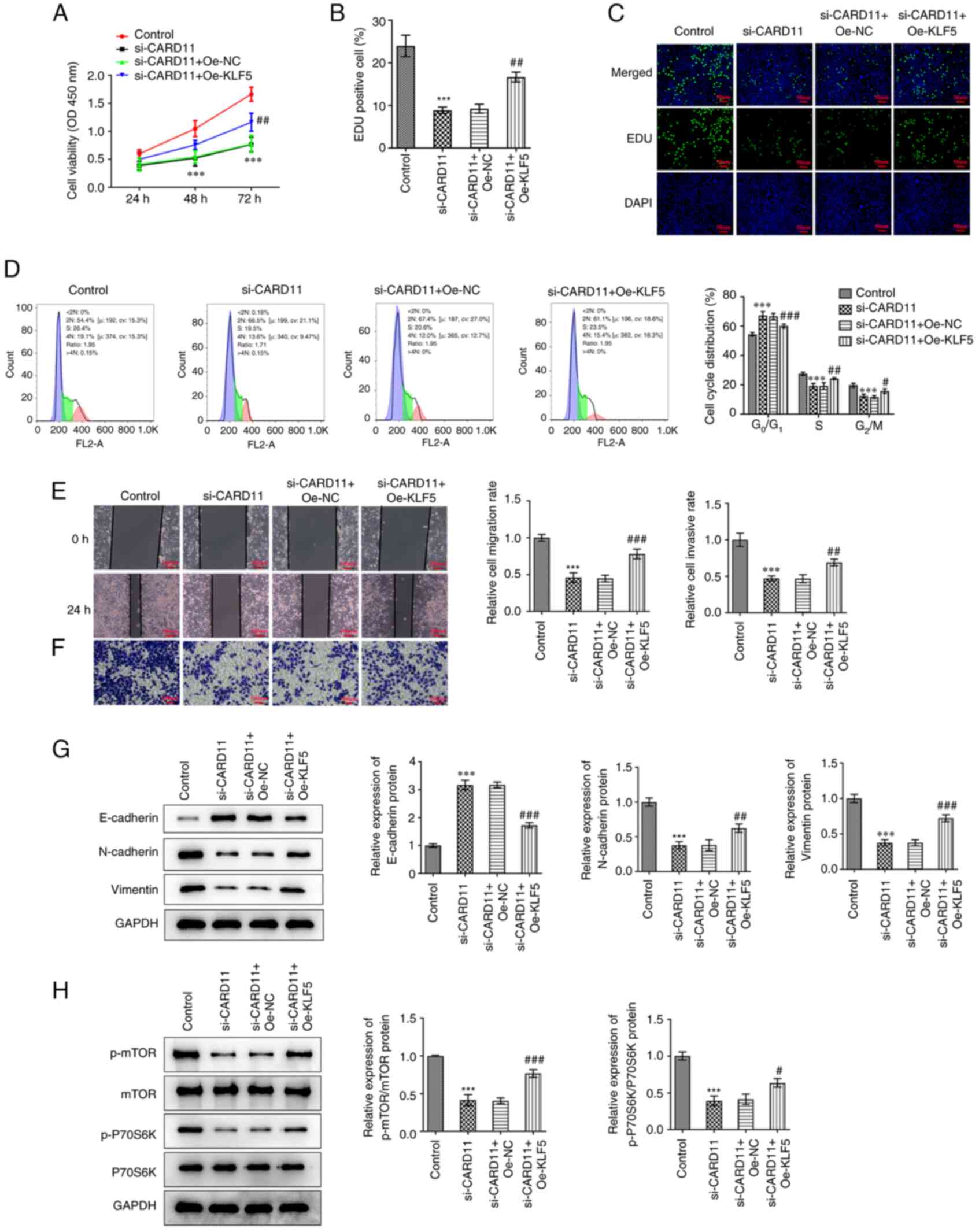
KLF5 regulates CARD11 to promote
malignant progression of gastric cancer perhaps by activating the
mTOR pathway
Subsequently, KLF5 was overexpressed to study the
mechanism of KLF5 regulation on CARD11 in the progression of
gastric cancer. It was found that KLF5 overexpression improved the
viability and proliferation of AGS cells transfected with si-CARD11
(Fig. 5A-C). The proportion of
cells at the G0/G1 phase was decreased and that of the
cells corresponding to the S and G2/M phases was
increased in AGS cells transfected with si-CARD11 and oe-KLF5
(Fig. 5D). Meanwhile, KLF5
overexpression improved the migration and invasion of AGS cells
transfected with si-CARD11 (Fig.
5E and F). Additionally, KLF5
overexpression suppressed the expression of E-cadherin, while it
promoted the expression levels of N-cadherin and vimentin in AGS
cells transfected with si-CARD11 (Fig.
5G). Interference of CARD11 expression downregulated the
expression levels of p-mTOR and p-P70S6K, which were activated by
KLF5 overexpression (Fig. 5H).
Above findings demonstrated that KLF5 regulated CARD11 to promote
malignant progression of gastric cancer might by activating the
mTOR pathway.
Discussion
At present, the CARD11 protein in mammals has been
studied extensively. The CARD11 protein is not only a member of the
membrane-associated guanylate kinase (MAGUK) family of enzymes, but
also a member of the CARD-MAGUK (CARMA) protein family, which
exists in the cytoplasm of the host cell. It is especially abundant
in hematopoietic cells and immune organs and is the only member of
the MAGUK family specifically expressed in lymphocytes (15). Members of the CARMA family were
originally discovered using bioinformatics methods based on the
CARD functional domain and named CARD11 (CARMA1), CARD14 (CARMA2)
and CARD10 (CARMA3) (16-18).
CARMA1 is highly expressed in specific tumor cell lines, such as
the chronic myelogenous leukemia cell K562, acute promyelocytic
leukocytosis HL-60 and Burkitt's lymphoma cell Raji (16). In addition, CARMA1 can activate
NF-κB through the CBM complex to cause excessive cell
proliferation, invasion and metastasis and tumor angiogenesis,
which is an important cause of autoimmune diseases or of lymphatic
hematopoietic system tumors (19-21).
Furthermore, high CARD11 expression in uveal melanoma is associated
with poor overall survival and CARD11 is associated with autophagy,
cell senescence and apoptosis (22). EMT is a cellular process by which
epithelial cells gain a mesenchymal phenotype through specific
changes in gene expression (23).
A number of studies have found that the occurrence of EMT
biological behavior plays an important role in the metastasis of
malignant tumors (24-26).
The epithelial cells undergoing EMT lose epithelial
characteristics, such as loss of E-cadherin expression and gain
mesenchymal features, such as overexpression of vimentin and
N-cadherin (27). The present
study found that high CARD11 expression in tissues of patients with
gastric cancer was predicted in the ENCORI database and CARD11
expression was increased in gastric cancer cells. Downregulation of
CARD11 expression suppressed the proliferation, invasion, migration
and EMT of gastric cancer cells and induced their cell cycle
arrest.
However, the regulatory models that mediate CARD11
expression and the mechanisms by which it modulates global gene
expression have not been well characterized in gastric cancer
cells. In the present study, a positive correlation between the
KLF5 transcription factor and CARD11 was noted by analyzing
HumanTFDB. The results derived from ChIP and luciferase reporter
assays confirmed high enrichment of KLF5 in the promoter of CARD11.
It was shown that KLF5 could activate CARD11 expression via an
interaction with its promoter. KLF5 is a basic transcription
element binding protein 2 in eukaryotes and a zinc finger protein
transcription factor (28). It can
regulate the tissue and time specificity of gene expression by
activating or inhibiting the transcription of target genes and
serves an important role in cell proliferation, differentiation and
apoptosis (28-30).
KLF5 expression is markedly upregulated in gastric cancer tissues
and knockdown of its expression suppresses proliferation and
arrested gastric cancer cell cycle at the G0/G1 phase
(31). Upregulation of KLF5
expression attenuated the function of crocin to promote the
migration, invasion and EMT of gastric cancer cells (32). Depletion of KLF5 expression reduces
gastric cancer proliferation in vitro and in vivo
(33). The present study further
indicated that KLF5 expression was increased in gastric cancer
cells and that its upregulation weakened the function of CARD11
interference to promote the migration, invasion and EMT; it also
decreased cell cycle arrest of gastric cancer cells.
CARD11 is activated by phosphorylation and forms the
CBM signalosome with the two downstream signaling molecules Bcl-10
and MALT1 (34-36).
Following the formation of CBM, the MALT1 protein recruits the
downstream signaling molecules to activate NF-κB, JNK and the mTOR
signaling pathways (35,37-39).
It was hypothesized that CARD11 may regulate the mTOR signaling
pathway. mTOR and its upstream PI3K-protein kinase B (Akt)
constitutes the PI3K/Akt/mTOR signaling pathway. This pathway is
highly activated in gastric cancer and regulates cell
proliferation, apoptosis, transcription, translation, metabolism
and other important cell biological processes in the occurrence and
development of gastric cancer (40). Lang et al (41) also found that the expression of
p-mTOR in gastric cancer was closely related to lymphatic
infiltration. Another study showed significant differences in the
expression of p-mTOR in gastric cancer tissues of different TNM
stages, whereas the positive expression of the p-mTOR protein in
gastric cancer tissues of stage III-IV is significantly higher than
that noted in tissues of stage I-II (42). The present study observed that
interference of CARD11 expression suppressed the mTOR expression in
gastric cancer cells, which was reversed by upregulation of KLF5
expression.
In conclusion, the present study demonstrated that
KLF5-mediated CARD11 promoted the proliferation, migration and
invasion of gastric cancer cells possibly by activating the mTOR
pathway. To the best of the authors' knowledge, the present study
is the first to demonstrate the suppressive role of CARD11
silencing in gastric cancer and the mechanism related to KLF5.
Overall, these findings may provide efficient therapeutic target
for the treatment of gastric cancer. Nevertheless, the present
study has a limitation. The present study only discussed the
regulatory effect of CARD11 and KLF5 on the progression of gastric
cancer cells. In vivo experiments involving transgenic
animals and the in-depth investigation of CARD11 and mTOR pathway
will be used in future investigations to support the conclusion
obtained in the present study.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
QL conceived and designed the current study. QL, SL
and ZL performed the experiments and QL, HX and WZ performed the
data analysis. QL wrote the manuscript. ZL and HX confirm the
authenticity of all the raw data. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Machlowska J, Baj J, Sitarz M, Maciejewski
R and Sitarz R: Gastric cancer: Epidemiology, risk factors,
classification, genomic characteristics and treatment strategies.
Int J Mol Sci. 21(4012)2020.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249.
2021.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Wang H, Guo W, Hu Y, Mou T, Zhao L, Chen
H, Lin T, Li T, Yu J, Liu H and Li G: Superiority of the 8th
edition of the TNM staging system for predicting overall survival
in gastric cancer: Comparative analysis of the 7th and 8th editions
in a monoinstitutional cohort. Mol Clin Oncol. 9:423–431.
2018.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Feng F, Liu J, Wang F, Zheng G, Wang Q,
Liu S, Xu G, Guo M, Lian X and Zhang H: Prognostic value of
differentiation status in gastric cancer. BMC Cancer.
18(865)2018.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Zhao B, Mei D, Lv W, Lu H, Bao S, Lin J
and Huang B: Clinicopathologic features, survival outcome, and
prognostic factors in gastric cancer patients 18-40 years of age. J
Adolesc Young Adult Oncol. 9:514–521. 2020.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Desjardins M, Arjunaraja S, Stinson JR,
Dorjbal B, Sundaresan J, Niemela J, Raffeld M, Matthews HF, Wang A,
Angelus P, et al: A Unique heterozygous CARD11 mutation combines
pathogenic features of both gain- and loss-of-function patients in
a four-generation family. Front Immunol. 9(2944)2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Juilland M and Thome M: Holding all the
CARDs: How MALT1 controls CARMA/CARD-dependent signaling. Front
Immunol. 9(1927)2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Jattani RP, Tritapoe JM and Pomerantz JL:
Cooperative control of caspase recruitment domain-containing
protein 11 (CARD11) signaling by an unusual array of redundant
repressive elements. J Biol Chem. 291:8324–8336. 2016.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Kou Z, Mao M, Liu H, Wang X, Wang Z, Gu Z,
Lang T, Nie Y, Wang Y, Huang Q, et al: CARD11 is a novel target of
miR-181b that is upregulated in chronic lymphocytic leukemia.
Biomark Med. 15:623–635. 2021.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Zhu L, Guo Q, Lu X, Zhao J, Shi J, Wang Z
and Zhou X: CTD-2020K17.1, a novel long non-coding RNA, promotes
migration, invasion, and proliferation of serous ovarian cancer
cells in vitro. Med Sci Monit. 24:1329–1339. 2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Watt SA, Purdie KJ, den Breems NY, Dimon
M, Arron ST, McHugh AT, Xue DJ, Dayal JH, Proby CM, Harwood CA, et
al: Novel CARD11 mutations in human cutaneous squamous cell
carcinoma lead to aberrant NF-κB regulation. Am J Pathol.
185:2354–2363. 2015.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Cheng Z, Liu G, Huang C and Zhao X: KLF5
activates lncRNA DANCR and inhibits cancer cell autophagy
accelerating gastric cancer progression. NPJ Genom Med.
6(75)2021.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Chen W, Zhang J, Fu H, Hou X, Su Q, He Y
and Yang D: KLF5 is activated by gene amplification in gastric
cancer and is essential for gastric cell proliferation. Cells.
10(1002)2021.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Ramadas RA, Roche MI, Moon JJ, Ludwig T,
Xavier RJ and Medoff BD: CARMA1 is necessary for optimal T cell
responses in a murine model of allergic asthma. J Immunol.
187:6197–6207. 2011.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Bertin J, Wang L, Guo Y, Jacobson MD,
Poyet JL, Srinivasula SM, Merriam S, DiStefano PS and Alnemri ES:
CARD11 and CARD14 are novel caspase recruitment domain
(CARD)/membrane-associated guanylate kinase (MAGUK) family members
that interact with BCL10 and activate NF-kappa B. J Biol Chem.
276:11877–11882. 2001.PubMed/NCBI View Article : Google Scholar
|
|
17
|
McAllister-Lucas LM, Jin X, Gu S, Siu K,
McDonnell S, Ruland J, Delekta PC, Van Beek M and Lucas PC: The
CARMA3-Bcl10-MALT1 signalosome promotes angiotensin II-dependent
vascular inflammation and atherogenesis. J Biol Chem.
285:25880–25884. 2010.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Wang S, Li Y, Hu YH, Song R, Gao Y, Liu
HY, Shu HB and Liu Y: STUB1 is essential for T-cell activation by
ubiquitinating CARMA1. Eur J Immunol. 43:1034–1041. 2013.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Park Y, Jin HS and Liu YC: Regulation of T
cell function by the ubiquitin-specific protease USP9X via
modulating the Carma1-Bcl10-Malt1 complex. Proc Natl Acad Sci USA.
110:9433–9438. 2013.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Qu C, Liu Y, Kunkalla K, Singh RR, Blonska
M, Lin X, Agarwal NK and Vega F: Trimeric G protein-CARMA1 axis
links smoothened, the hedgehog receptor transducer, to NF-κB
activation in diffuse large B-cell lymphoma. Blood. 121:4718–4728.
2013.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Blonska M and Lin X: CARMA1-mediated
NF-kappaB and JNK activation in lymphocytes. Immunol Rev.
228:199–211. 2009.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Shi X, Xia S, Chu Y, Yang N, Zheng J, Chen
Q, Fen Z, Jiang Y, Fang S and Lin J: CARD11 is a prognostic
biomarker and correlated with immune infiltrates in uveal melanoma.
PLoS One. 16(e0255293)2021.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Chen L, Yang QC, Li YC, Yang LL, Liu JF,
Li H, Xiao Y, Bu LL, Zhang WF and Sun ZJ: Targeting CMTM6
suppresses stem cell-like properties and enhances antitumor
immunity in head and neck squamous cell carcinoma. Cancer Immunol
Res. 8:179–191. 2020.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Hung JJ, Kao YS, Huang CH and Hsu WH:
Overexpression of Aiolos promotes epithelial-mesenchymal transition
and cancer stem cell-like properties in lung cancer cells. Sci Rep.
9(2991)2019.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Deng L, Zou J, Su Y, Wang M and Zhao L:
Resveratrol inhibits TGF-β1-induced EMT in gastric cancer cells
through Hippo-YAP signaling pathway. Clin Transl Oncol.
24:2210–2221. 2022.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Odero-Marah V, Hawsawi O, Henderson V and
Sweeney J: Epithelial-mesenchymal transition (EMT) and prostate
cancer. Adv Exp Med Biol. 1095:101–110. 2018.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Dong D, Na L, Zhou K, Wang Z, Sun Y, Zheng
Q, Gao J, Zhao C and Wang W: FZD5 prevents epithelial-mesenchymal
transition in gastric cancer. Cell Commun Signal.
19(21)2021.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Gao Y, Ding Y, Chen H, Chen H and Zhou J:
Targeting Krüppel-like factor 5 (KLF5) for cancer therapy. Curr Top
Med Chem. 15:699–713. 2015.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Marrero-Rodríguez D, la Cruz HA,
Taniguchi-Ponciano K, Gomez-Virgilio L, Huerta-Padilla V,
Ponce-Navarrete G, Andonegui-Elguera S, Jimenez-Vega F,
Romero-Morelos P, Rodriguez-Esquivel M, et al: Krüppel like factors
family expression in cervical cancer cells. Arch Med Res.
48:314–322. 2017.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Hart GT, Hogquist KA and Jameson SC:
Krüppel-like factors in lymphocyte biology. J Immunol. 188:521–526.
2012.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Chen P, Qian XK, Zhang YF, Sun XG, Shi XJ
and Gao YS: KLF5 promotes proliferation in gastric cancer via
regulating p21 and CDK4. Eur Rev Med Pharmacol Sci. 24:4224–4231.
2020.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Zhou Y, Xu Q, Shang J, Lu L and Chen G:
Crocin inhibits the migration, invasion, and epithelial-mesenchymal
transition of gastric cancer cells via miR-320/KLF5/HIF-1α
signaling. J Cell Physiol. 234:17876–17885. 2019.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Chia NY, Deng N, Das K, Huang D, Hu L, Zhu
Y, Lim KH, Lee MH, Wu J, Sam XX, et al: Regulatory crosstalk
between lineage-survival oncogenes KLF5, GATA4 and GATA6
cooperatively promotes gastric cancer development. Gut. 64:707–719.
2015.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Juilland M and Thome M: Role of the
CARMA1/BCL10/MALT1 complex in lymphoid malignancies. Curr Opin
Hematol. 23:402–409. 2016.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Lork M, Staal J and Beyaert R:
Ubiquitination and phosphorylation of the CARD11-BCL10-MALT1
signalosome in T cells. Cell Immunol. 340(103877)2019.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Roche MI, Ramadas RA and Medoff BD: The
role of CARMA1 in T cells. Crit Rev Immunol. 33:219–243.
2013.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Gross O, Grupp C, Steinberg C, Zimmermann
S, Strasser D, Hannesschläger N, Reindl W, Jonsson H, Huo H,
Littman DR, et al: Multiple ITAM-coupled NK-cell receptors engage
the Bcl10/Malt1 complex via Carma1 for NF-kappaB and MAPK
activation to selectively control cytokine production. Blood.
112:2421–2428. 2008.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Wegener E and Krappmann D:
CARD-Bcl10-Malt1 signalosomes: Missing link to NF-kappaB. Sci STKE.
2007(pe21)2007.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Wegener E, Oeckinghaus A, Papadopoulou N,
Lavitas L, Schmidt-Supprian M, Ferch U, Mak TW, Ruland J,
Heissmeyer V and Krappmann D: Essential role for IkappaB kinase
beta in remodeling Carma1-Bcl10-Malt1 complexes upon T cell
activation. Mol Cell. 23:13–23. 2006.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Sun D, Toan X, Zhang Y, Chen Y, Lu R, Wang
X and Fang J: Mammalian target of rapamycin pathway inhibition
enhances the effects of 5-aza-dC on suppressing cell proliferation
in human gastric cancer cell lines. Sci China C Life Sci.
51:640–647. 2008.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Lang SA, Gaumann A, Koehl GE, Seidel U,
Bataille F, Klein D, Ellis LM, Bolder U, Hofstaedter F, Schlitt HJ,
et al: Mammalian target of rapamycin is activated in human gastric
cancer and serves as a target for therapy in an experimental model.
Int J Cancer. 120:1803–1810. 2007.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Xu DZ, Xia LP, Lin TY, Cai MY, Fan XJ,
Zhan YQ, Zhou ZW and Li W: Expression and clinical significance of
p-mTOR in gastric cancer. J Sun Yat-Sen Univ (Med Sci). 30:304–307.
2009.
|