Introduction
Venous thromboembolism (VTE) and its complications
are common causes of death worldwide; 30% of patients with VTE die
<30 days after diagnosis (1).
Additionally, one-third of the remaining 70% of patients experience
relapse within 10 years and have long-term complications, including
deep vein thrombosis post-thrombotic and post-pulmonary embolism
syndrome, chronic thromboembolic pulmonary hypertension in
pulmonary embolism and death (2,3).
Pro- and antithrombotic factors exist in the human body and an
imbalance between these, such as decreases in the levels of
antithrombotic factors and/or increases in the levels of
prothrombotic factors, may serve key roles in thrombosis (4). However, the mechanisms underlying
this imbalance remain unclear. To predict and diagnose VTE, it is
necessary to understand the mechanisms underlying its pathogenesis
and to construct accurate and effective diagnosis and prediction
models.
Vascular endothelial cells (VECs) serve key roles in
VTE pathogenesis. Previous studies have reported that VTE causes
VEC injury, the accumulation of inflammatory substances, blood
hypercoagulation and aggravated thromboembolism, which lead to
severe health consequences (5-7).
Damage to VECs may disrupt the integrity of blood vessels and lead
to bleeding, which affects therapeutic efficacy in VTE (8). In addition to damage caused by the
accumulation of inflammatory substances, lactate (LA) may serve an
important role in VEC damage (9,10).
LA metabolism is regulated by LA metabolism-related molecules
(LMRMs). VECs take up glucose from peripheral blood and, via the
catalytic activity of various enzymes, such as glycogen and
pyruvate kinase, convert glucose to tricarboxylic acid. This serves
as an energy source for cells and pyruvate can also be converted to
LA by lactic acid dehydrogenase A (LDHA) (11-14).
However, LA does not provide energy to cells but must first be
converted by LDHB to pyruvate, which can then be used as a
substrate in the tricarboxylic acid cycle (15). LA in VECs is primarily excreted
through monocarboxylic acid transporter (MCT) 1(12) and Stabenow et al (16) reported that an increase in LA
levels promotes VEC aging, which may be associated with increased
conversion of pyruvate to LA by LDHA. Franczyk et al
(17) suggested that abnormal LA
metabolism may be associated with VTE. Additionally, VTE can lead
to glucose and oxygen deficiency in local VECs, which can cause
changes in levels of metabolic substances, including LA, which
serves an important role in VTE (17,18).
However, the roles of LA metabolism and LMRMs in the mechanisms
underlying VTE remain unclear.
Therefore, the present study screened differentially
expressed LMRMs (DE-LMRMs) in patients with VTE and constructed a
disease prediction model for VTE to the end of clarifying the roles
of LA metabolism and LMRMs in VTE pathogenesis. Further, the
oxygen-glucose deprivation (OGD) model of VECs and an in
vitro model of thromboembolism (19,20)
were used to verify the expression of the DE-LMRMs. The present
study may provide a novel future target for VTE diagnosis and
treatment.
Materials and methods
Data acquisition, preprocessing and
screening of DE-LMRMs
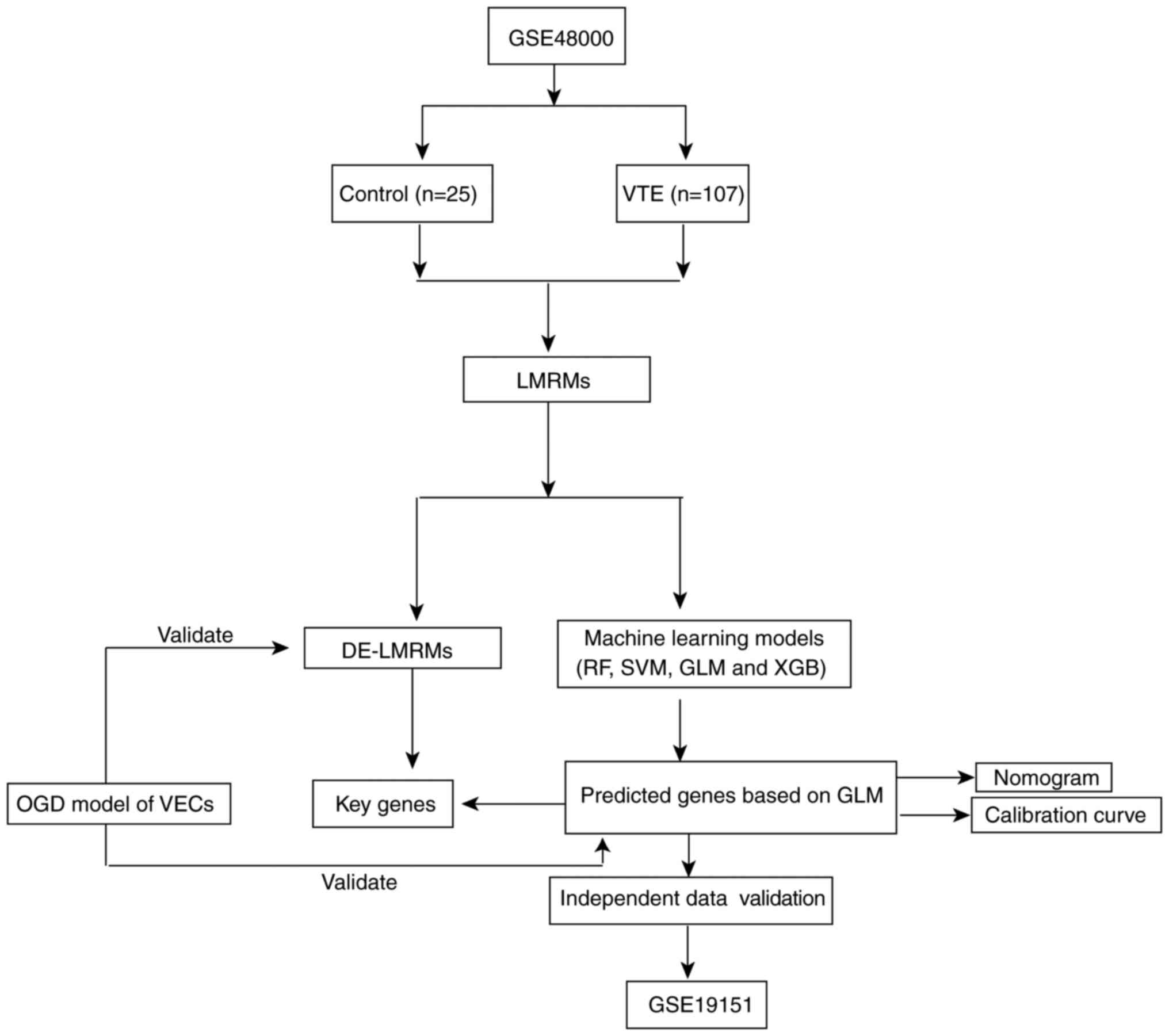
The datasets were downloaded from the Gene
Expression Omnibus, which is an open-access database with gene
chips and high-throughput sequencing datasets (ncbi.nlm.nih.gov/geo). The downloaded datasets
included GSE48000 (accession no. GPL10558) and GSE19151 (accession
no. GPL571; Fig. 1). GSE48000
training set comprised 25 peripheral blood samples from healthy
individuals (control) and 107 peripheral blood samples from
patients with VTE (VTE group). The validation set GSE19151
comprised 63 peripheral blood samples from healthy individuals and
70 samples from patients with VTE. The datasets were pre-processed
using Perl programming language. Furthermore, LMRMs were obtained
from the MsigDB database (version 7.0; gsea-msigdb.Org/gsea/msigdb/). As described by Li
et al (21), the ‘limma’ R
package was used to screen DE-LMRMs from the GSE48000 dataset and
the ‘ggpubr’ and ‘pheatmap’ R packages were used to generate box
plots and heatmaps, respectively. Finally, ‘corrplot’ R package was
used for correlation analysis(Pearson's coefficient) of the
DE-LMRMs. Perl programming language (version 5.30.0.1, URL:
https://www.perl.org/) and ‘RCircos’ package were
used to determine the location of the LMRMs on chromosomes.) and R
programming language (version 4.1.3, URL: https://www.r-project.org/) was employed to conduct
this study.
Construction of predictive model using
machine learning methods
Based on screened LMRMs, the ‘caret’ R package was
used to build machine learning models, namely, the random forest
(RF), support vector machine (SVM) and generalized linear model
(GLM) and eXtreme Gradient Boosting (XGB) (21). The ‘DALEX’ R package was used to
interpret the four models and generate the residual distribution of
each model in the test set. The ‘pROC’ R package was used to
visualize the area under the receiver operating characteristic
(ROC) curve (AUC). Next, the optimal machine learning model was
identified. The model with highest AUC was considered the optimal
model. The top five variables were considered key predictor genes
associated with VTE. ROC curve analysis of the GSE48000 dataset was
performed to verify the diagnostic value of the model.
Construction and validation of the
nomogram model
Using the ‘rms’ R package, nomogram models were
constructed to evaluate VTE clusters. Each predictor had a
corresponding score and ‘total score’ represented the sum of the
individual scores of all the predictors. Calibration curve and
decision curve analysis (DCA) were used to estimate the predictive
power of the nomogram model.
Establishment of the OGD model
The OGD model is an in vitro thromboembolic
model (19,20). Human umbilical (HU)VECs were
purchased from Shanghai Anwei Biotechnology Co., Ltd. (cat. no.
HUVEC-SV40T). Mycoplasma testing was performed on HUVECs and the
cell line was authenticated using immunofluorescence. Cells were
cultured in RPMI-1640 medium (cat. no. 22400097) with 10% fetal
bovine serum (cat. no. 12483020) and 1% penicillin-streptomycin
(cat. no. 15140122; all Gibco; Thermo Fisher Scientific, Inc.) in a
humidified incubator at 37˚C with 5% CO2 and 95% air for
7 days. Cells (1.5x105/ml) were transferred to a 6-well
plate in glucose-free DMEM (cat. no. 119660-25; Gibco; Thermo
Fisher Scientific, Inc.) and cultured for 24 h in a humidified
incubator at 37˚C with 5% CO2 and 95% N2.
Cell morphology was observed using a light microscope
(magnification, x200; cat. no. D-35578; Leica Microsystems
GmbH).
MTT analysis of VEC survival rate
VEC survival was analyzed as described by Li et
al (22) using the MTT kit
(cat. no. AR1156; Wuhan Boster Biotechnology, Ltd.). MTT staining
solution (10 µl) was added to each well and incubated at 37˚C for 4
h. Next, 100 µl formazan as added to each well and samples were
incubated at 37˚C for 4 h. Absorbance was measured at 570 nm using
an enzyme-labeled instrument.
Detection of LA levels
Total protein was extracted from VECs using a
protein extraction kit (cat. no. SD-001/SN-002; Invent
Biotechnologies Inc.). BCA assay kit (cat. no. P0012S; Beyotime
Institute of Biotechnology) was used to evaluate total protein
concentration levels. LA content assay kit (cat. no. BC2235;
Beijing Solarbio Science & Technology Co., Ltd.) was used to
measure the LA levels, as previously described (23,24).
Western blotting analysis
Samples (20 µg protein/lane) were obtained as
aforementioned and separated using SDS-PAGE (12.5%) and transferred
onto nitrocellulose (NC) membranes (MilliporeSigma) using wet
transfer cell (Bio-Rad Mini-Protean 1658001, Bio-Rad Laboratories,
Inc.). NC membranes were blocked with 5% BSA blocking buffer (cat.
no. SW3015, Solarbio Science & Technology Co., Ltd.) for 1 h at
25˚C and incubated overnight with primary antibodies at 4˚C. The
primary antibodies were as follows: Anti-monocarboxylic acid
transporter (MCT)1 (cat. no. ab85021; 1:1,000; Abcam), anti-embigin
(EMB; cat. no. ab170927; 1:1,000: Abcam), anti-MCT2 (cat. no.
ab81262; 1:400; Abcam), anti-LDHB (cat. no. ab264358; 1:2,000;
Abcam), anti-solute carrier family 5 member 12 (SLC5A12; cat. no.
sc-515141; 1:800; Santa Cruz Biotechnology, Inc.), anti-SLC16A8
(cat. no. D262213; 1:1,000; Sangon Biotech Co., Ltd.) and
anti-GAPDH (cat. no. D110016; 1:10,000; Sangon Biotech Co., Ltd.).
NC membranes were incubated with horseradish peroxidase-conjugated
Affinipure goat anti-rabbit IgG (H+L) antibody (cat. no. SA00001-2;
1:8,000; Proteintech Group, Inc.) for 1 h at 25˚C. Further,
immunoreactive bands were visualized using the Omni-ECL™ Femto
Light Chemiluminescence kit (cat. no. SQ201; EpiZyme) and
ChemiScope6000 (Clinx) visualization system. Band intensities were
quantified used ImageJ software (version: 1.8.0, National
Institutes of Health). The total protein levels were normalized to
GAPDH.
Statistical analysis
All statistical analysis was performed using SPSS
software (version 25.0; IBM Corp.). Data are expressed as the mean
± standard deviation (n=6). Unpaired independent sample t-test was
performed to analyze differences between groups. P<0.05 was
considered to indicate a statistically significant difference.
Results
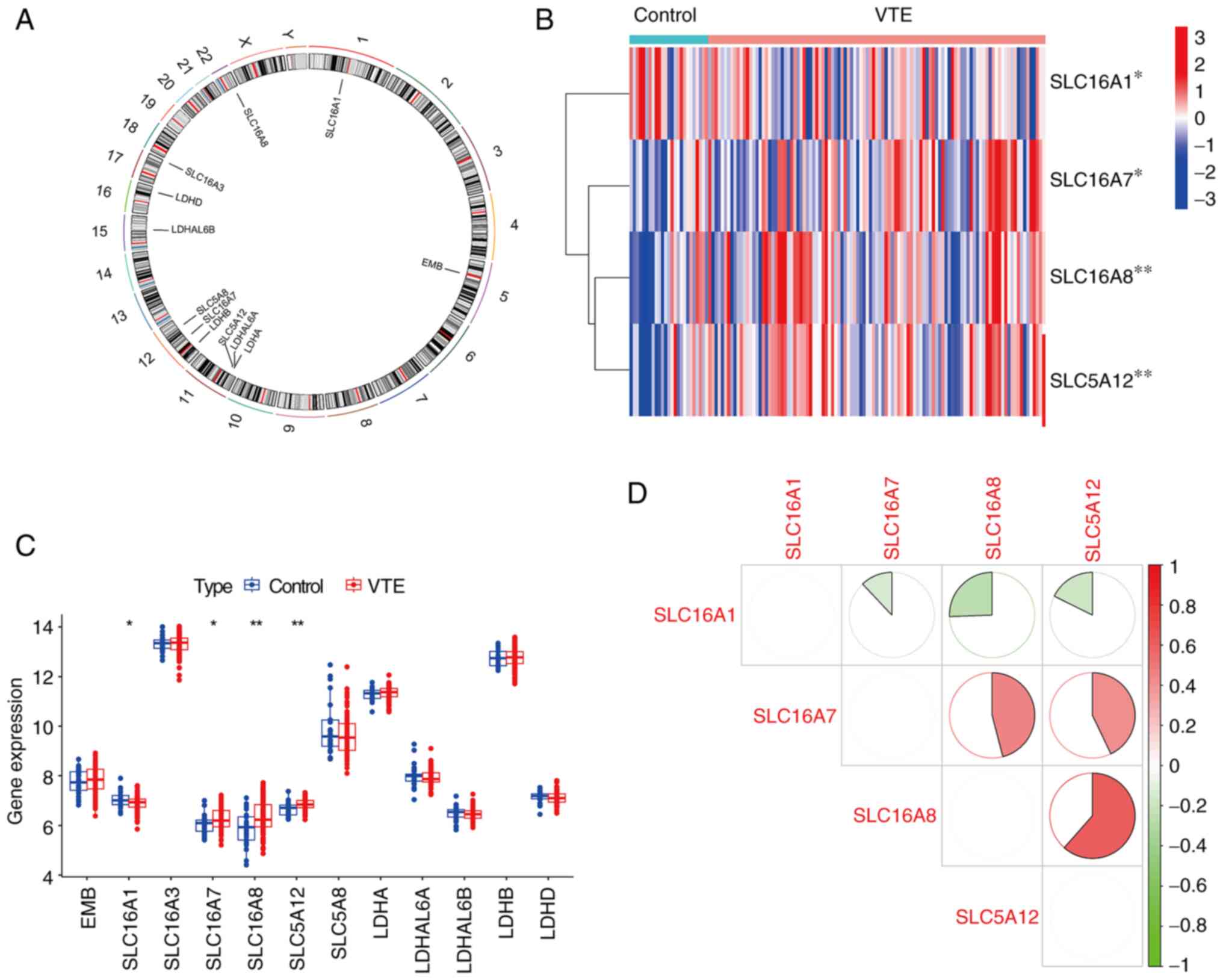
Identification of DE-LMRMs in VTE
The location of the LMRMs on chromosomes was
determined (Fig. 2A). The ‘limma’
package in R was used to screen DE genes from the dataset(GSE48000)
(21). Compared with the control,
four DE-LMRMs (SLC16A1, SLC16A7), SLC16A8 and SLC5A12) were
identified in the VTE group. Specifically, SLC16A1 demonstrated
decreased expression, whereas SLC16A7, SLC16A8 and SLC5A12
demonstrated increased expression in the VTE group compared with
the control group (Fig. 2B and
C). Correlation analysis was used
to analyze correlations between the four DE-LMRMs. Significant
positive correlations were found between SLC16A7 and SCL16A8,
SLC16A7 and SCL5A12, SLC16A8 and SCL5A12. Significant negative
correlations were found SLC16A1 and SLC16A7, SCL16A1 and SLC16A8,
SLC16A1 and SLC5A12 (Fig. 2D).
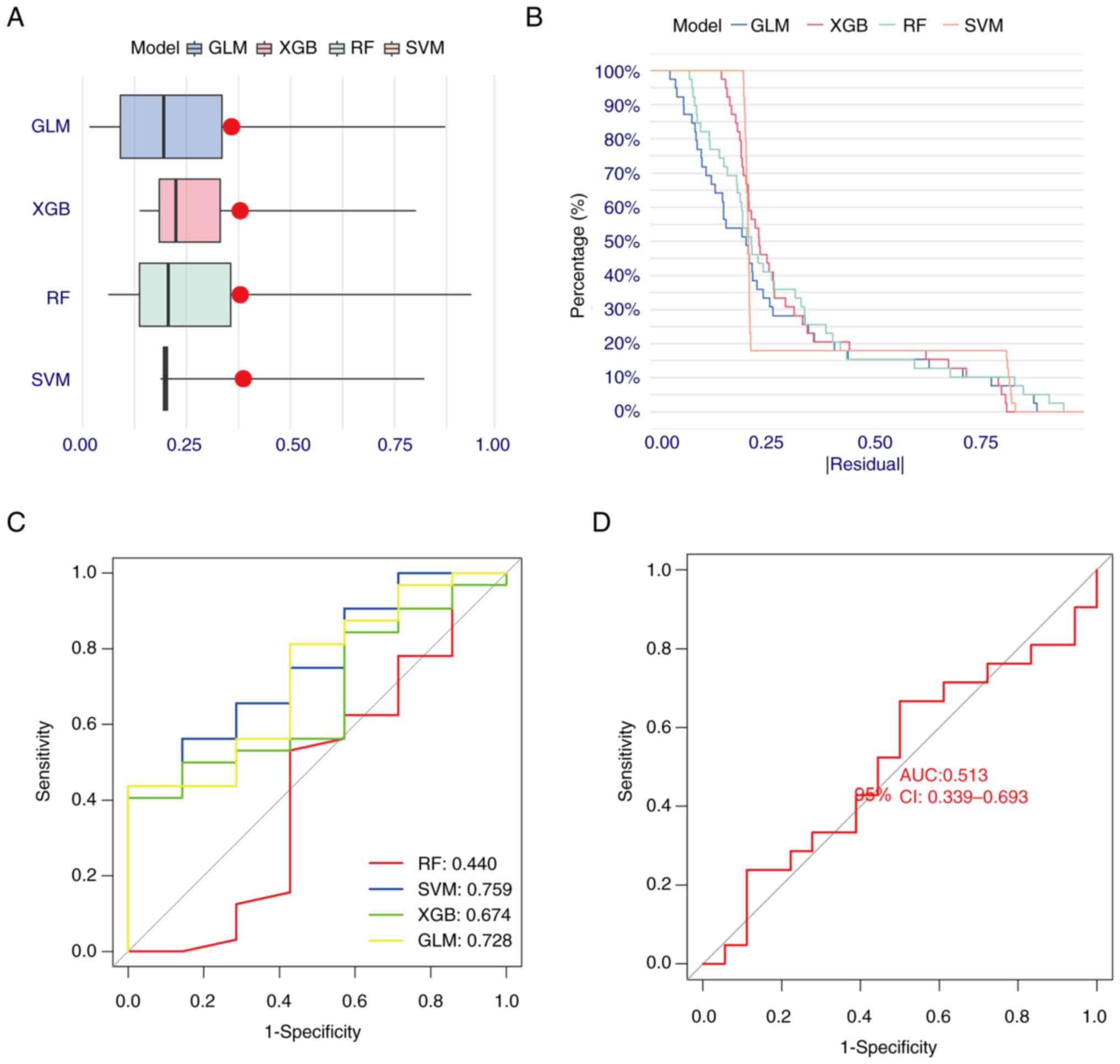
Construction and assessment of machine
learning models
A total of four machine learning models (RF, SVM,
GLM and XGB) were constructed based on expression levels of LMRMs.
GLM and XGB machine learning models demonstrated relatively low
residuals (Fig. 3A and B). Discriminative performances of the
four machine learning algorithms were evaluated by generating ROC
curves based on five-fold cross-validation in the GSE48000 training
dataset (Fig. 3C). The AUCs were
as follows: RF=0.440, SVM=0.759, XGB=0.674 and GLM=0.728. Based on
the residual and AUC values, the GLM machine learning model was
optimal with respect to distinguishing patients with VTE. The five
most important genes (EMB, LDHB, SLC16A1, SLC5A12 and SLC16A8)
determined by this model were selected as predictor genes for
further analysis. Finally, the GSE19151 dataset was used to verify
the accuracy of the machine learning model. In the GSE152532
dataset, the ROC curve for the GLM gene prediction model performed
well, with AUC=0.513 (Fig.
3D).
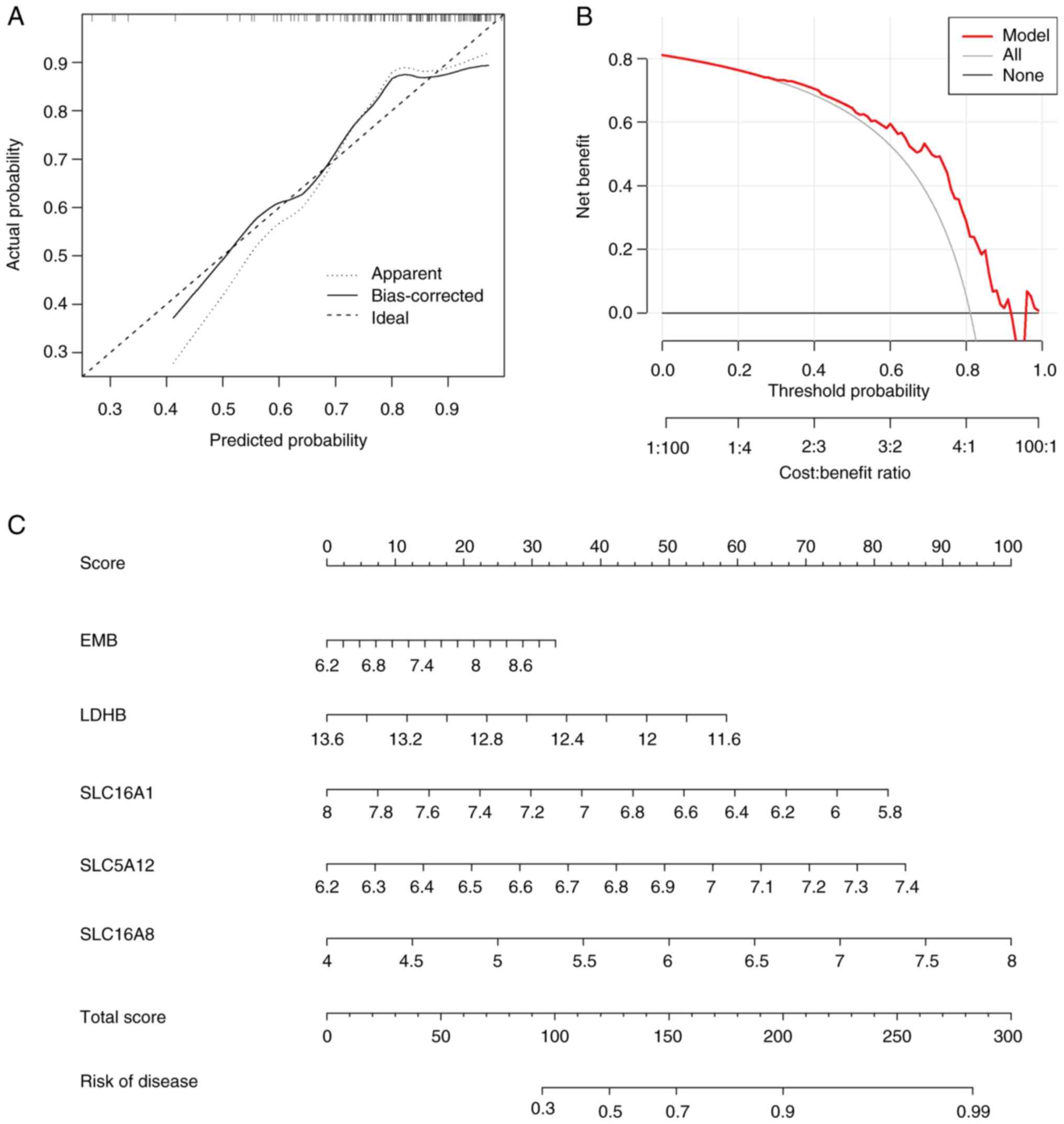
Construction of the nomogram
model
A nomogram integrates multiple prediction indicators
based on multi-factor regression analysis (27). A nomogram model was constructed to
estimate VTE risk (Fig. 4A). The
calibration curve and DCA were used to evaluate the prediction
efficiency of the nomogram model. Based on the calibration curve,
the error between the actual and predicted risks for the VTE
cluster was low (Fig. 4B). DCA
indicated that the nomogram was accurate (Fig. 4C). To facilitate the clinical use
of this diagnostic model, a nomogram was constructed. Based on the
actual measured values of expression levels of the five DE-LMRMs in
the blood of patients with VTE, it was possible to find these
DE-LMRMs on the corresponding scale in the nomogram and project to
the point scale on top to read the value for each variant. The
total number of points was obtained by summing the individual
points and the risk probability for a patient with VTE could be
estimated using the bottom scale by projecting the total.
Survival rate, LA levels and
expression of LMRMs in the OGD model
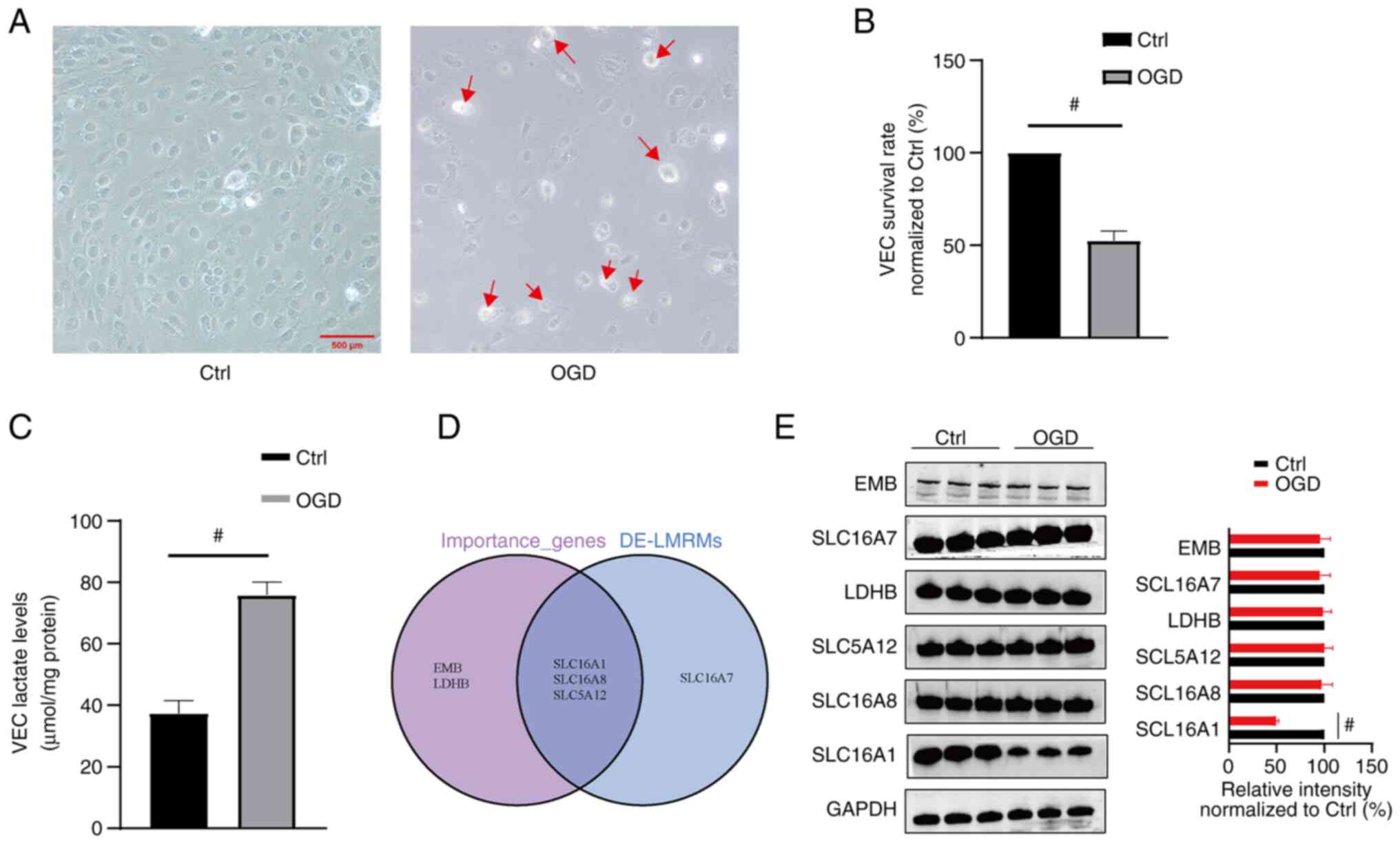
Compared with the control, the OGD model showed a
lower number of VECs and these VECs appeared swollen and ruptured
(Fig. 5A). MTT is a cell viability
assay that involves conversion of the water-soluble yellow dye MTT
to insoluble purple formazan by the action of mitochondrial
reductase (28). MTT assay
demonstrated a significant decrease in the survival rate of the
VECs in the OGD compared with the control. Compared with control
group, the survival rate significantly decreased to 52.49±5.17% in
the OGD group (Fig. 5B).
Furthermore, compared with the control, the OGD VECs demonstrated
significantly higher LA levels (37.38±4.18 vs. 75.90±4.21 µmol/mg
protein, respectively; Fig. 5C).
Based on the genes predicted using the GLM model and DE-LMRMs,
three key molecules (SLC16A1, SLC5A12 and SLC16A8) were identified
(Fig. 5D). Additionally, western
blotting to detect the protein expression levels of EMB, LDHB,
SLC16A1, SCL16A7, SLC5A12 and SLC16A8 in VECs demonstrated that the
protein expression of SLC16A1 significantly decreased to
49.69±2.55% in the OGD group (Fig.
5E), while the protein expression levels of the other five
molecules demonstrated no significant change (Fig. 5E).
Discussion
Although it has previously been reported that an
imbalance between prothrombotic and antithrombotic factors serves a
key role in VTE pathogenesis (4),
the associated mechanisms remain unclear. It has been suggested
that an imbalance in levels of several metabolic substances in the
blood may also be associated with VTE pathogenesis (17). Among these, LA serves an important
role in VEC damage (9,10). VECs obtain glucose from blood and
convert it to ATP to supply cells with energy (11-14).
VTE can cause ischemia and hypoxia of local blood vessels that can
result in glucose and oxygen deficiency in VECs, which may lead to
energy metabolism disorder. If these are not corrected, cell damage
can occur (22). It has also been
reported that in energy metabolism disorder, the accumulation of
large amounts of LA serves a key role in cell function and
structural damage (9).
Additionally, intracellular LA, which is primarily
regulated by LMRMs, is associated with LA production, consumption
and transport (11-14).
In the present study, LMRMs from the MsigDB database were screened
and DE-LMRMs were identified by comparing gene expression between
healthy individuals and patients with VTE. A total of four DE-LMRMs
were identified in the VTE group, with SLC16A1 demonstrating
decreased expression levels, whereas SLC16A7, SLC16A8 and SLC5A12
demonstrated increased expression. Reportedly, these DE-LMRMs are
primarily responsible for LA transport (12,26),
which suggests that abnormal LA transport may be involved in VTE
pathogenesis. Machine learning models have previously been used to
predict prevalence of a number of diseases, with lower error rates
and superior results compared with conventional logistic regression
(27). Machine learning models,
such as RF, SVM, GLM and XGB, demonstrate clinical relevance in
disease prediction (21). In the
present study, machine learning models based on the expression of
the LMRMs were constructed. After comparing AUC values
corresponding to the four models, GLM was identified as the optimal
model and its verification using external data indicated that it
demonstrated relatively good accuracy in predicting VTE. The five
predicted genes in GLM were EMB, LDHB, SLC16A1, SLC5A12 and
SLC16A8. DCA indicated that the nomogram could potentially provide
a basis for clinical decision-making EMB is a companion protein
located on the cell membrane that promotes localization of MCT1 to
the plasma membrane and alters transport of LA by MCT1(28). LDH is a homologous or
heterotetrametric enzyme with two subunits, LDHA and LDHB, which
are encoded by different genes and have different chromosomal
locations. LDHA is located on chromosome 11, while LDHB is
primarily located on chromosome 12 (13,29).
LDHA preferentially converts pyruvate to LA, whereas LDHB converts
LA to pyruvate (15,30,31).
The present study demonstrated that LDHB was an important gene for
predicting VTE, which suggested that intracellular LA conversion
may be involved in VTE pathogenesis.
VTE pathogenesis is a complex process and its
simulation using in vivo as well in vitro models
remains challenging. In vivo models of venous thrombosis are
primarily established via ferric chloride administration (32,33);
in in vitro models, in which cells are deprived of glucose
and oxygen, the cells are in a state of hypoxia and sugar
deficiency (19,20,34).
VTE also causes local blood vessel ischemia and hypoxia, which
results in ECs in these blood vessels being in a state of glucose
and oxygen deprivation. Hence, the OGD model only partially
simulates thromboembolism (19,20).
In the present study, OGD was used to verify the association
between LA, LMRMs and VTE. Light microscopy demonstrated a decrease
in number of VECs and edema in the OGD model compared with control
cells. A significant decrease in the survival rate of VECs in the
OGD group compared with the control was shown and the intracellular
LA levels increased significantly. This suggested that cell
survival was negatively associated with intracellular LA.
Additionally, more obvious cell damage and a lower cell survival
rate was observed at higher LA levels. According to the
GLM-predicted genes and DE-LMRMs, three molecules, SLC16A1, SLC16A7
and SLC16A8, were identified as key in VTE pathogenesis. Western
blotting was employed to detect the expression levels of the
proteins encoded by DE-LMRMs and GLM-predicted genes. The
expression of SLC16A1 was significantly decreased in the OGD group,
consistent with the prediction results. Further, MCT1, which is
encoded by SLC16A1, is primarily expressed in VECs (12) and decreased SLC16A1 expression may
limit excretion of intracellular LA, which results in an increase
in intracellular LA levels, cell acidification and impaired cell
function (35). The expression
levels of proteins encoded by EMB, LDHB, SCL16A7, SLC5A12 and
SLC16A8 did not change significantly in the OGD compared with the
control group. EMB, a companion protein of MCT1, primarily
regulates the expression of MCT1 on cell membrane (28). The expression of LDHB did not
change significantly, which suggested that LA conversion to
pyruvate was not affected in the OGD group. SLC16A7, which encodes
the MCT2 protein, is primarily expressed in the brain (36). SLC5A12, which encodes
sodium-coupled monocarboxylate transporter 2 (SMCT2), is primarily
expressed in the kidney (37) and
SLC16A8, which encodes MCT3, is primarily expressed in the retina
(38). The aforementioned reports
suggest that EMB, LDHB, SLC16A7, SLC5A12 and SLC16A8 do not serve
key roles in LA metabolism in VECs.
The present study had a number of limitations. The
in vitro OGD model cannot completely simulate VTE,
therefore, it is necessary to collect clinical samples and build an
in vivo model for further studies on VTE.
In the present study, it was demonstrated that LMRMs
may participate in VTE pathogenesis. Thus, a prediction model was
developed and showed a certain degree of accuracy. Model-predicted
results were verified using an in vitro model, which
confirmed that an increase in intracellular LA levels may be
associated with a decrease in SLC16A1 expression. In summary,
LMRMs, particularly SLC16A1, may serve key roles in VTE
pathogenesis and be a potential target for VTE diagnosis and
treatment.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by Guangxi Key Research
and Development Plan (grant. no. 2017AB45033)
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZQ wrote the manuscript, performed the experiments
and analyzed the data. JC performed cellular experiments. JZ and HL
contributed to the design of this study and involved in proof
reading and editing. QC contributed to acquisition of data and data
interpretation. HL and QC confirm the authenticity of all the raw
data. ZQ and QC provided the funding and supervised the study. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Battinelli EM, Murphy DL and Connors JM:
Venous thromboembolism overview. Hematol Oncol Clin North Am.
26:345–367, ix. 2012.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Bartholomew JR: Update on the management
of venous thromboembolism. Cleve Clin J Med. 84 (Suppl 3):39–46.
2017.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Klok FA, van der Hulle T, den Exter PL,
Lankeit M, Huisman MV and Konstantinides S: The post-PE syndrome: A
new concept for chronic complications of pulmonary embolism. Blood
Rev. 28:221–226. 2014.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Palta S, Saroa R and Palta A: Overview of
the coagulation system. Indian J Anaesth. 58:515–523.
2014.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Torres C, Matos R, Morais S, Campos M and
Lima M: Soluble endothelial cell molecules and circulating
endothelial cells in patients with venous thromboembolism. Blood
Coagul Fibrinolysis. 28:589–595. 2017.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Pilard M, Ollivier EL, Gourdou-Latyszenok
V, Couturaud F and Lemarié CA: Endothelial cell phenotype, a major
determinant of venous thrombo-inflammation. Front Cardiovasc Med.
9(864735)2022.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Poredos P and Jezovnik MK: The role of
inflammation in venous thromboembolism and the link between
arterial and venous thrombosis. Int Angiol. 26:306–311.
2007.PubMed/NCBI
|
|
8
|
Barsh GS, Molina-Ortiz P, Orban T, Martin
M, Habets A, Dequiedt F and Schurmans S: Rasa3 controls turnover of
endothelial cell adhesion and vascular lumen integrity by a
Rap1-dependent mechanism. PLoS Genet. 14(e1007195)2018.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Yang K, Fan M, Wang X, Xu J, Wang Y, Gill
PS, Ha T, Liu L, Hall JV, Williams DL and Li C: Lactate induces
vascular permeability via disruption of VE-cadherin in endothelial
cells during sepsis. Sci Adv. 8(eabm8965)2022.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Yang K, Holt M, Fan M, Lam V, Yang Y, Ha
T, Williams DL, Li C and Wang X: Cardiovascular dysfunction in
covid-19: Association between endothelial cell injury and lactate.
Front Immunol. 13(868679)2022.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Mamun AA, Hayashi H, Yamamura A, Nayeem MJ
and Sato M: Hypoxia induces the translocation of glucose
transporter 1 to the plasma membrane in vascular endothelial cells.
J Physiol Sci. 70(44)2020.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Shao H and Li S: A new perspective on HIV:
Effects of HIV on brain-heart axis. Front Cardiovasc Med.
10(1226782)2023.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Edwards YH, Povey S, LeVan KM, Driscoll
CE, Millan JL and Goldberg E: Locus determining the human
sperm-specific lactate dehydrogenase, LDHC, is syntenic with LDHA.
Dev Genet. 8:219–232. 1987.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Mdluli K, Booth MP, Brady RL and Rumsby G:
A preliminary account of the properties of recombinant human
glyoxylate reductase (GRHPR), LDHA and LDHB with glyoxylate, and
their potential roles in its metabolism. Biochim Biophys Acta.
1753:209–216. 2005.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Koukourakis MI, Giatromanolaki A, Sivridis
E, Bougioukas G, Didilis V, Gatter KC and Harris AL: Tumour and
Angiogenesis Research Group. Lactate dehydrogenase-5 (LDH-5)
overexpression in non-small-cell lung cancer tissues is linked to
tumour hypoxia, angiogenic factor production and poor prognosis. Br
J Cancer. 89:877–885. 2003.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Stabenow LK, Zibrova D, Ender C, Helbing
DL, Spengler K, Marx C, Wang ZQ and Heller R: Oxidative glucose
metabolism promotes senescence in vascular endothelial cells.
Cells. 11(2213)2022.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Franczyk B, Gluba-Brzózka A, Ławiński J,
Rysz-Górzyńska M and Rysz J: Metabolomic profile in venous
thromboembolism (VTE). Metabolites. 11(495)2021.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Lim CS, Kiriakidis S, Sandison A, Paleolog
EM and Davies AH: Hypoxia-inducible factor pathway and diseases of
the vascular wall. J Vasc Surg. 58:219–230. 2013.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Li WQ, Qin ZS, Chen S, Cheng D, Yang SC,
Choi YMM, Chu B, Zhou WH and Zhang ZJ: Hirudin alleviates acute
ischemic stroke by inhibiting NLRP3 inflammasome-mediated
neuroinflammation: In vivo and in vitro approaches. Int
Immunopharmacol. 110(108967)2022.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Wang Z, Liu P, Hu M, Lu S, Lyu Z, Kou Y,
Sun Y, Zhao X, Liu F and Tian J: Naoxintong restores ischemia
injury and inhibits thrombosis via COX2-VEGF/ NFκB signaling. J
Ethnopharmacol. 270(113809)2021.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Li S, Long Q, Nong L, Zheng Y, Meng X and
Zhu Q: Identification of immune infiltration and
cuproptosis-related molecular clusters in tuberculosis. Front
Immunol. 14(1205741)2023.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Li S, Zheng Y, Long Q, Nong J, Shao H,
Liang G and Wu F: Drug-drug interactions between propofol and ART
drugs: Inhibiting neuronal activity by affecting glucose
metabolism. CNS Neurosci Ther: Aug 31, 2023 (Epub ahead of
print).
|
|
23
|
Xie X, Hu Y, Ye T, Chen Y, Zhou L, Li F,
Xi X, Wang S, He Y, Gao X, et al: Therapeutic vaccination against
leukaemia via the sustained release of co-encapsulated anti-PD-1
and a leukaemia-associated antigen. Nat Biomed Eng. 5:414–428.
2020.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Wang S, Zhou L, Ji N, Sun C, Sun L, Sun J,
Du Y, Zhang N, Li Y, Liu W, et al: Targeting acyp1-mediated
glycolysis reverses lenvatinib resistance and restricts
hepatocellular carcinoma progression. Drug Resist Updat,.
69(100976)2023.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Kumar P, Nagarajan A and Uchil PD:
Analysis of cell viability by the MTT assay. Cold Spring Harb
Protoc. 2018:2018.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Sivaprakasam S, Bhutia YD, Yang S and
Ganapathy V: Short-chain fatty acid transporters: Role in colonic
homeostasis. Compr Physiol. 8:299–314. 2017.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Zhao X, Lu Y, Li S, Guo F, Xue H, Jiang L,
Wang Z, Zhang C, Xie W and Zhu F: Predicting renal function
recovery and short-term reversibility among acute kidney injury
patients in the ICU: Comparison of machine learning methods and
conventional regression. Ren Fail. 44:1327–1338. 2022.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Xu B, Zhang M, Zhang B, Chi W, Ma X, Zhang
W, Dong M, Sheng L, Zhang Y, Jiao W, et al: Embigin facilitates
monocarboxylate transporter 1 localization to the plasma membrane
and transition to a decoupling state. Cell Rep.
40(111343)2022.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Li SS, Luedemann M, Sharief FS, Takano T
and Deaven LL: Mapping of human lactate dehydrogenase-A -B and -C
genes and their related sequences: the gene for LDHC is located
with that for LDHA on chromosome 11. Cytogenet Cell Genet.
48:16–18. 1988.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Markert CL, Shakle JB and Whitt GS:
Evolution of a gene. Multiple genes for LDH isozymes provide a
model of the evolution of gene structure, function and regulation.
Science. 189:102–114. 1975.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Swiderek K and Paneth P: Differences and
similarities in binding of pyruvate and L-lactate in the active
site of M4 and H4 isoforms of human lactate dehydrogenase. Arch
Biochem Biophys. 505:33–41. 2011.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Srivastava AK, Kalita J, Haris M, Gupta RK
and Misra UK: Radiological and histological changes following
cerebral venous sinus thrombosis in a rat model. Neurosci Res.
65:343–346. 2009.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Song W, Ci H, Tian G, Zhang Y and Ge X:
Edoxaban improves venous thrombosis via increasing hydrogen sulfide
and homocysteine in rat model. Mol Med Rep. 16:7706–7714.
2017.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Zhao Z, Wu C, He X, Zhao E, Hu S, Han Y,
Wang T, Chen Y, Liu T and Huang S: Microrna let-7f alleviates
vascular endothelial cell dysfunction via targeting HMGA2 under
oxygen-glucose deprivation and reoxygenation. Brain Res.
1772(147662)2021.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Benjamin D, Robay D, Hindupur SK, Pohlmann
J, Colombi M, El-Shemerly MY, Maira SM, Moroni C, Lane HA and Hall
MN: Dual inhibition of the lactate transporters MCT1 and MCT4 is
synthetic lethal with metformin due to NAD+ depletion in cancer
cells. Cell Rep. 25:3047–3058.e4. 2018.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Yu X, Zhang R, Wei C, Gao Y, Yu Y, Wang L,
Jiang J, Zhang X, Li J and Chen X: MCT2 overexpression promotes
recovery of cognitive function by increasing mitochondrial
biogenesis in a rat model of stroke. Anim Cells Syst (Seoul).
25:93–101. 2021.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Srinivas SR, Gopal E, Zhuang L, Itagaki S,
Martin PM, Fei YJ, Ganapathy V and Prasad PD: Cloning and
functional identification of slc5a12 as a sodium-coupled
low-affinity transporter for monocarboxylates (SMCT2). Biochem J.
392:655–664. 2005.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Daniele LL, Sauer B, Gallagher SM, Pugh EN
Jr and Philp NJ: Altered visual function in monocarboxylate
transporter 3 (slc16a8) knockout mice. Am J Physiol Cell Physiol.
295:C451–C457. 2008.PubMed/NCBI View Article : Google Scholar
|