Introduction
Metabolic abnormalities, particularly imbalances in
macrophage polarization, are critical factors in the pathogenesis
of several diseases, including inflammation, autoimmunity and
metabolic disorders (1,2). Under various stimuli, macrophages
display remarkable plasticity, switching between pro-inflammatory
M1 and anti-inflammatory M2 phenotypes (3,4).
When lipopolysaccharide (LPS) and interferon (IFN) activate M1
macrophages, they release pro-inflammatory cytokines such tumor
necrosis factor (TNF), interleukin (IL)-1 and IL-6 that cause
tissue damage and inflammation (5). On the other hand, M2 macrophages can
be induced by IL-4 cytokines and secrete anti-inflammatory factors
such as IL-10 and TGF-β, promoting tissue repair and resolution of
inflammation (6). However,
imbalances in M1/M2 macrophage transitions can lead to severe
inflammatory responses and oxidative stress, posing significant
obstacles to disease treatment and prognosis (7). This process involves interactions
between macrophage polarization, metabolic reprogramming, reactive
oxygen species (ROS) and inflammation (8,9).
Therefore, the study of macrophage polarization is essential for
understanding the pathological mechanisms of metabolic
diseases.
Metabolic reprogramming is a core process of
macrophage polarization, with M1 macrophages relying on glycolysis
for rapid energy production and M2 macrophages using oxidative
phosphorylation (OXPHOS) for more sustained energy supply (10). Uncoupling protein 2 (UCP2) is a
member of the mitochondrial anion carrier protein family located in
the inner mitochondrial membrane that can reduce ROS production by
dissipating the proton gradient, thereby affecting cellular
metabolism and energy balance (11,12).
Previous studies have suggested that UCP2 may play a critical role
in regulating macrophage phenotypic transitions and may be involved
in coordinating macrophage glycolysis, OXPHOS and ROS generation
(13-15).
However, the exact mechanisms by which UCP2 regulates macrophage
polarization, glycolysis and inflammation remain unclear.
The NF-κB signaling pathway is an important
regulator of immune and inflammatory responses, cell survival and
proliferation (16,17). Its activation can increase the
expression of glycolytic enzymes such as hexokinase 2 (HK2),
phosphofructokinase (PFK) and lactate dehydrogenase A (LDHA),
thereby promoting glycolysis (18,19).
In addition, research has shown that NF-κB signaling mediated
inflammatory responses can inhibit peroxisome
proliferator-activated receptor γ coactivator 1-α (PGC-1α),
potentially leading to reduced activity of nuclear respiratory
factor 1 (NRF1), a critical factor in maintaining mitochondrial
homeostasis, which in turn may result in decreased mitochondrial
biogenesis and impaired oxidative metabolism (20,21).
The present study aimed to elucidate the role of
UCP2 in modulating macrophage polarization by examining its effects
on M1/M2 homeostasis, glycolysis, mitochondrial function, cytokine
and chemokine secretion and glycolytic enzyme expression. These
findings may provide new insights into the molecular mechanisms
underlying UCP2-mediated macrophage polarization and explore the
potential therapeutic implications of targeting UCP2 in
inflammatory and immune-mediated diseases.
Materials and methods
Cell culture and macrophage
differentiation
Peripheral blood mononuclear cells from six healthy
volunteers from the People's Hospital of Ningxia Hui Autonomous
Region (approval number 2022319) were isolated by density gradient
centrifugation and CD14+ magnetic bead isolation (Thermo
Fisher Scientific, Inc.) following the guidelines of the
Declaration of Helsinki. Isolated monocytes were cultured in RPMI
1640 medium (Gibco; Thermo Fisher Scientific, Inc.), supplemented
with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.)
and 50 ng/ml macrophage colony-stimulating factor (Invitrogen;
Thermo Fisher Scientific, Inc.). The cells were incubated at 37˚C
in a humidified atmosphere with 5% CO2 for seven days to
induce differentiation into M0 macrophages.
Macrophage polarization
Post differentiation, for M1 macrophage
polarization, M0 macrophages were treated with a combination of 100
ng/ml LPS (MilliporeSigma) and 20 ng/ml IFN-γ (MilliporeSigma) for
48 h at 37˚C. The M0 macrophages were also treated with 20 ng/ml
IL-4 (MilliporeSigma) for 48 h at 37˚C to achieve M2 polarization.
The treatment medium was replaced every 24 h to maintain effective
concentrations of the cytokines. After polarization, cells were
analyzed by flow cytometry to confirm successful polarization, as
described below.
Lentivirus production and
infection
Lentivirus for UCP2 overexpression and knockdown was
produced using a 3rd generation lentiviral generation system in
293T cells. For transfection, 10 µg lentiviral plasmid was used in
combination with the lentiviral packaging plasmid psPAX2 and the
envelope plasmid pMD2.G at a ratio of 4:3:3, respectively. The
cells were co-transfected with the appropriate vector, either the
UCP2 cDNA cloned into a pLVX-IRES-ZsGreen1 vector for
overexpression (ov-UCP2) (Shanghai GenePharma Co., Ltd.) or short
hairpin RNA (shRNA) for UCP2 knockdown (cat. no. EHU007971;
MilliporeSigma), the sequence of which is: Sense,
5'-GAGACAUUGGCCUGAAAUCGUCUUCAAGAGAGACGAUUUCAGGCCAAUGUCU-3' and
antisense,
5'-GCUAAAGUCCGGUUACAGAUCUCUCUUGAAGAUCUGUAACCGGACUUUAGC-3',
integrated into a PLKO.1 vector. As a negative control for both
overexpression (ov-NC) and knockdown (sh-NC) of UCP2, cells were
co-transfected with an empty pLVX-IRES-ZsGreen1 or PLKO.1 vector.
Transfections were performed using Lipofectamine 3000 (Invitrogen;
Thermo Fisher Scientific, Inc.), according to the manufacturer's
instructions, along with the lentivirus packaging plasmids pMD2.G
and psPAX2. Lentivirus-containing supernatants were harvested 48 to
72 h after transfection and centrifuged at 100,000 x g for 2
h at 4˚C. Lentiviral particles were resuspended with PBS and added
to the medium at a multiplicity of infection of 50, supplemented
with 4 µg/ml of polybrene (Beyotime Institute of Biotechnology) to
enhance the efficiency of infection. After a subsequent incubation
of 48 h, cells were subjected to puromycin (Beyotime Institute of
Biotechnology) selection at a concentration of 2 µg/ml for 48 h. A
lower concentration (1 µg/ml) of puromycin was used for
maintenance. The cells were then incubated for a further 72 h under
normal conditions before being used for subsequent experimental
analyses.
Flow cytometry assay
Using flow cytometry, ROS levels and macrophage
surface markers were examined. CD68-phycoerythrin (PE) (dilution,
1:500; cat. no. 12-0689-41), CD86-allophycocyanin (dilution, 1:500;
cat. no. 17-0869-42) and CD206-PE (dilution, 1:500; cat. no.
12-2069-42) antibodies (Invitrogen; Thermo Fisher Scientific, Inc.)
were used to label the cells, which were then examined using a BD
FACSCanto II flow cytometer (BD Biosciences). Cells were exposed to
the fluorescent probe 2',7'-dichlorodihydrofluorescein diacetate
(MilliporeSigma) before being examined by flow cytometry (BD
FACSDiva software v9.0; BD Biosciences) to detect ROS. The flow
cytometry assay was repeated three times.
Enzyme-linked immunosorbent assay
(ELISA)
Culture supernatants were collected, and the levels
of TNF-α (cat. no. SEKH-0047), IL-1β (cat. no. SEKH-0002), IL-6
(cat. no. SEKH-0013), IL-10 (cat. no. SEKH-0018), TGF-β (cat. no.
SEKH-0316), C-X-C motif chemokine ligand (CXCL)2 (cat. no.
SEKH-0066), CXCL9 (cat. no. SEKH-0245), CXCL10 (cat. no.
SEKH-0070), C-C motif chemokine ligand (CCL)17 (cat. no.
SEKH-0315), CCL22 (cat. no. SEKH-0239) and CCL24 (cat. no.
SEKH-0063) were quantified using commercial ELISA kits (Beijing
Solarbio Science & Technology Co., Ltd.) according to the
manufacturer's instructions. ELISA assay was repeated three
times.
Assay of cellular ATP levels
An ATP detection kit (cat. no. BC0305; Beijing
Solarbio Science & Technology Co., Ltd.) was used according to
the manufacturer's instructions. Sample preparation included the
use of lysis buffer (Beyotime Institute of Biotechnology) for cell
and tissue samples to ensure proper cell lysis at 4˚C. After
centrifugation at 800 x g for 10 min at 4˚C, the supernatants were
collected and mixed with the ATP detection working solution
prepared. The chemiluminescence generated by the reaction was
measured using an enzyme-labeled instrument (Thermo Fisher
Scientific, Inc.) to determine the concentration of ATP in the
samples. The assay was repeated three times.
Reverse transcription quantitative PCR
(RT-qPCR)
TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) was used to extract total RNA from
macrophages according to the manufacturer's instructions. A
NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Inc.)
was used to measure the RNA concentration and purity for 5 sec at
21˚C. For cDNA synthesis, the SuperScript IV First-Strand Synthesis
System (Invitrogen; Thermo Fisher Scientific, Inc.) was used
according to the manufacturer's protocol. This comprises
SuperScript IV reverse transcriptase, the accompanying 5X SSIV
Buffer, 10 mM dNTP mix, and both oligo(dT)20 and random hexamer
primers. The Applied Biosystems®7500 Real-Time PCR
equipment (Thermo Fisher Scientific, Inc.) was utilized for the
RT-qPCR analysis using the SYBR® Premix Ex Taq™ II kit
(Takara Bio, Inc.). The PCR conditions were as follows: 95˚C for 32
sec, followed by 40 cycles of 95˚C for 6 sec and 62˚C for 32 sec.
The 2-ΔΔCq method (22)
was used to calculate the levels of UCP2, HK2, PFK, LDHA, PGC-1 and
NRF1 expression after normalizing them to the housekeeping gene
GAPDH. The sequences of each primer utilized in this investigation
were created by Shanghai GenePharma Co., Ltd., and they are all
presented in Table I. RT-qPCR
assay was repeated three times.
 | Table IPrimer sequences for reverse
transcription-quantitative PCR. |
Table I
Primer sequences for reverse
transcription-quantitative PCR.
| Primers | Sequences
(5'-3') |
|---|
| HK2 F |
GCTTGCCTACTTCTTCACG |
| HK2 R |
TTTCTCCATCTCCTTGCG |
| PFK F |
ACAGAAGCCTTGGTCTAACAC |
| PFK R |
GGAGAGTTGGAGGAATCAGTAG |
| LDHA F |
TTCAGCCCGATTCCGTTAC |
| LDHA R |
AGACACCAGCAACATTCATTCC |
| PGC-1α F |
CACCAGCCAACACTCAGCTA |
| PGC-1α R |
GTGTGAGGAGGGTCATCGTT |
| NRF1 F |
AAACGGCCTCATGTATTTGAGT |
| NRF1 R |
TAACGTGGCTCGAAGTTTCCG |
| UCP2 F |
TCTGATGCCCAGAGAAGGGA |
| UCP2 R |
AACTCTGCCGGAATAGGCAC |
| GAPDH F |
GCTCATTTGCAGGGGGGAG |
| GAPDH R |
GTTGGTGGTGCAGGAGGCA |
Western blotting
Cells were lysed with lysis buffer (Beyotime
Institute of Biotechnology), and protein concentrations in the
extracts were determined using a bicinchoninic acid protein assay
kit (Beyotime Institute of Biotechnology) according to the
manufacturer's instructions. Equal quantities of protein (20
µg/lane) were separated by SDS-PAGE (10%) and transferred onto
nitrocellulose membranes (Cytiva) after macrophages were lysed
using RIPA buffer (Beyotime Institute of Biotechnology). The
membranes were treated with primary antibodies for an overnight
period at 4˚C after being blocked with 5% BSA in Tris-buffered
saline containing 0.1% Tween-20 for 1 h at 21˚C. The primary
antibodies utilized in this investigation were all from Abcam and
were anti-UCP2 (dilution, 1:1,000; cat. no. ab97931), anti-p65
(dilution, 1:1,000; cat. no. ab16502), anti-phospho-p65 (dilution,
1:2,000; cat. no. ab86299), and anti-GAPDH (dilution, 1:2,000; cat.
no. ab181602). Membranes were cleaned with 10% tris-buffered
saline-0.1% Tween-20 (v/v; Beijing Solarbio Science &
Technology Co., Ltd.) before being incubated for 1 h at 21˚C with
the Goat Anti-Rabbit IgG (HRP) secondary antibody (1:5,000; cat.
no. ab205718; Abcam). SuperSignal West Pico PLUS Chemiluminescent
Substrate (Thermo Fisher Scientific, Inc.) was used to see protein
bands, and a ChemiDoc XRS+ Imaging System (Bio-Rad Laboratories,
Inc.) was used to capture images of bands. ImageJ (version 1.48;
National Institutes of Health) densitometric analysis was performed
to analyze the bands. Western blotting was repeated three
times.
Metabolic assays
The extracellular acidification rate and
O2 consumption rate was assessed using a Seahorse XF96
Extracellular Flux Analyzer (Agilent Technologies, Inc.) in
accordance with the manufacturer's instructions to examine
glycolysis and oxidative phosphorylation in macrophages. Sequential
injections of glucose, oligomycin and 2-deoxyglucose (2-DG) were
used to evaluate the function of the glycolytic system of the
cells, while sequential injections of oligomycin, carbonyl
cyanide-4-(trifluoromethoxy)phenylhydrazone, and a combination of
antimycin A and rotenone were used to assess the function of the
mitochondrial system. The metabolic assay was repeated three
times.
Statistical analysis
Data from at least three separate experiments are
presented as mean ± standard deviation. One-way ANOVA analysis of
variance was used for statistical comparisons, and Tukey's multiple
comparison post hoc test was performed after that. P<0.05 was
considered to indicate a statistically significant difference.
GraphPad Prism 9.0 software (GraphPad Software, Inc.; Dotmatics)
was used for all statistical analyses.
Results
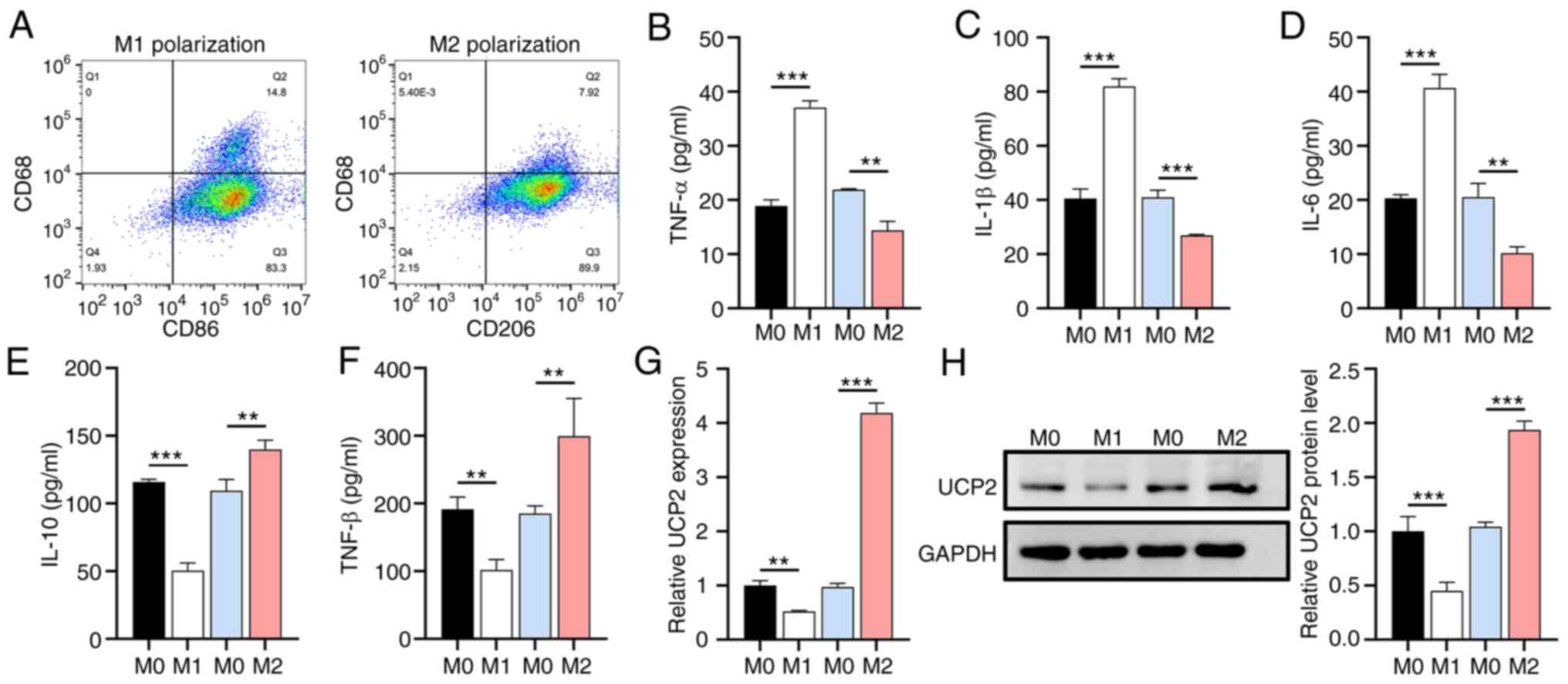
M1 polarization suppresses UCP2
expression
To confirm the successful polarization of primary
human M0 macrophages into M1 and M2 macrophages, subtype-specific
surface markers and cytokines were analyzed. The flow cytometry
results showed that LPS and IFN-γ induction increased the M1
macrophage population (CD68+CD86+), whereas
IL-4 induction increased the M2 macrophage population
(CD68+CD206+) (Fig. 1A). The ELISA results demonstrated
that, under the impetus of LPS and IFN-γ, M0 macrophages
facilitated an elevation in proinflammatory cytokines TNF-α, IL-1β,
and IL-6 secretion by 96.12, 102.39 and 100.49% respectively,
whilst experiencing a reduction of 34.25, 34.20 and 50.32%
correspondingly under the influence of IL-4 (Fig. 1B-D). Contrariwise, the
anti-inflammatory cytokines IL-10 and TGF-β diminished by 56.49 and
46.73%, respectively, under the stimulus of LPS and IFN-γ, yet they
exhibited an increment of 27.90 and 61.70%, respectively, under the
inducement of IL-4 (Fig. 1E and
F). Subsequent analysis of M1 or
M2 macrophage polarization on endogenous expression levels of UCP2
using RT-qPCR and western blotting revealed a significant increase
in UCP2 expression in M2 macrophages and a decrease in M1
macrophages (Fig. 1G and H), suggesting a regulatory role for UCP2
in macrophage polarization.
UCP2 overexpression decreases
glycolysis and ATP synthesis and promotes OXPHOS and ROS
To elucidate the metabolic effects of UCP2 on
macrophages, all macrophage subtypes were infected with lentivirus
expressing UCP2 to generate primary human macrophages
overexpressing UCP2. Flow cytometry results indicated that,
compared with the M0 ov-NC groups, there was an increase in the
proportion of CD86+ cells in the M1 ov-NC group, while
the proportion of CD206+ cells was decreased. By
contrast, the M2 ov-NC group exhibited the opposite trend compared
with the M1 ov-NC group. Furthermore, following UCP2
overexpression, compared with the M0, M1 and M2 ov-NC groups, the
proportions of CD86+ cells in the M0, M1 and M2 groups,
respectively, were significantly reduced, while the proportions of
CD206+ cells were increased. This suggested that UCP2
increased the proportion of M2 macrophages (CD206+
cells) and decreased the proportion of M1 macrophages
(CD86+ cells), regardless of their original subtype
(Fig. 2A). M1 macrophages
exhibited increased ROS levels and glycolytic rates and decreased
OXPHOS levels compared with M0, whereas M2 macrophages showed the
opposite trend (Fig. 2B-D).
Furthermore, UCP2 overexpression significantly reduced glycolytic
rates and increased OXPHOS levels in all macrophage subtypes,
accompanied by a decrease in ATP synthesis (Fig. 2E), expression of key glycolytic
enzymes such as HK2, PFK and LDHA (Fig. 2F-H), and an increase in expression
of mitochondrial biogenesis markers PGC-1α and NRF1 (Fig. 2I and J).
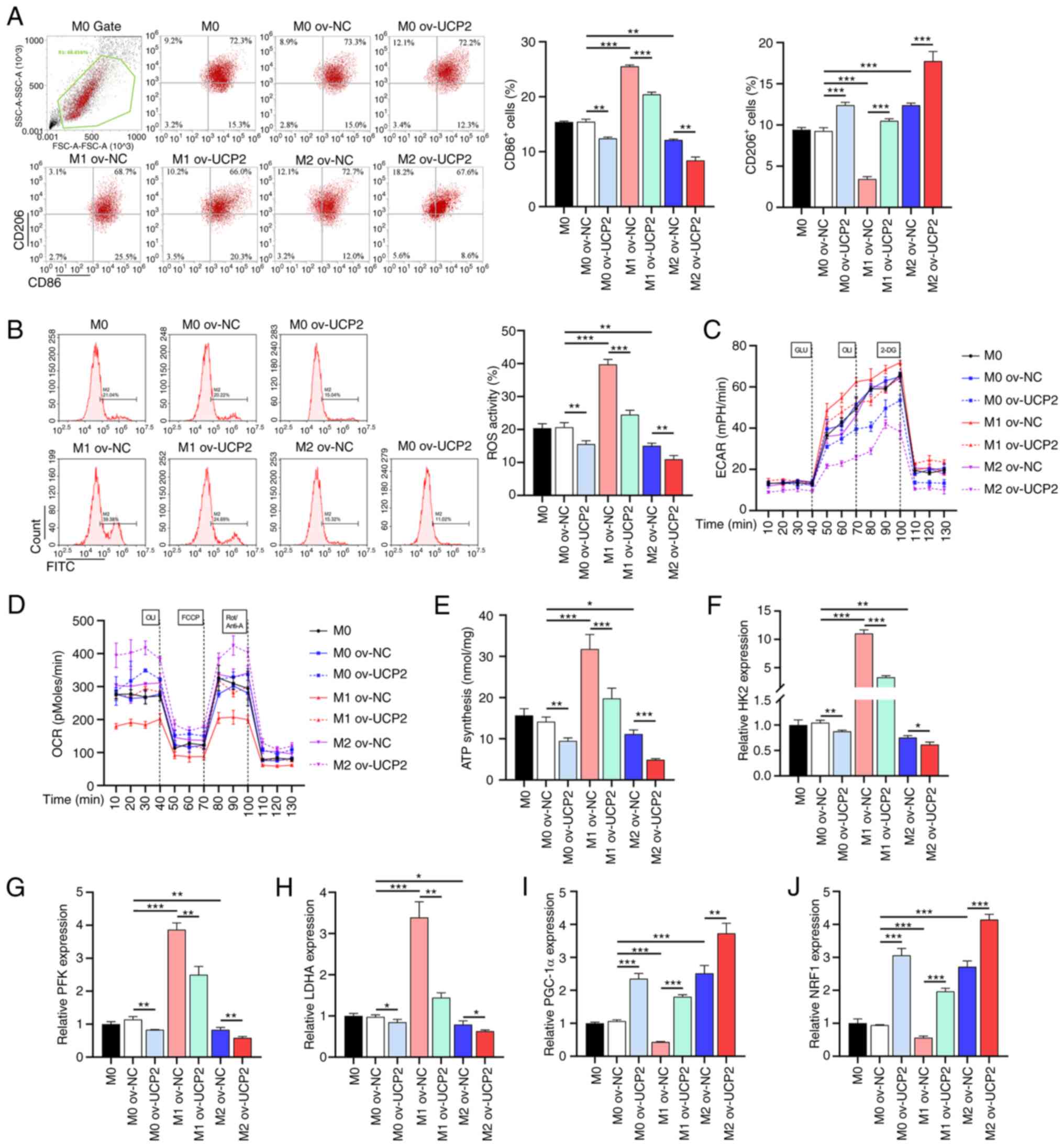 | Figure 2UCP2 overexpression alters macrophage
metabolism and polarization. (A) Flow cytometry analysis of
M1-to-M2 macrophage conversion with UCP2 overexpression (n=3). (B)
ROS levels, (C) ECAR and (D) OCR in macrophages with UCP2
overexpression (n=3). (E) ATP synthesis in macrophages with UCP2
overexpression (n=3). RT-qPCR analysis of key glycolytic enzymes
(F) HK2, (G) PFK and (H) LDHA in UCP2-overexpressing macrophages
(n=3). RT-qPCR analysis of mitochondrial biogenesis markers (I)
PGC-1α and (J) NRF1 in UCP2-overexpressing macrophages (n=3).
*P<0.05, **P<0.01,
***P<0.001. RT-qPCR, reverse transcription
quantitative PCR; ROS, reactive oxygen species; OXPHOS, oxidative
phosphorylation; UCP2, uncoupling protein 2; HK2, hexokinase 2;
PFK, phosphofructokinase; LDHA, lactate dehydrogenase A; PGC-1α,
peroxisome proliferator-activated receptor g coactivator 1-α; NRF1,
nuclear respiratory factor 1; ov-, overexpression; NC, negative
control; ECAR, extracellular acidification rate; OCR, O2
consumption rate. |
UCP2 overexpression regulates
macrophage cytokine and chemokine secretion profiles through
inhibition of NF-κB signaling
Given the impact of UCP2 on macrophage polarization,
the effect of UCP2 regulation on cytokine and chemokine secretion
was further investigated. UCP2 overexpression in all macrophage
subtypes resulted in a significant decrease in pro-inflammatory
chemokines (CXCL2, CXCL9 and CXCL10) and cytokines (TNF-α, IL-1β
and IL-6) (Fig. 3A-F) compared
with the M2 ov-NC group, whereas anti-inflammatory chemokines
(CCL17, CCL22, and CCL24) and cytokines (IL-10 and TGF-β) were
significantly increased (Fig.
3G-K). The RT-qPCR results revealed an elevation in UCP2
expression by 279.45% within the ov-UCP2 group of M0 macrophages
when compared against the ov-NC group. Additionally, M1
polarization contributed to a 48.94% decrease in UCP2 expression
compared with the M0 ov-NC group, while M2 polarization stimulated
an increase of 305.85%. Within M1 macrophages, UCP2 expression in
the UCP2 group saw an increase of 323.21% as compared with the
ov-NC group. In M2 macrophages, the UCP2 group reported an increase
of UCP2 expression by 53.76% when compared with the ov-NC group
(Fig. 3L). Western blotting
illuminated notable fluctuations in protein levels within M0
macrophages, relative to the ov-NC group. Specifically, UCP2
overexpression resulted in a 47.72% increase in UCP2 protein levels
and a 14.40% decrease in the p-p65/p65 ratio, respectively,
compared with the M0 ov-NC group. Furthermore, M1 polarization
instigated a 43.36% reduction and a 36.53% increase in UCP2
expression and the p-p65/p65 ratio, respectively. Conversely, M2
polarization induced a 27.81% increase in UCP2 expression and a
decrease of 17.44% in the p-p65/p65 ratio. Within the ambit of M1
macrophages, the UCP2 group exhibited a 92.21% increase in UCP2
protein levels and a drop of 10.07% in the p-p65/p65 ratio compared
with the ov-NC group. Similarly, within M2 macrophages, the UCP2
group exhibited a 36.27% increase in UCP2 and a reduction of 16.64%
in the p-p65/p65 ratio compared with the ov-NC group (Fig. 3M).
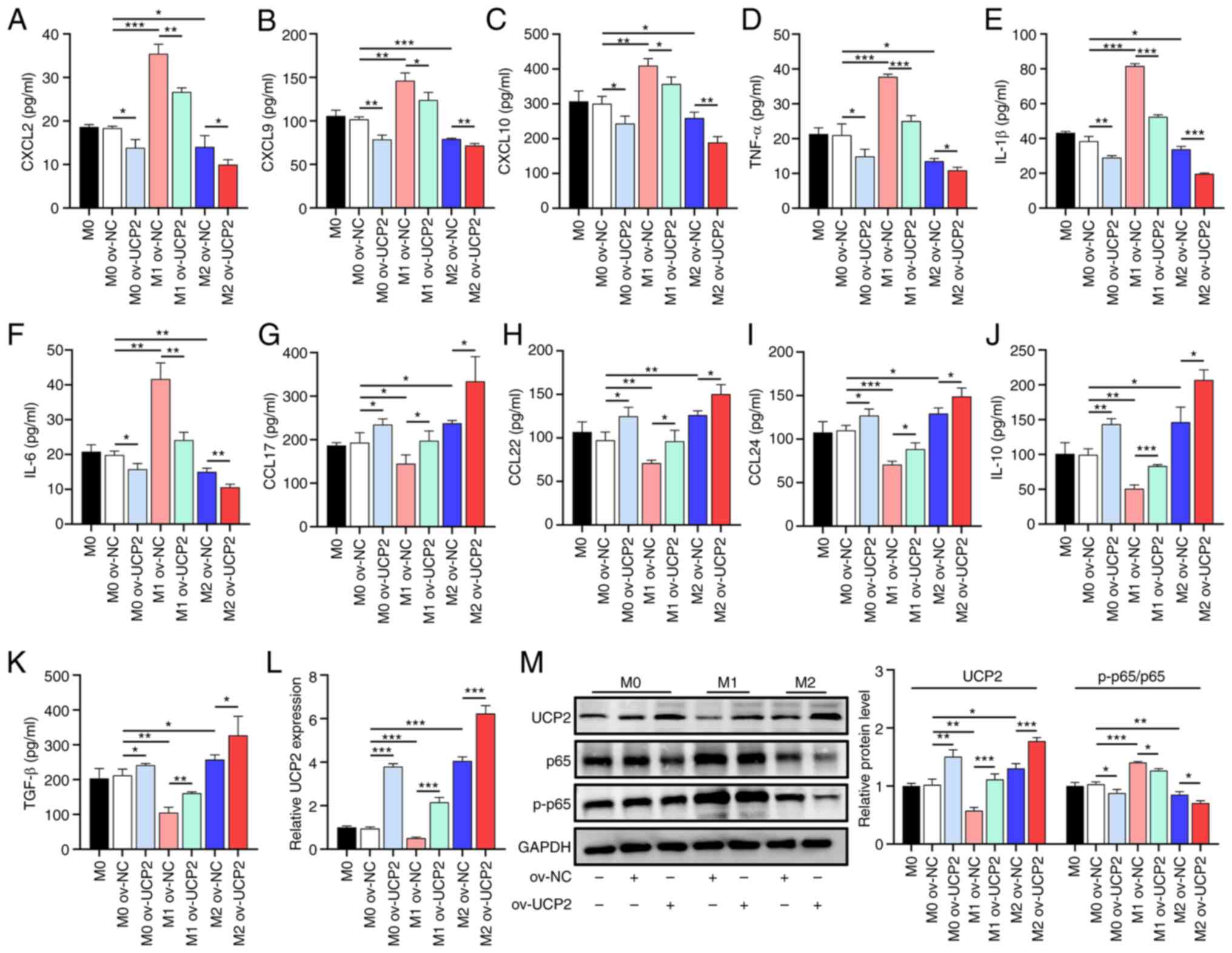 | Figure 3UCP2 overexpression modulates
macrophage cytokine and chemokine secretion through NF-κB signaling
inhibition. ELISA results showing changes in pro-inflammatory (A)
CXCL2, (B) CXCL9, (C) CXCL10, (D) TNF-α, (E) IL-1β (F) and IL-6 and
anti-inflammatory (G) CCL17, (H) CCL22, (I) CCL24, (J) IL-10 and
(K) TGF-β chemokine and cytokine secretion with UCP2 overexpression
(n=3). (L) RT-qPCR analysis of UCP2 mRNA expression in macrophages
with UCP2 overexpression (n=3). (M) Western blotting of UCP2
protein levels and the ratio of p-p65/p65 in macrophages with UCP2
overexpression (n=3). *P<0.05,
**P<0.01, ***P<0.001. RT-qPCR, reverse
transcription quantitative polymerase chain reaction; CXCL, C-X-C
motif chemokine ligand; CCL, C-C motif chemokine ligand; p65, NF-κB
p65 subunit; UCP2, uncoupling protein 2; ov-, overexpression; NC,
negative control; p-, phosphorylated. |
UCP2 knockdown exacerbates
inflammation and oxidative stress by promoting macrophage
glycolysis through lentivirus-mediated approach
To further confirm the role of UCP2 in macrophage
inflammation and oxidative stress, a lentivirus-mediated UCP2
knockdown experiment in M2 macrophages was performed. The RT-qPCR
results confirmed that sh-UCP2 vector could effectively inhibit the
expression of UCP2 in M0 macrophages with an inhibition efficiency
of 82.94% (Fig. 4A). UCP2
knockdown was observed to increase the rate of glycolysis and to
decrease the level of OXPHOS in M2 macrophages. These effects were
significantly attenuated in the presence of the glycolysis
inhibitor 2-DG (Fig. 4B and
C). In addition, increased
glycolysis caused by UCP2 knockdown resulted in increased
production of pro-inflammatory cytokines (Fig. 4D-H) and pro-inflammatory chemokines
(Fig. 4I-N), but was attenuated by
the effect of 2-DG. In addition, sh-UCP2 was observed to contribute
to a 66.06% increase in ROS accumulation, but an 18.46% decrease in
response to 2-DG (Fig. 4O). These
results demonstrated the effect of UCP2 on M1/M2 rebalancing, and
thus on reducing inflammation and oxidative stress, is mediated
through an effect on macrophage glycolysis.
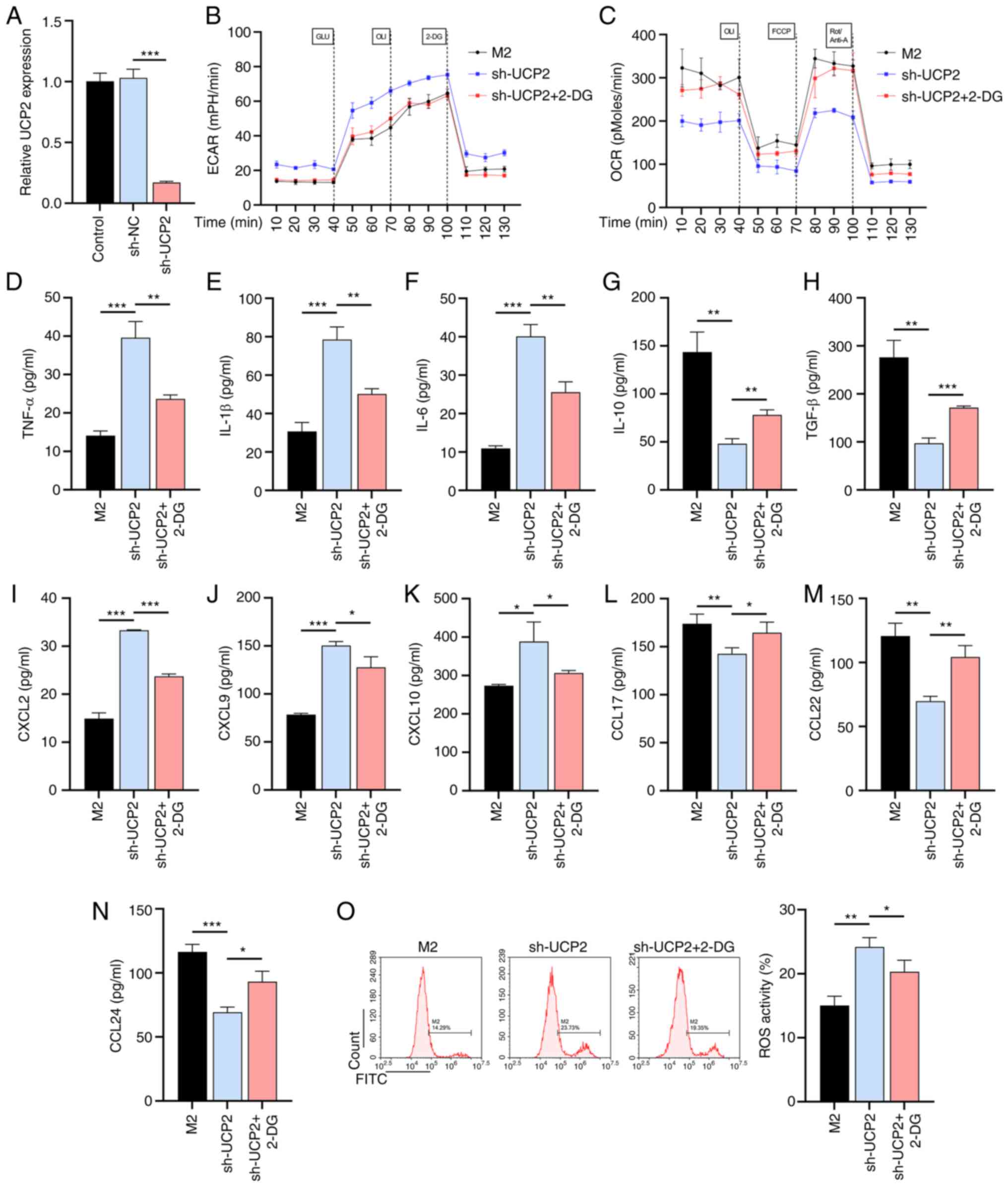 | Figure 4UCP2 knockdown exacerbates
inflammation and oxidative stress by promoting macrophage
glycolysis. (A) RT-qPCR to verify the validity of the sh-UCP2
vector (n=3). (B) Glycolysis and (C) OXPHOS levels in M2
macrophages with UCP2 knockdown and 2-DG treatment (n=3). ELISA
results of pro-inflammatory cytokines (D) TNF-α, (E) IL-1β, (F)
IL-6, (G) IL-10 and (H) TGF-β production in UCP2-knockdown M2
macrophages with or without 2-DG treatment (n=3). ELISA results of
production of pro-inflammatory chemokines (I) CXCL2, (J) CXCL9, (K)
CXCL10, (L) CCL17, (M) CCL22 and (N) CCL24 in UCP2-knockdown M2
macrophages with or without 2-DG treatment (n=3). (O) ROS
production in UCP2-knockdown M2 macrophages with or without 2-DG
treatment (n=3). *P<0.05, **P<0.01,
***P<0.001. 2-DG, 2-deoxy-D-glucose; OXPHOS,
oxidative phosphorylation; ROS, reactive oxygen species; IL,
interleukin; TNF-α, tumor necrosis factor alpha; CXCL, C-X-C motif
chemokine ligand; CCL, C-C motif chemokine ligand; UCP2, uncoupling
protein 2; sh-, short hairpin; ECAR, extracellular acidification
rate; OCR, O2 consumption rate. |
Discussion
Metabolic abnormalities are important factors
contributing to various diseases, in particular the M1/M2
macrophage imbalance they cause, which is a major obstacle to
improving disease outcomes as it can lead to severe inflammatory
responses (23). The present study
investigated the interplay between macrophage polarization,
glycolysis, metabolic reprogramming, ROS and inflammation, focusing
on the role of UCP2 in coordinating these processes.
Consistent with previous research (24), the present study observed that M1
macrophages identified by specific markers (CD86+) had
increased secretion of pro-inflammatory cytokines (TNF-α, IL-1β and
IL-6) and decreased UCP2 expression, whereas M2 macrophages
(CD206+) had increased secretion of anti-inflammatory
cytokines (IL-10 and TGF-β) and upregulated UCP2 expression. This
suggested that UCP2 may be a key regulator of macrophage phenotypic
shifts. As a member of the mitochondrial anion carrier protein
family, UCP2 is located in the inner mitochondrial membrane and can
reduce ROS production by dissipating the proton gradient (25). Under the influence of
lentivirus-mediated UCP2 overexpression, all macrophage subtypes
exhibited a shift from M1 to M2 polarization, accompanied by
reduced ROS production, possibly due to decreased glycolysis
(26). The present study confirmed
that UCP2 overexpression resulted in decreased glycolysis and key
glycolytic enzymes (including HK2, PFK and LDHA) in macrophages
with increased OXPHOS. M1 macrophage polarization is induced by LPS
and IFN-γ, which are activators of glycolysis (27), suggesting that UCP2 acts as an
inhibitor of glycolysis (28). ATP
is generated by OXPHOS, the central system of cellular metabolism
(29). UCP2 allows protons to leak
back across the membrane without passing through ATP synthase,
thereby reducing the efficiency of ATP production (30). The present study revealed that
UCP2-induced metabolic reprogramming was accompanied by a slight
decrease in ATP synthesis and upregulation of mitochondrial
biogenesis markers (PGC-1α and NRF1), suggesting that UCP2 may act
as a metabolic regulator in macrophages. As reported by Rius-Perez
et al (31),
inflammation-mediated inhibition of PGC-1α may lead to reduced
activity of NRF1, a critical factor in maintaining mitochondrial
homeostasis.
Notably, the present study showed that UCP2
overexpression resulted in decreased secretion of pro-inflammatory
cytokines and chemokines and increased secretion of
anti-inflammatory cytokines. This regulation may be achieved
through ROS-mediated modulation of signaling pathways involved in
cytokine and chemokine expression, such as NF-κB (32,33),
and by influencing the metabolic processes controlling their
production (34). The current
study subsequently confirmed an increase in p65 protein and
phosphorylation in M1 macrophages and a decrease in M2 macrophages,
with UCP2 promoting reduced p65 protein levels and
dephosphorylation in both subtypes.
To further investigate the role of UCP2 in
macrophage inflammation and oxidative stress, the present study
used a lentivirus-mediated approach to downregulate UCP2 expression
in M2 macrophages. Consistent with previous research (35,36),
UCP2 downregulation resulted in increased glycolysis rates,
decreased OXPHOS levels and increased production of
pro-inflammatory cytokines and chemokines, along with increased ROS
generation. These results further supported the importance of UCP2
in macrophage function. The observed effects were attenuated by
treatment with the glycolysis inhibitor 2-DG, suggesting that the
effects of UCP2 on macrophage polarization and inflammation are
mediated through the regulation of glycolysis. The findings might
have implications for cancer therapy, where tumor-associated
macrophages predominantly exhibit an M2 phenotype that supports
tumor progression (37). In such
circumstances, downregulation of UCP2 might tilt the balance back
toward an M1 state, possibly helping to combat tumor growth
(38). In general, overexpression
of UCP2 reduced glycolysis and increased OXPHOS levels in
macrophages. This led to activation of the NF-κB signaling pathway,
which ultimately triggered inflammation (Fig. 5). Therefore, understanding the
molecular mechanisms underpinning UCP2's regulation of macrophage
polarization, could lead to the development of small molecule
inhibitors or activators, which may serve as novel therapeutic
agents.
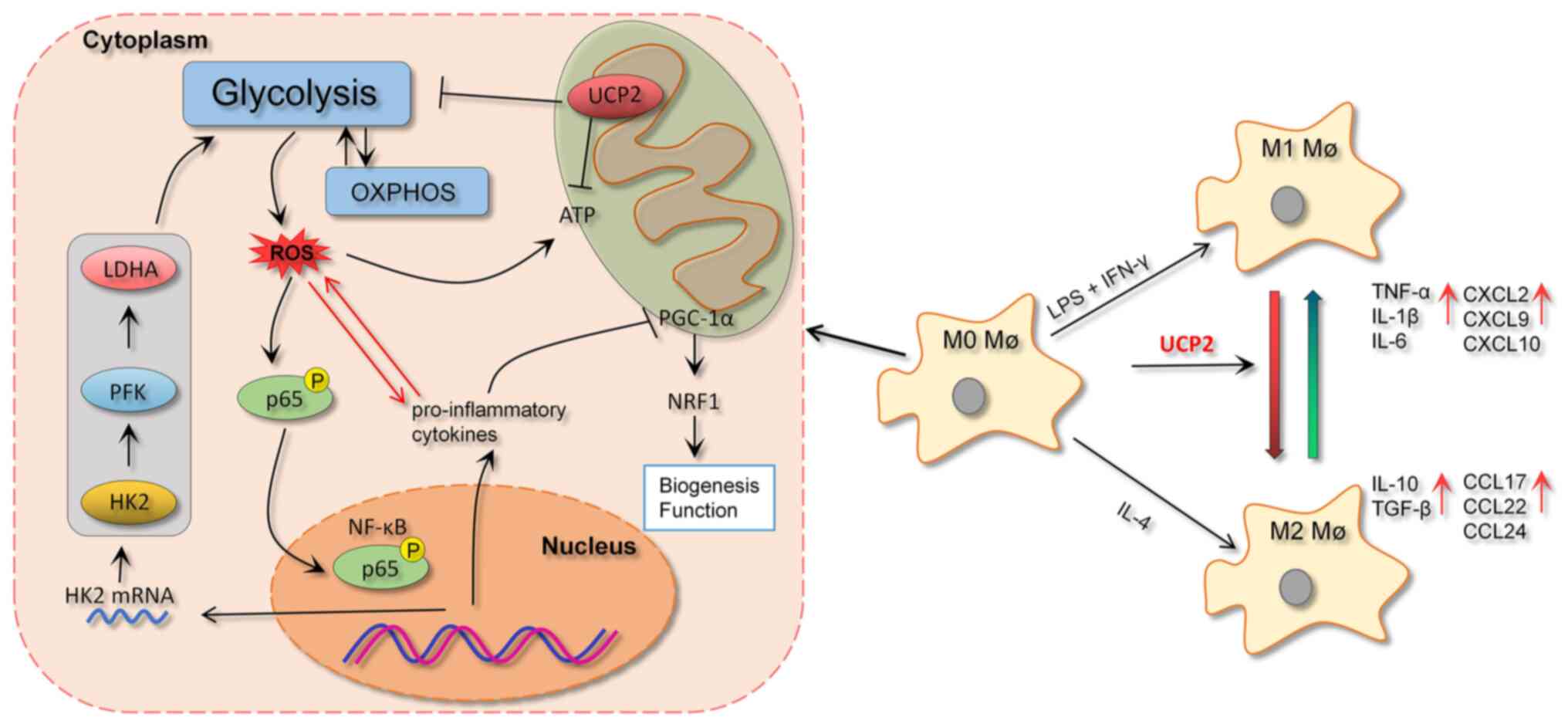 | Figure 5UCP2, by promoting the transformation
of M1 macrophages to M2 macrophages, effectively suppresses
glycolysis within the macrophages. This action subsequently leads
to a reduction in the production of ROS. Furthermore, by minimizing
the nuclear translocation of p65, UCP2 also diminishes the
secretion of pro-inflammatory cytokines and chemokines, thus
serving a pivotal role in modulating inflammation. OXPHOS,
oxidative phosphorylation; ROS, reactive oxygen species; IL,
interleukin; TNF-α, tumor necrosis factor alpha; CXCL, C-X-C motif
chemokine ligand; CCL, C-C motif chemokine ligand; UCP2, uncoupling
protein 2; HK2, hexokinase 2; PFK, phosphofructokinase; LDHA,
lactate dehydrogenase A; PGC-1α, peroxisome proliferator-activated
receptor g coactivator 1-α; NRF1, nuclear respiratory factor 1; P,
phosphorylated; LPS, lipopolysaccharide; IFN-γ, interferon γ; Mø,
macrophage. |
The present study had several limitations. First,
the mechanisms of UCP2 in specific metabolic diseases were not
investigated. Second, numerous non-coding RNAs, such as miRNAs, may
play critical regulatory roles in UCP2 expression, which were not
investigated in this study. Third, due to the lack of commercially
available products that can directly stimulate glycolysis, this
study lacked additional rescue experiments to further confirm the
inhibitory effect of UCP2 overexpression on glycolysis in M2
macrophages. Fourth, further in vivo studies are needed to
further determine the physiological relevance of the present
findings, as the complexity of the in vivo system may affect
macrophage behavior in ways not reflected in vitro. Finally,
the present study recognized the limitation of not employing
single-cell metabolic analyses such as the scMetabolism tool
(39). Such an approach could
potentially provide additional insights into the metabolic
heterogeneity of individual macrophages and further corroborate our
findings. Future research could investigate potential
pharmacological interventions, such as drugs that enhance or
inhibit UCP2 expression or activity. These limitations represent
key areas for future investigation and potential therapeutic
targets.
In conclusion, the current study demonstrated the
important role of UCP2 in regulating macrophage polarization,
metabolism, inflammation and oxidative stress through its effects
on glycolysis. These findings provide valuable insights into the
molecular mechanisms of UCP2 function in macrophages and highlight
the potential for developing novel therapeutic strategies targeting
UCP2 for macrophage-driven inflammatory and metabolic diseases. By
elucidating the importance of UCP2 in macrophage function and its
potential implications in immune regulation and inflammation, the
present research lays the groundwork for further investigation and
potential clinical applications.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LL and DZ designed the experiments. QJ and TM
performed the experiments and drafted the manuscript. LL, XM and YY
collected and analyzed the data. LL and DZ confirm the authenticity
of all the raw data. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
the People's Hospital of Ningxia Hui Autonomous Region (approval
no. 2022316). Written informed consent was obtained from
patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Mahroum N, Alghory A, Kiyak Z, Alwani A,
Seida R, Alrais M and Shoenfeld Y: Ferritin-from iron, through
inflammation and autoimmunity, to COVID-19. J Autoimmun.
126(102778)2022.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Li C, Xu MM, Wang K, Adler AJ, Vella AT
and Zhou B: Macrophage polarization and meta-inflammation. Transl
Res. 191:29–44. 2018.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Ross EA, Devitt A and Johnson JR:
Macrophages: The good, the bad, and the gluttony. Front Immunol.
12(708186)2021.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Viola A, Munari F, Sanchez-Rodriguez R,
Scolaro T and Castegna A: The metabolic signature of macrophage
responses. Front Immunol. 10(1462)2019.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Zhu L, Zhao Q, Yang T, Ding W and Zhao Y:
Cellular metabolism and macrophage functional polarization. Int Rev
Immunol. 34:82–100. 2015.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Shapouri-Moghaddam A, Mohammadian S,
Vazini H, Taghadosi M, Esmaeili SA, Mardani F, Seifi B, Mohammadi
A, Afshari JT and Sahebkar A: Macrophage plasticity, polarization,
and function in health and disease. J Cell Physiol. 233:6425–6440.
2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Wang S, Liu G, Li Y and Pan Y: Metabolic
reprogramming induces macrophage polarization in the tumor
microenvironment. Front Immunol. 13(840029)2022.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Marrocco A and Ortiz LA: Role of metabolic
reprogramming in pro-inflammatory cytokine secretion from LPS or
silica-activated macrophages. Front Immunol.
13(936167)2022.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Peace CG and O'Neill LA: The role of
itaconate in host defense and inflammation. J Clin Invest.
132(e148548)2022.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Mouton AJ, Li X, Hall ME and Hall JE:
Obesity, hypertension, and cardiac dysfunction: novel roles of
immunometabolism in macrophage activation and inflammation. Circ
Res. 126:789–806. 2020.PubMed/NCBI View Article : Google Scholar
|
|
11
|
van Dierendonck X, Sancerni T,
Alves-Guerra MC and Stienstra R: The role of uncoupling protein 2
in macrophages and its impact on obesity-induced adipose tissue
inflammation and insulin resistance. J Biol Chem. 295:17535–17548.
2020.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Steen KA, Xu H and Bernlohr DA: FABP4/aP2
regulates macrophage redox signaling and inflammasome activation
via control of UCP2. Mol Cell Biol. 37:e00282–e00216.
2017.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Esteves P, Pecqueur C, Ransy C, Esnous C,
Lenoir V, Bouillaud F, Bulteau AL, Lombes A, Prip-Buus C, Ricquier
D and Alves-Guerra MC: Mitochondrial retrograde signaling mediated
by UCP2 inhibits cancer cell proliferation and tumorigenesis.
Cancer Res. 74:3971–3982. 2014.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Emre Y, Hurtaud C, Karaca M, Nubel T,
Zavala F and Ricquier D: Role of uncoupling protein UCP2 in
cell-mediated immunity: How macrophage-mediated insulitis is
accelerated in a model of autoimmune diabetes. Proc Natl Acad Sci
USA. 104:19085–19090. 2007.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Nishio K, Qiao S and Yamashita H:
Characterization of the differential expression of uncoupling
protein 2 and ROS production in differentiated mouse
macrophage-cells (Mm1) and the progenitor cells (M1). J Mol Histol.
36:35–44. 2005.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Hayden MS and Ghosh S: NF-κB in
immunobiology. Cell Res. 21:223–244. 2011.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Lawrence T: The nuclear factor NF-kappaB
pathway in inflammation. Cold Spring Harb Perspect Biol.
1(a001651)2009.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Wang S, Yu H, Gao J, Chen J, He P, Zhong
H, Tan X, Staines KA, Macrae VE, Fu X, et al: PALMD regulates
aortic valve calcification via altered glycolysis and
NF-κB-mediated inflammation. J Biol Chem.
298(101887)2022.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Kooshki L, Mahdavi P, Fakhri S, Akkol EK
and Khan H: Targeting lactate metabolism and glycolytic pathways in
the tumor microenvironment by natural products: A promising
strategy in combating cancer. Biofactors. 48:359–383.
2022.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Liu H, Zhu S, Han W, Cai Y and Liu C: DMEP
induces mitochondrial damage regulated by inhibiting Nrf2 and
SIRT1/PGC-1alpha signaling pathways in HepG2 cells. Ecotoxicol
Environ Saf. 221(112449)2021.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Zhang Z, Zhang X, Meng L, Gong M, Li J,
Shi W, Qiu J, Yang Y, Zhao J, Suo Y, et al: Pioglitazone inhibits
diabetes-induced atrial mitochondrial oxidative stress and improves
mitochondrial biogenesis, dynamics, and function through the
PPAR-γ/PGC-1α signaling pathway. Front Pharmacol.
12(658362)2021.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Kim MJ, Lee H, Chanda D, Thoudam T, Kang
HJ, Harris RA and Lee IK: The role of pyruvate metabolism in
mitochondrial quality control and inflammation. Mol Cells.
46:259–267. 2023.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Tsai CF, Chen GW, Chen YC, Shen CK, Lu DY,
Yang LY, Chen JH and Yeh WL: Regulatory effects of quercetin on
M1/M2 macrophage polarization and oxidative/antioxidative balance.
Nutrients. 14(67)2021.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Forte M, Bianchi F, Cotugno M, Marchitti
S, Stanzione R, Maglione V, Sciarretta S, Valenti V, Carnevale R,
Versaci F, et al: An interplay between UCP2 and ROS protects cells
from high-salt-induced injury through autophagy stimulation. Cell
Death Dis. 12(919)2021.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Yu J, Shi L, Lin W, Lu B and Zhao Y: UCP2
promotes proliferation and chemoresistance through regulating the
NF-κB/β-catenin axis and mitochondrial ROS in gallbladder cancer.
Biochem Pharmacol. 172(113745)2020.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Meng L, Lu C, Wu B, Lan C, Mo L, Chen C,
Wang X, Zhang N, Lan L, Wang Q, et al: Taurine antagonizes
macrophages M1 polarization by mitophagy-glycolysis switch blockage
via dragging SAM-PP2Ac transmethylation. Front Immunol.
12(648913)2021.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Ji R, Chen W, Wang Y, Gong F, Huang S,
Zhong M, Liu Z, Chen Y, Ma L, Yang Z, et al: The warburg effect
promotes mitochondrial injury regulated by uncoupling protein-2 in
septic acute kidney injury. Shock. 55:640–648. 2021.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Ahmed ST, Craven L, Russell OM, Turnbull
DM and Vincent AE: Diagnosis and treatment of mitochondrial
myopathies. Neurotherapeutics. 15:943–953. 2018.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Jia JJ, Zhang X, Ge CR and Jois M: The
polymorphisms of UCP2 and UCP3 genes associated with fat
metabolism, obesity and diabetes. Obes Rev. 10:519–526.
2009.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Rius-Perez S, Torres-Cuevas I, Millan I,
Ortega AL and Perez S: PGC-1 α, inflammation, and oxidative stress:
An integrative view in metabolism. Oxid Med Cell Longev.
2020(1452696)2020.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Morgan MJ and Liu ZG: Crosstalk of
reactive oxygen species and NF-κB signaling. Cell Res. 21:103–115.
2011.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Korbecki J, Kojder K, Barczak K, Siminska
D, Gutowska I, Chlubek D and Baranowska-Bosiacka I: Hypoxia alters
the expression of CC chemokines and CC chemokine receptors in a
tumor-a literature review. Int J Mol Sci. 21(5647)2020.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Morrissey SM, Zhang F, Ding C,
Montoya-Durango DE, Hu X, Yang C, Wang Z, Yuan F, Fox M, Zhang HG,
et al: Tumor-derived exosomes drive immunosuppressive macrophages
in a pre-metastatic niche through glycolytic dominant metabolic
reprogramming. Cell Metab. 33:2040–2058 e2010. 2021.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Hu S, Feng J, Wang M, Wufuer R, Liu K,
Zhang Z and Zhang Y: Nrf1 is an indispensable redox-determining
factor for mitochondrial homeostasis by integrating
multi-hierarchical regulatory networks. Redox Biol.
57(102470)2022.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Ohkouchi S, Block GJ, Katsha AM, Kanehira
M, Ebina M, Kikuchi T, Saijo Y, Nukiwa T and Prockop DJ:
Mesenchymal stromal cells protect cancer cells from ROS-induced
apoptosis and enhance the Warburg effect by secreting STC1. Mol
Ther. 20:417–423. 2012.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Li D, Zhang Q, Li L, Chen K, Yang J, Dixit
D, Gimple RC, Ci S, Lu C, Hu L, et al: beta2-microglobulin
maintains glioblastoma stem cells and induces M2-like polarization
of tumor-associated macrophages. Cancer Res. 82:3321–3334.
2022.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Luby A and Alves-Guerra MC: UCP2 as a
cancer target through energy metabolism and oxidative stress
control. Int J Mol Sci. 23(15077)2022.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Wu Y, Yang S, Ma J, Chen Z, Song G, Rao D,
Cheng Y, Huang S, Liu Y, Jiang S, et al: Spatiotemporal immune
landscape of colorectal cancer liver metastasis at single-cell
level. Cancer Discov. 12:134–153. 2022.PubMed/NCBI View Article : Google Scholar
|