Introduction
Cervical cancer is the fourth most common malignancy
and the leading cause of cancer-associated death in women worldwide
(1,2). In the Gulf Cooperation Council
States, which include Bahrain, Kuwait, Oman, Saudi Arabia, Qatar
and the United Arab Emirates, cervical cancer is the ninth most
prevalent female malignancy and the number of cases in these
countries is expected to increase in the coming years (3). Several risk factors have been linked
to cervical cancer, such as human papillomavirus (HPV), early age
of sexual intercourse, multiple sexual partners, oral contraceptive
use and smoking (4-6).
However, since the isolation of HPV 6 DNA from human genital warts
in 1980(7), HPV has been
identified as the major causative agent for the development of
cervical cancer. The majority of cases of cervical cancer are
caused by persistent HPV infection, involving 15 genotypes of
high-risk HPV (hr-HPV) (8). In
these cases, HPV vaccination has been proven to be safe and
effective as a primary preventative strategy for cervical
intraepithelial neoplasia (CIN) and cervical cancer (9). A secondary preventative method
includes regular screening and prompt treatment for precancerous
lesions (9,10). The primary methods of screening for
cervical cancer include a Pap smear, visual inspection with acetic
acid and Lugol's iodine, liquid-based cytology and HPV testing
(10). In addition to
cytology-based tests, HPV testing is a pivotal method for cervical
cancer screening, allowing clinicians to assess cells for infection
with hr-HPV (10). The use of
cytology-based testing and HPV testing together or simultaneously
has assisted in guiding women who are at risk of developing
cervical cancer but has also led to an increase in the number of
referral cases to colposcopy clinics, in particular, cases with
low-grade cytological scores and hr-HPV infection (11,12).
Several countries have adopted the use of cytology-based testing
and HPV testing, including Australia, The Netherlands, United
Kingdom, Canada and Turkey (11,12).
In addition to HPV-positive cervical cancer, studies
have shown that ~5% of cervical cancer cases are not associated
with HPV infection (13-16).
Thus, the new World Health Organization Female Genital Tumors
classification subdivided cervical squamous and adenocarcinomas
into HPV-associated and HPV-independent tumors (17). HPV-negative cervical cancer
represents a biologically different subset of tumors with a
distinct pathogenic pathway and a poorer prognosis than
HPV-positive cervical cancer (18). Furthermore, HPV-negative and
HPV-positive cervical cancers show differences in molecular
profiles and no specific therapeutic strategies have been developed
based on HPV status (19).
Dynamic genetic alterations are the primary
causative mechanisms in the initiation and progression of human
cancer (20). In addition to
nuclear changes, the role of mitochondrial DNA (mtDNA) has been
widely investigated in various types of cancers, including cervical
cancer (21-24).
The mitochondria generate the majority of cellular energy through
the oxidative phosphorylation (OXPHOS) system and are also involved
in other cellular functions, such as the regulation of cell death
and generation of reactive oxygen species (ROS). The human mtDNA is
a 16.6 kb circular double-stranded DNA that contains 37 genes
coding for proteins essential for cellular respiration and normal
mitochondrial function (25).
mtDNA is present in multiple copies per cell, ranging from
1,000-10,000, and the mtDNA copy number varies by cell type
(26). Several factors make the
mtDNA particularly susceptible to oxidative stress, including its
proximity to the electron transport chain, lack of histone
protection and reduced DNA repair capacity (27). Thus, mtDNA is a particularly
susceptible target of ROS, and this may result in mutations or copy
number alterations (27). Changes
in mtDNA copy number can potentially lead to a decrease in
mitochondrial function with increased ROS production (27,28).
Variation in the mtDNA copy number has been reported in a wide
range of pathological conditions, such as neurodegenerative
diseases (29,30), autoimmune diseases (31) and different types of cancer
(32,33). Certain previous studies have
indicated an association between mtDNA copy number and the
development of cervical cancer. Sun et al (34) reported an increase in mtDNA copy
number in exfoliated cervical cells of women who tested positive
for HPV and hypothesized an association between mtDNA copy number
and cervical carcinogenesis. A study by Warowicka et al
(23) showed an association
between increased mtDNA copy number (and mtDNA mutations) and
cervical cancer development. However, these studies focused only on
HPV-positive cervical cancer cases, and thus far, no studies have
investigated the mtDNA copy number in HPV-negative cervical cancer.
Therefore, the aim of the present study was to examine the changes
in mtDNA copy number among HPV-positive and HPV-negative cervical
cancer cases. Knowledge regarding mtDNA copy number alterations in
HPV-positive and HPV-negative cervical cancer may assist in
understanding the role of mtDNA in the pathogenesis of the
disease.
Patients and methods
Sample collection
A total of 287 ThinPrep cervical samples were
collected from female patients who attended the Obstetrics and
Gynecology outpatient clinic at the Maternity Hospital (Al-Sabah
Health Area, Kuwait) and Mubarak Al Kabeer Hospital (Jabriya,
Kuwait) between January 2022 and January 2023, and the samples were
analyzed in the hospitals' Cytology Laboratory. Informed consent
was obtained from all participants and the study was approved by
the Health Science Center Ethics Committee at Kuwait University,
Kuwait and the Ministry of Health the Standing Committee for
Coordination of Health and Medical Research, Kuwait (no.
VDR/EC/3746). Samples were prepared and results were reported using
The Bethesda System 2014 guidelines (35). Samples with abnormal cytological
results (n=143) included the following: Squamous cell carcinoma
(SCC), high-grade squamous intraepithelial lesion (HSIL) and
low-grade squamous intraepithelial lesion (LSIL). Samples with SCC
and HSIL were categorized together as the SCC/HSIL group. Follow-up
histological information for cases with cervical abnormalities was
recorded when possible. Histological reports of SCC/HSIL included
the following diagnosis: CIN grade 1 (CIN1), CIN2 and CIN3. Cases
with LSIL with two sequentially abnormal cytology reports were also
referred for histological analysis. The median age of the cases
group was 39 years (age range, 19-79 years).
Control samples (n=144) were without any
intraepithelial lesions or malignancy. The HPV status (HPV-positive
and HPV-negative) for both cases and controls was determined by
genotyping. The median age of the control group was 38.5 years (age
range, 19-64 years).
DNA extraction
DNA was isolated from cervical cells using the MagNA
Pure LC DNA Kit I (Roche Diagnostics GmbH), based on magnetic-bead
technology, and it was performed using an automated MagNA Pure LC
Instrument (Roche Diagnostics GmbH). In brief, the sample was mixed
with a lysis/binding buffer for DNA lysis. Proteinase K was added
to complete the digestion of protein. Magnetic Glass Particles
(MGPs) were added for DNA binding to the surface and unbound
substances were discarded by several washing steps. The DNA was
eluted using a low salt buffer and MGPs. The DNA samples were
checked for purity using a NanoDrop 1000 system (Thermo Fisher
Scientific, Inc.) and the DNA concentration was determined using a
Qubit 3.0 Fluorometer (Thermo Fisher Scientific, Inc.).
Detection of HPV in cervical
samples
HPV DNA in cervical samples was detected by reverse
transcription-quantitative (q)PCR using two different sets of
primers, MY09/MY11 (MY09 reverse, 5'-CGTCCMARRGGAWACTGATC-3' and
MY11 forward, 5'-GCMCAGGGWCACAAYAATGG-3') and Gp5+/Gp6+ (Gp5+
forward, 5'-TTTGTTACTGTGGTAGATACTAC-3' and Gp6+ reverse,
5'-GAAAAATAAACTGTAAATCATATTC-3') (custom design; Thermo Fisher
Scientific, Inc.), as described previously (36). Samples with positive PCR
amplification results were subjected to a conventional PCR assay
then Sanger-based sequencing analysis was carried out using
MY09/MY11 and Gp5+/Gp6+ universal primers. The sample results were
analysed using in-house Sequencing Analysis Software version 3.7
(Applied Biosystems; Thermo Fisher Scientific, Inc.). The HPV
genotypes were identified using the BLASTn Software version 2.13.0
(http://www.ncbi.nlm.nih.gov/blast/htlm) and the Los
Almos Data National Laboratory Theoretical Biology and Biophysics
HPV database (https://pave.niaid.nih.gov/).
A total of 17 different HPV genotypes were detected
in the samples, including 11 hr and six low-risk (lr) genotypes.
hr-HPV included the following genotypes: HPV16, HPV18, HPV31,
HPV33, HPV35, HPV45, HPV53, HPV56, HPV58, HPV66 and HPV73. lr-HPV
included the following genotypes: HPV6, HPV11, HPV83, HPV90, HPV102
and HPV106. Only hr-HPV cases were included in the further
analysis. Representative Sanger sequencing chromatograms of an
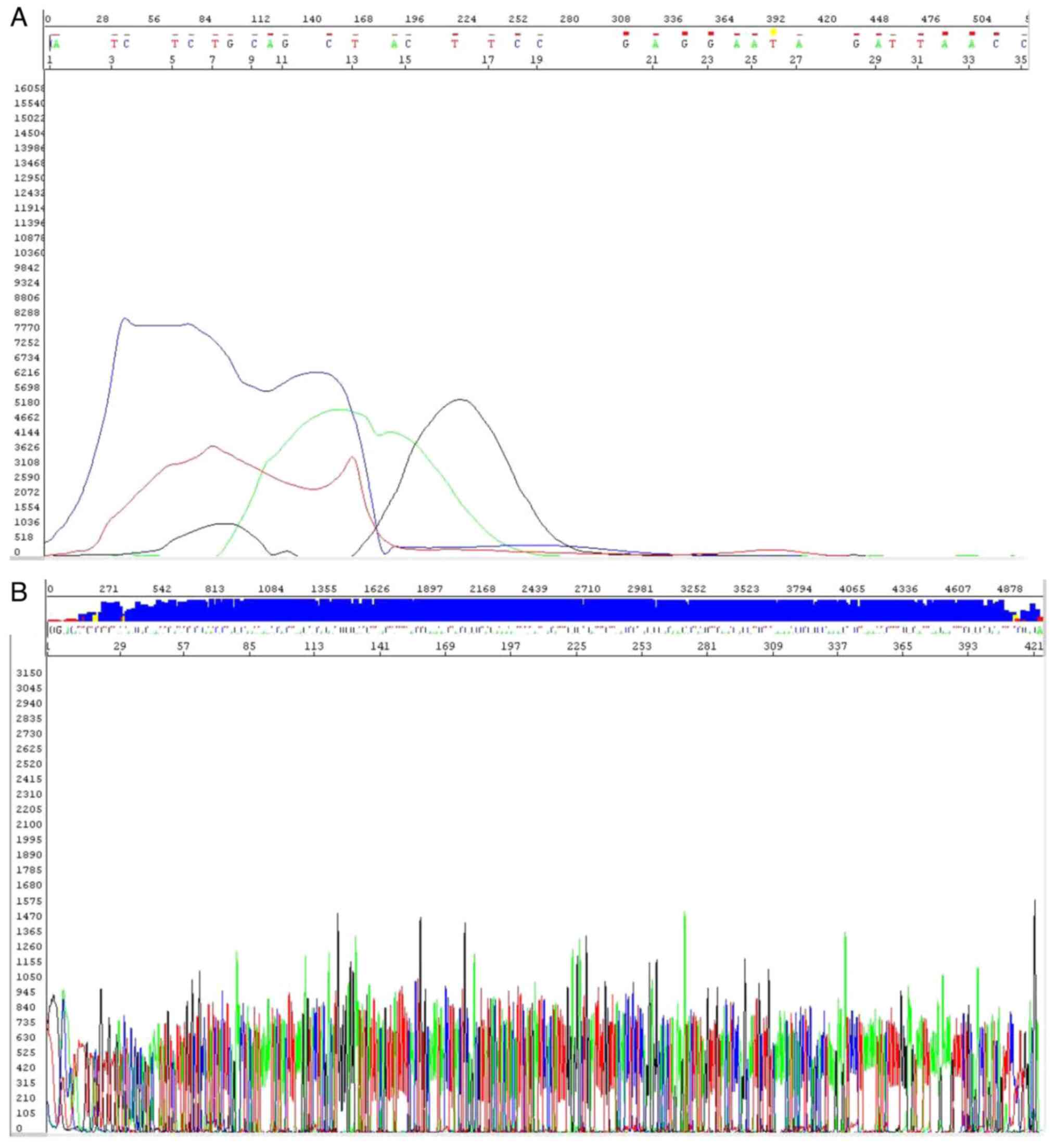
HPV-negative and HPV16-positive sample are provided in Fig. 1A and B, respectively.
Determination of mtDNA copy
number
qPCR was used to determine the mtDNA copy number
relative to nuclear DNA (nDNA) in cervical exfoliated cells. The
mtDNA-encoded human NADH dehydrogenase subunit 2 (mt-ND2) was used
as a target gene and the nuclear β2-macroglobulin (β2-M) was used
as a reference gene. Amplification was performed using the
following primers: mt-ND2 forward, 5'-CACAGAAGCTGCCATCAAGTA-3' and
reverse, 5'-CCGGAGAGTATATTGTTGAAGAG-3'; β2-M forward,
5'-CCAGCAGAGAATGGAAAGTCAA-3' and reverse,
5'-TCTCTCTCCATTCTTCAGTAAGTCAACT-3'. The qPCR was set up according
to the manufacturer's instruction and optimized for >95%
amplification efficiency using the SYBR1 Green PCR MasterMix
(Applied Biosystems; Thermo Fisher Scientific, Inc.) as described
previously (37). Reactions were
run in duplicate on a 7900HT Fast Real-Time PCR System (Applied
Biosystems; Thermo Fisher Scientific, Inc.) and a non-template
control was included in each run. The following thermocycling
program was used: 1 cycle of 50˚C for 2 min, 1 cycle of 95˚C for 10
min, and 30 cycles of 95˚C for 15 sec and 60˚C for 1 min. The
relative mtDNA copy number was calculated from the quantification
cycle (Cq) values of the target and reference genes using the
2-∆∆Cq method (38).
Statistical analysis
Statistical analysis was performed using SPSS
version 20.0 (IBM Corp.). The distribution of data was assessed
using a Kolmogorov-Smirnov test. Continuous variables were compared
between cases and controls using a Student's t-test. The
χ2 test was used to determine statistical differences
between categorical variables. Multiple groups were compared by
using one-way ANOVA followed by Tukey's post-hoc test. P<0.05
was considered to indicate a statistically significant difference.
Graphs were plotted using GraphPad Prism version 5.0 (GraphPad
Software; Dotmatics). The data are presented as the mean ± standard
deviation (SD) or n (%).
Results
Demographic and clinical
characteristics of study subjects
The demographic and clinical characteristics of the
study subjects, including 143 cases with cervical abnormalities and
144 controls, are presented in Table
I. There was no statistically significant difference in the
respective mean ± SD and median age of cases (38.2±10.4; 39 years)
and controls (36.9±10.3; 38.5 years; P=0.16). The distribution of
the HPV genotypes, which included hr-HPV positive and HPV-negative,
was also assessed. In the group of cases with cervical
abnormalities, 49 (34%) were hr-HPV positive and 94 (66%) were
HPV-negative. In the control group, 44 (31%) were hr-HPV positive
and 100 (69%) were HPV-negative. There were no significant
differences between the cases and controls among hr-HPV positive
subjects (P>0.05). In terms of cytological diagnosis, 28 (20%)
of the cases had SCC/HSIL and 115 (80%) had LSIL. SCC/HSIL cases,
as well as LSIL cases with two sequential abnormal cytological
reports, were further analysed based on the histological diagnosis.
Histology diagnoses were available for 56 cases, including 18 (32%)
CIN2/3 and 38 (68%) CIN1. Among the 28 SCC/HSIL cases, 18 (64%) had
CIN2/3 and 10 (36%) had CIN1, and among the 115 LSIL cases 28 (24%)
had CIN1.
 | Table IDemographic and clinical
characteristics of study groups. |
Table I
Demographic and clinical
characteristics of study groups.
| Variables | Cases (n=143) | Control
(n=144) | P-value |
|---|
| Age, years | 38.2±10.4 | 36.9±10.3 | 0.16 |
| HPV status | | | 0.47a |
|
Positive | 49(34) | 44(31) | |
|
Negative | 94(66) | 100(69) | |
| Cytology
diagnosis | 143 | n/a | |
|
SCC/HSIL | 28(20) | n/a | |
|
LSIL | 115(80) | n/a | |
| Histology
diagnosis | 56 | n/a | |
|
CIN2/3 | 18(32) | n/a | |
|
CIN1 | 38(68) | n/a | |
mtDNA copy number in squamous
intraepithelial lesions and controls
In the overall analysis, the mtDNA copy number was
compared between cases with cervical abnormalities and controls.
Cases were categorized based on the cytological diagnosis into
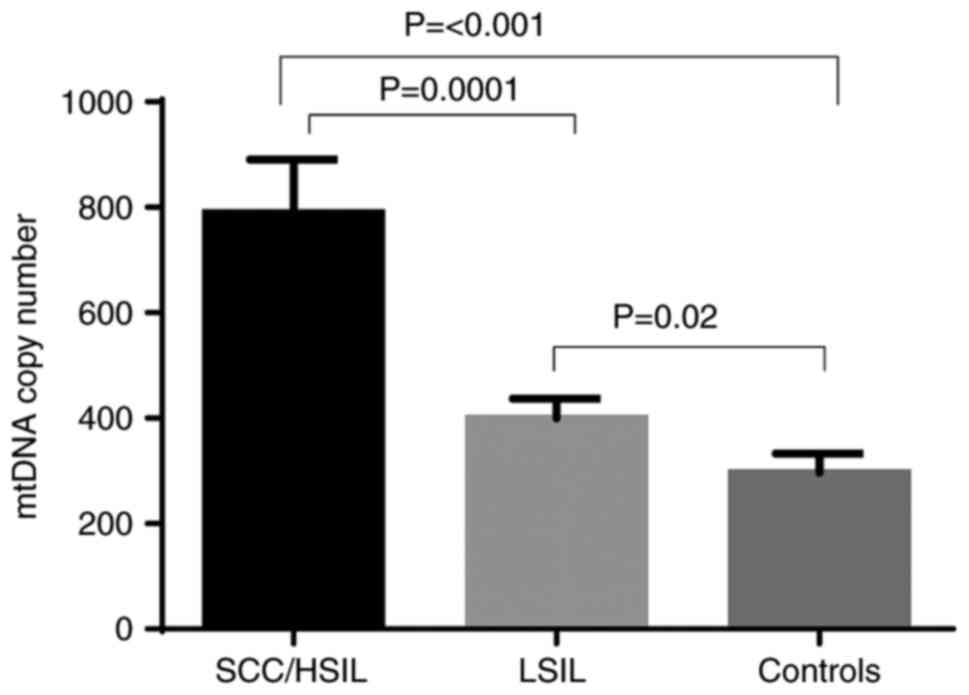
SCC/HSIL and LSIL. As presented in Fig. 2, the mtDNA copy number was
significantly higher in SCC/HSIL cases (788.8±102) and LSIL cases
(399.4±37.5) compared with the controls (296.6±36.4; P<0.001 and
P=0.02, respectively). Furthermore, cases with SCC/HSIL had a
significantly higher mtDNA copy number compared with the cases with
LSIL (P=0.0001).
mtDNA copy number in squamous
intraepithelial lesions and controls based on HPV status
In subsequent analyses, SCC/HSIL and LSIL cases and
controls were subdivided based on HPV status into hr-HPV-positive
and HPV-negative. The mtDNA copy number was evaluated in these
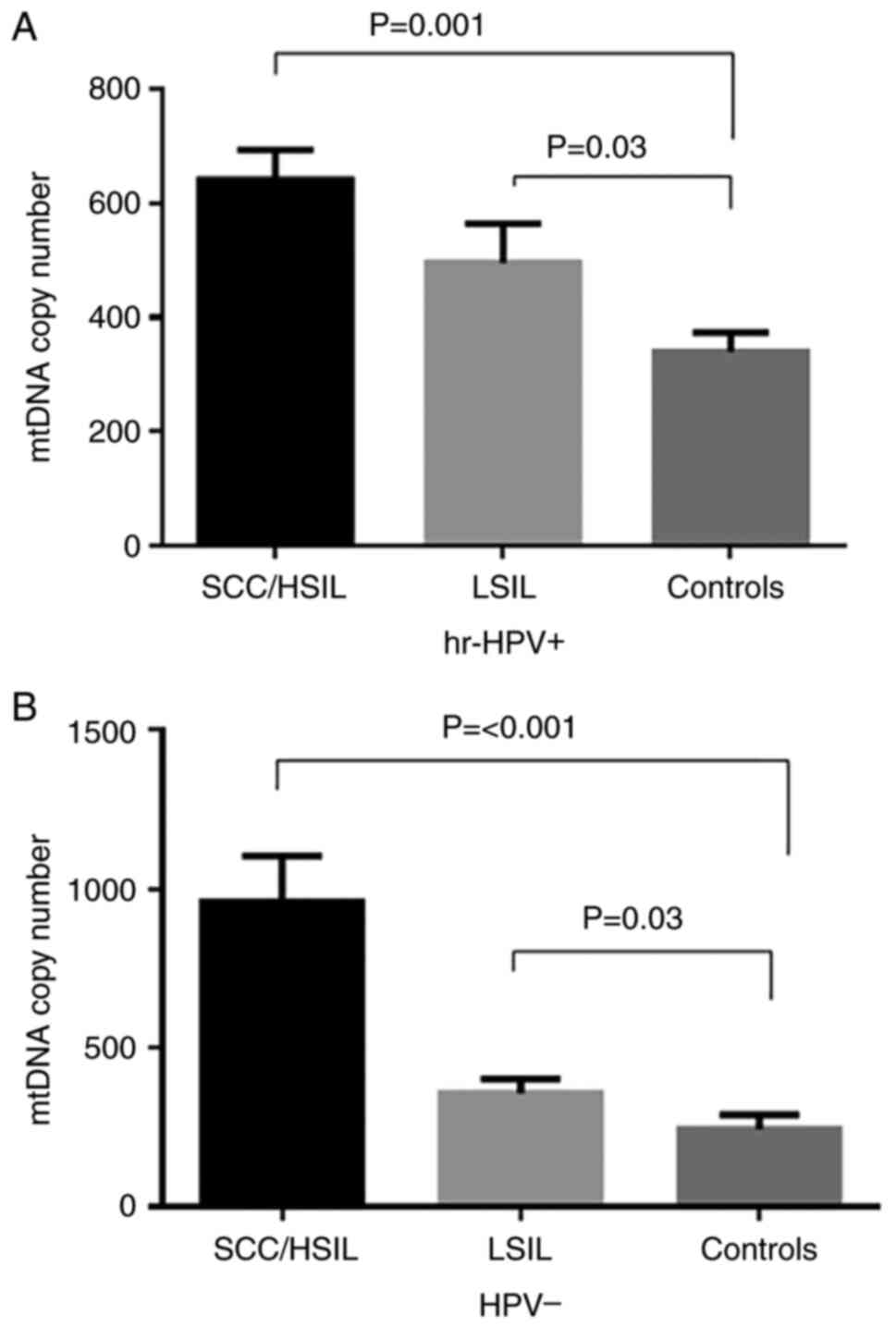
groups. In the hr-HPV-positive samples (Fig. 3A), the mtDNA copy number was
significantly higher in SCC/HSIL cases (642.1±53.0) and LSIL cases
(496.4±69.0) compared with the controls (339.0±34.0; P<0.001 and
P=0.03, respectively). Similar results were observed in the
HPV-negative samples. A significant increase in mtDNA copy number
was observed in SCC/HSIL cases (958.0±145.5) and LSIL cases
(358.7±44.0) compared with the controls (244.7±46.0; P<0.001;
Fig. 3B).
mtDNA copy number in squamous
intraepithelial lesions of different histological stages
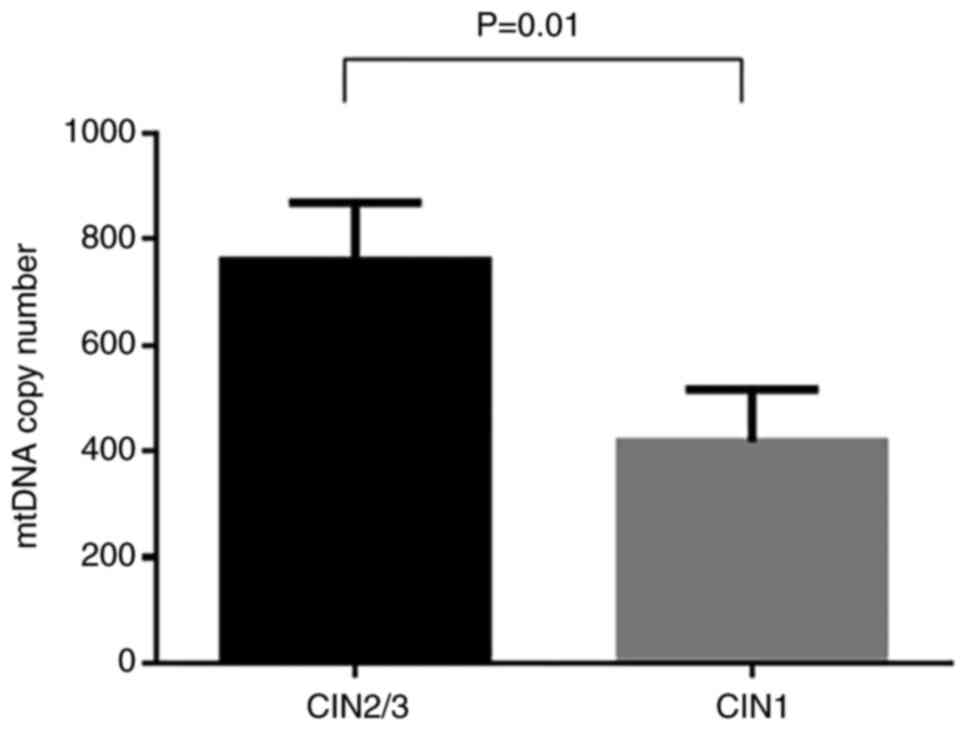
The mtDNA copy number in cases with squamous
intraepithelial lesions based on their histological diagnosis
(CIN1, CIN2 and CIN3) was determined. In the overall analysis
(Fig. 4), the mtDNA copy number
was significantly higher in CIN2/CIN3 cases (759.6±110.0) compared
with the CIN1 cases (417.8±99.0; P=0.01).
mtDNA copy number in squamous
intraepithelial lesions of different histological stages based on
HPV status
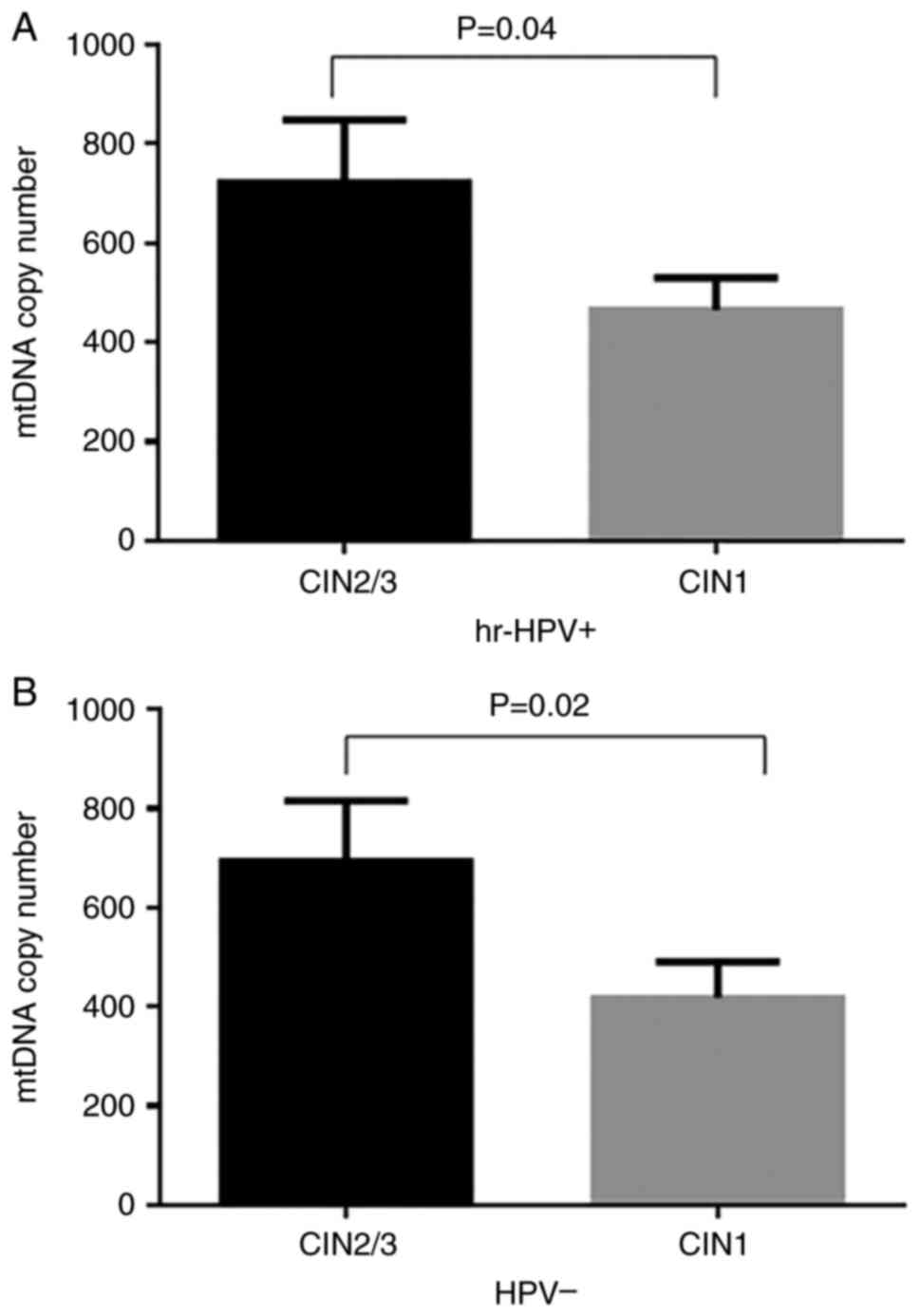
Subsequent stratification analyses were performed by
dividing CIN2/CIN3 and CIN1 cases based on HPV status into
hr-HPV-positive and HPV-negative. Subsequently, the mtDNA copy
number was compared between the groups. In hr-HPV-positive samples
(Fig. 5A), there was a significant
increase in the mtDNA copy number in cases with CIN2/CIN3
(722.0±127.0) compared to CIN1 (465.0±65.0; P=0.04). In
HPV-negative samples (Fig. 5B),
there was also a significant increase in the mtDNA copy number in
cases with CIN2/CIN3 (693.0±123.0) compared to cases with CIN1
(418.0±73.0; P=0.02).
Discussion
Mitochondria, the critical organelles involved in
various cellular functions, have their own multicopy genome
(mtDNA). The regulation of the mtDNA copy number is significant for
maintaining the cellular energy needs; thus, the mtDNA copy
number is considered as an indicator of mitochondrial activity and
function (39). Alterations in the
mtDNA copy number have been reported in several human diseases such
as Parkinson's disease, rheumatoid arthritis, multiple sclerosis
and cancer (29-31,40).
In particular, the role of mitochondria in tumor promotion and
development has been widely investigated, and an altered mtDNA copy
number has been shown to impact numerous cellular pathways
associated with cancer. Alteration of mtDNA copy number may cause
elevation of mtDNA oxidative stress and disruption of mtDNA gene
expression. As a result, the overall mtDNA functions may be
affected, including the OXPHOS system, ROS production, signal
transduction, cell apoptosis and cell growth. Therefore,
disturbances in the OXPHOS system may result in a reduction of
intracellular ATP, which triggers glycolysis to compensate for the
total ATP. Thus, the aberrant mtDNA copy number may reduce the rate
of mitochondrial biogenesis and normal cellular function that
eventually trigger tumorigenesis (41).
Previous studies have reported an increase or
decrease in mtDNA copy number in different types of cancer
(32,33,41),
suggesting that the effects of alterations in mtDNA copy number may
be cancer-specific, dependent on the energy needs of the specific
cancer tissue. Biologically, a decrease in mtDNA copy number may
result in a reduction in OXPHOS capacity, which triggers a
compensatory increase in glycolysis, resulting in the disruption of
cellular functions. A decrease in mtDNA copy number also promotes
resistance to apoptosis of cancer cells and increases their
sensitivity to chemotherapeutic drugs (42). Conversely, a high mtDNA copy number
has been proposed as a marker for oxidative stress and impairment
of aerobic pathways involved in the molecular mechanisms of
carcinogenesis (27). In previous
studies, an increase in mtDNA copy number was shown to be
associated with the risk (31) and
development (23) of cervical
cancer. However, these studies focused only on cervical cancer
cases who tested positive for HPV, the most common causative agent
of cervical carcinogenesis (7,8). To
date, no studies have explored the role of mtDNA copy number in
HPV-negative cervical cancer, which occurs in ~5% of cases
(13-16),
to the best of our knowledge. No HPV-negative cervical cancer cases
were included in the present study and all four squamous cervical
carcinoma cases were HPV-infected, this could be explained by the
small sample size of abnormal cases in this study, which could be
avoided in future studies. The inclusion of only HPV-positive
carcinoma cases is in line with previous mtDNA copy number studies
(22,23,34).
In the present study, the changes in mtDNA copy number in
HPV-positive and HPV-negative cases with cervical abnormalities and
controls were assessed.
In the overall analysis, the mtDNA copy number in
cases with cervical abnormalities categorized based on cytological
diagnosis into SCC/HSIL or LSIL, was compared with that in the
controls. It was found that the mtDNA copy number was significantly
higher in cases with SCC/HSIL and LSIL compared to the controls.
Furthermore, cases with SCC/HSIL displayed a significantly
increased mtDNA copy number compared with cases with LSIL.
Stratification of SCC/HSIL and LSIL cases based on HPV status into
hr-HPV+ and HPV-indicated that the mtDNA copy number was
significantly higher in cases with SCC/HSIL compared to LSIL cases
with and without HPV infection. These results suggest that the
increase in mtDNA copy number was not influenced by HPV status in
patients with different types of cervical abnormalities. In
addition, the mtDNA copy number was compared among abnormal
cervical cases with different stages. When these cases were
stratified based on histological diagnosis into CIN2/CIN3 and CIN1,
the mtDNA copy number was significantly higher in cases with
CIN2/CIN3 compared to CIN1. Stratification of CIN2/CIN3 and CIN1
cases based on HPV status showed that the mtDNA copy number was
significantly elevated in CIN2/CIN3 cases compared to CIN1 cases
with and without HPV infection. These results suggested that a
higher mtDNA copy number may be associated with the progression of
cervical cancer, regardless of the HPV status.
These results of a higher mtDNA copy number in cases
with cervical abnormalities are in agreement with those in previous
studies, which reported an elevated mtDNA copy number in women with
cervical cancer who tested positive for hr-HPV (34) and in SCC/HSIL compared to LSIL
(23), whereas a low mtDNA copy
number was reported in 20 cervical cancer tissues compared to 10
cervicitis samples (22). The
small sample size in the study by Kabekkodu et al (22) may explain the discrepancy between
their findings and the findings of other studies, including the
present study.
The results of the present study extend on the
previous above-mentioned findings of an elevated mtDNA copy number
in HPV-positive cervical cancer cases (23,34),
indicating that the mtDNA copy number in cervical abnormalities
without HPV infection was also higher.
HPV consists of a circular, double-stranded DNA of
~8 Kb pairs in size and encodes six early genes (E1, E2, E4, E5, E6
and E7) responsible for DNA maintenance, replication and
transcription, as well as two late genes (L1 and L2) that
constitute the viral capsid (4).
Following HPV infection, the early genes E1, E2, E4, E5, E6 and E7
are expressed and the viral DNA replicates from free DNA in the
basal cells at the cervix and integrates into the host genome. As
the infection progresses, upregulation of the E6 and E7 oncogenes
occurs (43), and these two viral
oncoproteins are necessary for malignant conversion (44). Studies have indicated that
oncoproteins E6 and E7 of hr-HPV may induce a chronic oxidative
stress response that increases the susceptibility to DNA damage
(45,46). A previous report by Warowicka et
al (23) showed higher mtDNA
copy number and increased ROS generation during cervical cancer
development.
Therefore, HPV infection may contribute to an
increase in mtDNA copy number in cervical cancer. Conversely, ROS
and free radicals contribute to changes in the mtDNA copy number
and mtDNA integrity in human cells, and the increased mtDNA copy
number is considered a biomarker of oxidative stress (27). Therefore, the observed increase in
mtDNA copy number in the absence of HPV infection may suggest that
oxidative stress associated with environmental exposure to
pollutants, tobacco, smoke or radiation also has an influence on
the elevated mtDNA copy number. In particular, the increased mtDNA
copy number has been suggested as an adaptive response mechanism to
compensate for oxidatively damaged mtDNA and energy deficiency. The
mtDNA is preferentially clonally amplified by making more
mitochondria and mtDNA to meet the energy demand of cells (27,28).
These observations suggest that an increase in mtDNA copy number in
cases with cervical abnormalities with and without HPV infection
may be a consequence of impaired mitochondrial function.
A long-term follow-up study revealed that ~40% of
CIN2 lesions progress to cervical cancer (47). In addition, women diagnosed with
HSIL and CIN2 may have either an early progression that requires
surgical intervention or a productive hr-HPV infection that may
regress without treatment (48).
Therefore, early detection is highly beneficial to prevent the
progression of CIN2 lesions to cervical cancer. The current
protocol followed by the Ministry of Health in Kuwait is a referral
of women primarily diagnosed with cytologically abnormal lesions
and positive hr-HPV infection for colposcopy. Applying this
protocol to the present study, 49 women (34%) with hr-HPV positive
results and abnormal cytology will be referred to colposcopy,
including cases with LSIL. Nevertheless, the positive predictive
value of HPV testing is <50% for high-grade lesions (49). Furthermore, there is no suitable
screening test for HPV-negative cervical cancer and the
identification of these cases is essential for the proper
management of patients. The mtDNA copy number has been proposed as
a potential biomarker in several diseases, including different
types of cancers, such as breast cancer, colorectal cancer,
cervical cancer and head and neck cancer (33,50,51).
Despite the small sample size in the present study,
the results of increased mtDNA copy number in cases with cervical
abnormalities may suggest a potential utility of the mtDNA copy
number as a biomarker, particularly for HPV-negative cases. Further
validation of these results in a larger sample size may highlight a
novel avenue for the clinical utility of mtDNA copy number as a
biomarker for the early detection of cervical lesions that are at a
high risk of progression to cervical cancer.
In conclusion, the current study investigated, for
the first time, changes in mtDNA copy number in cases of cervical
abnormalities and controls with both hr-HPV-positive and
HPV-negative status. The results of the present study revealed that
the mtDNA copy number was higher in SCC/HSIL and LSIL cases
compared to controls, as well as in SCC/HSIL cases compared to LSIL
cases, and the increase in mtDNA copy number was not influenced by
the HPV status in different types of cervical abnormalities. These
results also showed an increase in mtDNA copy number in cases with
CIN2/CIN3 compared to CIN1 for both hr-HPV positive and
HPV-negative, suggesting that a higher mtDNA copy number may be
involved in the progression of cervical cancer regardless of HPV
status. The mtDNA copy number thus deserves further investigation
for its role in cervical cancer and as a disease biomarker in a
larger cohort.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets generated and/or analyzed during the
current study are not publicly available due to ethical restriction
rules but are available from the corresponding author on reasonable
request.
Authors' contributions
RAA, MSA and SAW conceived the study. MSA and MA
developed the methodology. MSA and RAA performed data analysis.
MSA, SAW and RAA confirmed the authenticity of all the raw data.
RAA and SAW provided resources. RAA, MSA and MR curated data. MSA
and RAA wrote the manuscript. RAA, SAW and MSA revised the
manuscript. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
The study was conducted according to the guidelines
of the Declaration of Helsinki and approved by the Health Science
Center Ethics Committee at Kuwait University (Jabriya, Kuwait) and
the Ministry of Health the Standing Committee for Coordination of
Health and Medical Research (Safat, Kuwait) and registered under
no. VDR/EC/3746. Written informed consent was obtained from all
subjects involved in the study and all patients gave permission for
the use of the remaining cytology material for research
purposes.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Mattiuzzi C and Lippi G: Cancer
statistics: A comparison between World Health Organization (WHO)
and Global Burden of Disease (GBD). Eur J Public Health.
30:1026–1027. 2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Alkhalawi E, Al-Madouj A and Al-Zahrani A:
Cervical cancer incidence and trends among nationals of the gulf
cooperation council states, 1998-2012. Gulf J Oncolog. 1:7–13.
2019.PubMed/NCBI
|
|
4
|
zur Hausen H: Papillomaviruses and cancer:
From basic studies to clinical application. Nat Rev Cancer.
2:342–350. 2002.PubMed/NCBI View
Article : Google Scholar
|
|
5
|
Zhang S, Xu H, Zhang L and Qiao Y:
Cervical cancer: Epidemiology, risk factors and screening. Chin J
Cancer Res. 32:720–728. 2020.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Roura E, Castellsague X, Pawlita M,
Travier N, Waterboer T, Margall N, Bosch FX, de Sanjosé S, Dillner
J, Gram IT, et al: Smoking as a major risk factor for cervical
cancer and pre-cancer: Results from the EPIC cohort. Int J Cancer.
135:453–466. 2014.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Gissmann L and zur Hausen H: Partial
characterization of viral DNA from human genital warts
(Condylomata acuminata). Int J Cancer. 25:605–609.
1980.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Schiffman M, Castle PE, Jeronimo J,
Rodriguez AC and Wacholder S: Human papillomavirus and cervical
cancer. Lancet. 370:890–907. 2007.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Bloem P and Ogbuanu I: Vaccination to
prevent human papillomavirus infections: From promise to practice.
PLoS Med. 14(e1002325)2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
US Preventive Services Task Force. Curry
SJ, Krist AH, Owens DK, Barry MJ, Caughey AB, Davidson KW, Doubeni
CA, Epling JW Jr, Kemper AR, et al: Screening for CERVICAL CANcer:
US preventive services task force recommendation statement. JAMA.
320:674–686. 2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Delpero E and Selk A: Shifting from
cytology to HPV testing for cervical cancer screening in Canada.
CMAJ. 194:E613–E615. 2022.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Aitken CA, van Agt HME, Siebers AG, van
Kemenade FJ, Niesters HGM, Melchers WJG, Vedder JEM, Schuurman R,
van den Brule AJC, van der Linden HC, et al: Introduction of
primary screening using high-risk HPV DNA detection in the Dutch
cervical cancer screening programme: A population-based cohort
study. BMC Med. 17(228)2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Blatt AJ, Kennedy R, Luff RD, Austin RM
and Rabin DS: Comparison of cervical cancer screening results among
256,648 women in multiple clinical practices. Cancer Cytopathol.
123:282–288. 2015.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Petry KU, Liebrich C, Luyten A, Zander M
and Iftner T: Surgical staging identified false HPV-negative cases
in a large series of invasive cervical cancers. Papillomavirus Res.
4:85–89. 2017.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Clifford GM, Smith JS, Plummer M, Munoz N
and Franceschi S: Human papillomavirus types in invasive cervical
cancer worldwide: A meta-analysis. Br J Cancer. 88:63–73.
2003.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Guan P, Howell-Jones R, Li N, Bruni L, de
Sanjosé S, Franceschi S and Clifford GM: Human papillomavirus types
in 115,789 HPV-positive women: A meta-analysis from cervical
infection to cancer. Int J Cancer. 131:2349–2359. 2012.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Hohn AK, Brambs CE, Hiller GGR, May D,
Schmoeckel E and Horn LC: 2020 WHO classification of female genital
tumors. Geburtshilfe Frauenheilkd. 81:1145–1153. 2021.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Arezzo F, Cormio G, Loizzi V, Cazzato G,
Cataldo V, Lombardi C, Ingravallo G, Resta L and Cicinelli E:
HPV-Negative cervical cancer: A narrative review. Diagnostics
(Basel). 11(952)2021.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Fernandes A, Viveros-Carreno D, Hoegl J,
Avila M and Pareja R: Human papillomavirus-independent cervical
cancer. Int J Gynecol Cancer. 32:1–7. 2022.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Yusoff AAM, Abdullah WSW, Khair SZNM and
Radzak SMA: A comprehensive overview of mitochondrial DNA 4977-bp
deletion in cancer studies. Oncol Rev. 13(409)2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Ksiezakowska-Lakoma K, Zyla M and
Wilczynski JR: Mitochondrial dysfunction in cancer. Prz
Menopauzalny. 13:136–144. 2014.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Kabekkodu SP, Bhat S, Mascarenhas R,
Mallya S, Bhat M, Pandey D, Kushtagi P, Thangaraj K, Gopinath PM
and Satyamoorthy K: Mitochondrial DNA variation analysis in
cervical cancer. Mitochondrion. 16:73–82. 2014.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Warowicka A, Kwasniewska A and
Gozdzicka-Jozefiak A: Alterations in mtDNA: A qualitative and
quantitative study associated with cervical cancer development.
Gynecol Oncol. 129:193–198. 2013.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Zhai K, Chang L, Zhang Q, Liu B and Wu Y:
Mitochondrial C150T polymorphism increases the risk of cervical
cancer and HPV infection. Mitochondrion. 11:559–563.
2011.PubMed/NCBI View Article : Google Scholar
|
|
25
|
DiMauro S and Wallace DC: Mitochondrial
DNA in Human Pathology. Raven Press, New York, NY, 1993.
|
|
26
|
Clay Montier LL, Deng JJ and Bai Y: Number
matters: Control of mammalian mitochondrial DNA copy number. J
Genet Genomics. 36:125–131. 2009.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Lee HC and Wei YH: Mitochondrial
biogenesis and mitochondrial DNA maintenance of mammalian cells
under oxidative stress. Int J Biochem Cell Biol. 37:822–834.
2005.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Lee HC, Yin PH, Lu CY, Chi CW and Wei YH:
Increase of mitochondria and mitochondrial DNA in response to
oxidative stress in human cells. Biochem J. 348:425–432.
2000.PubMed/NCBI
|
|
29
|
Lee JW, Park KD, Im JA, Kim MY and Lee DC:
Mitochondrial DNA copy number in peripheral blood is associated
with cognitive function in apparently healthy elderly women. Clin
Chim Acta. 411:592–596. 2010.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Pyle A, Anugrha H, Kurzawa-Akanbi M,
Yarnall A, Burn D and Hudson G: Reduced mitochondrial DNA copy
number is a biomarker of Parkinson's disease. Neurobiol Aging.
38:216 e7–216 e10. 2016.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Svendsen AJ, Tan Q, Jakobsen MA,
Thyagarajan B, Nygaard M, Christiansen L and Mengel-From J: White
blood cell mitochondrial DNA copy number is decreased in rheumatoid
arthritis and linked with risk factors. A twin study. J Autoimmun.
96:142–146. 2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Reznik E, Miller ML, Senbabaoglu Y, Riaz
N, Sarungbam J, Tickoo SK, Al-Ahmadie HA, Lee W, Seshan VE, Hakimi
AA and Sander C: Mitochondrial DNA copy number variation across
human cancers. Elife. 5(e10769)2016.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Hu L, Yao X and Shen Y: Altered
mitochondrial DNA copy number contributes to human cancer risk:
Evidence from an updated meta-analysis. Sci Rep.
6(35859)2016.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Sun W, Qin X, Zhou J, Xu M, Lyu Z, Li X,
Zhang K, Dai M, Li N and Hang D: Mitochondrial DNA copy number in
cervical exfoliated cells and risk of cervical cancer among
HPV-positive women. BMC Womens Health. 20(139)2020.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Nayar R and Wilbur DC: The Pap test and
Bethesda 2014. Cancer Cytopathol. 123:271–281. 2015.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Al-Awadhi R, Chehadeh W and Kapila K:
Prevalence of human papillomavirus among women with normal cervical
cytology in Kuwait. J Med Virol. 83:453–460. 2011.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Alwehaidah MS, AlFadhli S and Al-Kafaji G:
Leukocyte mitochondrial DNA copy number is a potential non-invasive
biomarker for psoriasis. PLoS One. 17(e0270714)2022.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Filograna R, Mennuni M, Alsina D and
Larsson NG: Mitochondrial DNA copy number in human disease: The
more the better? FEBS Lett. 595:976–1002. 2021.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Al-Kafaji G, Bakheit HF, Alharbi MA,
Farahat AA, Jailani M, Ebrahin BH and Bakhiet M: Mitochondrial DNA
copy number in peripheral blood as a potential non-invasive
biomarker for multiple sclerosis. Neuromolecular Med. 22:304–313.
2020.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Abd Radzak SM, Mohd Khair SZN, Ahmad F,
Patar A, Idris Z and Mohamed Yusoff AA: Insights regarding
mitochondrial DNA copy number alterations in human cancer (Review).
Int J Mol Med. 50(104)2022.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Mei H, Sun S, Bai Y, Chen Y, Chai R and Li
H: Reduced mtDNA copy number increases the sensitivity of tumor
cells to chemotherapeutic drugs. Cell Death Dis.
6(e1710)2015.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Woodman CB, Collins SI and Young LS: The
natural history of cervical HPV infection: Unresolved issues. Nat
Rev Cancer. 7:11–22. 2007.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Bedell SL, Goldstein LS, Goldstein AR and
Goldstein AT: Cervical cancer screening: Past, present, and future.
Sex Med Rev. 8:28–37. 2020.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Williams VM, Filippova M, Filippov V,
Payne KJ and Duerksen-Hughes P: Human papillomavirus type 16 E6*
induces oxidative stress and DNA damage. J Virol. 88:6751–6761.
2014.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Marullo R, Werner E, Zhang H, Chen GZ,
Shin DM and Doetsch PW: HPV16 E6 and E7 proteins induce a chronic
oxidative stress response via NOX2 that causes genomic instability
and increased susceptibility to DNA damage in head and neck cancer
cells. Carcinogenesis. 36:1397–1406. 2015.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Buckley CH, Butler EB and Fox H: Cervical
intraepithelial neoplasia. J Clin Pathol. 35:1–13. 1982.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Leeman A, Del Pino M, Marimon L, Torné A,
Ordi J, Ter Harmsel B, Meijer CJLM, Jenkins D, Van Kemenade FJ and
Quint WGV: Reliable identification of women with CIN3+ using hrHPV
genotyping and methylation markers in a cytology-screened referral
population. Int J Cancer. 144:160–168. 2019.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Coquillard G, Palao B and Patterson BK:
Quantification of intracellular HPV E6/E7 mRNA expression increases
the specificity and positive predictive value of cervical cancer
screening compared to HPV DNA. Gynecol Oncol. 120:89–93.
2011.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Rai NK, Panjwani G, Ghosh AK, Haque R and
Sharma LK: Analysis of mitochondrial DNA copy number variation in
blood and tissue samples of metastatic breast cancer patients (A
pilot study). Biochem Biophys Rep. 26(100931)2021.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Chen N, Wen S, Sun X, Fang Q, Huang L, Liu
S, Li W and Qiu M: Elevated Mitochondrial DNA copy number in
peripheral blood and tissue predict the opposite outcome of cancer:
A meta-analysis. Sci Rep. 6(37404)2016.PubMed/NCBI View Article : Google Scholar
|