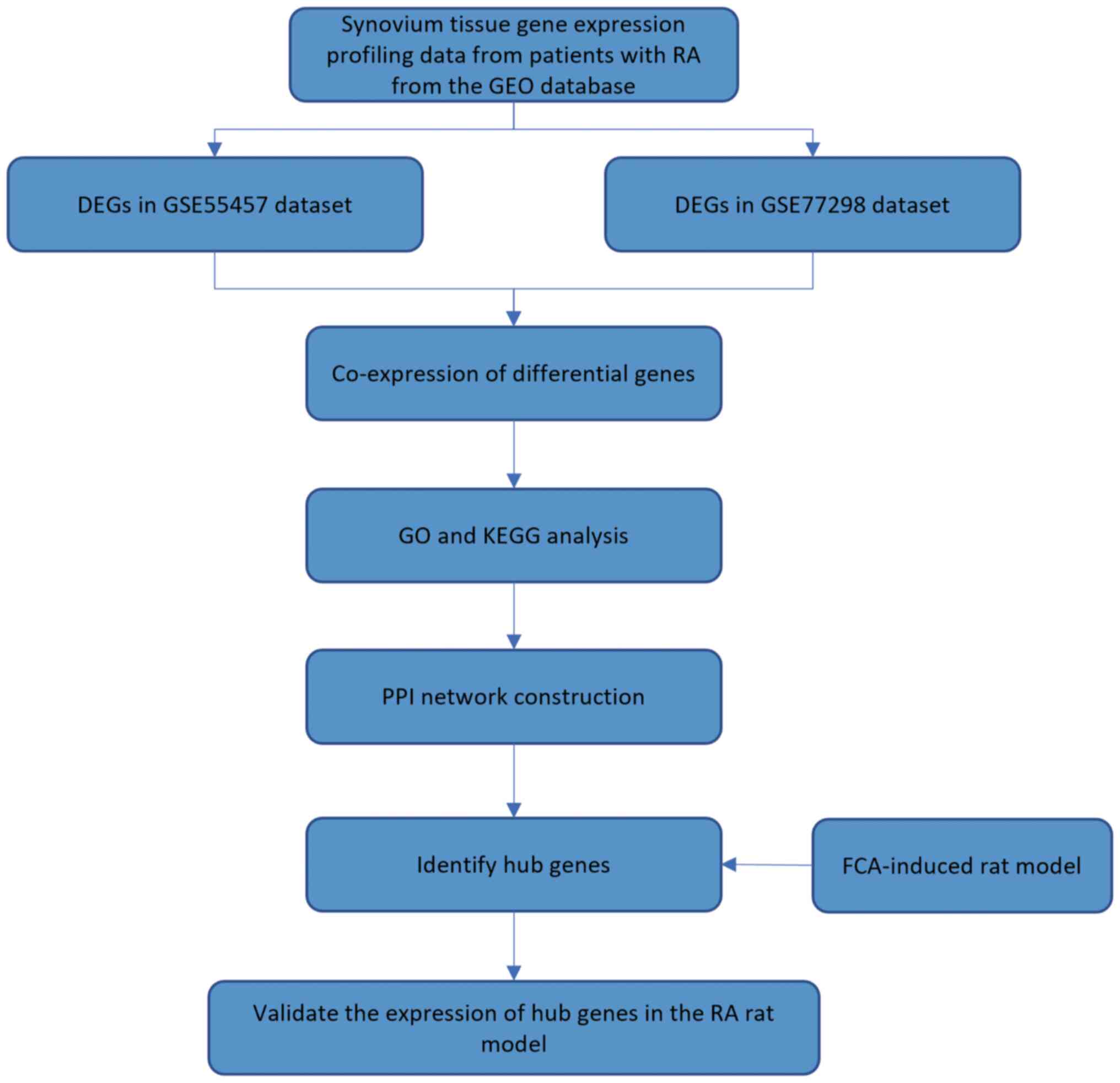
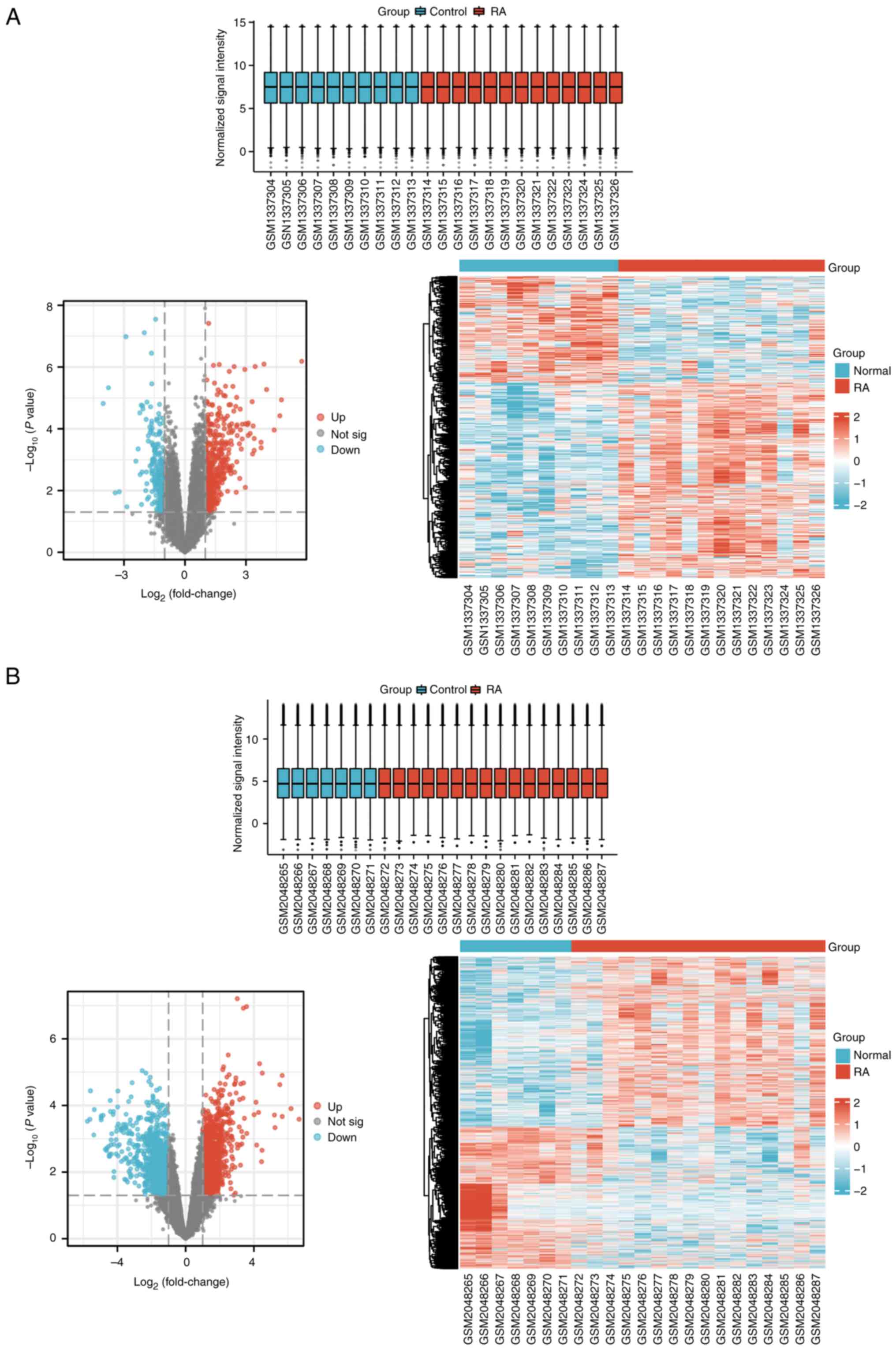
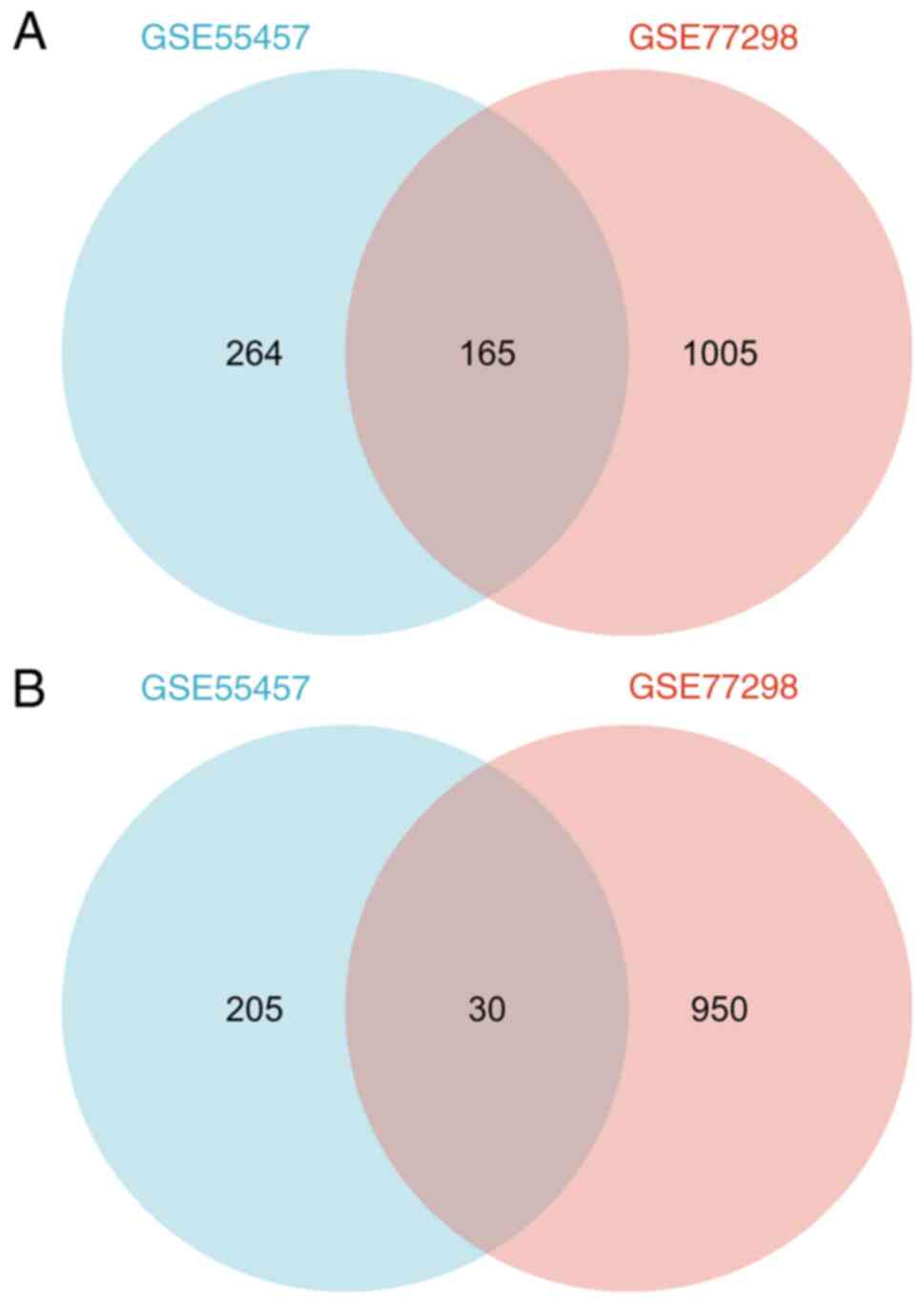
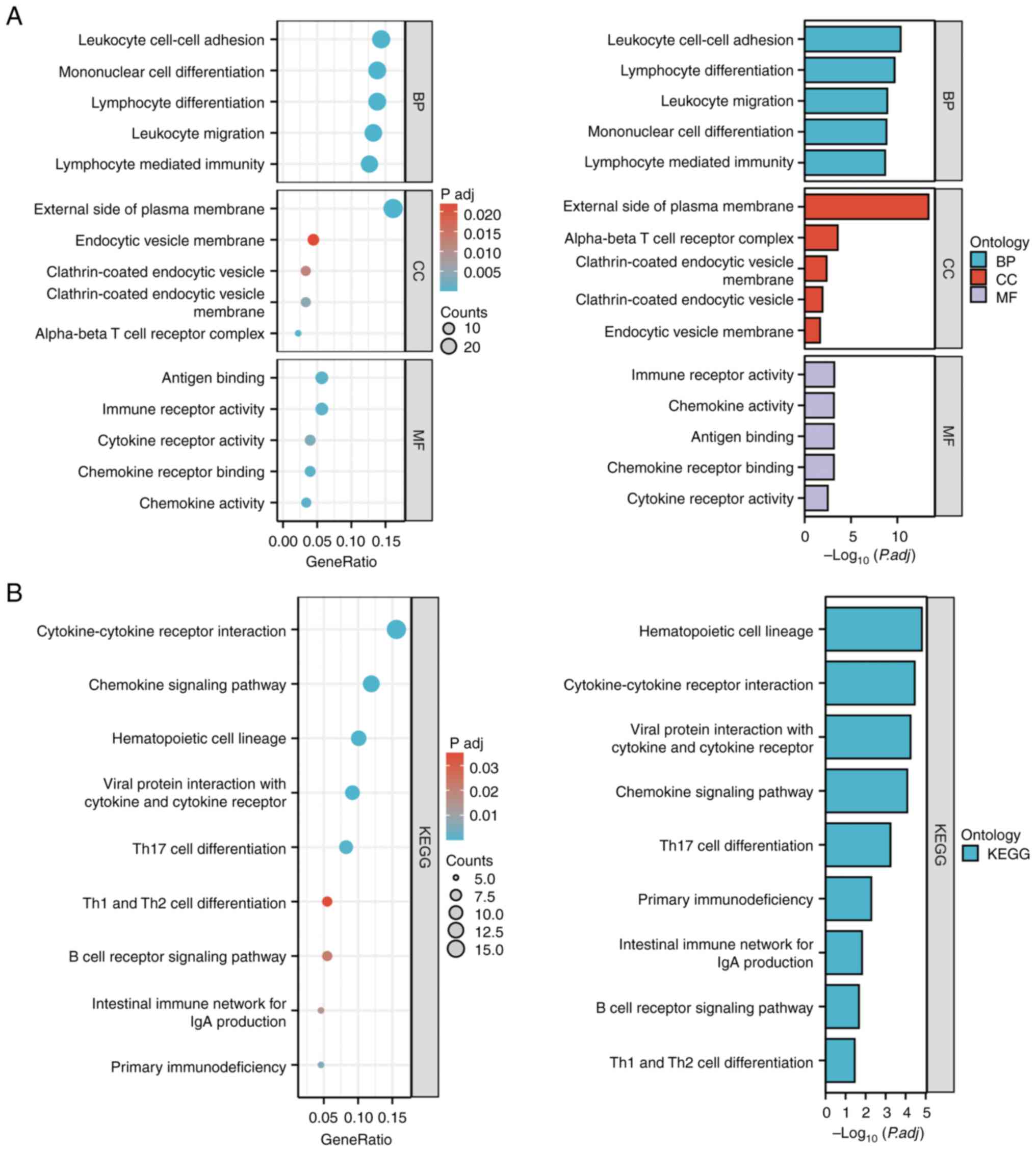
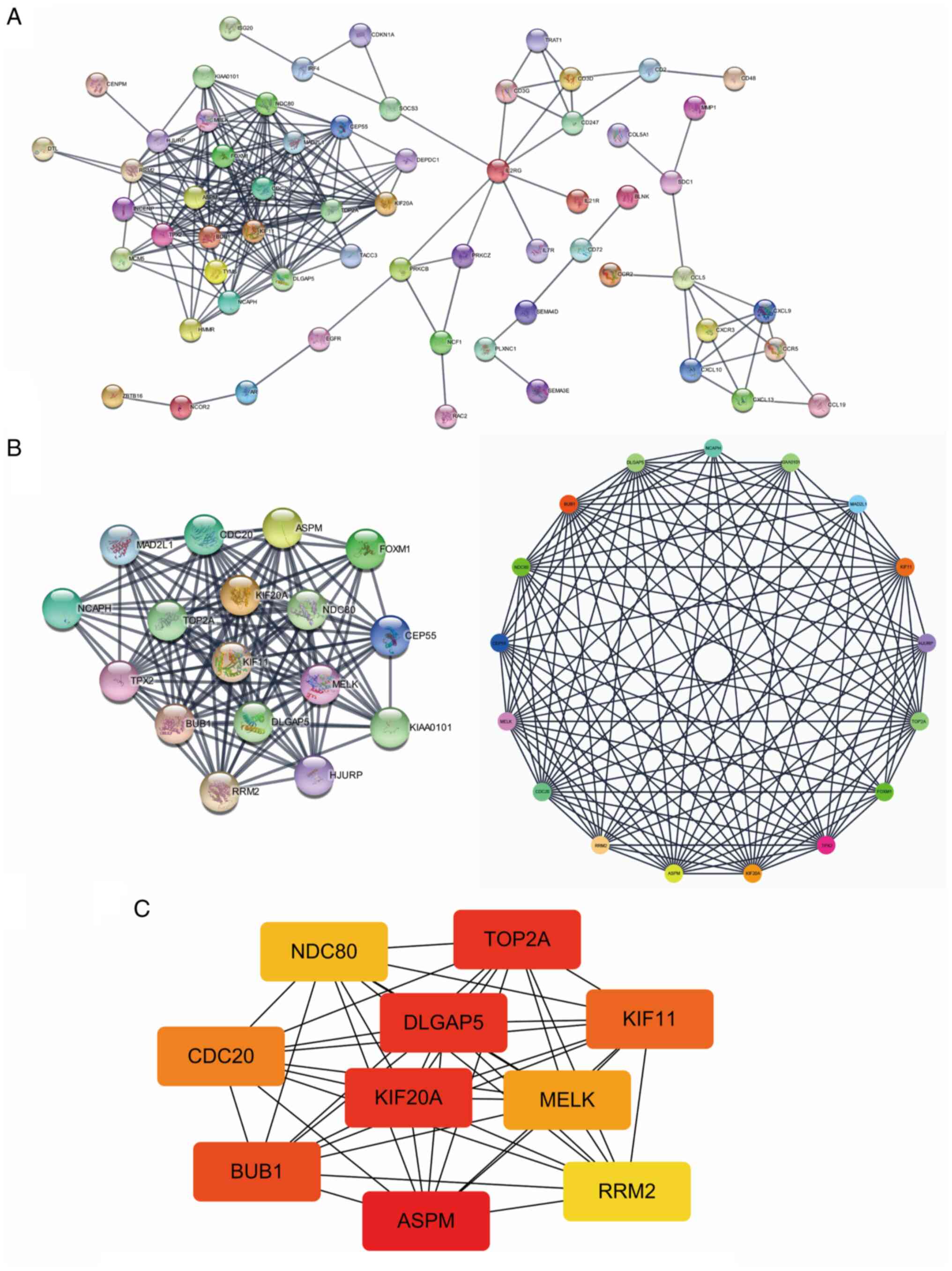
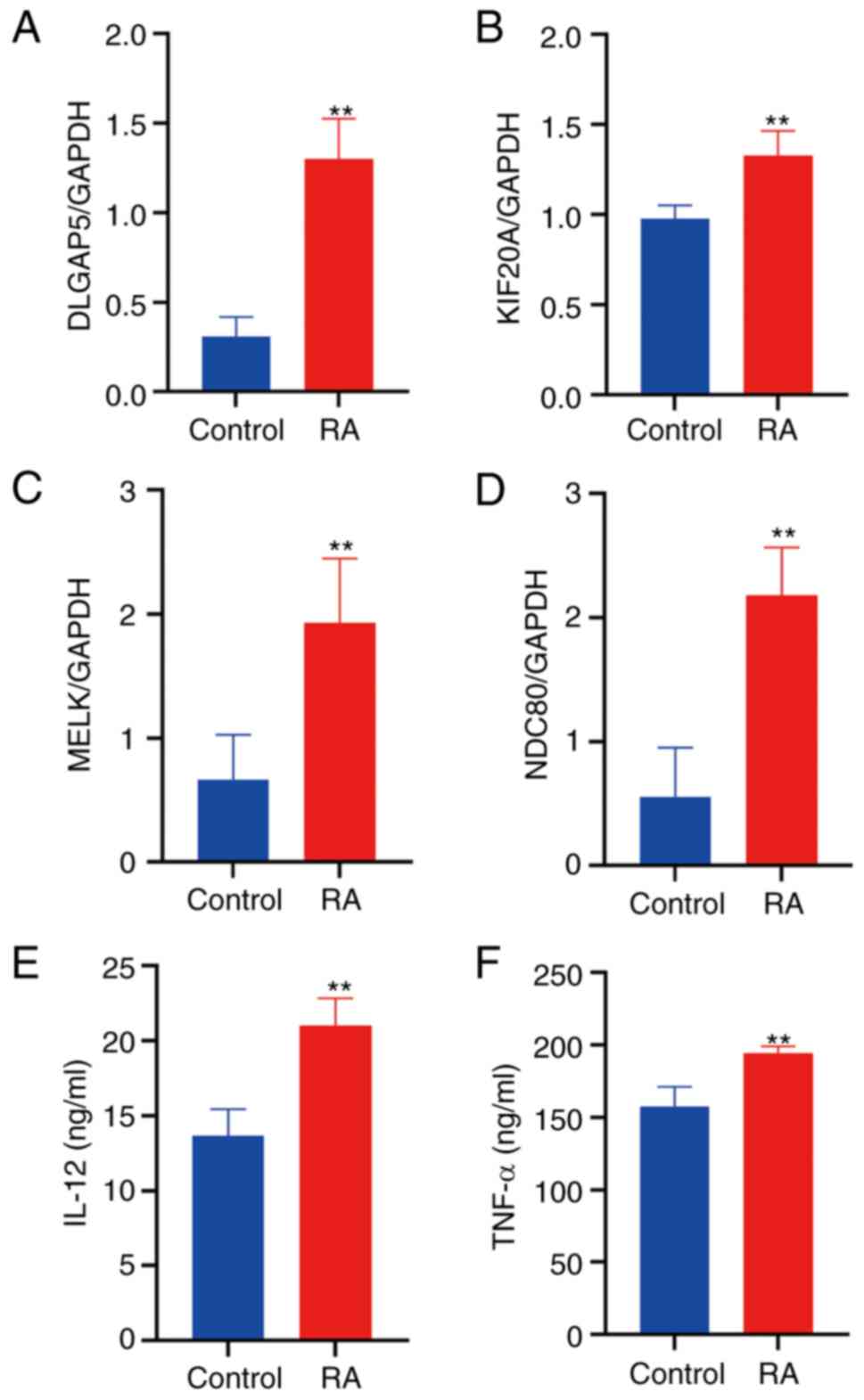
|
1
|
Giraud EL, Jessurun NT, van Hunsel FPAM,
van Puijenbroek EP, van Tubergen A, Ten Klooster PM and Vonkeman
HE: Frequency of real-world reported adverse drug reactions in
rheumatoid arthritis patients. Expert Opin Drug Saf. 19:1617–1624.
2020.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Smith MH and Berman JR: What is rheumatoid
arthritis? JAMA. 327(1194)2022.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Smolen JS, Aletaha D and McInnes IB:
Rheumatoid arthritis. Lancet. 388:2023–2038. 2016.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Gravallese EM and Firestein GS: Rheumatoid
arthritis-common origins, divergent mechanisms. N Engl J Med.
388:529–542. 2023.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Aletaha D and Smolen JS: Diagnosis and
management of rheumatoid arthritis: A review. JAMA. 320:1360–1372.
2018.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Woetzel D, Huber R, Kupfer P, Pohlers D,
Pfaff M, Driesch D, Häupl T, Koczan D, Stiehl P, Guthke R and Kinne
RW: Identification of rheumatoid arthritis and osteoarthritis
patients by transcriptome-based rule set generation. Arthritis Res
Ther. 16(R84)2014.PubMed/NCBI View
Article : Google Scholar
|
|
7
|
Broeren MG, de Vries M, Bennink MB, Arntz
OJ, Blom AB, Koenders MI, van Lent PL, van der Kraan PM, van den
Berg WB and van de Loo FA: Disease-regulated gene therapy with
anti-inflammatory interleukin-10 under the control of the CXCL10
promoter for the treatment of rheumatoid arthritis. Hum Gene Ther.
27:244–254. 2016.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Davis S and Meltzer PS: GEOquery: A bridge
between the gene expression omnibus (GEO) and BioConductor.
Bioinformatics. 23:1846–1847. 2007.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Smyth GK: Limma: Linear models for
microarray data. In: Bioinformatics and Computational Biology
Solutions using R and Bioconductor. Springer, New York, NY,
397-420, 2005.
|
|
10
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012.PubMed/NCBI View Article : Google Scholar
|
|
11
|
R Core Team: R: A language and environment
for statistical computing. R Foundation for Statistical Computing,
Vienna, Austria, 2023. https://www.R-project.org/.
|
|
12
|
Lu S, Yadav AK and Qiao X: Identification
of potential miRNA-mRNA interaction network in bone marrow T cells
of acquired aplastic anemia. Hematology. 25:168–175.
2020.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Liu S, Chen H, Ma W, Zhong Y, Liang Y, Gu
L, Lu X and Li J: Non-coding RNAs and related molecules associated
with form-deprivation myopia in mice. J Cell Mol Med. 26:186–194.
2022.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Chin CH, Chen SH, Wu HH, Ho CW, Ko MT and
Lin CY: cytoHubba: Identifying hub objects and sub-networks from
complex interactome. BMC Syst Biol. 8 (Suppl 4)(S11)2014.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Otasek D, Morris JH, Bouças J, Pico AR and
Demchak B: Cytoscape automation: Empowering workflow-based network
analysis. Genome Biol. 20(185)2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Szklarczyk D, Gable AL, Nastou KC, Lyon D,
Kirsch R, Pyysalo S, Doncheva NT, Legeay M, Fang T, Bork P, et al:
Correction to ‘The STRING database in 2021: Customizable
protein-protein networks, and functional characterization of
user-uploaded gene/measurement sets.’. Nucleic Acids Res.
49(10800)2021.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Li A, Zhang Z, Ru X, Yi Y, Li X, Qian J,
Wang J, Yang X and Yao Y: Identification of SLAMF1 as an
immune-related key gene associated with rheumatoid arthritis and
verified in mice collagen-induced arthritis model. Front Immunol.
13(961129)2022.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Zhong YM, Zhang LL, Lu WT, Shang YN and
Zhou HY: Moxibustion regulates the polarization of macrophages
through the IL-4/STAT6 pathway in rheumatoid arthritis. Cytokine.
152(155835)2022.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Chen Y, Liu K, Qin Y, Chen S, Guan G,
Huang Y, Chen Y and Mo Z: Effects of Pereskia aculeate Miller
petroleum ether extract on complete Freund's adjuvant-induced
rheumatoid arthritis in rats and its potential molecular
mechanisms. Front Pharmacol. 13(869810)2022.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Pan H, Guo R, Ju Y, Wang Q, Zhu J, Xie Y,
Zheng Y, Li T, Liu Z, Lu L, et al: A single bacterium restores the
microbiome dysbiosis to protect bones from destruction in a rat
model of rheumatoid arthritis. Microbiome. 7(107)2019.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Han C, Yang Y, Sheng Y, Wang J, Zhou X, Li
W, Guo L, Zhang C and Ye Q: Glaucocalyxin B inhibits cartilage
inflammatory injury in rheumatoid arthritis by regulating M1
polarization of synovial macrophages through NF-κB pathway. Aging
(Albany NY). 13:22544–22555. 2021.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Kumar H and Bot A: In this issue: Role of
immune cells and molecules in rheumatoid arthritis pathogenesis and
cancer immunotherapy. Int Rev Immunol. 37:127–128. 2018.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Zhang L, Zhong Y, Lu W, Shang Y, Guo Y,
Luo X, Chen Y, Luo K, Hu D, Yu H, et al: Moxibustion of Zusanli
(ST36) and Shenshu (BL23) alleviates the inflammation of rheumatoid
arthritis in rats through regulating macrophage migration
inhibitory factor/glucocorticoids signaling. J Tradit Chin Med (In
Chinese).
|
|
25
|
Su H, Su SY, Yang P, Guo YJ and Li J:
Progress of research on mechanism of acupuncture and moxibustion in
the treatment of rheumatoid arthritis. Zhen Ci Yan Jiu. 48:500–507.
2023.PubMed/NCBI View Article : Google Scholar : (In Chinese).
|
|
26
|
Zhong YM, Wu F, Luo XC, Chen Y, Ren JG,
Yang X, Ma WB and Zhou HY: Mechanism on moxibustion for rheumatoid
arthritis based on PD-1/PD-L1 signaling pathway. Zhongguo Zhen Jiu.
40:976–982. 2020.PubMed/NCBI View Article : Google Scholar : (In Chinese).
|
|
27
|
Chen Y, Li H, Luo X, Liu H, Zhong Y, Wu X
and Liu X: Moxibustion of Zusanli (ST36) and Shenshu (BL23)
alleviates cartilage degradation through RANKL/OPG signaling in a
rabbit model of rheumatoid arthritis. Evid Based Complement
Alternat Med. 2019(6436420)2019.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Krishna V, Yin X, Song Q, Walsh A,
Pocalyko D, Bachman K, Anderson I, Madakamutil L and Nagpal S:
Integration of the transcriptome and genome-wide landscape of BRD2
and BRD4 binding motifs identifies key superenhancer genes and
reveals the mechanism of bet inhibitor action in rheumatoid
arthritis synovial fibroblasts. J Immunol. 206:422–431.
2021.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Tang N, Dou X, You X, Shi Q, Ke M and Liu
G: Pan-cancer analysis of the oncogenic role of discs large homolog
associated protein 5 (DLGAP5) in human tumors. Cancer Cell Int.
21(457)2021.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Li K, Fu X, Wu P, Zhaxi B, Luo H and Li Q:
DLG7/DLGAP5 as a potential therapeutic target in gastric cancer.
Chin Med J. 135:1616–1618. 2022.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Zhang H, Liu Y, Tang S, Qin X, Li L, Zhou
J, Zhang J and Liu B: Knockdown of DLGAP5 suppresses cell
proliferation, induces G2/M phase arrest and apoptosis in ovarian
cancer. Exp Ther Med. 22(1245)2021.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Ren X, Chen X, Ji Y, Li L, Li Y, Qin C and
Fang K: Upregulation of KIF20A promotes tumor proliferation and
invasion in renal clear cell carcinoma and is associated with
adverse clinical outcome. Aging (Albany, NY). 12:25878–25894.
2020.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Wu C, Qi X, Qiu Z, Deng G and Zhong L: Low
expression of KIF20A suppresses cell proliferation, promotes
chemosensitivity and is associated with better prognosis in HCC.
Aging (Albany, NY). 13:22148–22163. 2021.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Zhang X, Wang J, Wang Y, Liu G, Li H, Yu
J, Wu R, Liang J, Yu R and Liu X: MELK inhibition effectively
suppresses growth of glioblastoma and cancer stem-like cells by
blocking AKT and FOXM1 pathways. Front Oncol.
10(608082)2020.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Ye J, Deng W, Zhong Y, Liu H, Guo B, Qin
Z, Li P, Zhong X and Wang L: MELK predicts poor prognosis and
promotes metastasis in esophageal squamous cell carcinoma via
activating the NF-κB pathway. Int J Oncol. 61(94)2022.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Tang B, Zhu J, Liu F, Ding J, Wang Y, Fang
S, Zheng L, Qiu R, Chen M, Shu G, et al: xCT contributes to
colorectal cancer tumorigenesis through upregulation of the MELK
oncogene and activation of the AKT/mTOR cascade. Cell Death Dis.
13(373)2022.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Xu Q, Ge Q, Zhou Y, Yang B, Yang Q, Jiang
S, Jiang R, Ai Z, Zhang Z and Teng Y: MELK promotes endometrial
carcinoma progression via activating mTOR signaling pathway.
EBioMedicine. 51(102609)2020.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Chen X, He Q, Zeng S and Xu Z:
Upregulation of nuclear division cycle 80 contributes to
therapeutic resistance via the promotion of autophagy-related
protein-7-dependent autophagy in lung cancer. Front Pharmacol.
13(985601)2022.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Chen J and Ünal E: Meiotic regulation of
the Ndc80 complex composition and function. Curr Genet. 67:511–518.
2021.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Huang J, Zheng M, Li Y, Xu D and Tian D:
DLGAP5 promotes gallbladder cancer migration and tumor-associated
macrophage M2 polarization by activating cAMP. Cancer Immunol
Immunother: Jul 8, 2023 (Epub ahead of print).
|
|
41
|
Zhang Z, Sun C, Li C, Jiao X, Griffin BB,
Dongol S, Wu H, Zhang C, Cao W, Dong R, et al: Upregulated MELK
leads to doxorubicin chemoresistance and M2 macrophage polarization
via the miR-34a/JAK2/STAT3 pathway in uterine leiomyosarcoma. Front
Oncol. 10(453)2020.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Chung SJ, Yoon HJ, Youn H, Kim MJ, Lee YS,
Jeong JM, Chung JK, Kang KW, Xie L, Zhang MR and Cheon GJ:
18F-FEDAC as a targeting agent for activated macrophages in DBA/1
mice with collagen-induced arthritis: Comparison with
18F-FDG. J Nucl Med. 59:839–845. 2018.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Park SY, Lee SW, Lee SY, Hong KW, Bae SS,
Kim K and Kim CD: SIRT1/adenosine monophosphate-activated protein
kinase α signaling enhances macrophage polarization to an
anti-inflammatory phenotype in rheumatoid arthritis. Front Immunol.
8(1135)2017.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Cutolo M, Campitiello R, Gotelli E and
Soldano S: The role of M1/M2 macrophage polarization in rheumatoid
arthritis synovitis. Front Immunol. 13(867260)2022.PubMed/NCBI View Article : Google Scholar
|