Introduction
Breast cancer is the most common malignant tumor in
women worldwide and is also the main cause of cancer-associated
mortality in women (1). The most
common treatments for breast cancer include surgery, chemotherapy,
hormone therapy and radiotherapy; however, breast cancer is highly
heterogeneous and these therapies are often ineffective for the
treatment of breast cancer metastasis (2). Breast cancer cell proliferation,
metastasis and malignant progression are the main causes of
cancer-related death (3).
Therefore, exploring new diagnostic biomarkers and molecular
targets is of great importance in the treatment of breast
cancer.
Acetyl-CoA carboxylase (ACC) is a biotin-dependent
enzyme, which can be used as a catalyst for CO2
carboxylation and conversion of acetyl-CoA to malonyl-CoA (4). ACC is highly enriched in adipogenic
tissues, and is under long term control at the transcriptional and
translational levels through targeted
phosphorylation/dephosphorylation of serine residues and sterol
conversion of citrate or palmitoyl-CoA (5). In recent years, numerous studies have
focused on the role of ACC in cancer. Zhan et al (6) identified ACC as an antitumor target
in colon cancer. Zhao et al (7) reported that the expression levels of
ACC in ovarian cancer stem cells were significantly increased. In
addition, ACC has been reported to be overexpressed in liver
cancer, where it can promote the proliferation of human hepatoma
HepG2 cells and the rat liver cell line BRL3A (8). ACC has been shown to be overexpressed
in gastric cancer, and ACC/phosphorylated-ACC has been considered a
potential target for cancer treatment (9). However, the role of ACC in breast
cancer remains unclear.
The present study aimed to determine the role of ACC
in the malignant progression of breast cancer proliferation and
metastasis. Lentiviral-mediated short hairpin RNA (shRNA) was used
to knock down the expression of ACC in two breast cancer cell lines
(MCF-7 and MDA-MB-231), and its functions were subsequently
explored. In addition, a nude mouse model of subcutaneous
metastatic tumors was established, and the results were compared
with the findings from breast cancer cells. It was hypothesized
that the ACC gene may affect cell viability and migration in the
malignant progression of breast cancer.
Materials and methods
Experimental animals and cells
A total of 12 female 4-week-old nude mice [Changzhou
Cavens Laboratory Animal Co., Ltd.; license no. SCXK (Su)
2016-0010], weighing ~20 g, were raised under the following
conditions: Temperature, 20-26˚C; humidity, 40-70%; with ad
libitum access to specific-pathogen-free grade animal feed and
sterilized drinking. Human MCF-7 (TCHu 74) and MDA-MB-231 (TCHu227)
triple-negative breast cancer cells were obtained from The Cell
Bank of Type Culture Collection of The Chinese Academy of Sciences.
The cells were cultured in high-glucose Dulbecco's modified Eagle's
medium (DMEM; cat. no. KGM12800S; Nanjing KeyGen Biotech Co.,
Ltd.), containing 10% fetal bovine serum (FBS; cat no. 10099-141;
Gibco; Thermo Fisher Scientific, Inc.) and 1%
penicillin-streptomycin (cat. no. P1400; Beijing Solarbio Science
& Technology Co., Ltd.) at 37˚C in a humidified atmosphere
containing 5% CO2. The interim cell line 293T and the
pLKO.1-EGFP Puro plasmid were purchased from Hanbio Biotechnology
Co., Ltd.
Main reagents and instruments
Polyethylene glycol (PEG)8000 (cat no. 81268) was
from MilliporeSigma. The Annexin V-FITC/PI Apoptosis Kit (cat. no.
AP101-100-kit) was purchased from MultiSciences (Lianke) Biotech,
Co., Ltd. Ultrapure RNA Kit (cat. no. CW0581M) was purchased from
CoWin Biosciences. HiScript II Q RT SuperMix for quantitative
polymerase chain reaction (qPCR) (+gDNA wiper) (cat. no. R223-01)
was from Vazyme Biotech Co., Ltd. 2X SYBR Green PCR Master Mix
(cat. no. A4004M) was purchased from Xiamen LifeInt Technology Co.,
Ltd. Radioimmunoprecipitation assay buffer (cat. no. C1053) was
obtained from Applygen Technologies Inc. Polyvinylidene difluoride
membranes (cat. no. IPVH00010) were purchased from MilliporeSigma.
Mouse monoclonal anti-GAPDH internal loading control antibody
(1:2,000; cat. no. TA-08), and goat anti-mouse IgG (H+L) HRP
conjugate (1:2,000; cat. no. ZB-2305) and goat anti-rabbit IgG
(H+L) HRP conjugate (1:2,000; cat. no. ZB-2301) secondary
antibodies were obtained from OriGene Technologies, Inc. The
following primary antibodies were used in the present study: Rabbit
anti N-cadherin (1:1,000; AF4039; Affinity Biosciences, Ltd.),
rabbit anti-vimentin (1:2,000; cat. no. 10366-1-ap), rabbit
anti-Bax (1:5,000; cat. no. 50599-2-lg) (both from Proteintech
Group, Inc.). Scott's Bluing Solution (cat. no. G1865) was obtained
from Beijing Solarbio Science & Technology Co., Ltd. The TUNEL
assay kit (cat. no. C1090) was purchased from Beyotime Institute of
Biotechnology. Minimum Essential Medium (MEM) high glucose complete
medium (cat no. KGM41500S) and Cell Counting Kit-8 (CCK-8) cell
proliferation detection kit (cat. no. KGA317) were obtained from
Nanjing KeyGen Biotech Co., Ltd. The NovoCyte™ flow cytometer
(NovoCyte 2060R) was from ACEA Bioscience, Inc. The fluorescence
PCR machine (CFX Connect™ Real-Time) and the ultra-sensitive
chemiluminescence imaging system (Chemi Doc™ XRS+) were from
Bio-Rad Laboratories, Inc. The automated microplate reader
(WD-2102B) and vertical protein electrophoresis system (DYY-6C)
were purchased from Beijing Liuyi Instrument Factory. An inverted
fluorescence microscope (MF53) was from Guangzhou Micro-shot
Technology Co., Ltd., and a CX41 fluorescence microscope was
obtained from Olympus Corp.
Selection of shRNA
According to the sequence of ACC, three shRNAs were
selected on a shRNA design website (https://www.sigmaaldrich.cn/CN/zh/semi-configurators/shrna?activeLink=selectClones)
and cloned into the pLKO.1-EGFP Puro plasmid. The 3rd generation
system was used for lentiviral transduction and the interim cell
line was 293T. Four plasmids (12 µg lentiviral plasmid + 12 µg
Lenti-Mix=24 µg total; Lenti-Mix was composed of pMDLg/pRRE :
pVSV-G : pRSV-Rev, 5:3:2) was added into a 5 ml Eppendorf (EP) tube
for transfection of 293T cells to produce lentiviral vectors.
Subsequently, 1,000 µl 0.25 M CaCl2 was added, and
gently mixed evenly with a pipette. Then, 1,000 µl 2X HBS was
added, gently mixed evenly with the pipette, and let stand at room
temperature for 10 min. Finally, the Lenti-DNA Mix transfection
system solution was added dropwise to the culture dish of the 293T
cells, the dish was gently agitated to mix well, and cultured in a
37˚C cell culture incubator containing 5% CO2.
A total of 8-10 h after transfection, the culture
medium was replaced with 20 ml prewarmed complete medium and
culture was continued. A total of 24 h after transfection,
observation was performed under a fluorescence microscope and
images were captured. Subsequently, 72 h after transfection, the
cell culture supernatant was collected, and centrifuged at 3,000 x
g and 4˚C for 10 min. Then, the supernatant was obtained, filtered
through a 0.45-µM filter and fully mixed with 5X PEG8000 lentivirus
concentration solution overnight at 4˚C. On the second day,
centrifugation was performed at 10,000 x g at 4˚C for 30 min, and
the lentiviral particles were resuspended in 1 ml serum-free DMEM,
aliquoted and stored at -80˚C.
The interference efficiencies of the shRNAs were
detected by qPCR. The shRNA with the best effect was selected for
further experiments. The sequences of the three ACC shRNAs are
listed in Table I.
 | Table IshRNA sequences. |
Table I
shRNA sequences.
| shRNA | Sequence, 5'-3' |
|---|
| ACC shRNA-1 |
TACAAGGGATACAGGTATTTA |
| ACC shRNA-2 |
TATGAGGTGGATCGGAGATTT |
| ACC shRNA-3 |
GTATGTTCGAAGGGCTTATAT |
| NC |
TAGGCTAGGCGTAGCTATAGC |
Cell transduction, stable screening
and grouping
MCF-7 and MDA-MB-231 cells were transduced with the
indicated lentiviruses. The cell culture medium was replaced with
serum-free medium (1 ml). The multiplicity of infection (MOI) was
3x108 TU/ml and the duration of transduction into cells
was 8-10 h. The time interval between transduction and subsequent
experimentation was 48 h. Puromycin (2.5 µg/ml) was added to the
stably expressed cells post-transduction and the medium was changed
every 2 days. If, under the CX41 fluorescence microscope, the
proportion of fluorescent cells was 100%, and they could be
subcultured and expanded normally, this indicated that the
screening of the stable transgenic strains was successful.
The cells were randomly allocated into the following
groups: i) Normal breast cancer cells (control group), ii) breast
cancer cells transduced with a negative control (NC) lentiviral
plasmid, and iii) breast cancer cells transduced with a shRNA ACC
lentiviral plasmid (shACC).
Reverse transcription (RT)-qPCR
TRIzon from the Ultrapure RNA Kit was used to
completely lyse the sample and extract RNA from the tumor tissue ;
RNA concentration and purity were then determined. cDNA was
synthesized from RNA using HiScript II Q RT SuperMix, according to
the manufacturer's protocol. Using cDNA as the template, qPCR was
performed using a fluorescence qPCR instrument; with GAPDH used as
the internal reference gene, the relative mRNA expression levels of
the genes of interest in each group were calculated. The reaction
system was as follows: 2X SYBR Green PCR Master Mix (10 µl), cDNA
(1 µl), upstream primer (0.4 µl), downstream primer (0.4 µl),
RNase-free ddH2O (8.2 µl). The reaction steps were as
follows: Pre-denaturation at 95˚C for 10 min; followed by 40 cycles
of denaturation at 95˚C at 10 sec, annealing at 58˚C for 30 sec and
extension at 72˚C for 30 sec. The primer sequences are shown in
Table II. GAPDH was used as the
internal reference, and the relative expression levels of ACC were
calculated according to the 2-ΔΔCq method (10).
 | Table IIPrimer information. |
Table II
Primer information.
| Primer name | Primer sequence,
5'-3' | Primer length,
nt | Product length,
bp | Annealing
temperature, ˚C |
|---|
| ACC | F:
GGACCCAGTCTACATCCACT | 20 | 194 | 57.65 |
| | R:
TATCGCTAATAACACCCTTCTCC | 23 | | |
| GAPDH | F:
TGACTTCAACAGCGACACCCA | 21 | 121 | 61.5 |
| | R:
CACCCTGTTGCTGTAGCCAAA | 21 | | |
Western blotting
The cell culture medium was removed from the culture
dish using a pipette. Subsequently, 100 µl cell lysis buffer was
added to each well and placed on ice for 20 min. The cells were
scraped to one side with a cell scraper and then transferred to EP
tubes, which were centrifuged at 13,523 x g at 4˚C for 10 min, and
the supernatant was used to assess total protein concentration
using a bicinchoninic acid kit. The proteins were then denatured,
loaded (50 µg/lane), separated by SDS-PAGE on 12% gels for 2 h, and
transferred to membranes at a constant current of 300 mA for 80
min. The membranes were incubated with 5% skimmed milk at room
temperature for 1.5 h. After blocking, they were incubated with the
primary antibodies at 4˚C overnight, and with the secondary
antibody at room temperature for 2 h. Drops of ECL luminescent
liquid were added to the membranes and exposed in the gel imaging
system. ImageJ software (version 1.8.0, National Institutes of
Health) was used to analyze the gray value of each band.
CCK-8 cell viability assay
The groups of cells were digested, resuspended,
counted and plated at a cell density of 5x103 cells/well
in a 96-well plate. The cells were then cultured for 48 h in MEM.
Subsequently, the culture medium was discarded and replaced with
the same medium (100 µl per well). CCK-8 reagent was added to each
well and placed in the incubator at 37˚C for 2 h. The absorbance of
each well was detected at a wavelength of 450 nm using a microplate
reader and the cell viability rate was calculated using the
following formula: Cell viability rate (%)=(experimental group cell
OD/control group OD) x100.
Apoptosis detected by flow
cytometry
Cells (1-3x106) were collected, added to
1 ml PBS, centrifuged at 300 x g at 4˚C for 3 min, and washed
twice. Binding buffer (5X) was diluted into 1X binding buffer with
double distilled water and 300 µl pre-cooled 1X binding buffer was
used to resuspend the cells. Subsequently, 5 µl Annexin V-FITC and
10 µl PI were added to each tube, mixed slightly and incubated at
room temperature for 10 min in the dark. Finally, 200 µl pre-cooled
1X binding buffer was added to each tube, mixed and apoptosis was
detected by the NovoCyte flow cytometer using the NovoExpress
software (version 1.5.6; Agilent Technologies, Inc.)
Cell migration detected by cell
scratch test
Once the cell density reached ≥90%, the scratch test
was performed. A 200-µl pipette tip was used to scratch each well,
the medium was discarded, the cells were washed three times with
PBS and the medium was replaced with serum-free medium. Images of
the wound were captured with an inverted fluorescence microscope
(MF53) at 0 h and after 24 h in an incubator at 37˚C. The scratch
width was measured at 0 and 24 h, and the cell migration rate was
calculated, as follows: Migration rate=migration distance/migration
time.
Establishment of a nude mouse tumor
model and grouping
Mice were randomly allocated into the following
experimental groups: i) Normal breast cancer cells (Control), ii)
breast cancer cells transduced with a NC lentiviral plasmid, and
iii) breast cancer cells transduced with shACC (n=4/group).
A nude mouse model of subcutaneous metastatic tumor
was established using MCF-7 breast cancer cells. Four nude mice in
each group were inoculated with MCF-7 breast cancer cells
(1x107 cells/mouse; 0.20 ml/mouse) subcutaneously into
the armpit. For mice in the NC and shACC groups, the breast cancer
cells were transduced with NC lentiviral plasmid or shACC,
respectively.
A total of 21 days after inoculation, the mice were
euthanized by CO2 inhalation (flow rate, 40% of the
chamber volume/min) and the tumor tissue was collected. The tumor
length and width were measured every 3 days after inoculation, and
the tumor volume was calculated according to the following formula:
Tumor volume=0.5 x long diameter x short diameter2.
Hematoxylin and eosin (H&E)
staining
The tissues were collected, fixed in 4%
paraformaldehyde at 4˚C for >24 h, dehydrated, paraffin embedded
and sliced into 4 µm sections, which were dewaxed and hydrated. The
sections were placed in distilled water, and were then stained in
hematoxylin aqueous solution for 3 min, differentiated with
hydrochloric acid ethanol differentiation solution for 15 sec,
slightly washed, bluing the hematoxylin stain with Scott's Bluing
Solution for 15 sec, and washed with running water. Subsequently,
the sections were stained with eosin for 3 min, washed with running
water, dehydrated, cleared, mounted and examined under a microscope
(CX43).
Apoptosis detected by TUNEL
staining
The tissues were fixed in 4% paraformaldehyde at 4˚C
for >24 h, dehydrated, paraffin-embedded and sectioned. The
paraffin-embedded sections (0.3 µm) were baked, dewaxed and
hydrated. The sections then underwent antigen retrieval by the
dropwise addition of Proteinase K working solution at 37˚C for 25
min, and were washed. A sufficient amount of TUNEL test solution
was added to each slide (2 µl TdT enzyme and 48 µl fluorescence
labeling solution for one sample) and incubated in the dark at 42˚C
for 1 h. The nucleus was counterstained by adding DAPI dropwise and
incubating for 3 min in the dark at room temperature, after which,
the slides were mounted with antifade mounting medium and observed
under a fluorescence microscope.
Statistical analysis
SPSS 19.0 software (IBM Corp.) was used for
statistical analysis. All experiments were repeated three times and
the quantitative results are expressed as the mean ± standard
deviation. One way ANOVA was used to compare the results among
multiple groups, and the least significant difference method was
used for pairwise comparison. P<0.05 was considered to indicate
a statistically significant difference. GraphPad Prism 5.0
(GraphPad Software; Dotmatics, Inc.) was used for graph generation
and ImageJ software (version 1.8.0; National Institutes of Health)
was used for gray value analysis.
Results
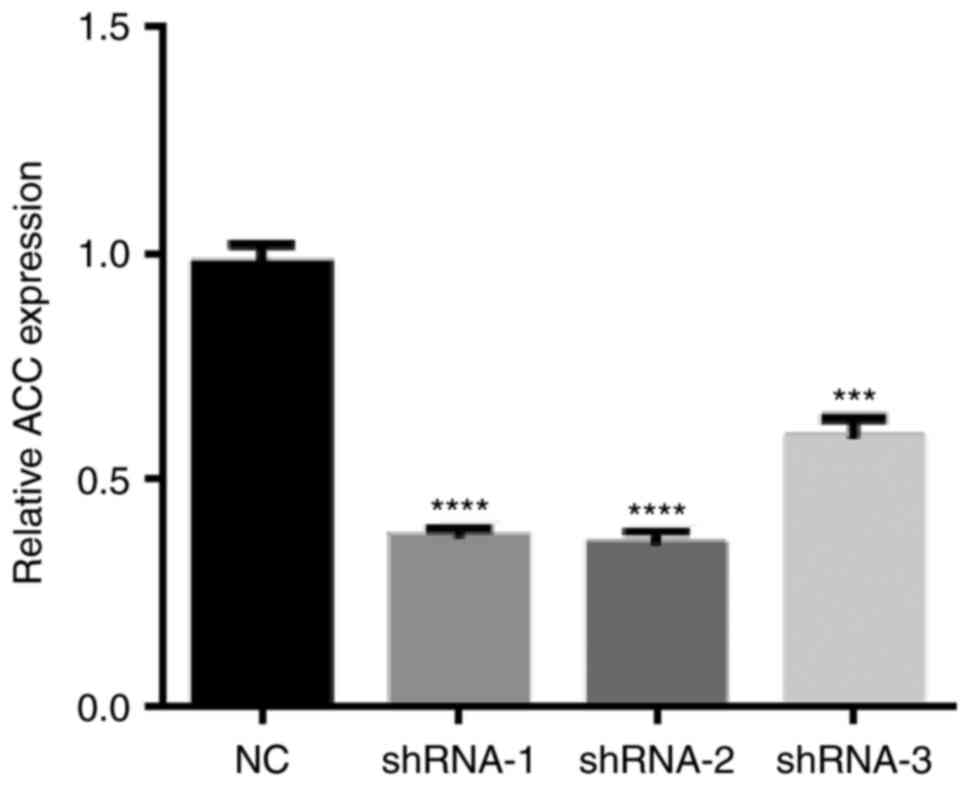
shACC selection
Compared with the NC group, shRNA-1 had the best
effect on ACC expression (Fig. 1);
therefore, this shRNA was selected for further experimentation.
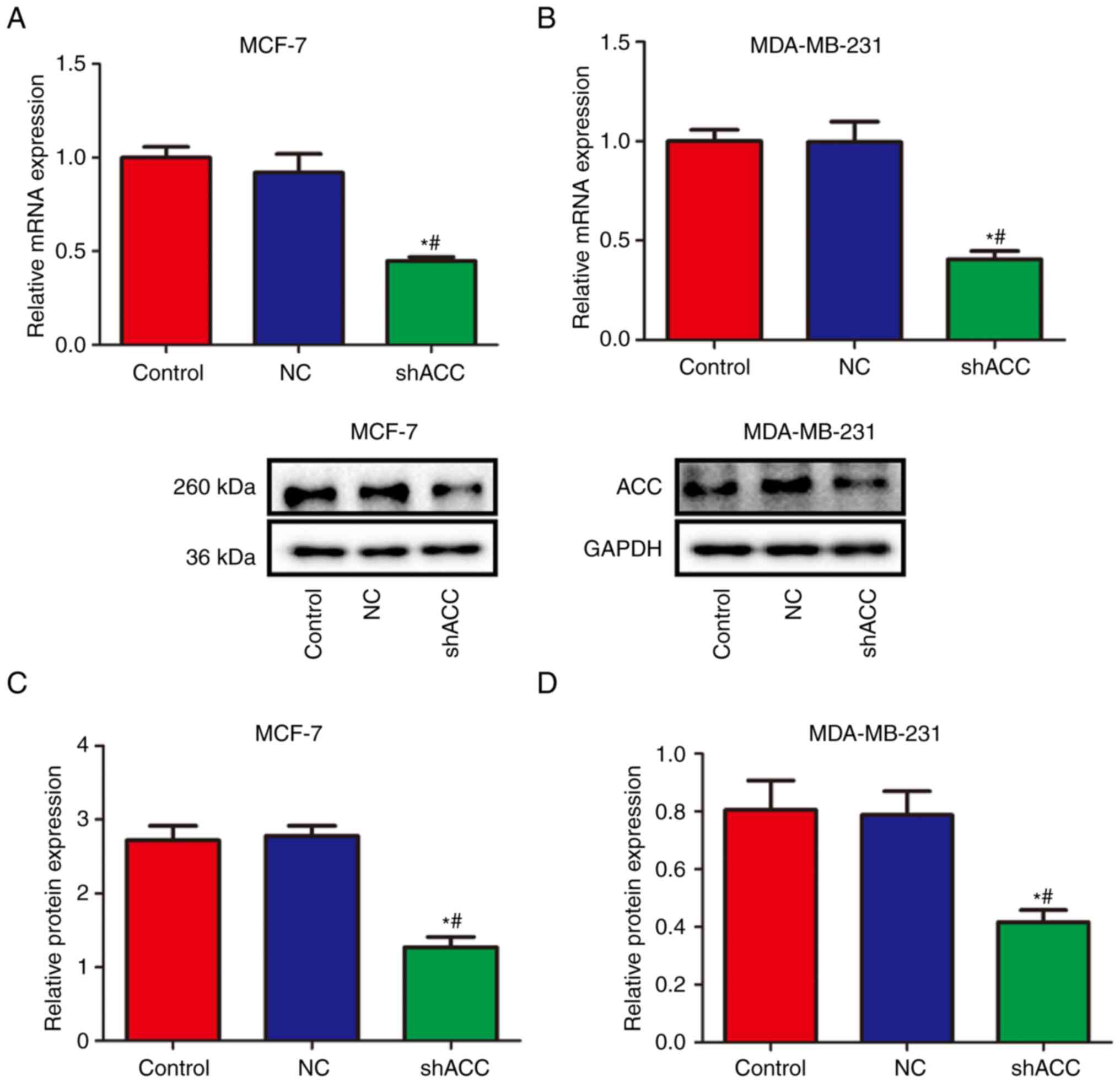
Effects of shACC on ACC
expression
Compared with those in the Control and NC groups,
the mRNA and protein expression levels of ACC were significantly
decreased in the shACC groups in both breast cancer cell lines
(Fig. 2). These findings suggested
that ACC gene interference was effective and successfully verified,
and subsequent experiments could be performed.
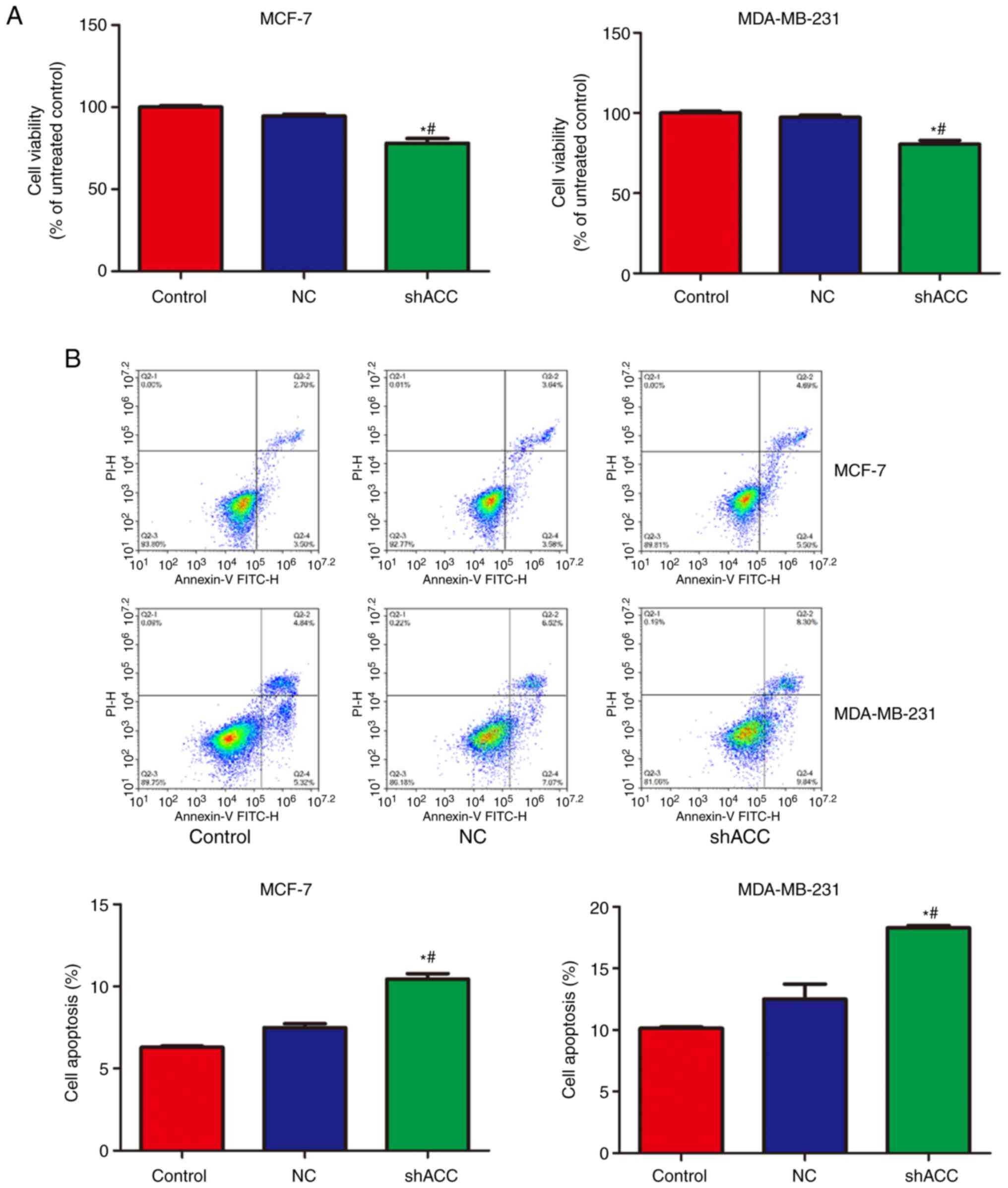
Effects of shACC on cell viability and
apoptosis
As shown in Fig. 3,
compared with that in the Control and NC groups, cell viability was
significantly decreased in the shACC groups of both breast cancer
cell lines, whereas apoptosis was significantly increased. These
findings indicated that ACC gene interference inhibited cell
viability and promoted apoptosis.
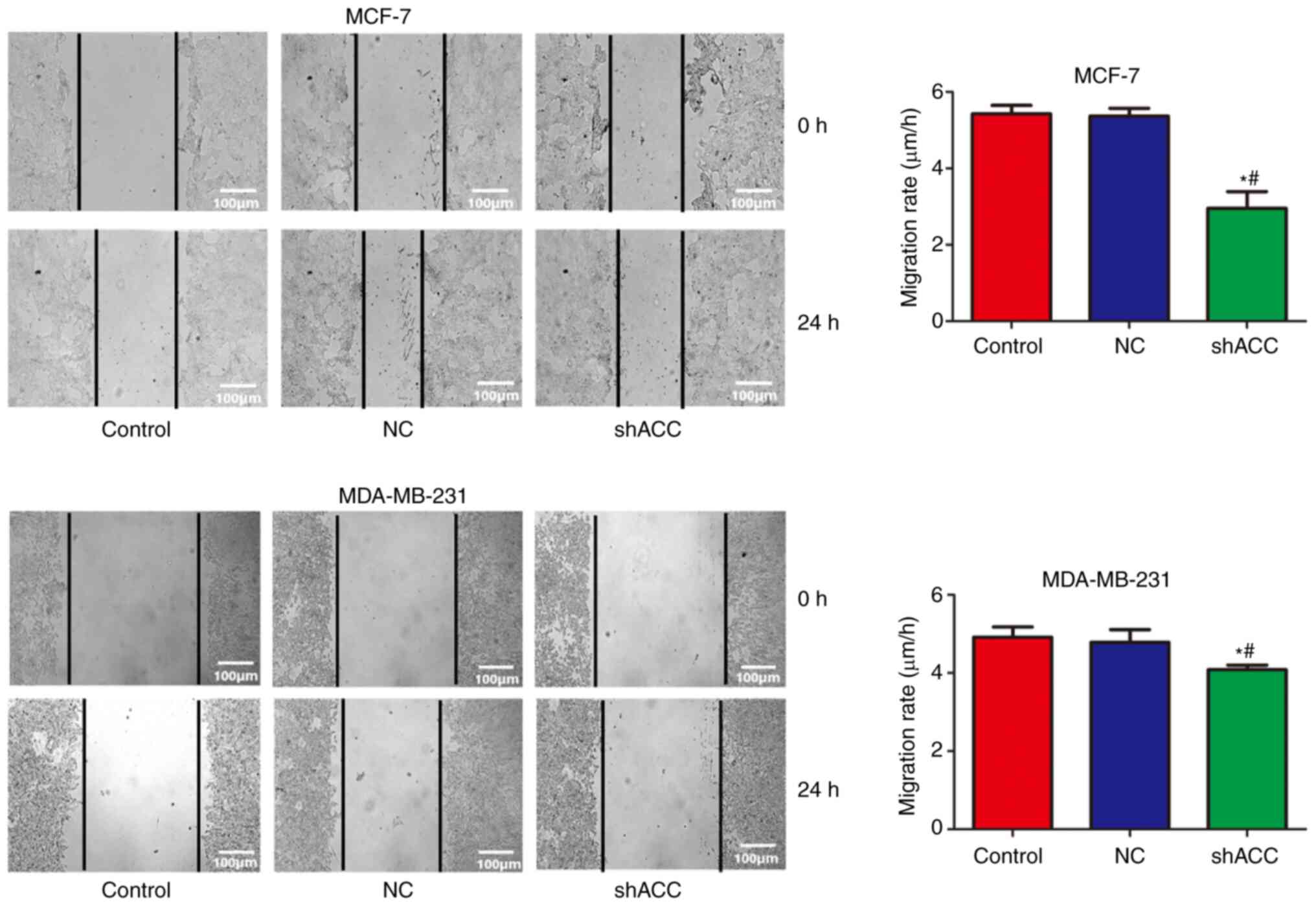
Effects of shACC on cell
migration
Compared with that in the Control and NC groups, the
cell migration rate was significantly decreased in the shACC groups
of both breast cancer cell lines (Fig.
4). These findings suggested that ACC gene interference
inhibited cell migration.
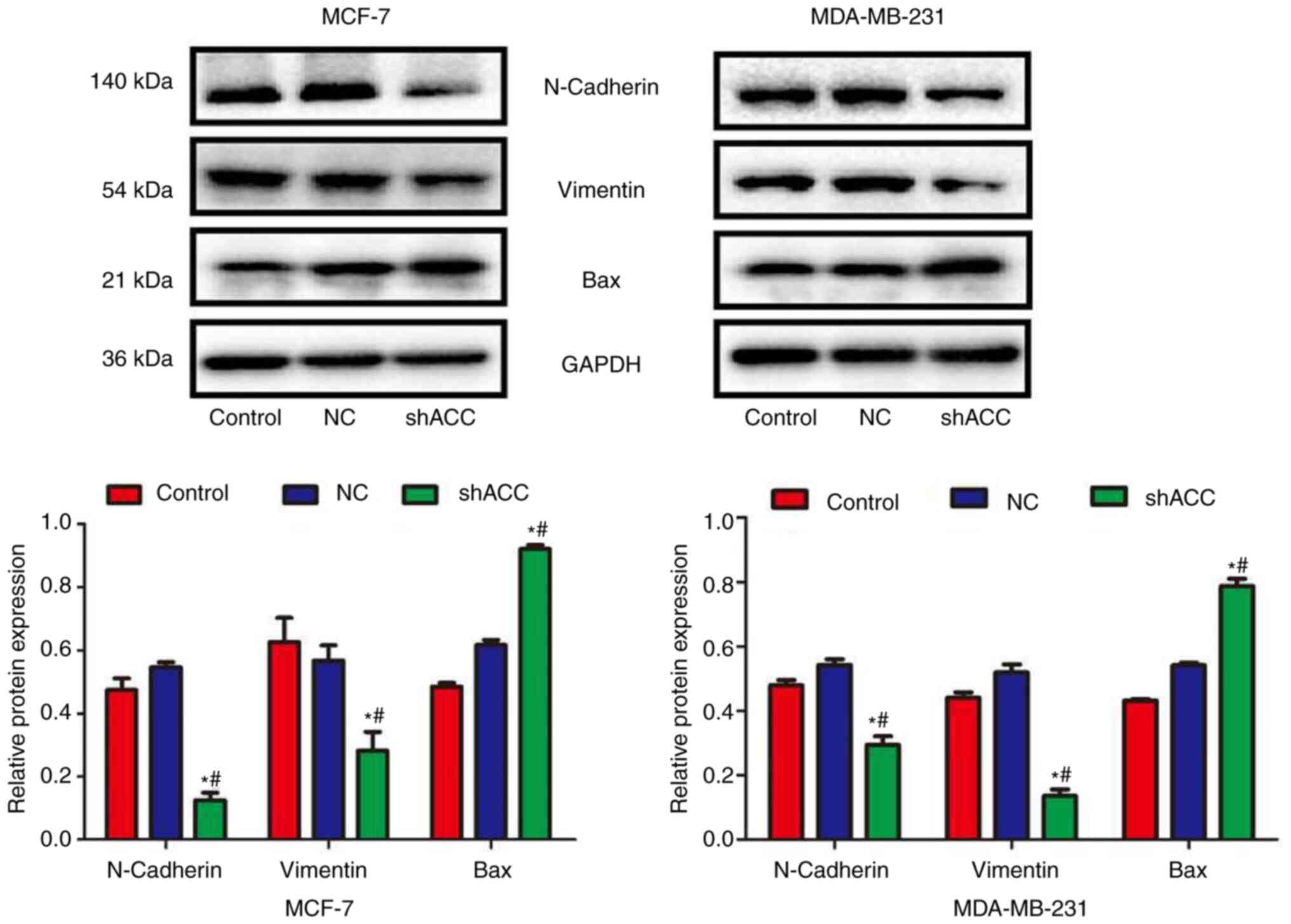
Effects of shACC on protein
expression
Compared with those in the Control and NC groups,
the protein expression levels of N-cadherin and Vimentin were
significantly decreased in the shACC groups of both breast cancer
cell lines, whereas Bax protein expression was significantly
increased (Fig. 5). These findings
indicated that ACC gene interference could affect the
epithelial-mesenchymal transition (EMT) of cells and the expression
of apoptosis-related proteins.
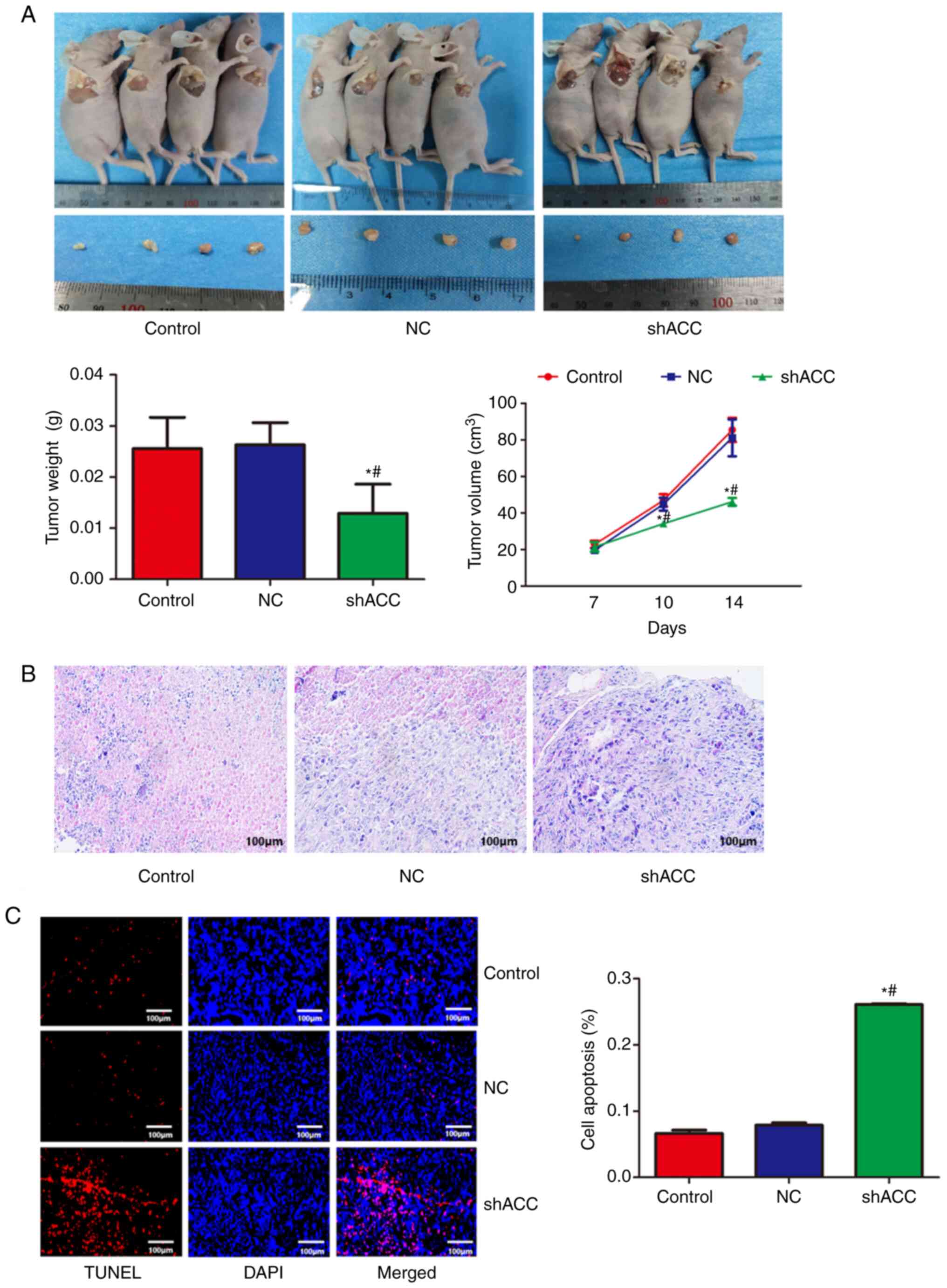
Effects of shACC on a MCF-7 xenograft
model
A nude mouse model of subcutaneous metastatic tumor
was established. As shown in Fig.
6A, compared with that in the Control and NC groups, the tumor
volume was significantly decreased in the shACC group,
demonstrating that ACC gene interference could reduce tumor volume
in nude mice. As shown in Fig. 6B,
the pathological findings showed that the breast cancer cells in
the Control and NC groups were flat and surrounded by antenna-like
extensions. By contrast, in the shACC group, the breast cancer
cells became round and smaller, decreased in number, with pyknotic
nuclei and apoptotic bodies observed, and chromatin agglutinated
into patches or crescent-shaped edges. As shown in Fig. 6C, compared with that in the Control
and NC groups, the apoptosis rate of the breast cancer cells in the
shACC group was significantly increased, suggesting that ACC gene
interference promoted apoptosis.
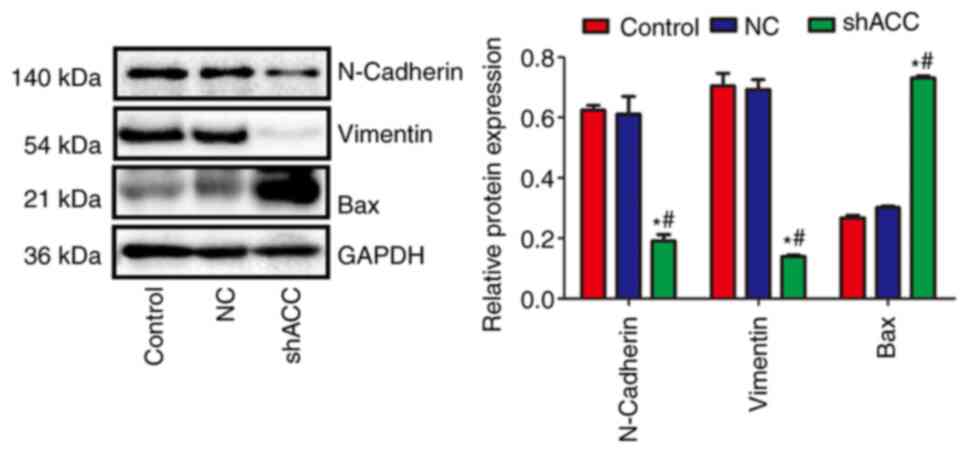
Effects of shACC on protein expression
in a MCF-7 xenograft model
Compared with those in the Control and NC groups,
the protein expression levels of N-cadherin and Vimentin were
significantly decreased in the shACC group, whereas the protein
expression levels of Bax were significantly increased (Fig. 7). This finding confirmed that ACC
gene interference also affected the EMT of cells and the expression
of apoptosis-related proteins in vivo.
Discussion
Notable progress has been made in understanding the
signaling network regulating breast cancer pathogenesis (11); however, due to various reasons,
such as tumor heterogeneity, drug resistance and drug-induced
toxicity (12), exploring
effective therapeutic targets remains a key issue that urgently
needs to be solved in breast cancer. Excessive cell proliferation
and metastasis of tumor cells are typical features of malignant
lesions. Compared with in normal human tissues, tumor cells prefer
to use exogenous lipids to promote growth and cell viability
(6). Previous studies have
reported that a variety of cancer cells contain high levels of
lipid-related enzymes (13),
including ACC (14), which can
significantly promote the malignant characteristics of high
proliferation and metastasis (15).
It has been shown that ACC is a valuable drug target
for the treatment of various metabolic diseases (4), including hepatic steatosis (16), nonalcoholic fatty liver disease,
metabolic syndrome (17), obesity
and hepatic insulin resistance (18). Previous studies have demonstrated
that ACC is also a potential cancer treatment target. In prostate
cancer (19) and liver cancer
(8) cells, knocking down ACC can
inhibit cancer proliferation and induce apoptosis without cytotoxic
effects on normal cells. The present study focused on two types of
breast cancer cell lines, MCF-7 and MDA-MB-231, as these two types
of cells are relatively representative cell lines of breast cancer.
After knocking down ACC gene expression, the viability of breast
cancer cells was decreased, whereas the apoptosis rate of cells was
increased, and the cell migration was significantly inhibited. The
results were consistent with those of previous studies (8,19).
In addition, knockdown of ACC gene expression inhibited the
formation and migration of nude mouse model tumors.
In order to understand the effect of ACC on the
malignant progression of MCF-7 and MDA-MB-231 cells, a CCK-8 assay
was used to detect the changes in cell viability after ACC
knockdown. The results showed that ACC knockdown could inhibit
cancer cell viability, and the results of flow cytometry also
revealed that ACC knockdown could induce breast cancer cell
apoptosis. EMT is the process of epithelial cells transitioning to
a mesenchymal state (20), which
is considered to be the main process of tumor metastasis (21). The molecular and cellular changes
observed in EMT are characterized by upregulation of specific
proteins, such as N-cadherin and vimentin, which are used as EMT
markers (22). It has been
reported that EMT may be related to the prognosis and
clinicopathological features of patients with breast cancer
(23). The overexpression of
mesenchymal cell marker factors and expression of EMT-related
transcription factors has been reported to promote metastasis in
breast cancer cells (24). In
order to further study the effect and mechanism of ACC on breast
cancer cells, a cell scratch test was performed to detect the
migratory ability of cells and western blotting was conducted to
determine the expression levels of EMT-related markers. The results
showed that ACC knockdown inhibited the migratory ability of breast
cancer cells and decreased the expression levels of N-cadherin and
vimentin. Therefore, ACC may inhibit the malignant progression of
breast cancer, and cell migration and metastasis, by affecting EMT.
In addition, the in vivo experiments revealed that knockdown
of ACC gene expression could inhibit the EMT process in tumors from
nude mice, which is consistent with the in vitro results.
Future studies may evaluate the expression of ACC in clinical
samples, as well as the association between ACC and survival of
patients with breast cancer.
In subsequent studies, a colony formation assay may
be performed to compare the findings with those of the CCK-8 assay.
Experiments with other shRNAs, to exclude off-target effects, and
comparison of ACC knockdown with ACC overexpression may also be
performed.
In conclusion, the findings of the present study
suggested that the ACC gene may affect cell viability and migration
in the malignant progression of breast cancer through the EMT
process. Targeting ACC expression could be used as a new target
therapy for breast cancer.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
MJ and LP designed the experiments. MJ, CY, SD and
BZ performed the experiments and analyzed the data. MJ wrote the
manuscript. LP revised the manuscript and supervised the
experiments. MJ and LP confirm the authenticity of all the raw
data. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of The Second Affiliated Hospital of Nanchang University
(approval no. 2021080401; Nanchang, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Dowsett M: Preoperative models to evaluate
endocrine strategies for breast cancer. Clin Cancer Res.
9:502S–510S. 2003.PubMed/NCBI
|
|
2
|
Youness RA, Gad AZ, Sanber K, Ahn YJ, Lee
GJ, Khallaf E, Hafez HM, Motaal AA, Ahmed N and Gad MZ: Targeting
hydrogen sulphide signaling in breast cancer. J Adv Res.
27:177–190. 2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Viedma-Rodríguez R, Baiza-Gutman L,
Salamanca-Gómez F, Diaz-Zaragoza M, Martínez-Hernández G, Ruiz
Esparza-Garrido R, Velázquez-Flores MA and Arenas-Aranda D:
Mechanisms associated with resistance to tamoxifen in estrogen
receptor-positive breast cancer (review). Oncol Rep. 32:3–15.
2014.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Chen L, Duan Y, Wei H, Ning H, Bi C, Zhao
Y, Qin Y and Li Y: Acetyl-CoA carboxylase (ACC) as a therapeutic
target for metabolic syndrome and recent developments in ACC1/2
inhibitors. Expert Opin Investig Drugs. 28:917–930. 2019.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Corbett JW: Review of recent acetyl-CoA
carboxylase inhibitor patents: Mid-2007-2008. Expert Opin Ther Pat.
19:943–956. 2009.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Zhan Y, Ginanni N, Tota MR, Wu M, Bays NW,
Richon VM, Kohl NE, Bachman ES, Strack PR and Krauss S: Control of
cell growth and survival by enzymes of the fatty acid synthesis
pathway in HCT-116 colon cancer cells. Clin Cancer Res.
14:5735–5742. 2008.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Zhao S, Cheng L, Shi Y, Li J, Yun Q and
Yang H: MIEF2 reprograms lipid metabolism to drive progression of
ovarian cancer through ROS/AKT/mTOR signaling pathway. Cell Death
Dis. 12(18)2021.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Ye B, Yin L, Wang Q and Xu C: ACC1 is
overexpressed in liver cancers and contributes to the proliferation
of human hepatoma Hep G2 cells and the rat liver cell line BRL 3A.
Mol Med Rep. 19:3431–3440. 2019.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Fang W, Cui H, Yu D, Chen Y, Wang J and Yu
G: Increased expression of phospho-acetyl-CoA carboxylase protein
is an independent prognostic factor for human gastric cancer
without lymph node metastasis. Med Oncol. 31(15)2014.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Zhou J, Chen C, Zhao X, Jiang T, Jiang Y,
Dai J and Chen J: Coding variants in the PCNT and CEP295 genes
contribute to breast cancer risk in Chinese women. Pathol Res
Pract. 225(153581)2021.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Miller EA, Pinsky PF, Heckman-Stoddard BM
and Minasian LM: Breast cancer risk prediction models and
subsequent tumor characteristics. Breast Cancer. 27:662–669.
2020.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Rysman E, Brusselmans K, Scheys K,
Timmermans L, Derua R, Munck S, Van Veldhoven PP, Waltregny D,
Daniëls VW, Machiels J, et al: De novo lipogenesis protects cancer
cells from free radicals and chemotherapeutics by promoting
membrane lipid saturation. Cancer Res. 70:8117–8126.
2010.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Schreurs M, Kuipers F and van der Leij FR:
Regulatory enzymes of mitochondrial beta-oxidation as targets for
treatment of the metabolic syndrome. Obes Rev. 11:380–388.
2010.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Harwood HJ Jr: Treating the metabolic
syndrome: Acetyl-CoA carboxylase inhibition. Expert Opin Ther
Targets. 9:267–281. 2005.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Savage DB, Choi CS, Samuel VT, Liu ZX,
Zhang D, Wang A, Zhang XM, Cline GW, Yu XX, Geisler JG, et al:
Reversal of diet-induced hepatic steatosis and hepatic insulin
resistance by antisense oligonucleotide inhibitors of acetyl-CoA
carboxylases 1 and 2. J Clin Invest. 116:817–824. 2006.PubMed/NCBI View
Article : Google Scholar
|
|
17
|
Kusunoki J, Kanatani A and Moller DE:
Modulation of fatty acid metabolism as a potential approach to the
treatment of obesity and the metabolic syndrome. Endocrine.
29:91–100. 2006.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Srivastava RA, Jahagirdar R, Azhar S,
Sharma S and Bisgaier CL: Peroxisome proliferator-activated
receptor-alpha selective ligand reduces adiposity, improves insulin
sensitivity and inhibits atherosclerosis in LDL receptor-deficient
mice. Mol Cell Biochem. 285:35–50. 2006.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Pang B, Zhang J, Zhang X, Yuan J, Shi Y
and Qiao L: Inhibition of lipogenesis and induction of apoptosis by
valproic acid in prostate cancer cells via the C/EBPα/SREBP-1
pathway. Acta Biochim Biophys Sin (Shanghai). 53:354–364.
2021.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Göbel A, Dell'Endice S, Jaschke N, Pählig
S, Shahid A, Hofbauer LC and Rachner TD: The role of inflammation
in breast and prostate cancer metastasis to bone. Int J Mol Sci.
22(5078)2021.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Tong D: Unravelling the molecular
mechanisms of prostate cancer evolution from genotype to phenotype.
Crit Rev Oncol Hematol. 163(103370)2021.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Barik GK, Sahay O, Behera A, Naik D and
Kalita B: Keep your eyes peeled for long noncoding RNAs: Explaining
their boundless role in cancer metastasis, drug resistance, and
clinical application. Biochim Biophys Acta Rev Cancer.
1876(188612)2021.PubMed/NCBI View Article : Google Scholar
|
|
23
|
González-Martínez S, Pérez-Mies B, Pizarro
D, Caniego-Casas T, Cortés J and Palacios J: Epithelial mesenchymal
transition and immune response in metaplastic breast carcinoma. Int
J Mol Sci. 22(7398)2021.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Lu Y, Ding Y, Wei J, He S, Liu X, Pan H,
Yuan B, Liu Q and Zhang J: Anticancer effects of traditional
chinese medicine on epithelial-mesenchymal transition (EMT) in
breast cancer: Cellular and molecular targets. Eur J Pharmacol.
907(174275)2021.PubMed/NCBI View Article : Google Scholar
|