Introduction
Cardiovascular diseases (CVDs) are life-threatening
conditions with high mortality and complication rates despite the
advances made in diagnostic and therapeutic approaches (1). The standard treatments for acute
myocardial infarction (AMI) are reperfusion or reoxygenation, which
limits the size of the infarct through percutaneous coronary
intervention (PCI) or thrombolytic therapy (2). However, myocardial reperfusion or
reoxygenation can cause further damage to the ischemic cardiac
tissues, resulting in ischemia-reperfusion injury (IRI) (3,4).
Several key pathological factors involved in IRI have been
identified, including the excessive production of ROS, regulated
cell death, Ca2+ overload, mitochondrial dysfunction,
and inflammatory responses. However, the exact biological process
and pathways related to IRI have not yet been elucidated (5). Thus, there is an urgent need to
elucidate the pathological mechanisms underlying IRI to develop
more effective pharmaceutical and other therapeutic strategies.
Ferroptosis is a novel type of programmed cell death
initiated by iron-dependent lipid peroxidation, which leads to the
condensation of mitochondrial membranes, and the reduction of
mitochondrial cristae (6). It is a
complex event mediated by multiple pathways and has been
increasingly implicated in CVD (7). A previous study showed that the
administration of deferoxamine during the reperfusion of isolated
rabbit hearts significantly decreased the free radical content and
protected heart tissue from IRI (8). Deferoxamine also reduced cardiac
function injury after coronary artery bypass grafting (9). Dysregulated iron homeostasis is a
leading cause of IRI, which increases the generation of hydroxyl
radicals (•OH) via the Fenton reaction, thereby triggering
ferroptosis, pyroptosis, necroptosis, and other forms of programmed
cell death (10). Although these
studies established the pathological role of ferroptosis in IRI,
the underlying mechanism and targets remain unclear (11).
The aim of the present study was to elucidate the
role of ferroptosis in the pathophysiology of IRI through
bioinformatics analysis and in vitro experiments. The
association between ferroptosis and IRI was confirmed and the
differentially expressed genes related to ferroptosis were
identified.
Materials and methods
Data collection
Gene expression datasets associated with myocardial
reperfusion injury were downloaded from GEO (https://www.ncbi.nlm.nih.gov/geo/) (12) using ‘ischemia-reperfusion’,
‘post-ischemia’, and ‘after ischemia’ as the key words. The
inclusion criteria for the microarray datasets were: i) Consisted
of cardiac tissue specimens, ii) included control and
ischemia/reperfusion samples, and iii) had an adequate sample size
(both IRI and normal samples >3). Accordingly, four microarray
datasets were obtained from GEO. The first two datasets were
GSE58486 (3 control and 6 IRI heart samples) and GSE160516 (4
control and 12 IRI heart samples), which were screened for
differentially expressed genes (DEGs) between the normal and IRI
heart samples), as well as for hub genes. GSE4105 (6 control and 6
IRI heart samples) was used for hub gene validation. GSE124176 (3
control and 3 IRI heart samples) was used for screening
differentially expressed miRNAs (DEmiRs) between the normal and IRI
heart samples. Ferroptosis-related genes (FRGs) were downloaded
from FerrDb (https://www.zhounan.org/ferrdb) (13).
Identification of DEGs, differentially
expressed ferroptosis-related genes (DEFRGs), and DEmiRs
The genes were first converted to the official gene
IDs based on the annotations from the corresponding platform. The
gene expression values were then transformed to a log2 format and
quantile normalization was performed. Identifying and extracting
DEGs using the limma package (version 3.48.0) (14) in R software (version 3.6.3)
(15), with |log2FC|≥1 and
P-values <0.05 as the thresholds for selection (where FC is fold
change). After removing repetitive genes, 388 genes were included
in the FRG set (Table SI). In
addition, 51 DEFRGs were obtained using the VennDiagram package
(version 1.7.3) (16) in R, and 8
DEmiRs were identified in GSE124176.
Gene set enrichment analysis
(GSEA)
GSEA was conducted on the 388 FRGs to evaluate the
functional correlation between ferroptosis and myocardial
reperfusion injury. Briefly, the FRGs in GSE58486 and GSE160516
were scored and ranked based on expression values to evaluate the
enrichment score (ES) and core genes. FRGs with a false discovery
rate (FDR) <0.25 and a P-value <0.05 were considered
significantly enriched.
Functional and pathway enrichment
analyses of DEFRGs
DEFRGs between the normal and IRI heart samples were
submitted to Data for Annotation, Visualization, and Integrated
Discovery (DAVID; david.ncifcrf.gov/) (17,18)
for Gene Ontology (GO) (19,20)
and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment
analyses (21). P<0.05 was
considered to indicate a statistically significant difference.
Construction of a protein-protein
interaction (PPI) network and hub gene identification
A PPI network of the DEFRGs was constructed using
the STRING database (https://string-db.org/) (22) and visualized in Cytoscape software
(version 3.8.2) (23). The hub
genes in DEFRGs were identified by Cytohubba, a Cytoscape plugin
based on the MCC algorithm (24).
Each DEFRG in the PPI network was allocated a value, and the genes
were ranked accordingly. The top 10 genes were screened as hub
genes.
Verification of hub genes in
GSE4105
The expression levels of the hub genes were
validated in the GSE4105 dataset. Comparisons between the control
and IRI samples were made using a Student's t-test and P<0.05
was considered to indicate a statistically significant
difference.
Prediction of target genes and
transcription factors of DEmiRs
The target mRNAs of the DEmiRs extracted from
GSE124176 were predicted using TargetScan (targetscan.org/), miRwalk (http://mirwalk.umm.uni-heidelberg.de/), and miRDB
(http://www.mirdb.org/) (25-27).
After removing the duplicate genes, the overlapping differentially
expressed target genes and DEFRGs were extracted using the
VennDiagram package in R. Potential transcription factors (TFs) for
key DEmiRs were screened using the TransmiR v2.0 database
(http://www.cuilab.cn/transmir) (28). The overlapping potential TFs and
DEFRGs were considered the key TFs involved in the miRNA-mRNA-TF
network.
Construction of the DEmiR-target
gene-TF regulatory network
A TF-miRNA-mRNA regulatory network was established
based on the key TFs, key DEmiRs, and target genes and visualized
using Cytoscape to identify the regulatory pathways.
H9c2 cell culture and treatment
Rat H9c2 cardiomyocytes, purchased from the Cell
Bank/Stem Cell Bank (Chinese Academy of Sciences, China) were
cultured in high-glucose DMEM (H-DMEM) (HyClone, Cytiva) containing
10% FBS (Gibco, Thermo Fisher Scientific, Inc.), 100 U/ml
penicillin and 100 µg/ml streptomycin (Beijing Solarbio Science
& Technology Co., Ltd.) at 37˚C under in a humidified incubator
supplied with 5% CO2 air and 21% O2. For the
experiments, the cells were treated with ferrostatin-1 (Fer-1,
MedChemExpress) freshly prepared in DMSO.
Establishment of the in vitro IRI
model
The cellular IRI model was established as described
previously (29). Briefly, H9c2
cells were subjected to anoxia for 3 h in a specialized medium (1
mM CaCl2, 10 mM KCI, 20 mM HEPES, 98.5 mM NaCl, 1.2 mM
MgSO4, 6 mM NaHCO3, 0.9 mM
NaH2PO4, and 40 mM sodium lactate, pH 6.8) in
a humidified incubator supplied with 95% N2 and 5%
CO2, and then reoxygenated for 2 h in a reoxygenation
medium (1 mM CaCl2, 5.5 mM glucose, 5 mM KCI, 20 mM
HEPES, 129.5 mM NaCl, 1.2 mM MgSO4, 20 mM
NaHCO3, and 0.9 mM NaH2PO4, pH
7.4) in a humidified incubator supplied with 95% O2 and
5% CO2. The cells were treated with 5 mM Fer-1 for 2 h
prior to anoxia/reoxygenation (A/R) to inhibit ferroptosis.
Measurement of cell viability
H9c2 cells were seeded in 96-well plates at a
density of 1x104 cells per well in 100 µl H-DMEM and
cultured for 24 h. Following treatment, cell viability was
determined using a Cell Counting Kit-8 assay (GlpBio Technology;
cat. no. #GK10001) and absorbance at 450 nm was assessed using a
Multiskan FC microplate reader (Thermo Fisher Scientific,
Inc.).
Measurement of total iron
concentration and malondialdehyde (MDA) levels
Treated H9c2 cells were harvested, sonicated, and
centrifuged at 12,000 g for 10 or 5 min at 4˚C to remove cell
debris. The quantity of MDA and iron in the supernatants were
analyzed using specific assay kits (Beyotime Institute of
Biotechnology, cat. no. #S01031M; Pulilai Gene Technology, cat. no.
#E1042), and the absorbance was measured at 530 and 560 nm,
respectively, using a Multiskan FC microplate reader.
Measurement of intracellular ROS
generation
Intracellular ROS generation was detected using an
ROS assay kit (Beyotime Institute of Biotechnology, cat. no.
#S0033M). Briefly, H9c2 cells were cultured with 10 µM DCFH-DA
solution in H-DMEM for 20 min at 37˚C in the dark and washed once
with H-DMEM. DCFH-DA fluorescence was detected using a fluorescence
Olympus IX73 microscope (Olympus Corporation) at an excitation
wavelength of 488 nm and an emission wavelength of 525 nm.
Transmission electron microscopy
(TEM)
Following the different treatments, H9c2 cells were
rapidly gathered and incubated in 2% glutaraldehyde for 2 h. Then,
the cells were observed by transmission electron microscopy after
washing, dehydration, embedding in Epon 812, sectioning (60 nm),
and stained with 2% uranyl acetate and 2.6% lead citrate for 8 min
at 37˚C. Finally, the Flameng scoring method was used to assess the
ultrastructural injury of the mitochondria (30).
Measurement of intracellular lipid
ROS
Following the different treatments, H9c2 cells were
collected and incubated with 10 µM C11-BODIPY581/591 (GlpBIO) in
the dark for 1 h at 37˚C and washed twice with PBS. The cells were
centrifuged at 400 g for 5 min at 25˚C and resuspended in PBS
supplemented with 10% FBS. The levels of lipid ROS were measured
using a Cytomics FC500 flow cytometer (Beckman Coulter, Inc.).
Increases in lipid ROS shifted the fluorescence emission peak from
~590 to ~510 nm, which reflected increased lipid ROS levels. The
flow cytometry data were analyzed using NovoExpress (v.6.2; Agilent
Technologies, Inc.).
Western blot analysis
Western blotting was performed as described in our
previous study (21).
Cardiomyocytes were lysed in RIPA lysis buffer (Beyotime Institute
of Biotechnology) containing 1% PMSF, and the protein concentration
was measured using the BCA protein assay kit (GLPBIO). 30 µg/lane
from each sample were loaded on a 10% SDS-gel, resolved by
SDS-PAGE, and then transferred to PVDF membranes. After blocking
with 5% non-fat dry milk in TBS with 0.1% Tween 20 (TBST) at room
temperature for 2.5 h, the membranes were incubated overnight with
anti-PTGS2 (ProteinTech Group, Inc.; cat. no. #12375-1-AP;
1:1,000), anti-GPX4 (ZENBIO; cat. no. #381958; 1:1,000), and
anti-β-actin (OriGene Technologies, Inc.; cat. no. #TA-09; 1:1,000)
antibodies at 4˚C. The membranes were then washed three times and
incubated with the secondary antibody (Beyotime Institute of
Biotechnology; cat. no. #A0208; 1:3,000) for 1.5 h at room
temperature. The protein bands were detected using an
ultra-high-sensitivity ECL kit (Beyotime Institute of
Biotechnology) and imaged using FluorChem FC3 (ProteinSimple). The
signal intensities of the bands were quantified using ImageJ 1.8.0
(National Institutes of Health) and normalized to that of the
respective β-actin band.
Reverse transcription-quantitative
(RT-q)PCR
RT-qPCR was performed as described previously
(19). Briefly, total RNA,
including miRNA, was isolated from H9c2 cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) and reverse-transcribed to cDNA using RevertAid™ (Thermo
Fisher Scientific, Inc.) after evaluating the quality and
concentration of RNA. RT were as follows: 42˚C for 1 h, 72˚C for 10
min. qPCR was performed using Power SYBR Green PCR MasterMix
(Thermo Fisher Scientific, Inc.) on an Applied Biosystems
StepOnePlus Real-time PCR system (Thermo Fisher Scientific, Inc.).
The RT-PCR process involved initial denaturation at 95˚C for 10
min, followed by thermocycling 95˚C for 15 sec, and 60˚C for 1 min,
repeated 40 cycles. Relative mRNA expression levels were calculated
using the 2-ΔΔCq method (31). All primers were synthesized by
Sangon Biotech Co., Ltd.; the sequences of the primers used in the
present study are shown in Table
SII.
Immune infiltration analysis
The expression data of GSE58486 and GSE160516 were
uploaded to Immune Cell Abundance Identifier (http://bioinfo.life.hust.edu.cn/web/ImmuCellAI/)
(32) to evaluate the infiltration
ratio of different immune cells. The infiltration ratio of 36 types
of immune cells was calculated using the ImmuCellAI algorithm and
compared between control and IRI samples using Student's t-test.
P<0.05 was considered to indicate a statistically significant
difference.
Prediction of potential therapeutic
drugs
The DSigDB database (http://tanlab.ucdenver.edu/DSigDB) was used to predict
potential therapeutic drugs for IRI, with an FDR of <0.05 and a
combined score of >2,000,000 as the cutoff criteria (33).
Statistical analysis
Data are presented as the mean ± standard
derivation. Statistical analysis was performed using GraphPad Prism
8.0 (GraphPad Software, Inc.) or the Sangerbox platform. An
unpaired two-tailed Student's t-test was used to compare
differences between two groups and a one-way ANOVA followed by a
Dunnett's post hoc multiple comparisons test was used to compare
differences between ≥3 groups. P<0.05 was considered to indicate
a statistically significant difference.
Results
Study protocol
The overall workflow of this study is shown in
Fig. 1.
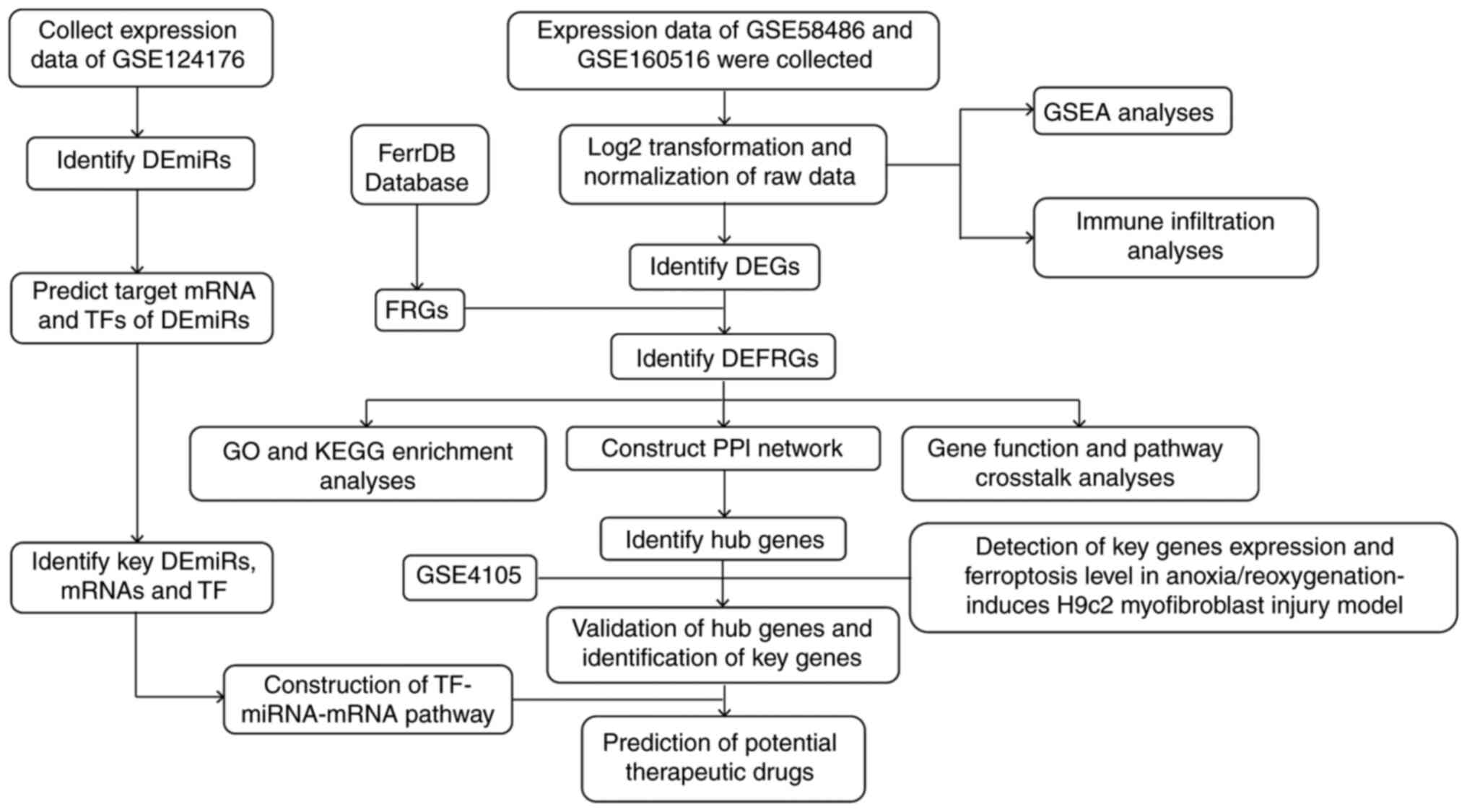 | Figure 1Overall workflow of the present
study. DEG, differentially expressed gene; miRNA, microRNA; DEmiR,
differentially expressed miRNA; TF, transcription factor; FRG,
ferroptosis related gene; DEFRG, differentially expressed FRG; GO,
Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; PPI,
protein-protein interaction; GSEA, gene set enrichment
analysis. |
Relationship between ferroptosis and
myocardial reperfusion
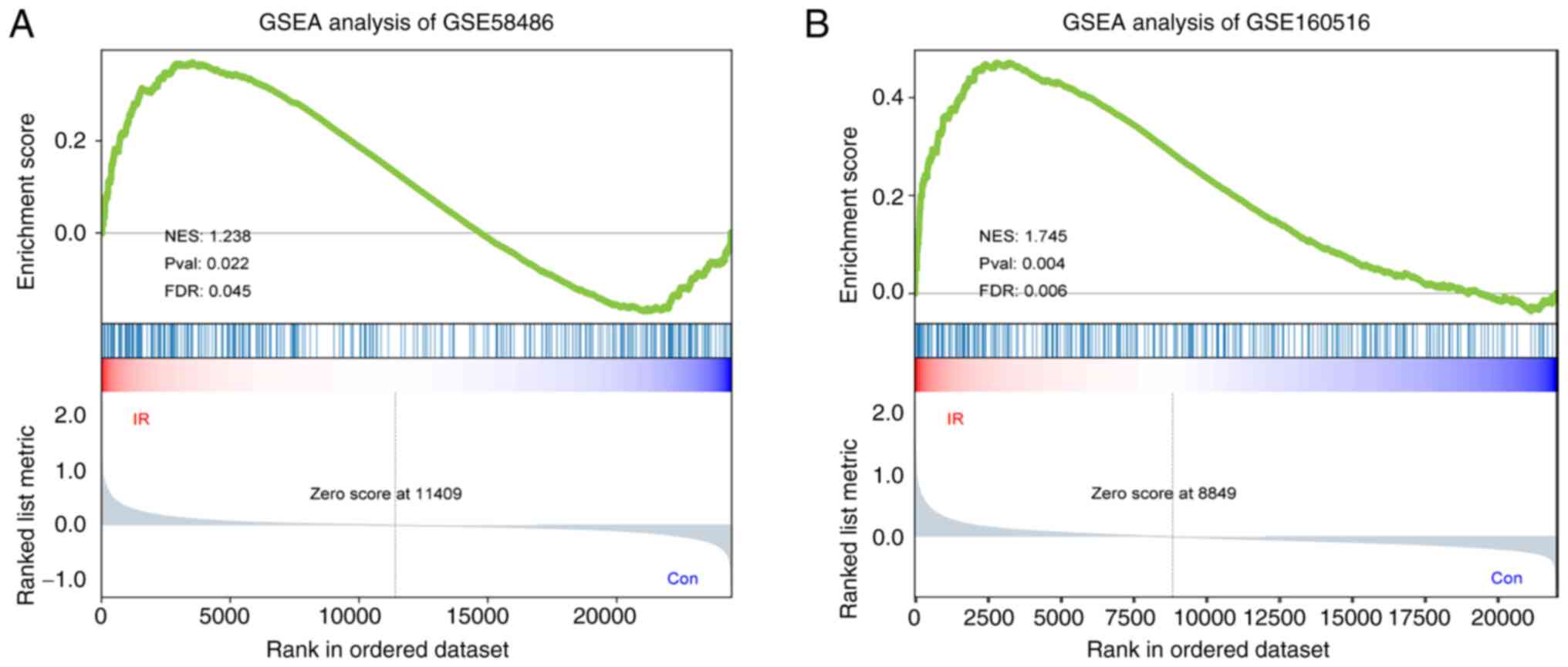
GSEA of the FRG dataset was used to determine
whether ferroptosis was a key pathophysiological factor in IRI. The
genes associated with ferroptosis were significantly enriched in
the GSE58486 and GSE160516 IRI samples with a normalized enrichment
score (NES) of >1 (nominal P-value <0.05 and FDR <0.25;
Fig. 2A and B), suggesting a relationship between IRI
and ferroptosis. A total of 87 and 84 genes were extracted from
GSE58486 and GSE160516, respectively, for GSEA and are listed in
Table SIII.
Screening of DEGs, DEmiRs, and
DEFRGs
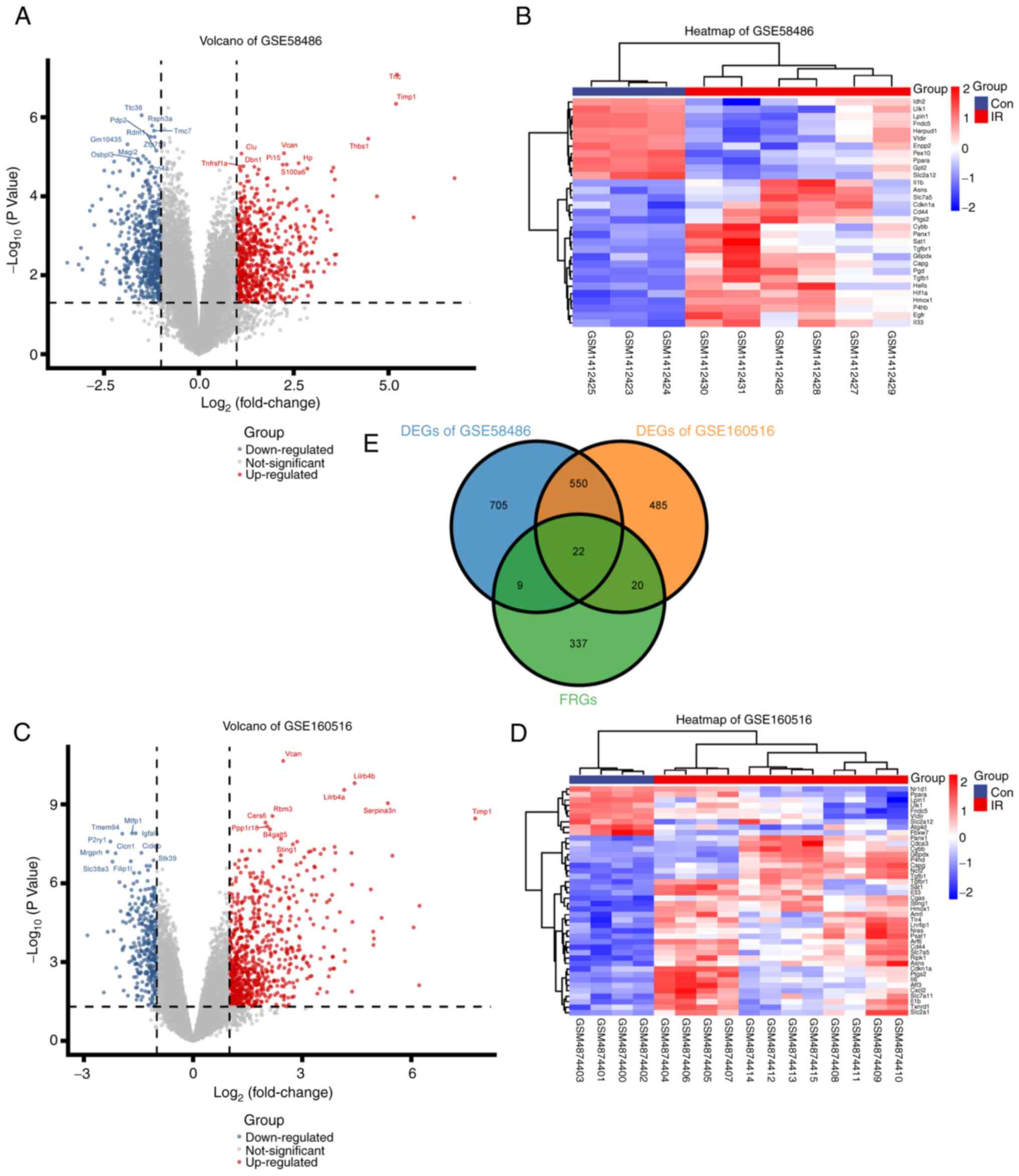
A total of 1,287 DEGs (673 upregulated and 614
downregulated) in GSE58486 and 1,079 DEGs (752 upregulated and 327
downregulated) in GSE160516 were screened (Fig. 3A and B). A total of eight DEmiRs (4
downregulated and 4 upregulated) were obtained from GSE124176
(Table I). Additionally, 51 DEFRGs
were identified based on the overlapping genes between the 388 FRGs
downloaded from FerrDb, and the DEGs screened from GSE58486 and
GSE160516 (Fig. 3E, Table SIV). The DEFRGs showed distinct
expression patterns in the normal and IRI samples (Fig. 3C and D).
 | Table IKey differentially expressed miRs
obtained from GSE124176. |
Table I
Key differentially expressed miRs
obtained from GSE124176.
| miRNA_ID | logFC | P-value |
|---|
| mmu-miR-23b-5p | -1.059027778 |
0.000560393c |
| mmu-miR-1940 | 1.042500000 |
0.001688678b |
|
mmu-miR-7030-5p | 1.022222222 |
0.001989443b |
| mmu-miR-706 | -1.084861111 |
0.005639392b |
| mmu-miR-582-5p | 2.275000000 |
0.009632056b |
|
mmu-let-7f-1-3p | -1.036805556 |
0.01466152a |
| mmu-miR-714 | -1.053055556 |
0.02414709a |
| mmu-miR-466j | 1.441666667 |
0.049095123a |
Functional and pathway enrichment
analysis of DEFRGs
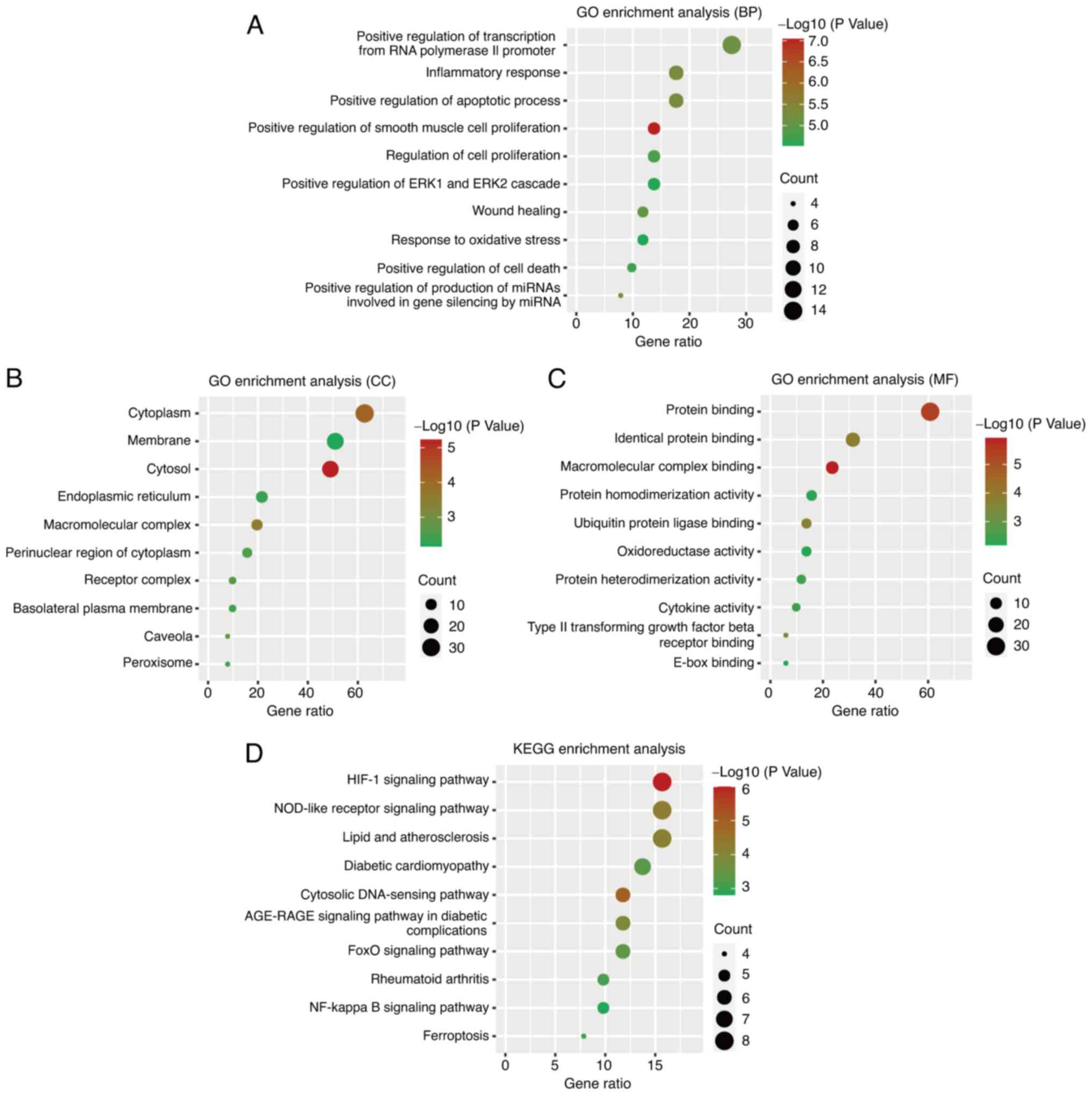
The DEFRGs were functionally annotated by GO
enrichment and KEGG pathway analyses. The GO terms were categorized
into biological process (BP), cellular component (CC), and
molecular function (MF). The DEFRGs were significantly enriched in
BP terms such as inflammatory response, positive regulation of the
apoptotic process, and positive regulation of transcription from
RNA polymerase II promoter (Fig.
4A). Cytoplasm, membrane, and cytosol were significantly
enriched CC terms (Fig. 4B), and
those related to the MF category were identical protein binding,
protein binding, and macromolecular complex binding (Fig. 4C). The pathways significantly
associated with DEFRGs included the hypoxia-inducible factor
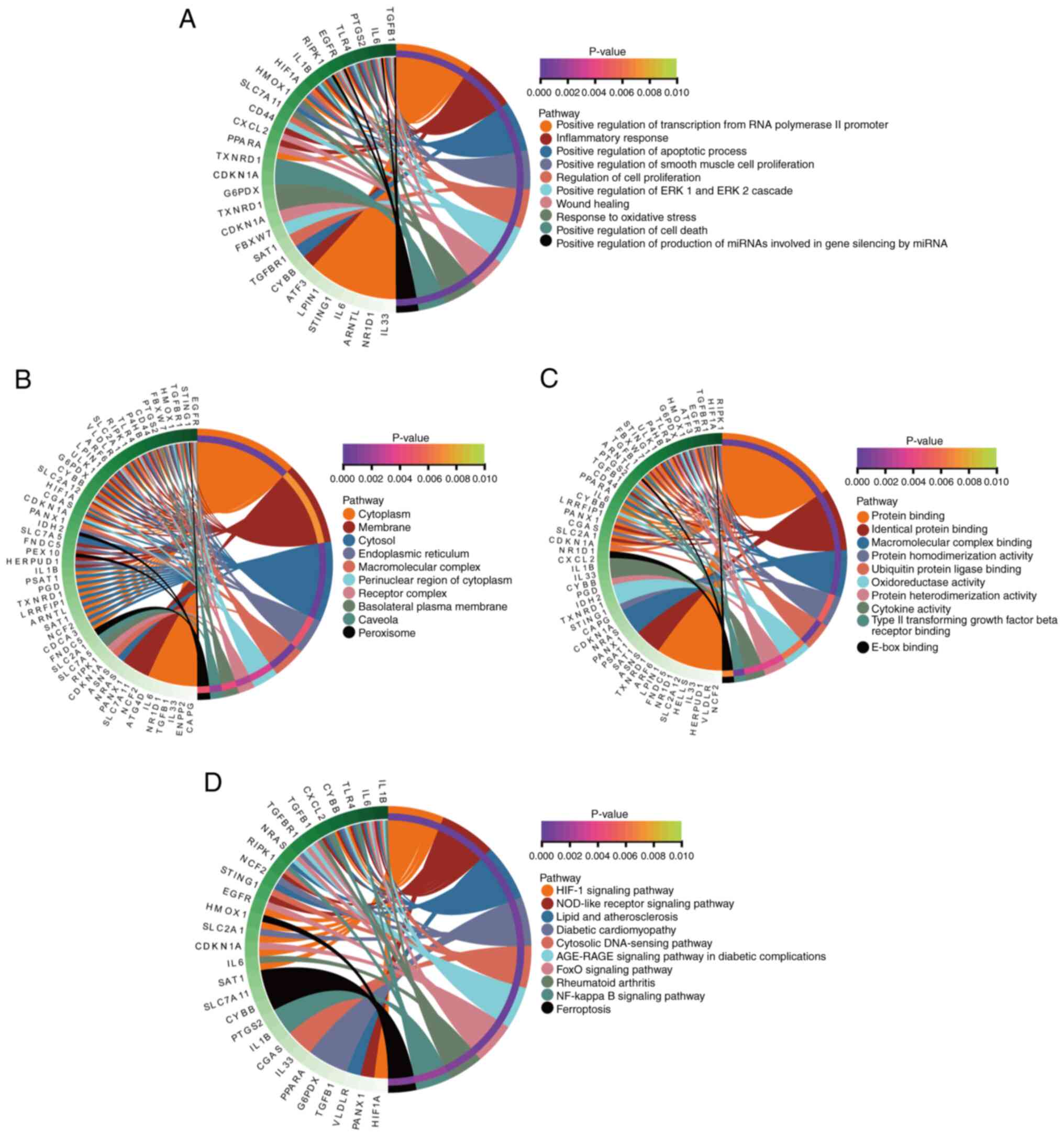
(HIF)-1, ferroptosis, and NF-κB signaling pathways (Fig. 4D). The interactions between DEFRGs
and the above functions and pathways were explored and found that
the regulation of DEFRGs during myocardial reperfusion injury may
involve multiple genes and pathways (Fig. 5).
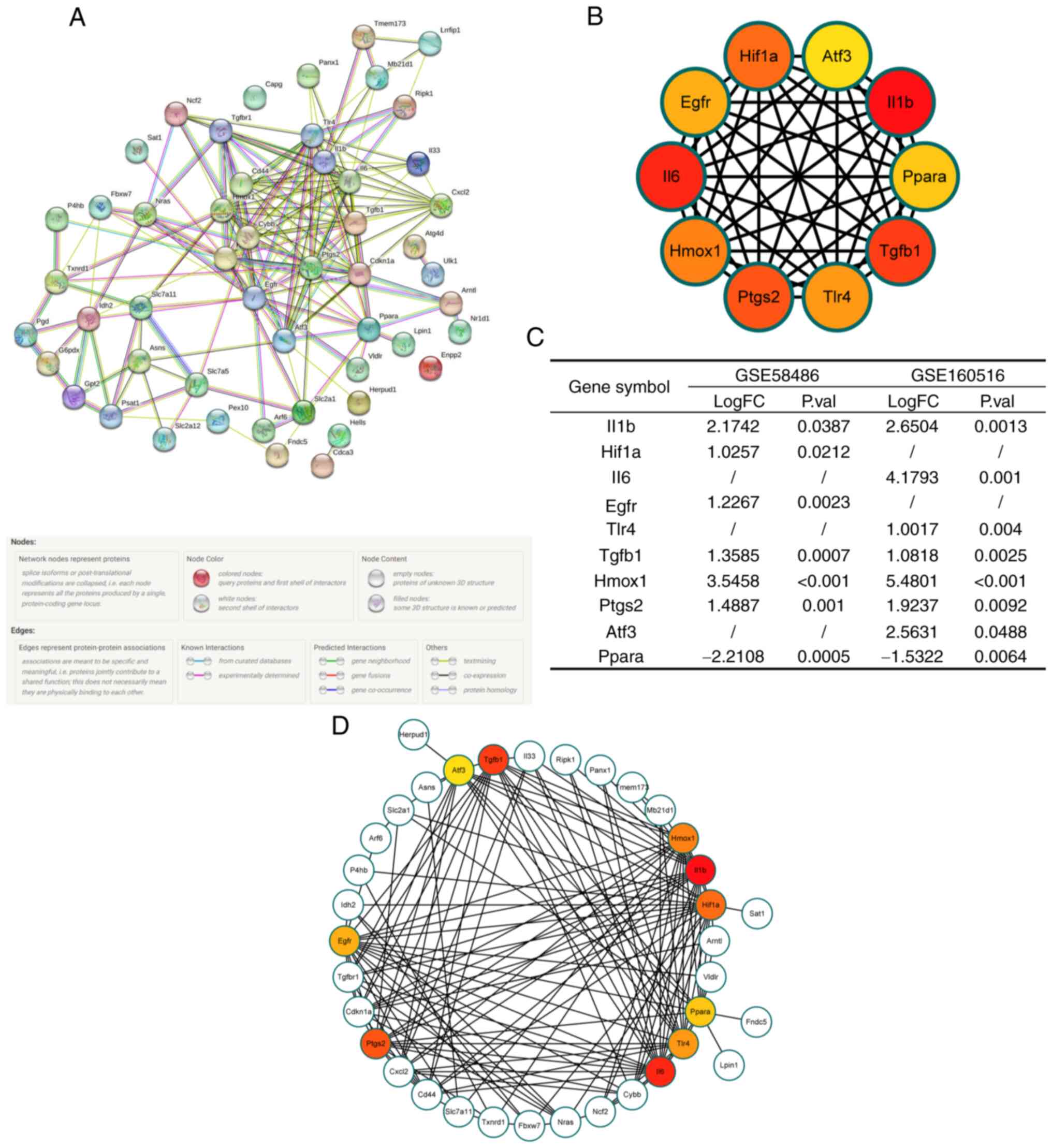
Construction of a PPI network and
extraction of hub genes
To further understand the interactions between these
DEFRGs, a PPI network was constructed using the STRING database
(Fig. 6A). The hub genes were
extracted using the Cytohubba plugin of Cytoscape and included
IL-1B, HIF-1A, IL-6, epidermal growth factor receptor (EGFR),
toll-like receptor (TLR)4, transforming growth factor (TGF)-B1,
heme oxygenase (HMOX)1, prostaglandin-endoperoxide synthase
(PTGS)2, activating transcription factor (ATF)3, and peroxisome
proliferator-activated receptor α (PPARα). The interactions between
the DEFRGs and hub genes are shown in Fig. 6B and D. The differential expression of hub
genes between normal and IRI hearts is shown in Fig. 6C.
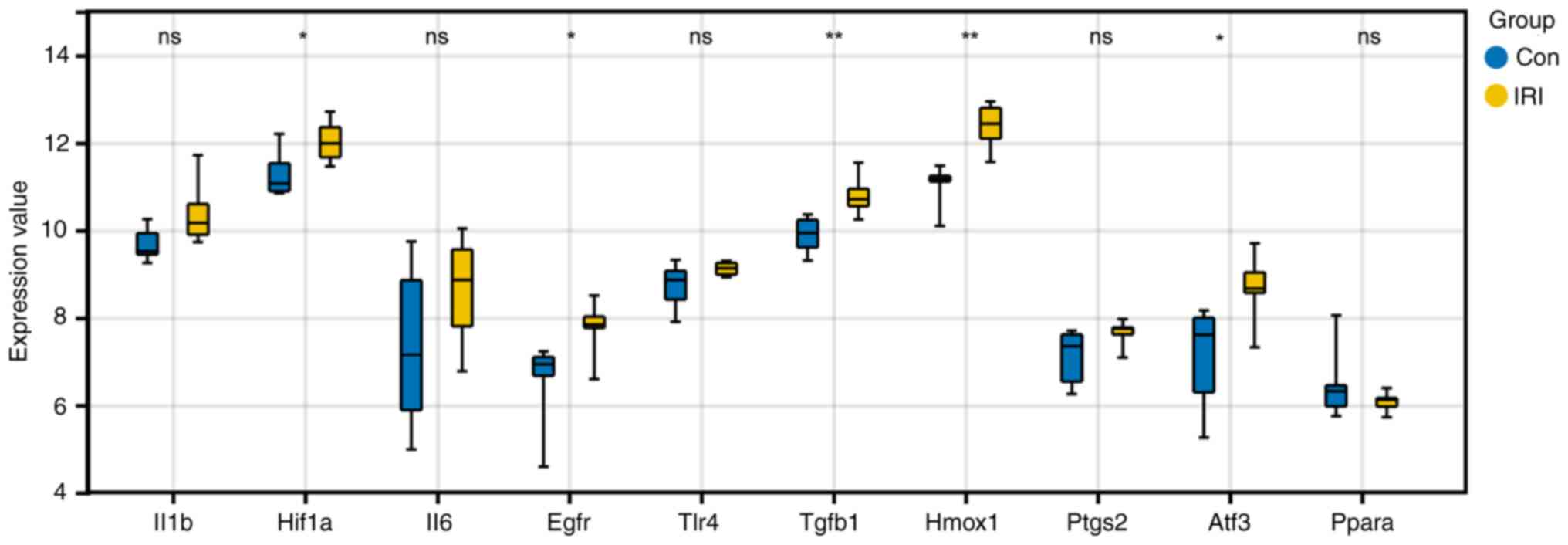
Verification of hub gene expression in
GSE4105
The differential expression of hub genes in the
GSE4105 dataset was analyzed to further clarify their role in IRI.
HIF-1A, EGFR, TGF-B1, HMOX1, and ATF3 showed similar expression
patterns in GSE4105 (Fig. 7) and
were markedly upregulated in the IRI samples compared to the
controls. Thus, these 5 genes were considered key FRGs involved in
myocardial reperfusion injury.
Ferroptosis is induced in
cardiomyocytes subjected to A/R injury in vitro
To validate the results of the bioinformatics
analysis, an A/R injury model was established to simulate IRI in
vitro and ferroptosis induction was explored. The viability of
H9c2 cells subjected to A/R was markedly decreased compared to that
of cells cultured under normoxic conditions. Interestingly,
pretreatment with the ferroptosis inhibitor Fer-1 protected the
cells against A/R injury (Fig.
8A). The levels of PTGS2 and GPX4 proteins were also analyzed,
as established markers of ferroptosis (34,35),
in the treated H9c2 cells. A/R treatment increased the expression
of PTGS2 and decreased that of GPX4, whereas Fer-1 neutralized the
effect of A/R and restored the levels of both proteins (Fig. 8B and C). Furthermore, cells subjected to A/R
showed a significant increase in the cytoplasmic levels of total
iron and MDA compared to the controls, which was attenuated by
Fer-1 pretreatment (Fig. 8E and
F). In addition, Fer-1
pretreatment alleviated A/R-induced increases in ROS levels
(Fig. 8D). Previous studies showed
that lipid peroxidation and extensive mitochondrial injury are key
signs of ferroptosis (34). The
mitochondria in A/R-induced H9c2 cells were significantly
distorted, and the Flameng scores were significantly increased;
Fer-1 pretreatment protected against such pathological changes
(Fig. 8G and H). As shown in Fig. 8I and J, the content of lipid ROS in the A/R
group was significantly increased compared to the control group,
and Fer-1 pretreatment abolished the effects induced by A/R
treatment. These findings suggested significant activation of the
ferroptosis program in cardiomyocytes in response to A/R injury.
The expression levels of HIF-1A, EGFR, TGF-B1, HMOX1, and ATF3, the
five key FRGs identified in bioinformatics analysis, were analyzed
in the treated cells. HIF-1A, EGFR, HMOX1, and ATF3 mRNA levels
increased significantly after A/R treatment and were downregulated
by Fer-1, indicating that these genes are key regulators of
ferroptosis during IRI (Fig.
8K-O).
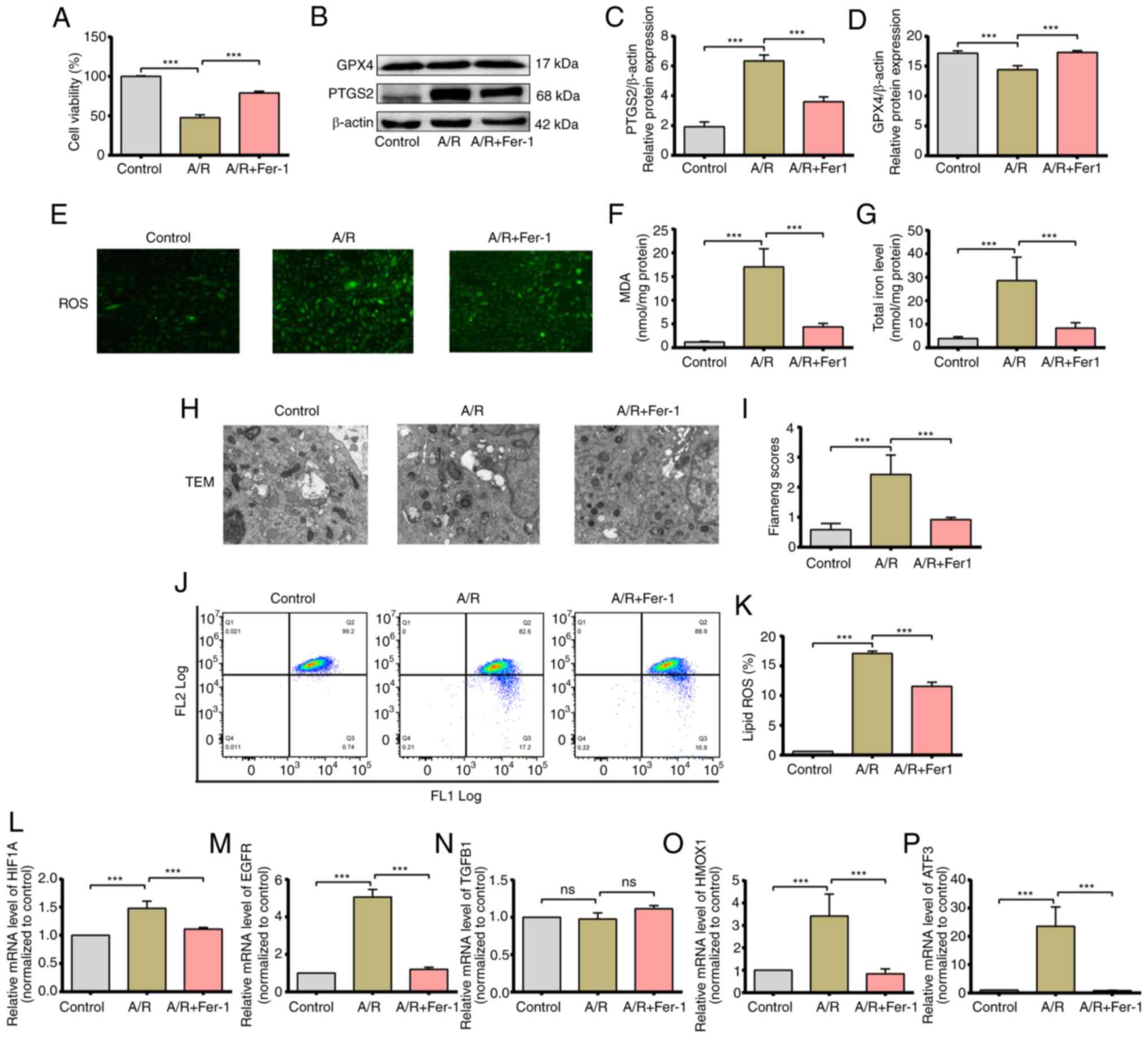 | Figure 8Activated ferroptosis and validation
of the expression of hub genes in the H9c2 A/R model, H9c2
myofibroblasts were exposed to anoxic conditions for 3 h and then
reoxygenated for 2 h in the after pretreatment with 5 mM Fer-1. (A)
Cell viability was measured using a CCK-8 assay. (B and C) Protein
expression levels of PTGS2 and GPX4. (D) Representative
fluorescence images of ROS in H9c2 myofibroblasts. (E) MDA levels
and (F) total iron levels in H9c2 myofibroblasts. (G and H) TEM
images and the FIameng scores of H9c2 cells. magnification, x8,000;
scale bar, 2 µm. (I and J) Lipid ROS levels of were assessed using
C11-BODIPY staining. (K-P) mRNA expression levels of hub genes.
***P<0.05. Data are presented as the mean ± SD of at
least three repeats. ROS, reactive oxygen species; MDA,
malondialdehyde; Fer-1, Ferrostatin-1; ns, not significant; A/R,
anoxia/reoxygenation; TEM, transmission electron microscope. |
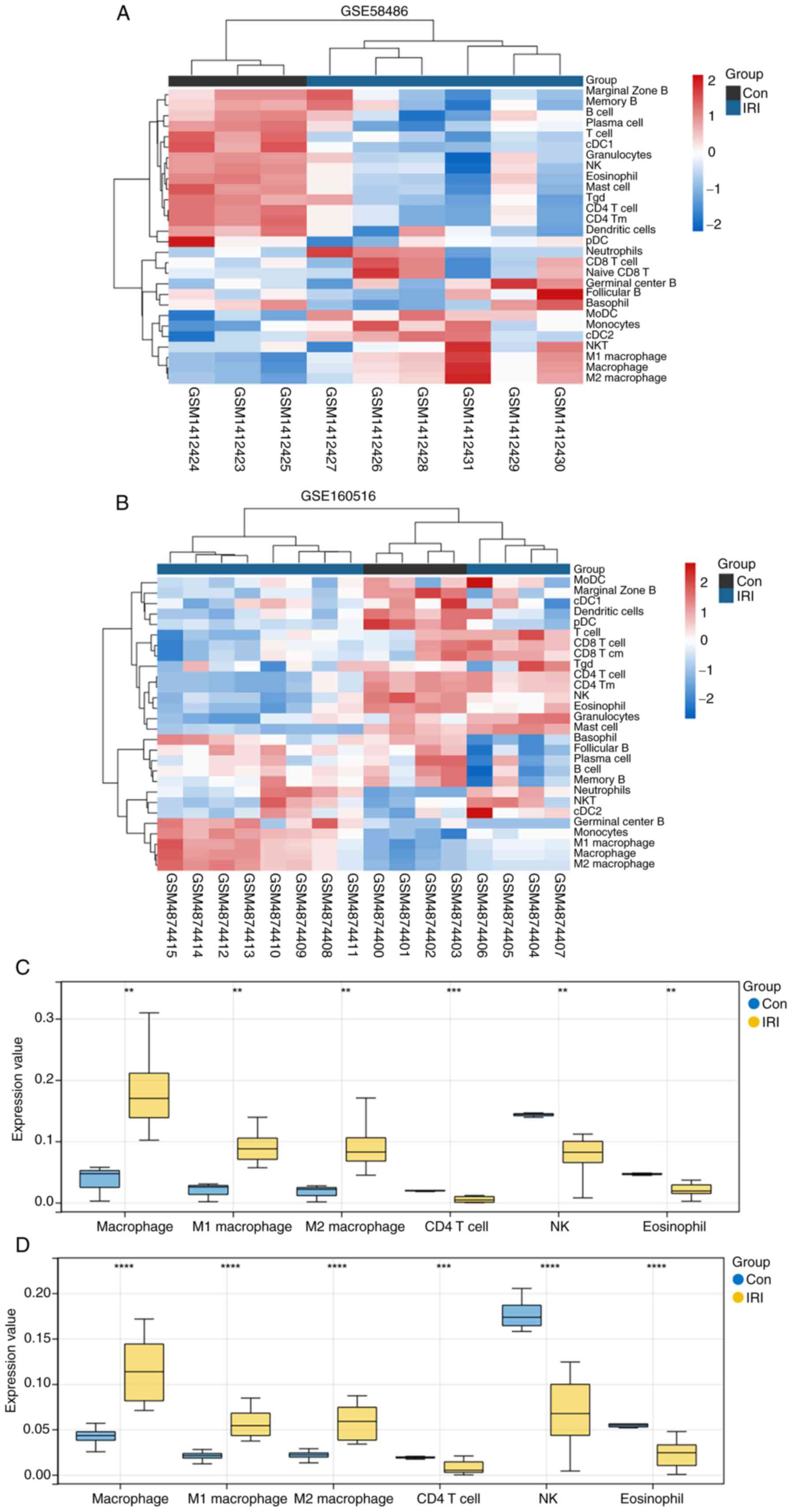
Immune infiltration analysis
Previous studies showed that ferroptosis is a form
of immune cell death (36,37). Therefore, the abundance of 36
immune cell types between IRI and control samples was compared
using the ImmuCellAI algorithm. Consistent with the clustered
heatmap results, immune infiltration was significantly different
between the IRI and control samples in the GSE58486 dataset,
whereas the differences were not obvious in the GSE160516 dataset
(Fig. 9A and C). However, the infiltration of M1 and M2
macrophages was markedly increased in the IRI samples in both
datasets.
Single Sample Gene Set Enrichment Analysis (ssGSEA)
results showed that the abundance of NK and CD4 T cells,
eosinophils, CD4 Tm cells, and Tregs was notably lower in the IRI
samples in both the GSE58486 and GSE160516 datasets (Fig. 9B and D). In addition, the IRI and control
sample infiltration scores evaluated by ssGSEA were significantly
different in GSE58486 (P<0.05) and GSE160516 (P<0.05).
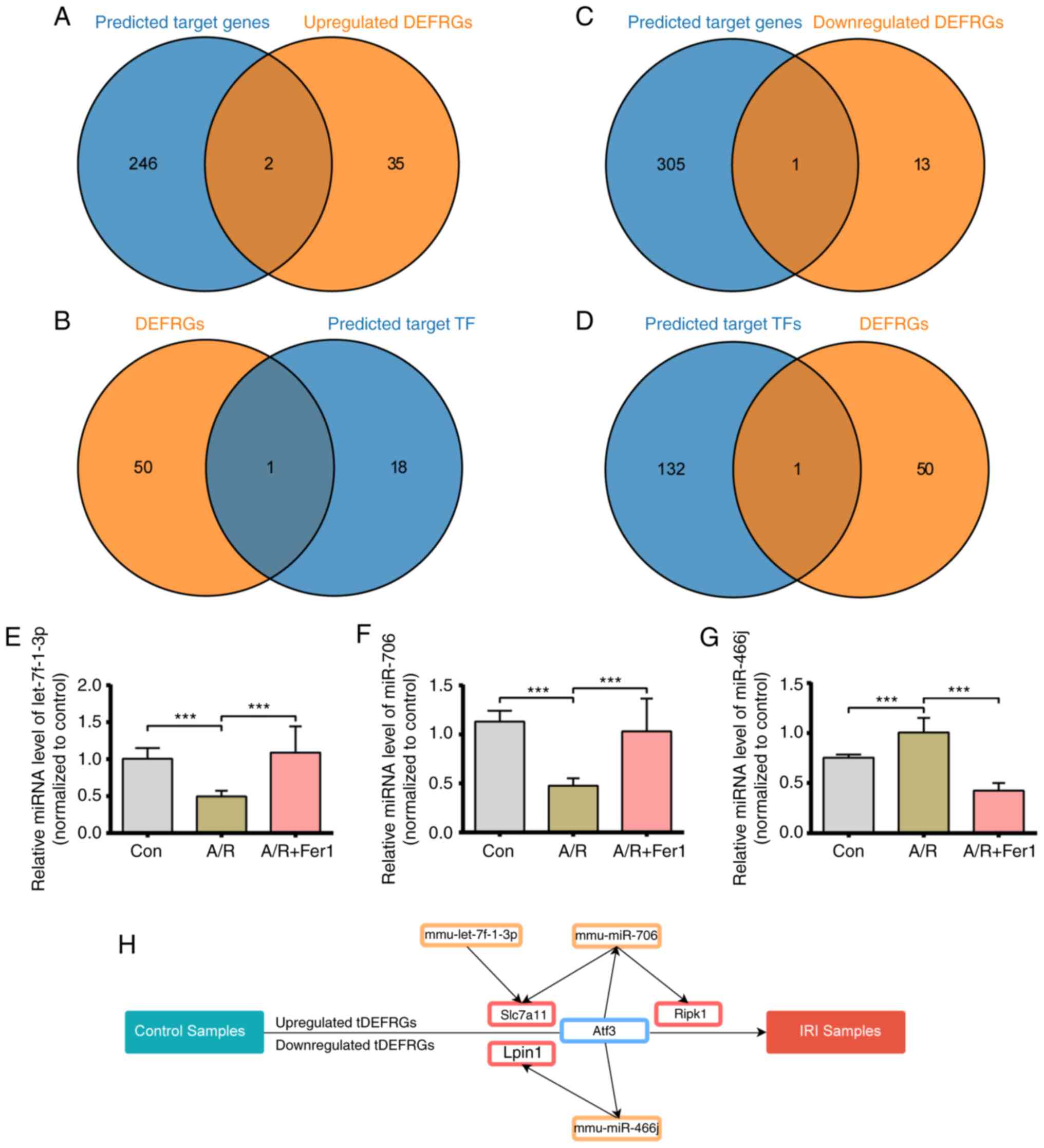
Prediction of the target genes and TFs
of DEmiRs
The target genes of DEmiRs were predicted using
miRwalk, TargetScan, and miRDB; 248 upregulated and 306
downregulated mRNAs that were common to all three databases were
screened. Two upregulated and one downregulated target mRNA
overlapped with the DEFRG set (Fig.
10A and C). Three DEmiRs,
including mmu-let-7f-3p, mmu-mir-706, and mmu-mir-466j, were the
putative regulators of these overlapping genes and, thus, were
considered key miRNAs involved in ferroptosis during myocardial
reperfusion injury. The target TFs of these key DEmiRs were then
screened in the TransmiR database, and ATF3 was identified as the
key TF for the DEFRGs (Fig. 10B
and D).
Establishment of TF-miRNA-mRNA
network
To further elucidate the role of key miRNAs in IRI,
the expression of mmu-let-7f-3p, mmu-mir-706, and mmu-mir-466j was
assessed in vitro. As shown in Fig. 10E, mmu-mir-466j miRNA levels were
significantly increased in the A/R group compared to the control
group and downregulated by Fer-1 pretreatment. The levels of
mmu-let-7f-3p and mmu-mir-706 miRNA were significantly reduced in
the A/R group, whereas Fer-1 pretreatment increased mmu-let-7f-3p
and mmu-mir-706 levels compared with the A/R group (Fig. 10F and G). A TF-miRNA-mRNA network was also
constructed. As shown in Fig.
10H, the network consisted of three key DEmiRs, three target
genes, and one TF.
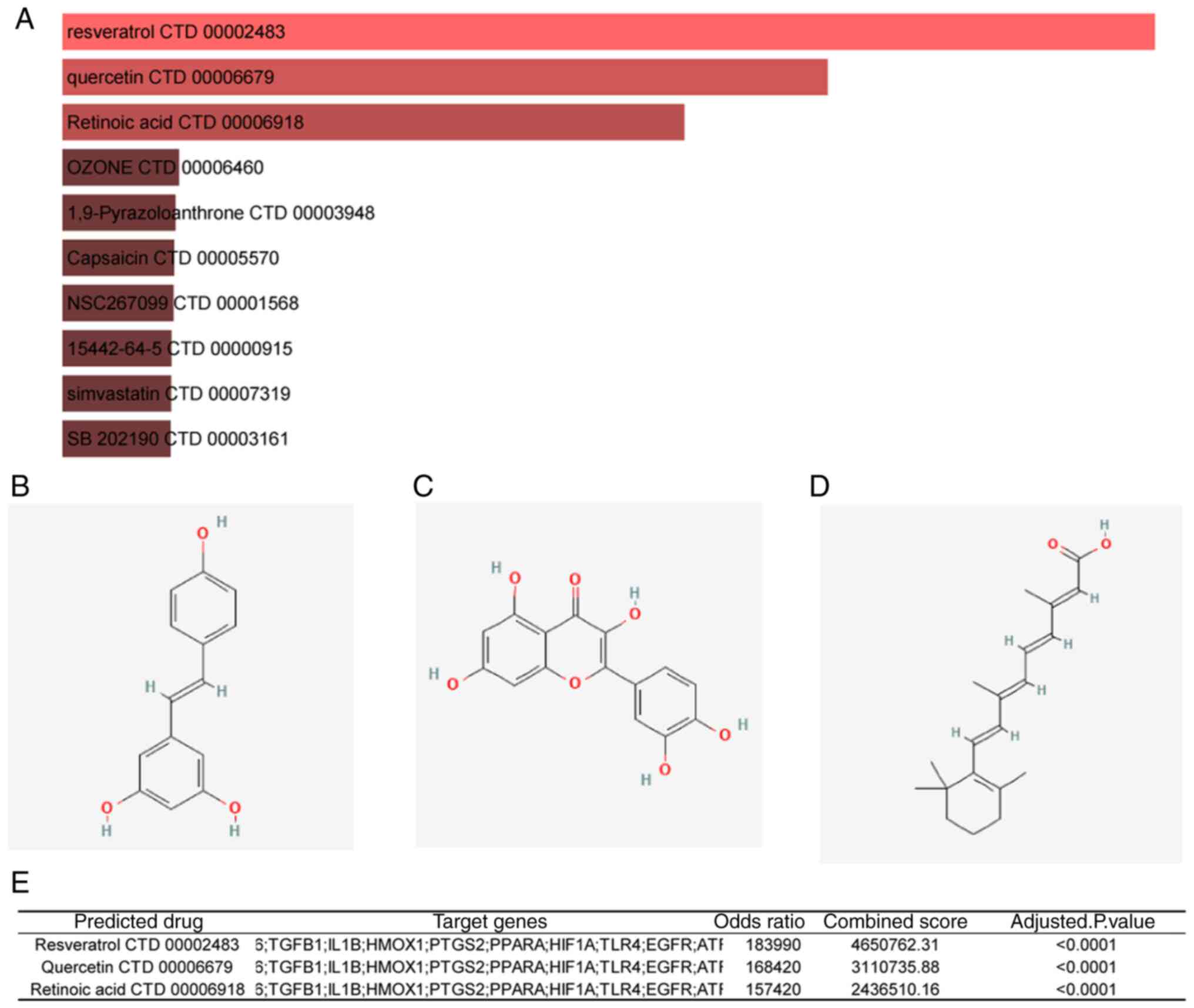
Targeted drug prediction of hub
genes
The DSigDB database was used to predict drugs
correlated with the hub genes, which can be potentially effective
against IRI. The candidate drugs were screened based on adjusted
P-values and combined scores. The top 10 predicted target drugs
ranked by combined scores are listed in Fig. 11A. As shown in Fig. 10E, the top three drugs
significantly correlated with the target genes (adjusted P-value
<0.0001, combined score >2x106) were resveratrol
(Fig. 11B, combined
score=4,650,762), quercetin (Fig.
11C, combined score=3,110,735), and retinoic acid (Fig. 11D, combined score=2,436,510).
Discussion
Ferroptosis, a novel type of programmed cell death
characterized by iron-induced lipid membrane peroxidation, is a
pathological factor in several types of cardiovascular diseases,
including heart failure, and acute aortic dissection (38-40).
However, the association between ferroptosis and IRI remains
unclear. Ischemic heart disease is often succeeded by myocardial
IRI, which is the result of the excessive production of free
radicals and cardiomyocyte apoptosis due to mitochondrial
dysfunction, calcium overload, and increased heat shock protein
levels (41). Several clinical
studies showed that iron overload was a critical factor involved in
incomplete left ventricular remodeling after IRI (42). Likewise, the inhibition of
glutaminolysis alleviated IRI by blocking ferroptosis (43). Based on these reports, it is
hypothesized that ferroptosis plays a significant role in the
development of IRI. Here, Fer-1 pretreatment could remarkably
reduce excess lipid ROS generation, iron accumulation, and
excessive mitochondrial injury induced by A/R treatment. Based on
the primary manifestations of ferroptosis, it was confirmed that
ferroptosis was involved in the pathological process of IRI. To
this end, the genes related to ferroptosis were screened and their
regulatory networks in IRI were explored through bioinformatics
analysis using multiple transcriptomic datasets of normal and
ischemic heart tissue.
Consistent with previous studies, several FRGs were
aberrantly expressed in the IRI samples, indicating that the
activation of ferroptosis is a key pathological factor (44). In addition, DEFRGs were
significantly enriched in GO terms related to ferroptosis and
oxidative stress, which is a major trigger of ferroptosis,
apoptosis, and the inflammatory response (45). Other GO terms associated with
DEFRGs included identical protein binding and macromolecular
complex binding, which may also affect the pathological process of
IRI. KEGG analysis revealed that the HIF-1 signaling pathway and
NOD-like receptor signaling pathway may mediate ferroptosis during
IRI, although neither pathway has yet been implicated in IRI.
To further clarify the mechanisms underlying
ferroptosis activation in IRI, a PPI network of DEFRGs was
constructed, and several hub genes were identified, of which
HIF1A, EGFR, HMOX1, and ATF3 were
validated experimentally in an in vitro A/R model. Previous
studies showed that these critical genes participate in the
regulation of ferroptosis (46-49).
Furthermore, the HIF1A/PTGS2 pathway aggravates ferroptosis and
mediates myocardial injury and inflammation after coronary
microembolization (50). By
contrast, the overexpression of ATF3 prevents the activation of
ferroptosis by erastin and RSL3 in cardiomyocytes (51). There are currently no reports on
the role of EGFR and HMOX1 in cardiovascular diseases in the
context of ferroptosis to the best of our knowledge. Since the
aforementioned genes also participate in other pathological
processes, they may regulate IRI through mechanisms independent of
ferroptosis. Thus, it is necessary to identify the connections
between ferroptosis and other biological processes in IRI, such as
autophagy. Although TGFB1 was screened as a hub gene, no changes in
its expression levels were found between the control and A/R
groups. It is possible that TGFB1 mediates ferroptosis during IRI
and is altered at the protein level.
As a non-coding RNA, miRNAs can regulate gene
expression post-transcriptionally by silencing target mRNAs
(52). A TF-miRNA-DEFRG network
was established to determine the possible regulatory pathways of
the DEFRGs; 3 key miRNAs were identified (mmu-miR-706,
mmu-let-7f-1-3p, and mmu-miR-466j) and were verified experimentally
using an in vitro A/R model, and the overlapping target
DEFRGs and TFs were SLC7A11, RIPK1, LIPIN1,
and ATF3. Whereas mmu-miR-706 inhibited cardiomyocyte
ferroptosis by suppressing PTGS2(53), mmu-7f-1-3p alleviated smoke-induced
bronchial and alveolar epithelial cell apoptosis via regulating
FOXO1(54). These potential
regulatory pathways may thus offer novel therapeutic targets for
the treatment of IRI, and thus warrant further investigation.
Previous studies have demonstrated that the
inflammatory response is crucial for the development of IRI, and
that any disruption in iron homeostasis affects immune cell
function, differentiation and death (55,56).
Consistent with this, notable differences in the immune
infiltration patterns of normal and IRI samples were observed.
While the abundance of M1 and M2 macrophages increased in ischemic
heart tissue, that of CD4+ T cells, NK cells, Tregs, eosinophils,
and CD4+ Tm cells decreased, consistent with previous studies
(57,58). These findings indicate the
possibility of crosstalk among the key DEFRGs and aberrant immune
responses in IRI, which need to be validated further.
Since the genes involved in pathological processes
are potential therapeutic targets (59), the potential drug candidates for
the management of IRI based on the ferroptosis-related hub genes
were predicted. Resveratrol, quercetin, and retinoic acid showed
the highest correlation with the target genes. Retinoic acid was
shown to attenuate myocardial injury and inhibit post-IRI
cardiomyocyte apoptosis by increasing ADAM10 expression (60). Quercetin exhibited therapeutic
effects in different H9c2 cardiomyoblast injury models induced by
lipopolysaccharide, doxorubicin, and A/R (61-64).
Resveratrol can also protect against IRI by inhibiting ferroptosis
and reducing oxidative stress (65). Nevertheless, further research is
needed to determine whether these compounds can alleviate IRI by
targeting ferroptosis.
Although the changes in expression levels of hub
DEFRGs were validated in another independent dataset, as well as in
an in vitro experimental model, the exact roles of these
genes in the pathological process of IRI requires further
clarification using in vivo experiments and molecular
assays. In addition, although animal models can simulate the
morphological signs of human IRI, they cannot recapitulate the
natural history and histological features, resulting in potentially
inaccurate and even contradictory conclusions. Thus, human tissue
samples need to be analyzed to validate our findings. Furthermore,
observing the role of the key genes in IRI through more precise
interference experiments is critical for the discovery of
meaningful therapeutic and diagnostic targets. Therefore, further
functional studies are needed to extend the results of the present
study to the clinic.
In conclusion, several dysregulated FRGs associated
with IRI were identified and experimentally validated, and the
aberrant immune infiltration in ischemic heart tissue was explored.
Resveratrol, quercetin, and retinoic acid were also identified as
potential candidate drugs for the management of IRI via targeting
of ferroptosis. The present study offers novel insight into the
role of ferroptosis in IRI, which can help to elucidate its
pathobiological mechanisms and identify novel diagnostic and
therapeutic targets.
Supplementary Material
Merged sets of ferroptosis-related
genes
Sequences of primers for HIF-1α,
HMOX1, EGFR, ATF3, and TGFB1.
Core genes extracted from the GSE58486
and GSE160516 datasets.
DEGFRs extracted from GSE58486 and
GSE160516
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the Natural Science
Foundation of Jiangxi (grant no. 20212BAB206021), Key Projects of
Jiangxi Natural Science Foundation (grant no. 20224ACB206002), the
National Natural Science Foundation of China (grant nos. 81860054
and 81960059), and the Major Discipline Academic and Technical
Leaders Training Program of Jiangxi Province (grant no.
20204BCJL23056).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author upon reasonable
request.
Authors' contributions
SQL, JCL, and HH conceived and designed the study.
JCL provided administrative support. SYL, ZHC, FJH, and YCW
collected the data. TH, WPY, JCL and HXZ analyzed the data. TH,
WPY, and YCW performed the experiments. JCL revised the manuscript.
SQL and JCL confirm the authenticity of all the raw data. All
authors have read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Mozaffarian D, Benjamin EJ, Go AS, Arnett
DK, Blaha MJ, Cushman M, de Ferranti S, Després JP, Fullerton HJ,
Howard VJ, et al: Heart disease and stroke statistics-2015 update:
A report from the American heart association. Circulation.
131:e29–e322. 2015.PubMed/NCBI View Article : Google Scholar
|
|
2
|
O'Gara PT, Kushner FG, Ascheim DD, Casey
DE Jr, Chung MK, de Lemos JA, Ettinger SM, Fang JC, Fesmire FM,
Franklin BA, et al: 2013 ACCF/AHA guideline for the management of
ST-elevation myocardial infarction: A report of the American
college of cardiology foundation/American heart association task
force on practice guidelines. Circulation. 127:e362–e425.
2013.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Heusch G and Gersh BJ: The pathophysiology
of acute myocardial infarction and strategies of protection beyond
reperfusion: A continual challenge. Eur Heart J. 38:774–784.
2017.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Eitel I, Stiermaier T, Rommel KP, Fuernau
G, Sandri M, Mangner N, Linke A, Erbs S, Lurz P, Boudriot E, et al:
Cardioprotection by combined intrahospital remote ischaemic
perconditioning and postconditioning in ST-elevation myocardial
infarction: The randomized LIPSIA CONDITIONING trial. Eur Heart J.
36:3049–3057. 2015.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Zhou L, Han S, Guo J, Qiu T, Zhou J and
Shen L: Ferroptosis-a new dawn in the treatment of organ
ischemia-reperfusion injury. Cells. 11(3653)2022.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Chen X, Li J, Kang R, Klionsky DJ and Tang
D: Ferroptosis: Machinery and regulation. Autophagy. 17:2054–2081.
2021.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Tang D, Chen X, Kang R and Kroemer G:
Ferroptosis: Molecular mechanisms and health implications. Cell
Res. 31:107–125. 2021.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Williams RE, Zweier JL and Flaherty JT:
Treatment with deferoxamine during ischemia improves functional and
metabolic recovery and reduces reperfusion-induced oxygen radical
generation in rabbit hearts. Circulation. 83:1006–1014.
1991.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Paraskevaidis IA, Iliodromitis EK,
Vlahakos D, Tsiapras DP, Nikolaidis A, Marathias A, Michalis A and
Kremastinos DT: Deferoxamine infusion during coronary artery bypass
grafting ameliorates lipid peroxidation and protects the myocardium
against reperfusion injury: Immediate and long-term significance.
Eur Heart J. 26:263–270. 2005.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Nakamura T, Naguro I and Ichijo H: Iron
homeostasis and iron-regulated ROS in cell death, senescence and
human diseases. Biochim Biophys Acta Gen Subj. 1863:1398–1409.
2019.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Ravingerová T, Kindernay L, Barteková M,
Ferko M, Adameová A, Zohdi V, Bernátová I, Ferenczyová K and Lazou
A: The molecular mechanisms of iron metabolism and its role in
cardiac dysfunction and cardioprotection. Int J Mol Sci.
21(7889)2020.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Edgar R, Domrachev M and Lash AE: Gene
expression omnibus: NCBI gene expression and hybridization array
data repository. Nucleic Acids Res. 30:207–210. 2002.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Zhou N and Bao J: FerrDb: A manually
curated resource for regulators and markers of ferroptosis and
ferroptosis-disease associations. Database (Oxford).
1(baaa021)2020.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43(e47)2015.PubMed/NCBI View Article : Google Scholar
|
|
15
|
R Core Team (2020). R: A language and
environment for statistical computing. R Foundation for Statistical
Computing, Vienna, Austria. URL https://www.R-project.org/.
|
|
16
|
Hanbo Chen (2022). VennDiagram: Generate
High-Resolution Venn and Euler Plots. R package version 1.7.3.
https://CRAN.R-project.org/package=VennDiagram.
|
|
17
|
Huang DW, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Huang DW, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The gene
ontology consortium. Nat Genet. 25:25–29. 2000.PubMed/NCBI View Article : Google Scholar
|
|
20
|
The Gene Ontology Resource: 20 years and
still GOing strong. Nucleic acids research. 47:D330–d338.
2019.PubMed/NCBI View Article : Google Scholar : doi:
10.1093/nar/gky1055.
|
|
21
|
Kanehisa M: A database for post-genome
analysis. Trends Genet. 13:375–376. 1997.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Szklarczyk D, Gable AL, Lyon D, Junge A,
Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork
P, et al: STRING v11: Protein-protein association networks with
increased coverage, supporting functional discovery in genome-wide
experimental datasets. Nucleic Acids Res. 47:D607–D613.
2019.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Chin CH, Chen SH, Wu HH, Ho CW, Ko MT and
Lin CY: CytoHubba: Identifying hub objects and sub-networks from
complex interactome. BMC Syst Biol. 8 (Suppl 4)(S11)2014.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Friedman RC, Farh KK, Burge CB and Bartel
DP: Most mammalian mRNAs are conserved targets of microRNAs. Genome
Res. 19:92–105. 2009.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Dweep H and Gretz N: MiRWalk2.0: A
comprehensive atlas of microRNA-target interactions. Nat Methods.
12(697)2015.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Wong N and Wang X: miRDB: An online
resource for microRNA target prediction and functional annotations.
Nucleic Acids Res. 43:D146–D152. 2015.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Tong Z, Cui Q, Wang J and Zhou Y: TransmiR
v2.0: An updated transcription factor-microRNA regulation database.
Nucleic Acids Res. 47:D253–D258. 2019.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Huang H, Lai S, Luo Y, Wan Q, Wu Q, Wan L,
Qi W and Liu J: Nutritional preconditioning of apigenin alleviates
myocardial ischemia/reperfusion injury via the mitochondrial
pathway mediated by notch1/hes1. Oxid Med Cell Longev.
2019(7973098)2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Flameng W, Borgers M, Daenen W and
Stalpaert G: Ultrastructural and cytochemical correlates of
myocardial protection by cardiac hypothermia in man. J Thorac
Cardiovasc Sur. 79:413–424. 1980.PubMed/NCBI
|
|
31
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Miao YR, Zhang Q, Lei Q, Luo M, Xie GY,
Wang H and Guo AY: ImmuCellAI: A unique method for comprehensive
T-cell subsets abundance prediction and its application in cancer
immunotherapy. Adv Sci (Weinh). 7(1902880)2020.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Yoo M, Shin J, Kim J, Ryall KA, Lee K, Lee
S, Jeon M, Kang J and Tan AC: DSigDB: Drug signatures database for
gene set analysis. Bioinformatics. 31:3069–3071. 2015.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Li Y, Cao Y, Xiao J, Shang J, Tan Q, Ping
F, Huang W, Wu F, Zhang H and Zhang X: Inhibitor of
apoptosis-stimulating protein of p53 inhibits ferroptosis and
alleviates intestinal ischemia/reperfusion-induced acute lung
injury. Cell Death Differ. 27:2635–2650. 2020.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Lillo-Moya J, Rojas-Solé C,
Muñoz-Salamanca D, Panieri E, Saso L and Rodrigo R: Targeting
ferroptosis against ischemia/reperfusion cardiac injury.
Antioxidants (Basel). 10(667)2021.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Chen X, Kang R, Kroemer G and Tang D:
Ferroptosis in infection, inflammation, and immunity. J Exp Med.
218(e20210518)2021.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Liao P, Wang W, Wang W, Kryczek I, Li X,
Bian Y, Sell A, Wei S, Grove S, Johnson JK, et al: CD8(+) T cells
and fatty acids orchestrate tumor ferroptosis and immunity via
ACSL4. Cancer Cell. 40:365–378.e6. 2022.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Zou HX, Qiu BQ, Lai SQ, Huang H, Zhou XL,
Gong CW, Wang LJ, Yuan MM, He AD and Liu JC: Role of
ferroptosis-related genes in Stanford type a aortic dissection and
identification of key genes: New insights from bioinformatic
analysis. Bioengineered. 12:9976–9990. 2021.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Fang X, Wang H, Han D, Xie E, Yang X, Wei
J, Gu S, Gao F, Zhu N, Yin X, et al: Ferroptosis as a target for
protection against cardiomyopathy. Proc Natl Acad Sci USA.
116:2672–2680. 2019.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Li W, Feng G, Gauthier JM, Lokshina I,
Higashikubo R, Evans S, Liu X, Hassan A, Tanaka S, Cicka M, et al:
Ferroptotic cell death and TLR4/Trif signaling initiate neutrophil
recruitment after heart transplantation. J Clin Invest.
129:2293–2304. 2019.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Chen Y, Yi X, Huo B, He Y, Guo X, Zhang Z,
Zhong X, Feng X, Fang ZM, Zhu XH, et al: BRD4770 functions as a
novel ferroptosis inhibitor to protect against aortic dissection.
Pharmacol Res. 177(106122)2022.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Liu X, Bian H, Dou QL, Huang XW, Tao WY,
Liu WH, Li N and Zhang WW: Ginkgetin alleviates inflammation,
oxidative stress, and apoptosis induced by hypoxia/reoxygenation in
H9C2 cells via caspase-3 dependent pathway. Biomed Res Int.
2020(1928410)2020.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Bulluck H, Rosmini S, Abdel-Gadir A, White
SK, Bhuva AN, Treibel TA, Fontana M, Ramlall M, Hamarneh A, Sirker
A, et al: Residual myocardial iron following intramyocardial
hemorrhage during the convalescent phase of reperfused
st-segment-elevation myocardial infarction and adverse left
ventricular remodeling. Circ Cardiovasc Imaging.
9(e004940)2016.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Gao M, Monian P, Quadri N, Ramasamy R and
Jiang X: Glutaminolysis and transferrin regulate ferroptosis. Mol
Cell. 59:298–308. 2015.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Wu X, Li Y, Zhang S and Zhou X:
Ferroptosis as a novel therapeutic target for cardiovascular
disease. Theranostics. 11:3052–3059. 2021.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Yu Y, Yan Y, Niu F, Wang Y, Chen X, Su G,
Liu Y, Zhao X, Qian L, Liu P and Xiong Y: Ferroptosis: A cell death
connecting oxidative stress, inflammation and cardiovascular
diseases. Cell Death Discov. 7(193)2021.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Li Y, Cao Y, Xiao J, Shang J, Tan Q, Ping
F, Huang W, Wu F, Zhang H and Zhang X: Inhibitor of
apoptosis-stimulating protein of p53 inhibits ferroptosis and
alleviates intestinal ischemia/reperfusion-induced acute lung
injury. Cell Death Differ. 27:2635–2650. 2020.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Liu S, Yan S, Zhu J, Lu R, Kang C, Tang K,
Zeng J, Ding M, Guo Z, Lai X, et al: Combination RSL3 treatment
sensitizes ferroptosis- and EGFR-inhibition-resistant HNSCCs to
cetuximab. Int J Mol Sci. 23(9014)2022.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Meng Z, Liang H, Zhao J, Gao J, Liu C, Ma
X, Liu J, Liang B, Jiao X, Cao J and Wang Y: HMOX1 upregulation
promotes ferroptosis in diabetic atherosclerosis. Life Sci.
284(119935)2021.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Wang L, Liu Y, Du T, Yang H, Lei L, Guo M,
Ding HF, Zhang J, Wang H, Chen X and Yan C: ATF3 promotes
erastin-induced ferroptosis by suppressing system Xc. Cell Death
Differ. 27:662–675. 2020.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Liu T, Shu J, Liu Y, Xie J, Li T, Li H and
Li L: Atorvastatin attenuates ferroptosis-dependent myocardial
injury and inflammation following coronary microembolization via
the Hif1a/Ptgs2 pathway. Front Pharmacol.
13(1057583)2022.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Liu H, Mo H, Yang C, Mei X, Song X, Lu W,
Xiao H, Yan J, Wang X, Yan J, et al: A novel function of ATF3 in
suppression of ferroptosis in mouse heart suffered
ischemia/reperfusion. Free Radic Biol Med. 189:122–135.
2022.PubMed/NCBI View Article : Google Scholar
|
|
53
|
van Wijk N, Zohar K and Linial M:
Challenging cellular homeostasis: Spatial and temporal regulation
of miRNAs. Int J Mol Sci. 23(16152)2022.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Gao F, Zhao Y, Zhang B, Xiao C, Sun Z, Gao
Y and Dou X: Suppression of lncRNA Gm47283 attenuates myocardial
infarction via miR-706/ Ptgs2/ferroptosis axis. Bioengineered.
13:10786–10802. 2022.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Han Z, Zhu Y, Cui Z, Guo P, Wei A and Meng
Q: MicroRNA Let-7f-1-3p attenuates smoke-induced apoptosis in
bronchial and alveolar epithelial cells in vitro by targeting
FOXO1. Eur J Pharmacol. 862(172531)2019.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Bacmeister L, Schwarzl M, Warnke S,
Stoffers B, Blankenberg S, Westermann D and Lindner D: Inflammation
and fibrosis in murine models of heart failure. Basic Res Cardiol.
114(19)2019.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Ni S, Yuan Y, Kuang Y and Li X: Iron
metabolism and immune regulation. Front Immunol.
13(816282)2022.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Fan Q, Tao R, Zhang H, Xie H, Lu L, Wang
T, Su M, Hu J, Zhang Q, Chen Q, et al: Dectin-1 contributes to
myocardial ischemia/reperfusion injury by regulating macrophage
polarization and neutrophil infiltration. Circulation. 139:663–678.
2019.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Sun K, Li YY and Jin J: A double-edged
sword of immuno-microenvironment in cardiac homeostasis and injury
repair. Signal Transduct Target Ther. 6(79)2021.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Huang M, Cai S and Su J: The pathogenesis
of sepsis and potential therapeutic targets. Int J Mol Sci.
20(5376)2019.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Zhu Z, Zhu J, Zhao X, Yang K, Lu L, Zhang
F, Shen W and Zhang R: All-trans retinoic acid ameliorates
myocardial ischemia/reperfusion injury by reducing cardiomyocyte
apoptosis. PLoS One. 10(e0133414)2015.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Li C, Zhang WJ and Frei B: Quercetin
inhibits LPS-induced adhesion molecule expression and oxidant
production in human aortic endothelial cells by p38-mediated Nrf2
activation and antioxidant enzyme induction. Redox Biol. 9:104–113.
2016.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Chen X, Peng X, Luo Y, You J, Yin D, Xu Q,
He H and He M: Quercetin protects cardiomyocytes against
doxorubicin-induced toxicity by suppressing oxidative stress and
improving mitochondrial function via 14-3-3γ. Toxicol Mech Methods.
29:344–354. 2019.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Tang L, Peng Y, Xu T, Yi X, Liu Y, Luo Y,
Yin D and He M: The effects of quercetin protect cardiomyocytes
from A/R injury is related to its capability to increasing
expression and activity of PKCε protein. Mol Cell Biochem.
382:145–152. 2013.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Li T, Tan Y, Ouyang S, He J and Liu L:
Resveratrol protects against myocardial ischemia-reperfusion injury
via attenuating ferroptosis. Gene. 808(145968)2022.PubMed/NCBI View Article : Google Scholar
|