Introduction
Allergic asthma is a common respiratory disease in
clinical practice. Its main pathogenesis involves the production of
specific IgE antibodies, chronic airway inflammation, airway
remodeling and airway hyperresponsiveness during the response of
the immune system to environmental allergens (1,2). The
most important link in the pathogenesis of asthma is allergic
sensitization through the activation of respiratory epithelial
cells (3). In recent years, the
incidence and prevalence of allergic asthma have rapidly increased,
particularly in low and middle income countries (4,5).
Respiratory viral infections are the most common cause of allergic
asthma deteriorations (6).
Rhinovirus (RV) is a single-stranded RNA (ssRNA)
virus of the picornavirus family, the most common human respiratory
virus and is considered to be the main pathogen causing asthma
deterioration (7,8). In children, 80% of asthma
deteriorations are caused by RV infections (9). It has been demonstrated that
cadherin-related family member 3 (CDHR3), a specific cell marker of
ciliated cells, is a risk gene for asthma deterioration induced by
RV in children (10).
Additionally, repeated RV infection can promote airway remodeling
by upregulating TNF superfamily member 14, IL-1β and TGF-β, even in
the absence of allergens (11).
Notably, double-stranded RNA (dsRNA) generated by RV in the process
of replication can also cause a strong immune response of the host
(12). Airway epithelial cells are
the primary target for RV and the first line of defense in the
immune response (13). A study has
shown that ssRNA infection in epithelial cells mainly activates the
interferon-induced antiviral signaling pathway with interferon
regulatory factor (IRF) 7(14).
The mechanism through which dsRNA infects and induces antiviral
immune responses in upper and lower respiratory tract epithelial
cells is similar; however, the induction of interferon-related
genes (such as IRF3, interferon-α/β receptor 1 and interferon β1)
is impaired in patients with asthma (15). RV-induced asthma deterioration is a
complex pathological process involving a large number of gene
expression changes and signaling pathways (7,12,14,15).
However, it is so far unknown, whether there is a common target and
regulatory network of rhinovirus single and double stranded RNA
inducing asthma deterioration.
Bioinformatics analysis is a powerful tool used to
explore the key genes and molecular regulatory networks in
pathogenesis (16). In the present
study, common differentially expressed genes (cDEGs) of asthma
deterioration induced by RV dsRNA and ssRNA were screened using
bioinformatics analysis. The functions of the proteins encoded by
the cDEGs and their interaction networks were analyzed using Gene
Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG),
and protein-protein interaction (PPI) analyses, respectively. Hub
genes were identified and validated. The hub genes and regulatory
networks identified in the present study may not only help
comprehend the molecular mechanism of RV-induced asthma
deterioration, but also serve as potential targets to improve
treatment.
Materials and methods
Clinical samples and ethics
A total of eight clinical samples (paired sample,
the comparison of the deterioration and recovery periods) were
collected from pediatric patients with RV-induced asthma
deterioration between May 2022 and August 2022. The patients
included five boys and three girls (mean age, 13 years). Sterile
cytology brushes were used to collect airway epithelium and
secretions from the posterior surface of the inferior turbinate
from patients at Wuhu Hospital of Traditional Chinese Medicine
(Wuhu, China). Collected brushes were immediately submerged in RLT
Plus lysis buffer (cat. no. 1053393; Qiagen GmbH) with
β-mercaptoethanol (cat. no. 444203; MilliporeSigma) and frozen at
-80˚C for subsequent experiments. The inclusion criteria for the
samples used in the present study were established by referring to
relevant literature (17): RV
nucleic acid test (+) using the human Rhv ELISA kit (cat. no.
YJ711802; Shanghai Enzyme-linked Biotechnology Co., Ltd.),
proportion of eosinophils >5% using blood cell analyzer
(XS-1800; Sysmex America, Inc.), bronchial dilation test (forced
expiratory volume of first second) >12% (MasterScreen IOS; Erich
Jaeger GmbH), chest tightness, cough and diffuse wheezes in both
lungs, and all other respiratory diseases ruled out. Samples were
collected from each of the 8 patients at both timepoints during the
asthma attacks and 14 days after the symptoms had disappeared with
treatment. The present study was approved by the Ethics Committee
of Wuhu Hospital of Traditional Chinese Medicine (approval no.
20220430; Wuhu, China).
Retrieval of datasets
In the present study, two datasets (GSE30326 and
GSE51392) were downloaded from the Gene Expression Omnibus (GEO)
database (https://www.ncbi.nlm.nih.gov/geo/) (14,15).
The characteristics of the patients in these two datasets are
presented in Table I. GSE30326
analyzed gene expression profiles in nasal lavage samples of 16
asthmatic children during acute RV ssRNA-induced deterioration and
after 7-14 days. The cellular composition of the acute samples was
macrophages (83.9±2.7%), neutrophils (12.3±2.5%), epithelial cells
(2.2±1.0%) and eosinophils (1.6±0.5%). The recovery period samples
contained macrophages (72.1±4.8%), epithelial cells (16.6±4.7%),
neutrophils (9.1±1.2%) and eosinophils (1.7±0.8%). In GSE30326, a
network of susceptible genes was constructed using the weighted
gene co-expression network analysis algorithm (14). The GSE51392 dataset was used to
identify microarray gene expression profiles of RV dsRNA-treated
nasal and bronchial epithelial cells from the same individual. A
total of 6 patients with asthma induced by RV dsRNA and 6 healthy
controls from the GSE51392 dataset (5 samples were involved in
allergic rhinitis and were excluded due to weak correlation with
this study) were selected as the study sample (15). The susceptibility genes in the
aforementioned two datasets were identified by moderated t
statistics, and genes that were significantly modulated at a false
discovery rate adjusted P<0.05 were used for further
experiments.
 | Table IPatient characteristics for the two
datasets (GSE30326 and GSE51392). |
Table I
Patient characteristics for the two
datasets (GSE30326 and GSE51392).
| GEO Accession | GSE30326 | GSE51392 |
|---|
| Subjects | 16 children | 17 adults |
| Mode of
pairing | Acute phase vs.
remission phase | Allergic asthma vs.
healthy controls |
| RNA type | Single-stranded
RNA | Double-stranded
RNA |
| RNA identification
method | Reverse
transcription-quantitative PCR | Poly(I:C), a
synthetic double-stranded RNA |
| Experimental
design | Differentially
expressed genes in respiratory epithelial cells between acute
exacerbation and remission were analyzed using a gene chip | Microarray
identification of double-stranded RNA-induced gene expression
profiles in respiratory epithelial cells |
| Sample source | Nasal cavity | Nasal cavity and
bronchi, paired samples |
| Predicted FEV1 at
remission, % | 103.4±10.9 | 109±11.0 |
| Allergen skin test
positive, % | 68.7 | 100 |
Screening and functional annotation of
cDEGs
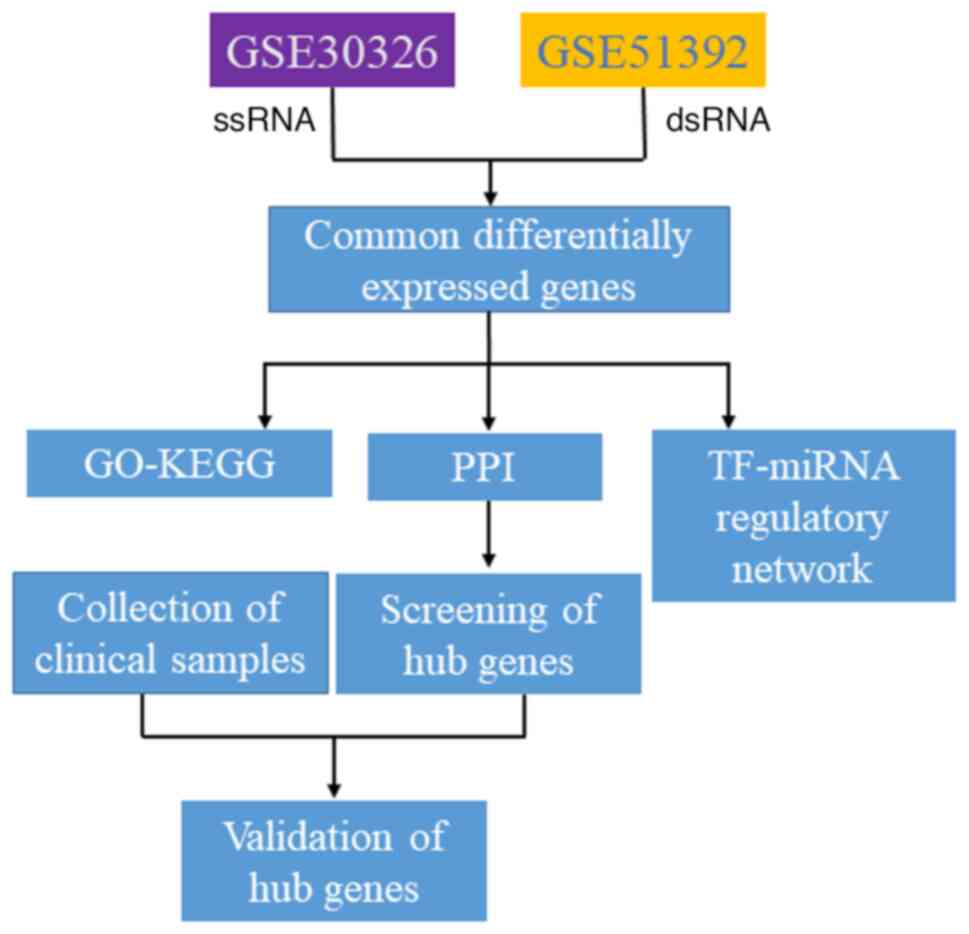
The flowchart of the present study is shown in
Fig. 1. First, cDEGs from the
GSE30326 and GSE51392 datasets were analyzed using GEO2Rweb
(https://www.ncbi.nlm.nih.gov/geo/geo2r). The cut-off
criteria were: Adjusted P<0.05 and log2-fold change
>1.5 or <-1.5. Second, the cDEGs were visualized using
ggplot2 (version 3.3.3; https://cran.r-project.org/mirrors.html), and
functional annotation was performed using GO and KEGG pathway
analysis [clusterProfiler (version 4.4.4) R package (version
4.2.1); http://bioconductor.org/packages/release/bioc/html/clusterProfiler.html].
Screening hub genes and key
module
The PPI network of DEGs was constructed using the
STRING database (https://cn.string-db.org/). PPIs were analyzed using
Cytoscape software (https://cytoscape.org/; version 3.8.2), and the hub
genes were evaluated based on the score of topological algorithms
in Cytoscape, including the BottleNeck, closeness, degree, density
of maximum neighborhood component, EcCentricity and maximal clique
centrality algorithms. In addition, the key modules from the PPI
network were analyzed using the Molecular Complex Detection plugin
in Cytoscape. The functional annotation of key module were
performed using GO-KEGG database.
Identification of candidate regulators
of hub genes
The regulators of hub genes [including transcription
factors (TFs), microRNAs (miRNAs/miRs) and drugs] were screened
through online resources, such as NetworkAnalyst (version 3.0;
https://www.networkanalyst.ca/) and
DSigDB (https://amp.pharm.mssm.edu/Enrichr/).
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from each sample using
TRNzol Universal Reagent (cat. no. DP424; Tiangen Biotech Co.,
Ltd.) according to the manufacturer's instructions. Chloroform was
added to separate the organic and inorganic phases, RNA was
precipitated with isopropanol and washed with 75% ethanol. The RNA
concentration (A260/280) was measured using a spectrophotometer
(Nanodrop; Thermo Fisher Scientific, Inc.). Next, 1 µg RNA was
reverse transcribed using HiScript II Q RT SuperMix (cat. no.
R222-01; Vazyme Biotech Co., Ltd.). Temperature protocol of reverse
transcription: 50˚C for 15 min; 85˚C for 5 sec. Quantification of
signal transducer and activator of transcription 1 (STAT1) and
interferon induced with helicase C domain 1 (IFIH1) expression was
performed using a Real-Time PCR Detection System (LightCycler 96;
Roche Diagnostics). Reactions were performed in three replicates
with 2 µl cDNA per reaction using the 2XSG Fast qPCR Master Mix
(cat. no. B639273; Sangon Biotech Co., Ltd.). Thermocycling
conditions were as follows: 3 min at 95˚C and 30 sec at 60˚C, for
40 cycles. The primers were: STAT1 forward,
5'-GCTTGACAATAAGAGAAAGG-3' and reverse,
5'-CGCTCTGCTGTCTCCGCTTCCACTCC-3'; IFIH1 forward,
5'-GTTGAAAAGGCTGGCTGAAAAC-3' and reverse,
5'-TCGATAACTCCTGAACCACTG-3'; and GAPDH forward,
5'-GAAGGTGAAGGTCGGAGTC-3' and reverse, 5'-GAAGATGGTGATGGGATTTCC-3'.
All primers were synthesized by TsingKe Biological Technology.
Relative gene expression was calculated using the
2-ΔΔCq method (18). GAPDH was used as the internal
reference.
Statistical analysis
SPSS (version 20; IBM Corp.) was utilized for
statistical analysis of three technical replicates per sample. The
paired t-test was used to compare differences in mRNA expression
levels. Data are shown as mean ± standard deviation. P<0.05 was
considered to indicate a statistically significant difference.
Results
Identification of cDEGs between
GSE51392 and GSE30326
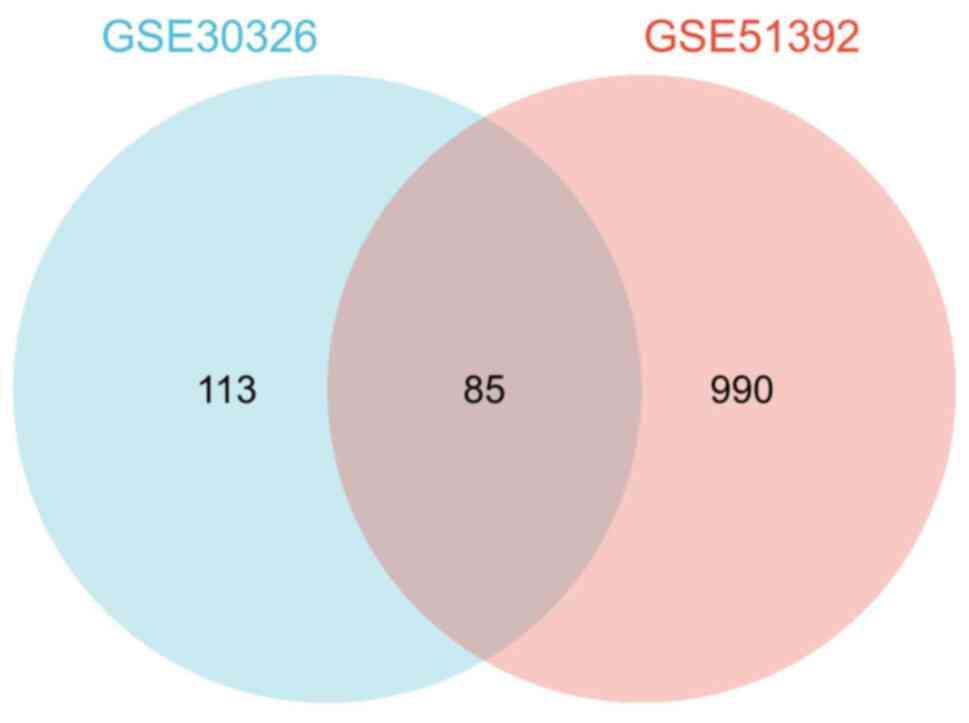
In the GSE51392 dataset, 1,075 DEGs were identified.
Among them, 337 genes were upregulated and 738 downregulated. In
the GSE30326 dataset, a total of 198 DEGs were identified and all
of them were downregulated. Of note, a total of 85 cDEGs were
identified in the aforementioned two datasets (Fig. 2).
Functional enrichment analysis of
cDEGs
To understand the functions of the cDEGs, GO and
KEGG pathway analysis was carried out using the Enrichr web server
and KEGG Mapper, respectively. GO analysis revealed enrichment for
‘response to virus’, ‘double-stranded RNA binding’ and
‘single-stranded RNA binding’. In addition, KEGG pathway analysis
revealed enrichment of cDEGs associated with ‘influenza A’,
‘hepatitis C’ and ‘NOD-like receptor signaling pathway’. More
detailed functional enrichment information of DEGs is presented in
Table II.
 | Table IIGO-KEGG enrichment analysis for
common differentially expressed genes. |
Table II
GO-KEGG enrichment analysis for
common differentially expressed genes.
| Terms | ID | Description | GeneRatio | Adjusted
P-value |
|---|
| GO | | | | |
|
BP | GO:0009615 | Response to
virus | 39/79 |
1.59x10-41 |
|
BP | GO:0051607 | Defense response to
virus | 35/79 |
4.89x10-40 |
|
BP | GO:0140546 | Defense response to
symbiont | 35/79 |
4.89x10-40 |
|
BP | GO:0002831 | Regulation of
response to biotic stimulus | 25/79 |
9.69x10-22 |
|
BP | GO:1903900 | Regulation of viral
life cycle | 17/79 |
3.54x10-18 |
|
MF | GO:0003725 | Double-stranded RNA
binding | 10/80 |
1.8x10-10 |
|
MF | GO:0003727 | Single-stranded RNA
binding | 7/80 |
1x10-5 |
|
MF | GO:0004842 | Ubiquitin-protein
transferase activity | 12/80 |
2.36x10-5 |
|
MF | GO:0019787 | Ubiquitin-like
protein transferase activity | 12/80 |
3.22x10-5 |
|
MF | GO:0003724 | RNA helicase
activity | 6/80 |
3.49x10-5 |
| KEGG | hsa05164 | Influenza A | 13/49 |
9.93x10-10 |
| KEGG | hsa05160 | Hepatitis C | 10/49 |
8.63x10-7 |
| KEGG | hsa04621 | NOD-like receptor
signaling pathway | 9/49 |
2.78x10-5 |
| KEGG | hsa05162 | Measles | 8/49 |
2.78x10-5 |
| KEGG | hsa04622 | RIG-I-like receptor
signaling pathway | 6/49 |
5.15x10-5 |
PPI network analysis
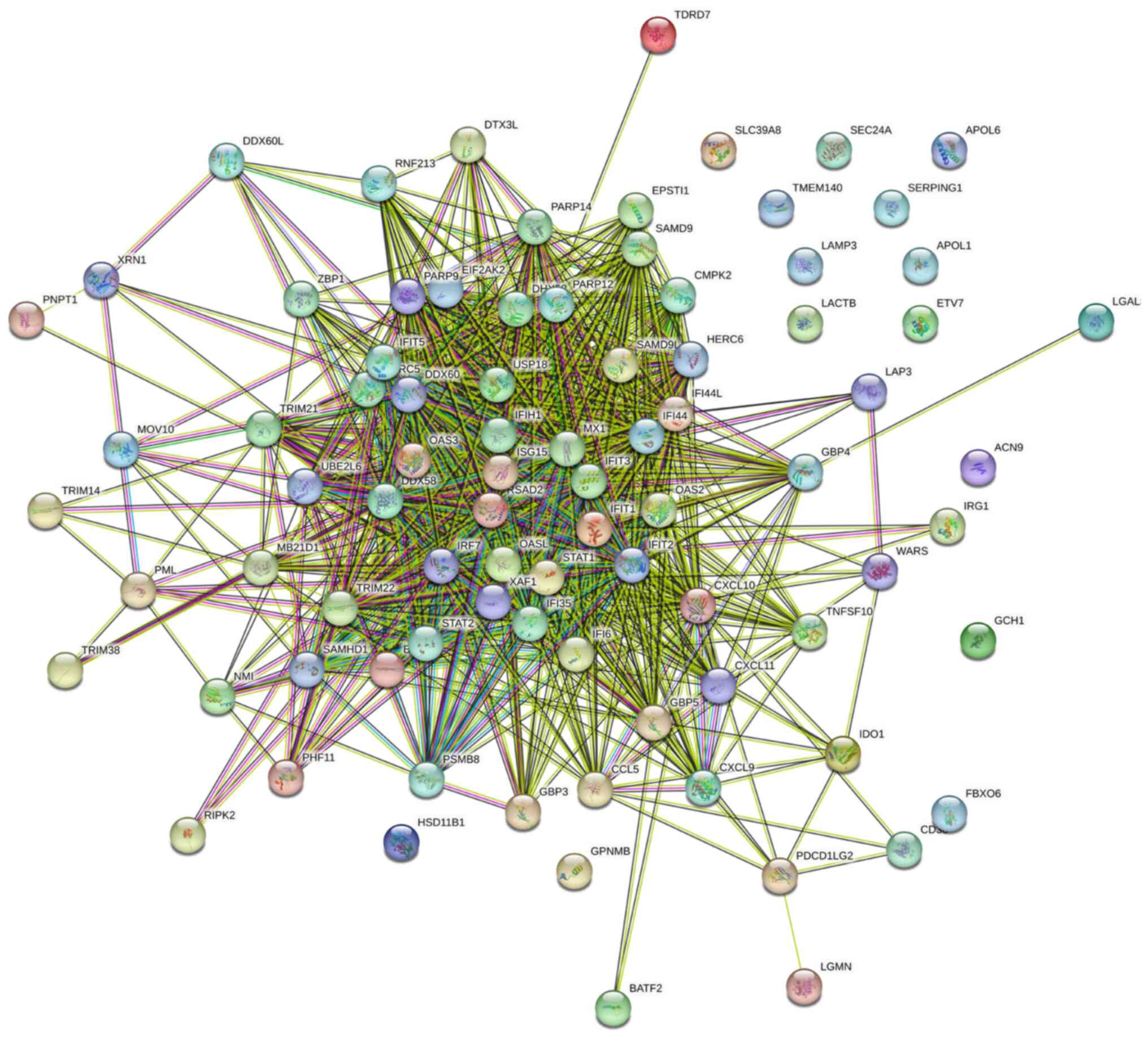
The 85 cDEGs were analyzed using the STRING online
tool and the result is shown in Fig.
3. The analyzed network had 85 nodes and 996 edges. The average
node degree was 23.4, and the average local clustering coefficient
was 0.648.
Detection of hub genes and module
analysis
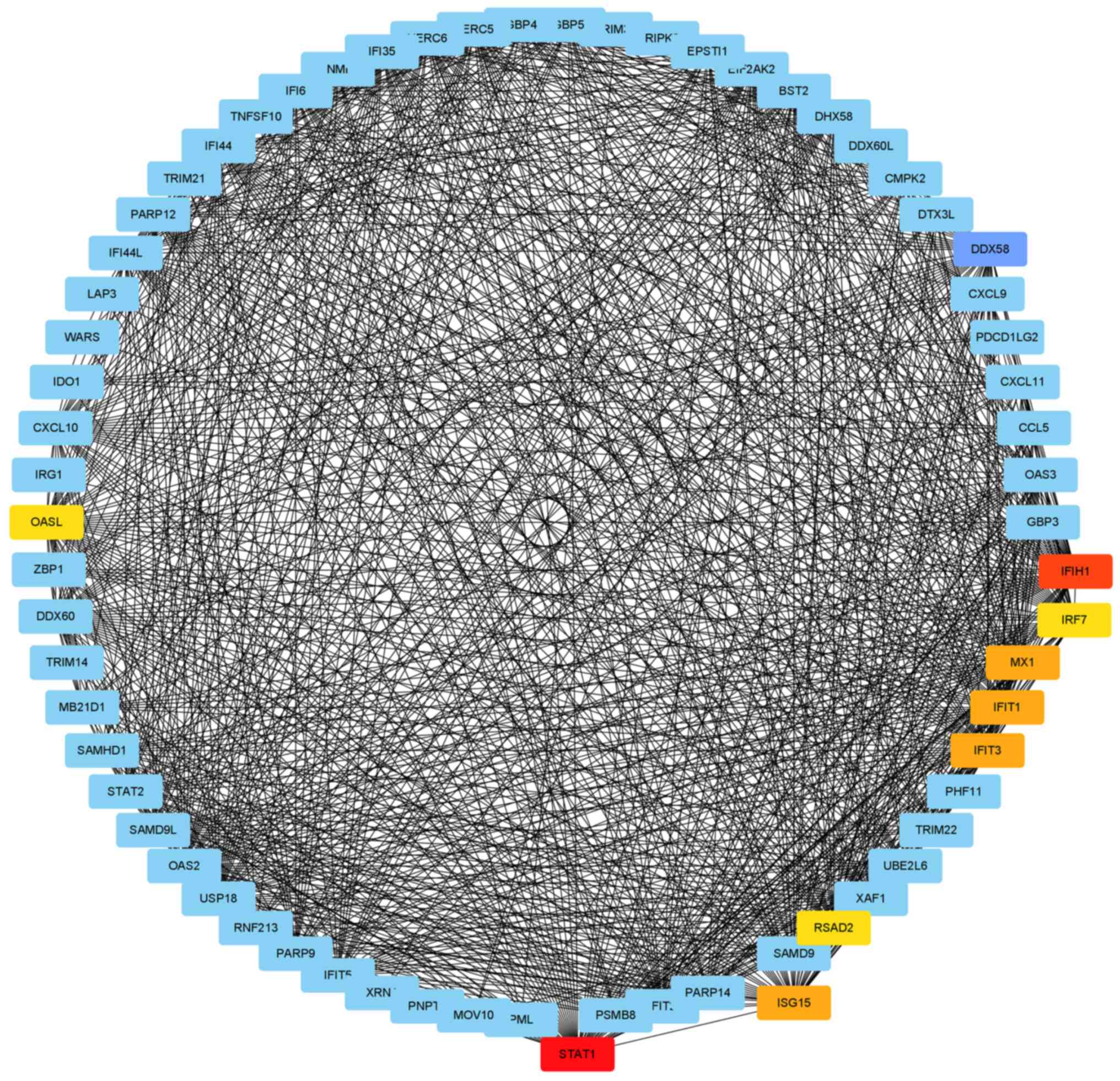
According to topology analysis, five cDEGs [STAT1,
IFIH1, IRF7, DExD/H box helicase 58 (DDX58) and
interferon-stimulating gene 15 (ISG15)] were considered to be hub
genes (Table III). The proteins
encoded by the hub genes have rich interactions with other
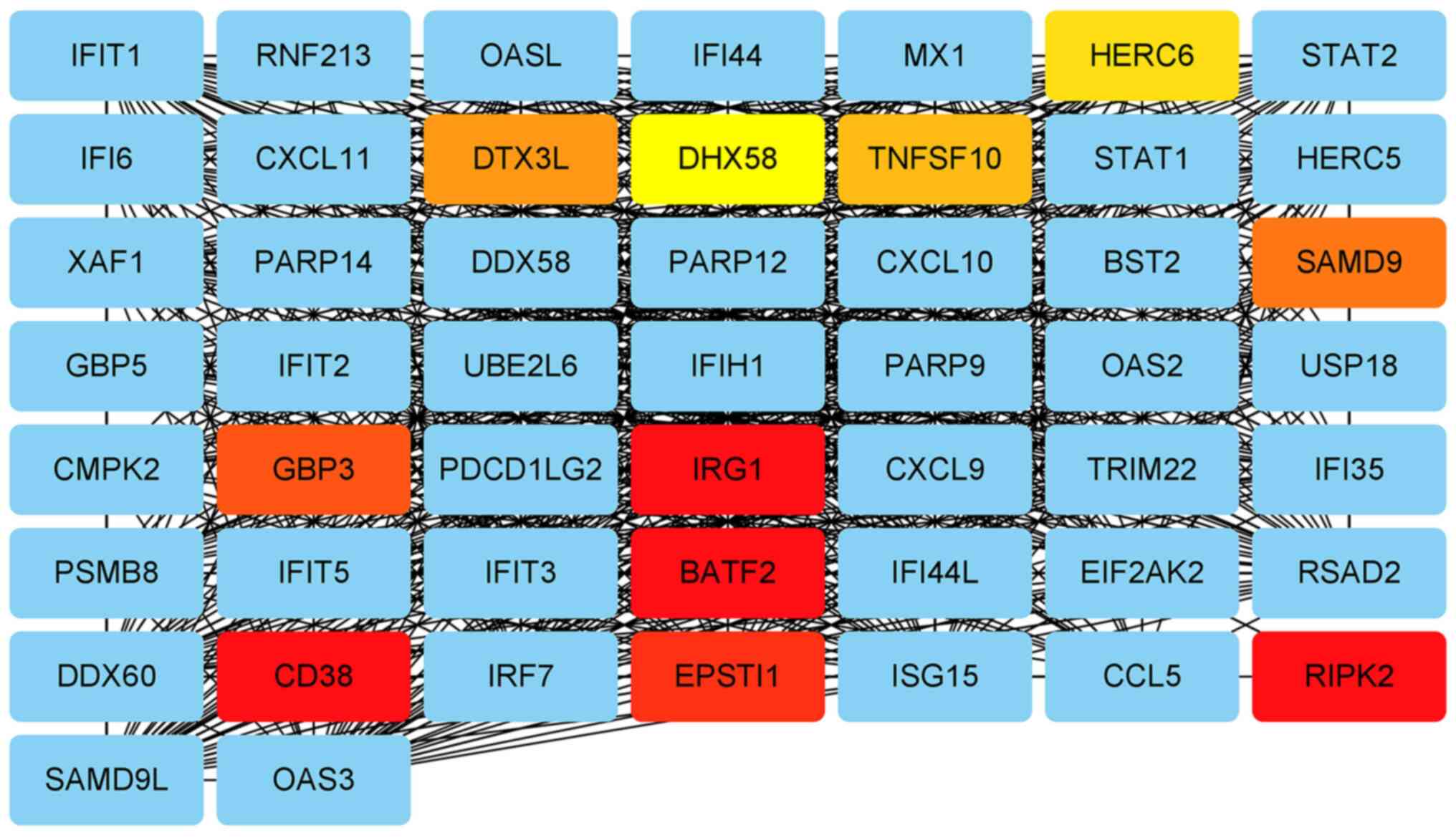
proteins, including 118 nodes and 1,531 edges (Fig. 4). Module analysis demonstrated that
immune-responsive gene 1 (IRG1), basic leucine zipper transcription
factor 2 (BATF2) and epithelial stromal interaction 1 were high
density modules and had significant interactions with hub genes
(Fig. 5). The genes in the module
were mainly enriched in ‘influenza A’, ‘Toll-like receptor
signaling pathway’, ‘coronavirus disease-COVID-19’, ‘hepatitis B’
and ‘herpes simplex virus 1 infection’ signaling pathways (Table IV).
 | Table IIIRanking of the top 5 genes based on 6
algorithms. |
Table III
Ranking of the top 5 genes based on 6
algorithms.
| Degree | MCC | DMNC | EcCentricity | Closeness | BottleNeck |
|---|
| STAT1 | IFIH1 | HERC6 | STAT1 | STAT1 | STAT1 |
| IFIH1 | RSAD2 | IFI6 | IFIH1 | IFIH1 | HELZ |
| IRF7 | IFIT3 | HERC5 | RSAD2 | IRF7 | PRKCA |
| DDX58 | STAT1 | EPSTI1 | IFI35 | ISG15 | XIAP |
| ISG15 | MX1 | SAMD9 | USP18 | DDX58 | MX1 |
 | Table IVAnalysis of the signaling pathways
associated with the genes within the key module. |
Table IV
Analysis of the signaling pathways
associated with the genes within the key module.
| ID | Description | Adjusted
P-value | Gene ID |
|---|
| hsa05164 | Influenza A |
6.8x10-6 |
OAS2/RSAD2/STAT1/IRF7/IFIH1 |
| hsa04620 | Toll-like receptor
signaling pathway |
7.2x10-5 |
CXCL11/CXCL9/STAT1/IRF7 |
| hsa05171 | Coronavirus
disease-COVID-19 |
0.1x10-3 |
OAS2/ISG15/STAT1/IFIH1 |
| hsa05161 | Hepatitis B | 0.004 |
STAT1/IRF7/IFIH1 |
| hsa05168 | Herpes simplex
virus 1 infection | 0.034 |
OAS2/STAT1/IRF7/IFIH1 |
TF regulatory network analysis of hub
genes
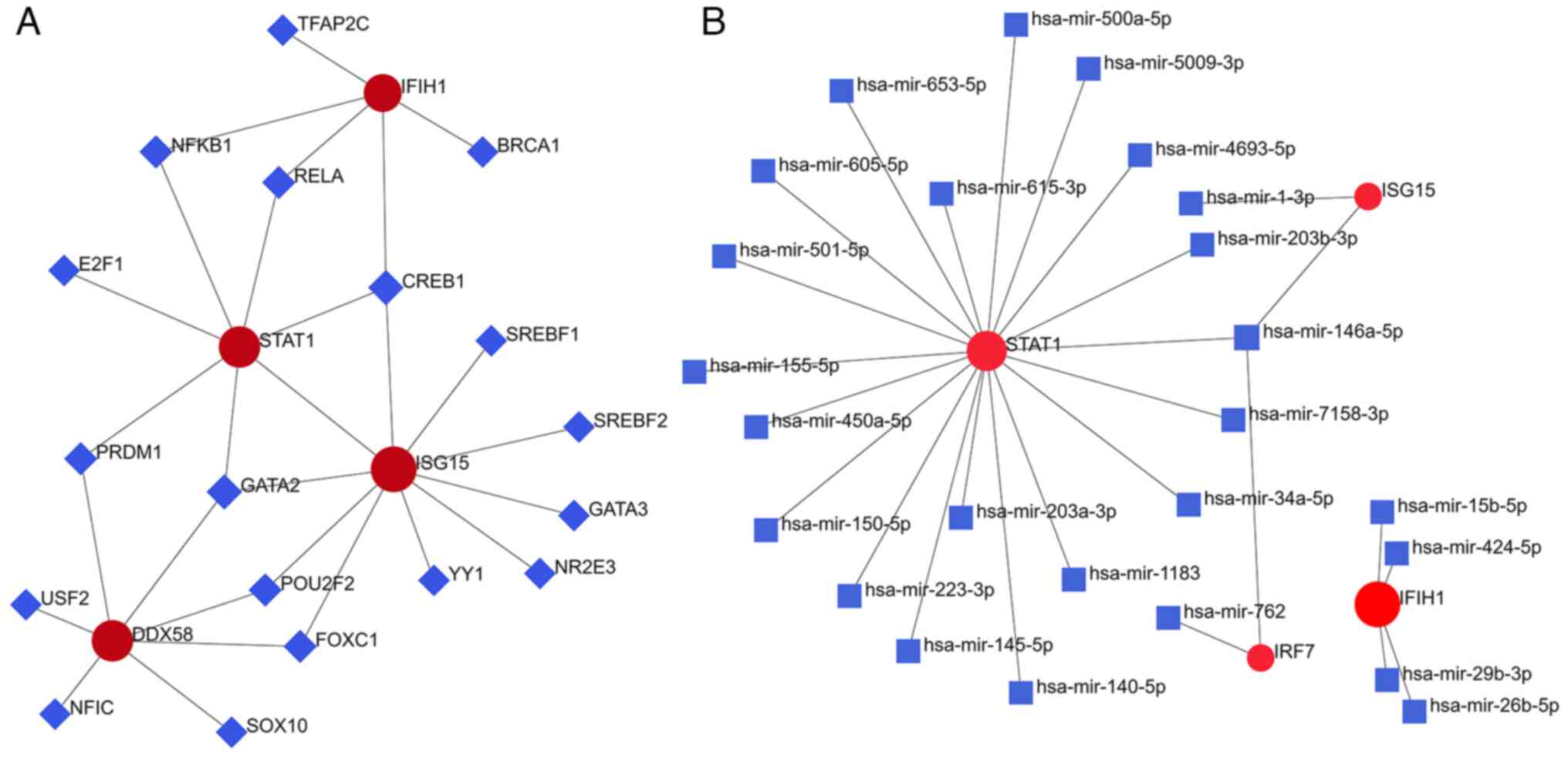
A total of 18 TFs critical for regulating the
expression of hub genes were identified using NetworkAnalyst. Among
them, GATA-binding factor 2 (GATA2) was found to regulate the
STAT1, DDX58 and ISG15 hub genes, and CAMP responsive element
binding protein 1 (CREB1) was found to regulate STAT1, IFIH1 and
ISG15 (Fig. 6A). In addition, 25
miRNAs critical for regulating the expression of hub genes were
identified. In the regulatory network, miR-146a-5p regulated the
STAT1, IRF7 and ISG15 hub genes. IFIH1 was mainly regulated by
miR-424-5p (Fig. 6B).
Screening of potential therapeutic
drugs
Enrichr Platform analysis revealed that suloctidil,
3'-Azido-3'-deoxythymidine, estradiol and acetaminophen were
potential therapeutic targets for cDEGs and hub genes (Table V).
 | Table VPotential drug components for
treatment of rhinovirus-induced asthma exacerbation. |
Table V
Potential drug components for
treatment of rhinovirus-induced asthma exacerbation.
| Component | Overlap | Adjusted
P-value | Odds ratio | Combined score | Target genes |
|---|
| Suloctidil | 38/141 |
6.85x10-57 | 146.1635 | 19919.95 | IFIT5, IFI6, IFIT1,
IFI44L, IFIT3, IFIT2, OASL, IFIH1, IFI44, EIF2AK2, ISG15, CXCL10,
CXCL11, AIM2, OAS2, OAS3, TFEC, IRF7, C19ORF66, CD69, XAF1, SP140L,
IRF9 |
|
3'-Azido-3'-deoxythymidine | 27/374 |
2.15x10-23 | 24.95654 | 1436.213 | SAMD9L, IFIT5,
IFI6, DDX60L, IFIT1, IFI44L, IFIT2, IFIH1, CASP5, LAMP3, C3AR1,
EPSTI1, TNFSF10, LGALS9, TRIM22, STAT1, MX1, ISG15 |
| Estradiol | 45/4336 |
6.29x10-8 | 3.809732 | 78.47791 | IFIT5, IFI6,
UBE2L6, DDX60L, IFIT1, TARP, IFI44L, IFIT3, IFIT2, IFIH1, DDX58,
STAT1, P2RY14, CYBB, IFI44, EIF2AK2, ISG15, NMI, PARP9, CXCL10,
CXCL11, OAS2, OAS3, IRF7, CMPK2, XAF1, HAMP, IRF9 |
| Acetaminophen | 43/4135 |
1.74x10-7 | 3.694254 | 71.97291 | IFI6, TMEM51,
UBE2L6, DDX60L, IFIT1, IFIT3, IFIT2, IFIH1, PSTPIP2, LAMP3, C3AR1,
TNFSF10, NBN, TRIM21, TRIM22, GBP4, TNS1, GBP3, RSAD2, DYNLT1,
DDX58, STAT1, SPHK1, PHF11, MX1, IFI44, EIF2AK2, NMI, RNASE2,
PARP9, PATL1, IRF9 |
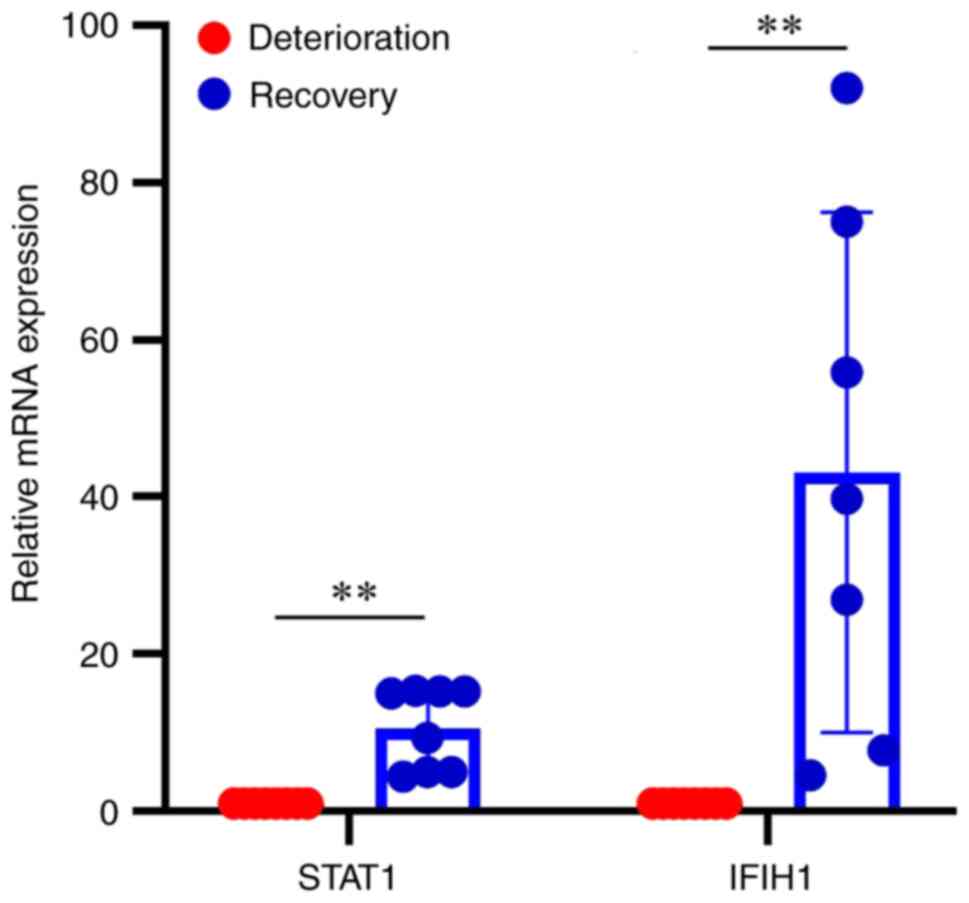
Validation of key hub genes
RT-qPCR was used to validate the mRNA expression of
the hub genes STAT1 and IFIH1, and it was revealed that the mRNA
expression levels of STAT1 were downregulated in asthma
deterioration samples compared with post-treatment recovery
samples, and the mRNA expression levels of IFIH1 exhibited the same
trend (Fig. 7; P<0.01).
Discussion
The two datasets involved in the present study
revealed that both STAT1 and IFIH1 were significantly downregulated
in respiratory epithelial cells infected with RV ssRNA or dsRNA.
Network analysis demonstrated that STAT1 and IFIH1 served important
roles in key modules as hub genes. Finally, it was found that the
expression levels of these two genes were significantly higher in
the recovery phase than in the deterioration phase in clinical
samples, suggesting that they can be used as potential therapeutic
targets to improve asthma deteriorations caused by RV ssRNA or
dsRNA. Studies have shown that STAT1 is an important TF that
maintains T helper 1 (Th1) cell development and serves an important
role in regulating asthma deterioration; low levels of STAT1
increase the risk of asthma attacks and virus susceptibility
(19,20). STAT1 is necessary to control the
replication of influenza A in vivo and serves a crucial role
in the Toll-like receptor (TLR) signaling pathway-mediated
inflammatory response (21,22).
In the course of COVID-19, STAT1 is dysfunctional and enhancing the
activity of STAT1 has a therapeutic effect (23). A study has shown that eosinophils
in the airway of allergic asthma enhanced the antiviral immune
response to influenza A by upregulating IFIH1 transcription
(24). IFIH1 can recognize and
bind to dsRNA produced in the process of viral replication
(25). TLR can recognize ssRNA in
the nucleus and bind with dsRNA and synergistically mediate
congenital antiviral immune response with IFIH1(26). Studies have shown that there are
two alleles of IFIH1 in the pathogenesis of COVID-19: The patients
with rs1990760/IFIH1 exhibit an attenuated inflammatory response
and improved outcomes, while patients with rs19907601/IFIH1 exhibit
poor prognosis (27,28). In addition, IFIH1/DDX58 is the
primary cytoplasmic sensor for the intracellular detection of viral
pathogen-associated molecular patterns of viral RNA (29). It is an important component in the
formation of cytoplasmic RNA-binding protein complement, which
contributes to innate antiviral immunity (30). IFIH1/DDX58 expression in epithelia
of the upper airway is higher in children than in adults, resulting
in stronger early innate antiviral immunity to severe acute
respiratory syndrome coronavirus 2 (SARS-CoV-2) infection in
children (31). Low IFIH1/DDX58
expression in patients with viral infection predicts poor prognosis
(32). These previous results were
consistent with those of the present study (that STAT1 and IFIH1
were downregulated during asthma deterioration).
In the two datasets involved in the present study,
IFIH1 is more downregulated than STAT1 during RV-induced asthma
deteriorations (14,15). This may be due to two reasons.
Firstly, asthma exhibits an imbalance of Th1/Th2 cells in cytology
and Th2 cells increase during asthma attacks (33), which reduces the generation of Th1
cytokine interferon, causing a deficiency in antiviral response and
increase in viral load in patients with asthma (34). Secondly, IgE antibodies eliminate
the biological activity of interferon α and reduce its
concentration and antiviral ability (35). GO-KEGG enrichment analysis showed
that cDEGs were mainly involved in ‘(defense) response to virus’,
‘regulation of viral life cycle’, ‘double-stranded RNA binding and
‘single-stranded RNA binding’, as well as in ‘influenza A’ and
‘NOD-like receptor signaling pathway’. A previous study has shown
that the nucleotide oligomerization domain (NOD)-like receptor
signaling pathway mediates airway epithelial inflammation and mucus
production, which is an important mechanism of RV-induced airway
remodeling (36). Dataset analysis
showed that the genes with ds/ssRNA binding functions [such as
DExD/H-box 60 like (DDX60L) and polyribonucleotide
nucleotidyltransferase 1 (PNPT1)] were significantly reduced
(14,15). DDX60L is an interferon-stimulated
gene product, which can effectively inhibit viral replication
(37). PNPT1 prevents dsRNA
formation (38). Based on the
above evidence, the present study hypothesized that low expression
of the aforementioned genes helps the host obtain a higher viral
load. A study has shown that the TRIM38 is an important member of
the antiviral network of the interferon family, which can activate
the promoter of interferon β (39). IRF7 is an important bridge
connecting interferon-mediated responses in virus-induced asthma
deteriorations (14). The present
study demonstrated that IFIH1 was significantly downregulated in
the acute phase of RV-induced asthma deterioration, which was
consistent with the aforementioned report (32) and further confirmed the role of the
interferon family in anti-rhinovirus infection.
TFs are the regulatory factors of gene expression
and are closely associated with the occurrence and development of
human diseases. In the present study, GATA2 and CREB1 were found to
be the most important TFs regulating hub genes. GATA2 is essential
for the survival and renewal of hematopoietic stem cells and
interacts with a variety of TFs (40). GATA2 deficiency can cause a variety
of immune cell disorders and increase virus susceptibility
(41). CREB1, a member of the
CREB/activating TF protein family, is involved in the
transcriptional regulation of various viruses, including hepatitis
B (42). A study has shown that
IL-6 mediated the interaction between CREB1 and STAT1 in adrenal
medulla chromaffin cells to regulate the release of cytokines and
inflammatory mediators (43).
Furthermore, the present study identified 25 miRNAs that regulate
hub genes, such as miR-146a-5p and miR-424-5p. In diabetic
nephropathy, miR-146a-5p can activate the STAT1 signaling pathway
and promote M2-type macrophage polarization to enhance
anti-inflammatory effects and improve renal function (44). In the course of dengue virus
infection, miR-146a-5p is considered to be an important molecule
regulating the expression of STAT1 and ISG15(45). Additionally, miR-424-5p serves an
important role in inhibiting hepatitis B virus (HBV) infection and
the progression of HBV-related hepatocellular carcinoma (46). In the present study, IFIH1
regulated by miR-424-5p (Fig. 6)
and enriched in the hepatitis B signaling pathway, which supports
the aforementioned report.
Our previous study demonstrated that the potential
of SARS-CoV-2 to induce asthma exacerbation was similar to that of
rhinovirus (47). Importantly, the
hub genes for asthma deterioration induced by both viruses were
nearly the same. This suggested that asthma deteriorations induced
by different respiratory viruses may have the same mechanism, which
provides a potential therapeutic target for asthma deteriorations
induced by mixed viral infections. In addition, the signaling
pathways associated with the cDEGs of the two dataset pairs also
overlapped (for example, ‘Influenza A’ and ‘NOD-like receptor
signaling pathway’ were identified in both studies). However, the
key modules of asthma deterioration induced by these two
respiratory viruses are different: The IRG1/BATF2 module was
dominated by RV, and the TRIM38/GBP3 module was dominated by
SARS-CoV-2(47).
In conclusion, in the present study, the cDEGs of RV
ssRNA and dsRNA-induced asthma deteriorations were screened using
the GSE51392 and GSE30326 datasets. On this basis, the mechanisms
of asthma deterioration induced by ssRNA and dsRNA were
systematically studied at the molecular, signaling pathway and
interaction network levels. GO-KEGG enrichment analysis and
identification of key modules showed that the hub genes STAT1,
IFIH1, IRF7, DDX58 and ISG15 are considered to serve an important
role in the progression of RV ssRNA and dsRNA-induced asthma
deterioration. At the pathway level, ‘influenza A’, and TLR
signaling pathway suggest that inflammatory factor expression and
antiviral effects serve a major role in innate immunity. From an
interaction network perspective, a key module with STAT1 as the
core was identified. Finally, the molecules and drugs that regulate
cDEGs (including hub genes) were analyzed and key hub genes (STAT1
and IFIH1) were verified using RT-qPCR. It is expected that the
aforementioned findings will contribute to an improved
understanding of the molecular mechanisms of RV-induced asthma
deterioration.
However, the present study had certain limitations.
First, the two datasets analyzed in the present study were from
different age groups, which may have influenced the results.
Secondly, clinical samples were collected from patients with
RV-induced asthma deteriorations for self-paired comparison, but a
control group of healthy children was not included.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the Science
Foundation of Wuhu Science and Technology Bureau (grant no.
WH2022226).
Availability of data and materials
Both datasets involved in this study are available
for download from the GEO database with the accession numbers of
GSE30326 and GSE51392 (https://www.ncbi.nlm.nih.gov/geo/).
Authors' contributions
QA and XZ conceived the idea, collected clinical
samples and wrote the manuscript. WG, YC and ZJ produced the
figures and analyzed data. HLu and HLi contributed to visualization
and performed qPCR. XZ contributed to overall editing and
supervision. All authors read and approved the final version of the
manuscript. QA and XZ confirm the authenticity of all the raw
data.
Ethics approval and consent to
participate
The present study was performed in line with the
principles of The Declaration of Helsinki. The patients and their
guardians provided written informed consent for participation in
the study. The present study was approved by the Ethics Committee
of Wuhu Hospital of Traditional Chinese Medicine (approval no.
20220430; Wuhu, China).
Patient consent for publication
All patients and their guardians provided written
informed consent for publication.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Helou DG, Shafiei-Jahani P, Lo R, Howard
E, Hurrell BP, Galle-Treger L, Painter JD, Lewis G, Soroosh P,
Sharpe AH and Akbari O: PD-1 pathway regulates ILC2 metabolism and
PD-1 agonist treatment ameliorates airway hyperreactivity. Nat
Commun. 11(3998)2020.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Cho JL, Ling MF, Adams DC, Faustino L,
Islam SA, Afshar R, Griffith JW, Harris RS, Ng A, Radicioni G, et
al: Allergic asthma is distinguished by sensitivity of
allergen-specific CD4+ T cells and airway structural cells to type
2 inflammation. Sci Transl Med. 8(359ra132)2016.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Zhu Z, Lee PH, Chaffin MD, Chung W, Loh
PR, Lu Q, Christiani DC and Liang L: A genome-wide cross-trait
analysis from UK Biobank highlights the shared genetic architecture
of asthma and allergic diseases. Nat Genet. 50:857–864.
2018.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Garcia-Marcos L, Asher MI, Pearce N,
Ellwood E, Bissell K, Chiang CY, El Sony A, Ellwood P, Marks GB,
Mortimer K, et al: The burden of asthma, hay fever and eczema in
children in 25 countries: GAN Phase I study. Eur Respir J.
60(2102866)2022.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Mortimer K, Reddel HK, Pitrez PM and
Bateman ED: Asthma management in low and middle income countries:
Case for change. Eur Respir J. 60(2103179)2022.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Toussaint M, Jackson DJ, Swieboda D,
Guedan A, Tsourouktsoglou TD, Ching YM, Radermecker C, Makrinioti
H, Aniscenko J, Bartlett NW, et al: Host DNA released by NETosis
promotes rhinovirus-induced type-2 allergic asthma exacerbation.
Nat Med. 23:681–691. 2017.PubMed/NCBI View
Article : Google Scholar
|
|
7
|
Michi AN, Love ME and Proud D:
Rhinovirus-Induced modulation of epithelial phenotype: Role in
Asthma. Viruses. 12(1328)2020.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Han M, Rajput C, Hinde JL, Wu Q, Lei J,
Ishikawa T, Bentley JK and Hershenson MB: Construction of a
recombinant rhinovirus accommodating fluorescent marker expression.
Influenza Other Respir Viruses. 12:717–727. 2018.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Niespodziana K, Stenberg-Hammar K,
Megremis S, Cabauatan CR, Napora-Wijata K, Vacal PC, Gallerano D,
Lupinek C, Ebner D, Schlederer T, et al: PreDicta chip-based high
resolution diagnosis of rhinovirus-induced wheeze. Nat Commun.
9(2382)2018.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Bochkov YA, Watters K, Ashraf S, Griggs
TF, Devries MK, Jackson DJ, Palmenberg AC and Gern JE:
Cadherin-related family member 3, a childhood asthma susceptibility
gene product, mediates rhinovirus C binding and replication. P Natl
Acad Sci USA. 112:5485–5490. 2015.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Mehta AK, Doherty T, Broide D and Croft M:
Tumor necrosis factor family member LIGHT acts with IL-1β and TGF-β
to promote airway remodeling during rhinovirus infection. Allergy.
73:1415–1424. 2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Wang Q, Nagarkar DR, Bowman ER, Schneider
D, Gosangi B, Lei J, Zhao Y, McHenry CL, Burgens RV, Miller DJ, et
al: Role of double-stranded RNA pattern recognition receptors in
rhinovirus-induced airway epithelial cell responses. J Immunol.
183:6989–6997. 2009.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Ganjian H, Rajput C, Elzoheiry M and
Sajjan U: Rhinovirus and innate immune function of airway
epithelium. Front Cell Infect Microbiol. 10(277)2020.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Bosco A, Ehteshami S, Panyala S and
Martinez FD: Interferon regulatory factor 7 is a major hub
connecting interferon-mediated responses in virus-induced asthma
exacerbations in vivo. J Allergy Clin Immunol. 129:88–94.
2012.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Wagener AH, Zwinderman AH, Luiten S,
Fokkens WJ, Bel EH, Sterk PJ and van Drunen CM: dsRNA-induced
changes in gene expression profiles of primary nasal and bronchial
epithelial cells from patients with asthma, rhinitis and controls.
Respir Res. 15(9)2014.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Zhu X, Tang LP, Mao J, Hameed Y, Zhang J,
Li N, Wu D, Huang Y and Li C: Decoding the Mechanism behind the
Pathogenesis of the Focal Segmental Glomerulosclerosis. Comput Math
Method Med. 2022(1941038)2022.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Wesolowska-Andersen A, Everman JL,
Davidson R, Rios C, Herrin R, Eng C, Janssen WJ, Liu AH, Oh SS,
Kumar R, et al: Dual RNA-seq reveals viral infections in asthmatic
children without respiratory illness which are associated with
changes in the airway transcriptome. Genome Biol.
18(12)2017.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Thompson EA, Sayers BC, Glista-Baker EE,
Shipkowski KA, Ihrie MD, Duke KS, Taylor AJ and Bonner JC: Role of
signal transducer and activator of transcription 1 in murine
allergen-induced airway remodeling and exacerbation by carbon
nanotubes. Am J Respir Cell Mol Biol. 53:625–636. 2015.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Kang YH, Biswas A, Field M and Snapper SB:
STAT1 signaling shields T cells from NK cell-mediated cytotoxicity.
Nat Commun. 10(912)2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Jewell NA, Cline T, Mertz SE, Smirnov SV,
Flano E, Schindler C, Grieves JL, Durbin RK, Kotenko SV and Durbin
JE: Lambda interferon is the predominant interferon induced by
influenza A virus infection in vivo. J Virol. 84:11515–11522.
2010.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Luu K, Greenhill CJ, Majoros A, Decker T,
Jenkins BJ and Mansell A: STAT1 plays a role in TLR signal
transduction and inflammatory responses. Immunol Cell Biol.
92:761–769. 2014.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Matsuyama T, Kubli SP, Yoshinaga SK,
Pfeffer K and Mak TW: An aberrant STAT pathway is central to
COVID-19. Cell Death Differ. 27:3209–3225. 2020.PubMed/NCBI View Article : Google Scholar
|
|
24
|
LeMessurier KS, Rooney R, Ghoneim HE, Liu
BM, Li K, Smallwood HS and Samarainghe AE: Influenza A virus
directly modulates mouse eosinophil responses. J Leukoc Biol.
108:151–168. 2020.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Mu X and Hur S: Immunogenicity of In
Vitro-Transcribed RNA. Acc Chem Res. 54:4012–4023. 2021.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Yan K, Zhu W, Yu L, Li N, Zhang X, Liu P,
Chen Q, Chen Y and Han D: Toll-like receptor 3 and RIG-I-like
receptor activation induces innate antiviral responses in mouse
ovarian granulosa cells. Mol Cell Endocrinol. 372:73–85.
2013.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Amado-Rodriguez L, Salgado Del Riego E,
Gomez de Ona J, López Alonso I, Gil-Pena H, Lopez-Martinez C,
Martin-Vicente P, Lopez-Vazquez A, Gonzalez Lopez A, Cuesta-Llavona
E, et al: Effects of IFIH1 rs1990760 variants on systemic
inflammation and outcome in critically ill COVID-19 patients in an
observational translational study. Elife. 11(e73012)2022.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Dieter C, de Almeida Brondani L, Lemos NE,
Schaeffer AF, Zanotto C, Ramos DT, Girardi E, Pellenz FM, Camargo
JL, Moresco KS, et al: Polymorphisms in ACE1, TMPRSS2, IFIH1,
IFNAR2, and TYK2 genes are associated with worse clinical outcomes
in COVID-19. Genes (Basel). 14(29)2022.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Wang P, Yang L, Cheng G, Yang G, Xu Z, You
F, Sun Q, Lin RT, Fikrig E and Sutton RE: UBXN1 interferes with
Rig-I-like receptor-mediated antiviral immune response by targeting
MAVS. Cell Rep. 3:1057–1070. 2013.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Pugh C, Kolaczkowski O, Manny A,
Korithoski B and Kolaczkowski B: Resurrecting ancestral structural
dynamics of an antiviral immune receptor: Adaptive binding pocket
reorganization repeatedly shifts RNA preference. BMC Evol Biol.
16(241)2016.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Loske J, Rohmel J, Lukassen S, Stricker S,
Magalhães VG, Liebig J, Chua RL, Thürmann L, Messingschlager M,
Seegebarth A, et al: Pre-activated antiviral innate immunity in the
upper airways controls early SARS-CoV-2 infection in children. Nat
Biotechnol. 40:319–324. 2022.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Selinger C, Tisoncik-Go J, Menachery VD,
Agnihothram S, Law GL, Chang J, Kelly SM, Sova P, Baric RS and
Katze MG: Cytokine systems approach demonstrates differences in
innate and pro-inflammatory host responses between genetically
distinct MERS-CoV isolates. BMC Genomics. 15(1161)2014.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Rachmiel M, Bloch O, Bistritzer T,
Weintrob N, Ofan R, Koren-Morag N and Rapoport MJ: TH1/TH2 cytokine
balance in patients with both type 1 diabetes mellitus and asthma.
Cytokine. 34:170–176. 2006.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Zhu J, Message SD, Mallia P, Kebadze T,
Contoli M, Ward CK, Barnathan ES, Mascelli MA, Kon OM, Papi A, et
al: Bronchial mucosal IFN-α/β and pattern recognition receptor
expression in patients with experimental rhinovirus-induced asthma
exacerbations. J Allergy Clin Immunol. 143:114–125 e4.
2019.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Gill MA, Bajwa G, George TA, Dong CC,
Dougherty II, Jiang N, Gan VN and Gruchalla RS: Counterregulation
between the FcepsilonRI pathway and antiviral responses in human
plasmacytoid dendritic cells. J Immunol. 184:5999–6006.
2010.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Liu T, Zhou YT, Wang LQ, Li LY, Bao Q,
Tian S, Chen MX, Chen HX, Cui J and Li CW: NOD-like receptor
family, pyrin domain containing 3 (NLRP3) contributes to
inflammation, pyroptosis, and mucin production in human airway
epithelium on rhinovirus infection. J Allergy Clin Immunol.
144:777–787 e9. 2019.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Grünvogel O, Esser-Nobis K, Reustle A,
Schult P, Müller B, Metz P, Trippler M, Windisch MP, Frese M,
Binder M, et al: DDX60L is an interferon-stimulated gene product
restricting hepatitis C virus replication in cell culture. J Virol.
89:10548–10568. 2015.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Wolin SL and Maquat LE: Cellular RNA
surveillance in health and disease. Science. 366:822–827.
2019.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Kane M, Zang TM, Rihn SJ, Zhang FW, Kueck
T, Alim M, Schoggins J, Rice CM, Wilson SJ and Bieniasz PD:
Identification of interferon-stimulated genes with antiretroviral
activity. Cell Host Microbe. 20:392–405. 2016.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Cohen JI: GATA2 deficiency and
epstein-barr virus disease. Front Immunol. 8(1869)2017.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Spinner MA, Sanchez LA, Hsu AP, Shaw PA,
Zerbe CS, Calvo KR, Arthur DC, Gu W, Gould CM, Brewer CC, et al:
GATA2 deficiency: A protean disorder of hematopoiesis, lymphatics,
and immunity. Blood. 123:809–821. 2014.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Zhao X, Fan H, Chen X, Zhao X, Wang X,
Feng YJ, Liu M, Li S and Tang H: Hepatitis B Virus DNA polymerase
restrains viral replication through the CREB1/HOXA distal
transcript antisense RNA Homeobox A13 Axis. Hepatology. 73:503–519.
2021.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Jenkins DE, Sreenivasan D, Carman F, Samal
B, Eiden LE and Bunn SJ: Interleukin-6-mediated signaling in
adrenal medullary chromaffin cells. J Neurochem. 139:1138–1150.
2016.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Zhang Y, Le X, Zheng S, Zhang K, He J, Liu
M, Tu C, Rao W, Du H, Ouyang Y, et al: MicroRNA-146a-5p-modified
human umbilical cord mesenchymal stem cells enhance protection
against diabetic nephropathy in rats through facilitating M2
macrophage polarization. Stem Cell Res Ther. 13(171)2022.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Xie LM, Yin X, Bi J, Luo HM, Cao XJ, Ma
YW, Liu YL, Su JW, Lin GL and Guo XG: Identification of potential
biomarkers in dengue via integrated bioinformatic analysis. PLoS
Negl Trop Dis. 15(e0009633)2021.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Chen Y, Li S, Wei Y, Xu Z and Wu X:
Circ-RNF13, as an oncogene, regulates malignant progression of
HBV-associated hepatocellular carcinoma cells and HBV infection
through ceRNA pathway of circ-RNF13/miR-424-5p/TGIF2. Bosn J Basic
Med Sci. 21:555–568. 2021.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Wei G, Yi C, Ziyun J, Hui L, Hui L and
Yilong X: Network-Based analysis of the genetic effects of
SARS-CoV-2 infection to patients with exacerbation of Virus-Induced
Asthma (VAE). Research Square: https://doi.org/10.21203/rs.3.rs-948407/v1.
|