Introduction
Bupleurum is a widely used traditional Chinese
medicine (1,2). Modern scientific research has
revealed Bupleurum to possess various beneficial properties,
including antitumor, anti-inflammatory, hepatoprotective,
anti-fibrotic, sedative and antiepileptic effects (3). Consequently, Bupleurum-associated
preparations have found extensive application in clinical
practice.
With >100 ancient prescriptions and >549
formulated preparations containing Bupleurum currently available,
such as Chaihu injections, Chaihushugan tablets, Xiaoyaowan
tablets, Chaihu oral solution and Chaihu granules, utilization of
Bupleurum-associated preparations in clinical practice continues to
expand in China and a number of other Asian countries such as Japan
and South Korea (4).
Combinations of the Dachaihu decoction (a type of
Chaihu preparation) with hypoglycaemic medicines (such as
metformin, liraglutide and insulin) have been used for the
treatment of type 2 diabetes mellitus (5), whereas combinations of the
Chaihushugan decoction with antiepileptic drugs (such as sodium
valproate, carbamazepine, lamotrigine, sodium valproate and
phenobarbital) have been used for the treatment of epilepsy
(6). In addition, the combination
of Xiaochaihu decoction (a type of Chaihu preparation) with
Entecavir has been used for the treatment of chronic viral
hepatitis B (7). However, the
increasing use of these combinations may result in potential drug
interactions. It has been previously reported that the combined use
of the Xiaochaihu decoction and cyclosporin A (CsA) can lead to an
increase in the blood concentration of CsA resulting in toxic
effects or therapeutic failure, though the specific mechanism
remains unclear (8).
The cytochrome P450 (CYP) family serves a crucial
role in the metabolism of a number of clinical drugs (9-11).
Among the various CYPs, CYP3A4 accounts for 60% of the total
protein of CYPs in the liver and is primarily responsible for
metabolizing ~50% of all drugs currently used in the clinic,
including antiviral drugs, calcium channel blockers, macrolides and
benzodiazepines (12). By
contrast, CYP1A2 constitutes ~13% of the total CYP content in the
liver and participates in the metabolism of 9% of all drugs
currently used in the clinic (13), including propranolol,
chlorpromazine, phenytoin, mexiletine, propanolol, fluoxetine,
verapamil and nitrendipine. Therefore, investigating the effects of
drugs on CYP3A4 and CYP1A2 is likely to yield useful theoretical
information and practical value.
The HepaRG cell line was first developed by Gripon
et al (14) in 2002 whilst
investigating hepatitis B virus-infected human liver cancer cell
line. In the presence of 2% DMSO, HepaRG cells can differentiate
into canaliculus-like and hepatocyte-like cells that can
functionally express CYP1A2, CYP2D6, CYP2C9, CYP2E1 and CYP3A4 to
resemble human liver cells (14-16).
Therefore, HepaRG cells have been proposed to be a viable model for
studying drug metabolism and interactions in vitro as a
substitute for human primary liver cells (15-17).
Previous studies have established a HepaRG-based model to
investigate drug metabolism and interactions. Specifically, studies
on the effects of saikosaponin (SS)-D on the mRNA and protein
expression levels of CYP1A2, CYP2D6 and CYP3A4, as well as the
relative enzyme activities in HepaRG cells have been conducted
(18,19).
Bupleurum-associated preparations exert
pharmacological effects through multiple targets and pathways in
the body, but the specific active components responsible for
regulating CYP expression remain to be fully determined. Total
saikosaponins (TSS) are triterpene saponins that are extracted
solely from Bupleurum and consist of >100 different types,
including SSA, SSB, SSC and SSD (20,21).
A number of pharmacological effects of Bupleuri Radix have been
directly associated with TSS (22). Several previous studies have
examined TSS metabolism in rats, identifying cetyl-SSE, saikogenin
A, saikogenin G, SSA, SSB2 and saikogenin C as major metabolites
(22-24).
Therefore, the present study used HepaRG cells as a
model to investigate the impact of TSS on the mRNA and protein
expression of CYP3A4 and CYP1A2, aiming to elucidate the underlying
basis for potential drug interactions involving preparations
containing TSS or Bupleurum. It is hoped that these findings will
provide a theoretical basis for rational combination therapy and
further research on drug metabolism involving TSS or Bupleurum.
Materials and methods
Reagents and cells
Undifferentiated HepaRG cells were purchased from
Shanghai Binsui Biotechnology Co., Ltd. (cat. no. C554). TSS
(purity, ≥70%) was purchased from Shanghai Yuanye Biotechnology
Co., Ltd. Omeprazole and rifampicin were acquired from Shanghai
Aladdin Biochemical Technology Co., Ltd. Other reagents used were
as follows: TB Green® Premix EX Taq™ Ⅱ (cat. no. RR820A;
Takara Biotechnology Co., Ltd.), PrimeScript™ IV 1st strand cDNA
Synthesis Mix (cat. no. 6215A; Takara Biotechnology Co., Ltd.),
CYP3A4 rabbit polyclonal antibody (cat. no. DF3586; Affinity
Biosciences), CYP1A2 rabbit polyclonal antibody (cat. no. AF5312;
Affinity Biosciences), GAPDH rabbit polyclonal antibody (cat. no.
10494-1-AP; ProteinTech Group, Inc.), HRP-conjugated Affinipure
Goat Anti-Rabbit IgG (cat. no. SA00001-2; ProteinTech Group, Inc.),
BCA protein assay kit (cat. no. CW0014S; CoWin Biosciences) and
Cell Counting Kit-8 (CCK8; cat. no. SC119-01; Seven Innovation
(Beijing) Biotechnology Co., Ltd.).
Cell culture and morphology
Undifferentiated HepaRG cells were cultured in RPMI
1640 medium (Gibco; Thermo Fisher Scientific, Inc.) containing 10%
FBS (Gibco; Thermo Fisher Scientific, Inc.), 1% antibiotics (100x
streptomycin-penicillin) and 50 µM hydrocortisone sodium
hemisuccinate (basal growth medium; Shanghai Aladdin Biochemical
Technology Co., Ltd.), incubated in a constant temperature
incubator at 37˚C, 5% CO2 and 95% humidity for 2 weeks.
Subsequently, the same culture medium supplemented with 2% DMSO
(differentiation medium) was added to the cells for a further 2
weeks at 37˚C (14). Images of
differentiated and undifferentiated cells were captured using an
inverted light microscope (Olympus CKX53; Olympus Corporation).
Differentiated HepaRG cells were seeded into 96-well plates or
6-well plates for follow-up experiments (14,25).
The differentiation medium was renewed every 2-3 days.
Cell viability assay
A toxicity analysis of TSS on HepaRG cells was
performed to determine a suitable range of concentrations for the
study regarding drug-drug interactions. Differentiated HepaRG cells
were seeded into 96-well plates at a density of 4.5x105
cell/cm2 and incubated for 12 h at 37˚C before being
treated with different concentrations of TSS dissolved in 0.1% DMSO
(0, 1, 5, 10, 15, 20, 30 and 40 µg/ml) for 72 h at 37˚C with 5%
CO2. Untreated cells that were incubated in the medium
served as a control and the medium served as a blank. Subsequently,
10 µl CCK8 solution (5 mg/ml) prepared in RPMI 1640 medium (without
FBS, antibiotics and hydrocortisone sodium hemisuccinate) was added
into each well. After 2 h of incubation at 37˚C, the
CCK8-containing medium was removed. The absorbance of each well was
detected by an enzyme-labelled instrument (iMARK; Bio-Rad
Laboratories, Inc.) at a wavelength of 490 nm.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
To characterize the differentiation of HepaRG cells,
differentiated HepaRG cells were seeded into 6-well plates at a
density of 4.5x105 cell/cm2 and incubated for
24 h at 37˚C, whereas undifferentiated cells were used as a
control. To investigate the effects of TSS on the mRNA expression
levels of CYP1A2 and CYP3A4, differentiated HepaRG cells were
treated with 50 µM rifampicin, 50 µM omeprazole or different
concentrations of TSS [0, 5, 10 or 15 µg/ml (this range was
selected based on the CCK8 results in order to achieve cell
viability)] and incubated for 72 h at 37˚C. Total RNA was extracted
from HepaRG cells using the RNAiso Plus reagent (Takara
Biotechnology Co., Ltd.) according to the manufacturer's protocol
and the total RNA concentration was quantified using a
spectrophotometer (A260/A280). Total RNA (500 ng) was reverse
transcribed using the PrimeScriptTM IV 1st strand cDNA
Synthesis Mix (cat. no. 6215A; Takara Biotechnology Co., Ltd.), and
the reverse transcription conditions were: 37˚C for 15 min, 85˚C
for 5 sec and 4˚C for 10 min. The complementary DNA (cDNA) obtained
from reverse transcription was diluted 10 times with RNase-free
distilled water. The TB Green® Premix EX
TaqTM Ⅱ (TIi RNaseH Plus) (cat. no. RR820A; Takara
Biotechnology Co., Ltd.) was used for quantitation with the CFX96
real-time PCR detection system (Bio-Rad Laboratories, Inc.). The
PCR amplification procedure was performed as follows: Sample
volume, 25 µl; initial denaturation, 95˚C for 30 sec; PCR
amplification (40 cycles), consisting of denaturation (95˚C for 5
sec) and annealing/extension (60˚C for 30 sec). Analysis of each
specimen for each target was repeated three times. Gene
transcription was analysed with the
2-ΔΔCq method (26) using GAPDH expression as a
reference. The primer sequences used are presented in Table I.
 | Table IPrimer sequences used for reverse
transcription-quantitative polymerase chain reaction. |
Table I
Primer sequences used for reverse
transcription-quantitative polymerase chain reaction.
| Gene | Forward
(5'-3') | Reverse
(5'-3') |
|---|
| CYP3A4 |
CTTCATCCAATGGACTGCATAAA |
TCCCAAGTATAACACTCTACACACACA |
| CYP1A2 |
ATGCTCAGCCTCGTGAAGAAC |
GTTAGGCAGGTAGCGAAGGAT |
| CYP2B6 |
TTCCTACTGCTTCCGTCTATC |
GTGCAGAATCCCACAGCTCA |
| GAPDH |
AGAAGGCTGGGGCTCATTTG |
AGGGGCCATCCACAGTCTTC |
Western blot analysis
The effects of TSS on the protein expression levels
of CYP3A4 and CYP1A2 in HepaRG cells were detected using western
blot analysis. The differentiated HepaRG cells were seeded in 6
well plates, incubated and treated as aforementioned. Total protein
was extracted from HepaRG cells using RIPA cell lysis buffer
containing proteinase inhibitor (Beijing Solarbio Science &
Technology Co., Ltd.). The protein concentration was measured using
a BCA assay kit (cat. no. CW0014S; CoWin Biosciences). Protein
samples (40 µg) were loaded per lane and separated on 10% sodium
dodecyl sulphate-polyacrylamide gels with electrophoresis and
transferred onto polyvinylidene difluoride membranes. The membranes
were blocked with 5% skimmed milk at room temperature for 2 h.
Subsequently, the membranes were incubated with primary antibodies
at 4˚C overnight and were then incubated with an HRP-conjugated
secondary antibody at room temperature for 1 h. The membranes were
washed three times with TBST, which contains 0.1% Tween20.
Chemiluminescent detection was performed using
electrochemiluminescence reagents (Nanjing KeyGen Biotech Co.,
Ltd.) with a ChemiDoc MP imaging system (Bio-Rad Laboratories,
Inc.).
The primary and secondary antibodies were used at
the following dilutions: Antibodies for GAPDH (1:5,000), CYP3A4
(1:1,000), CYP1A2 (1:2,000) and HRP-conjugated Affinipure Goat
Anti-Rabbit IgG (1:5,000). The protein expression levels were
quantified by comparing against the GAPDH bands using ImageJ
Software v1.6.0 (National Institutes of Health).
Statistical analysis
SPSS 29.0 (IBM Corp.) was used for statistical
analysis of all data, and the results are presented as the mean ±
standard deviation (n=3) as indicated in corresponding figure
legends. The Shapiro-Wilk normality test was used to test
distribution normality and the Levene test to assess variance
homogeneity. The comparison between groups were conducted using
one-way analysis of variance (ANOVA). When variances were found to
be homogeneous, multiple comparisons between groups were evaluated
using the Bonferroni post hoc test. In instances where the assessed
variances exhibited non-normality and/or unequal variances,
non-parametric statistical analysis via the Kruskal-Wallis test
with Dunn's multiple comparisons post hoc test was utilized. For
comparison between two groups, the unpaired Student's t-test was
performed. P<0.05 was considered to indicate a statistically
significant difference.
Results
Identification of differentiated
HepaRG cells
Microscopic examination of undifferentiated HepaRG
cells revealed the presence of numerous characteristic granular
epithelial cell colonies encircled by flat, well-cytoplasmic
cholangiocyte-like cells, as depicted in Fig. 1A-a. By contrast, differentiated
HepaRG cells exhibited distinct features under phase contrast
microscopy, with hepatocyte-like cells displaying a dense and
indefinite cytoplasm, epithelial-like cells with flat and
well-defined cytoplasm, and the presence of bile canalicular
structures, as illustrated in Fig.
1A-b. The mRNA expression levels of CYP3A4, CYP1A2 and CYP2B6
were significantly increased in the differentiated HepaRG cells
compared with those in the undifferentiated cells (Fig. 1B-D).
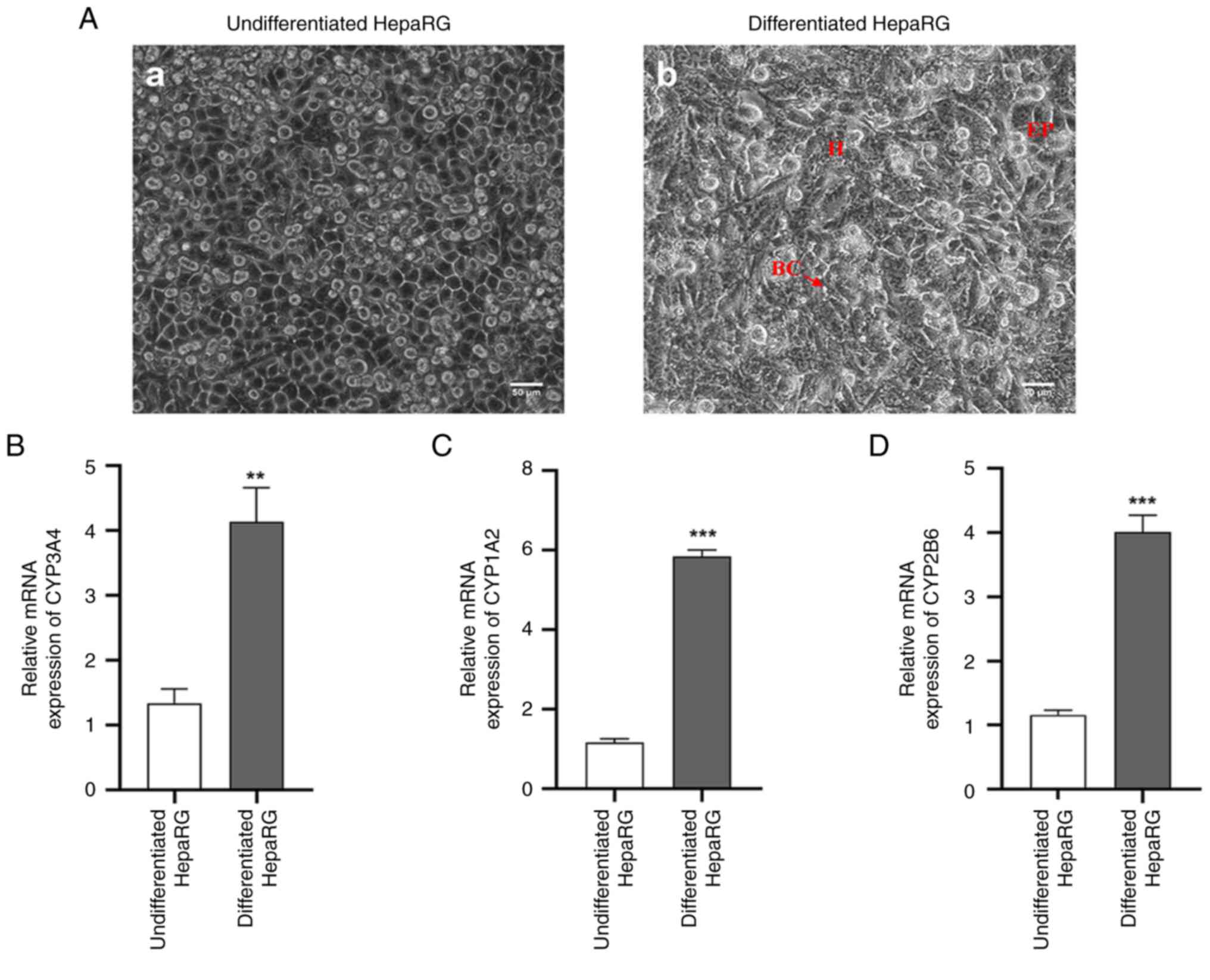 | Figure 1Identification of differentiated
HepaRG cells. (A-a) Phase contrast micrograph of undifferentiated
HepaRG cells, which indicated the formation of a large number of
typical granular epithelial cell colonies surrounded by flat and
well-cytoplasmic cholangiocyte-like cells. (A-b) Phase contrast
micrograph of differentiated HepaRG cells. Hepatocyte-like cells
with dense and indefinite cytoplasm (indicated by ‘H’),
epithelial-like cells with flat and well-defined cytoplasm
(indicated by ‘EP’), and bile canalicular structures (indicated by
‘BC’ and an arrow) can be observed. Expression levels of (B)
CYP3A4, (C) CYP1A2 and (D) CYP2B6 mRNA in HepaRG cells. Scale bars,
50 µm. **P<0.01 and ***P<0.001 vs.
undifferentiated HepaRG cells. For comparison between two groups,
the unpaired Student's t-test was used to test the statistical
significance (B, C and D). H, hepatocyte-like cells; EP,
epithelial-like cells; BC, bile canalicular structures; CYP,
cytochrome P450. |
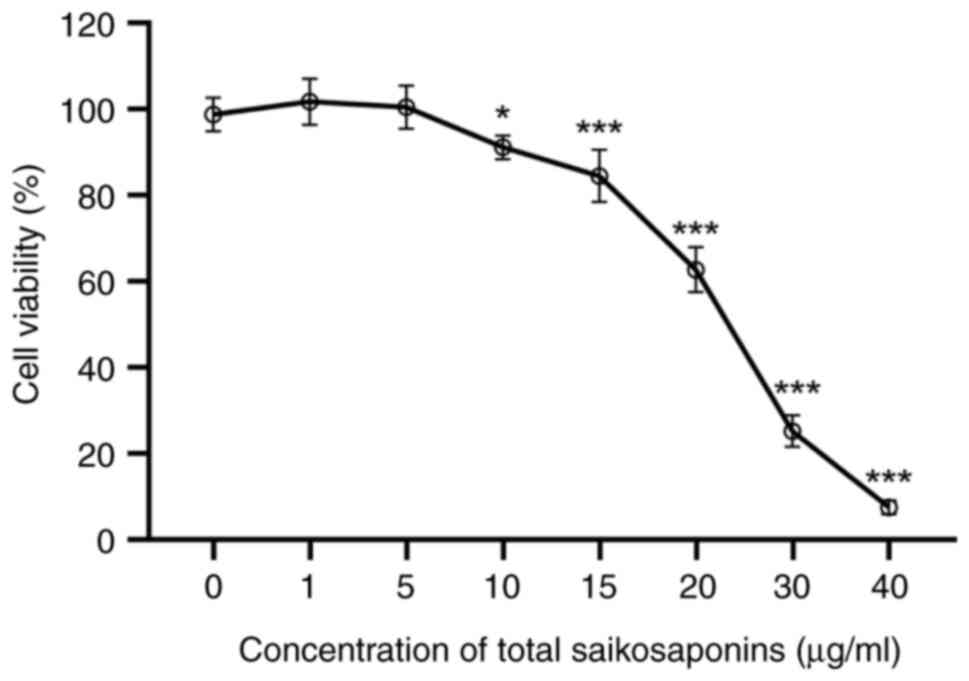
Cell viability analysis
Results of the in vitro cell viability assay
indicated that the HepaRG cell viability was >80% with TSS
concentrations in the range of 0-15 µg/ml, indicating no cytotoxic
effect (Fig. 2). This suggested
that TSS concentrations <15 µg/ml could be used for the
subsequent drug interaction experiments.
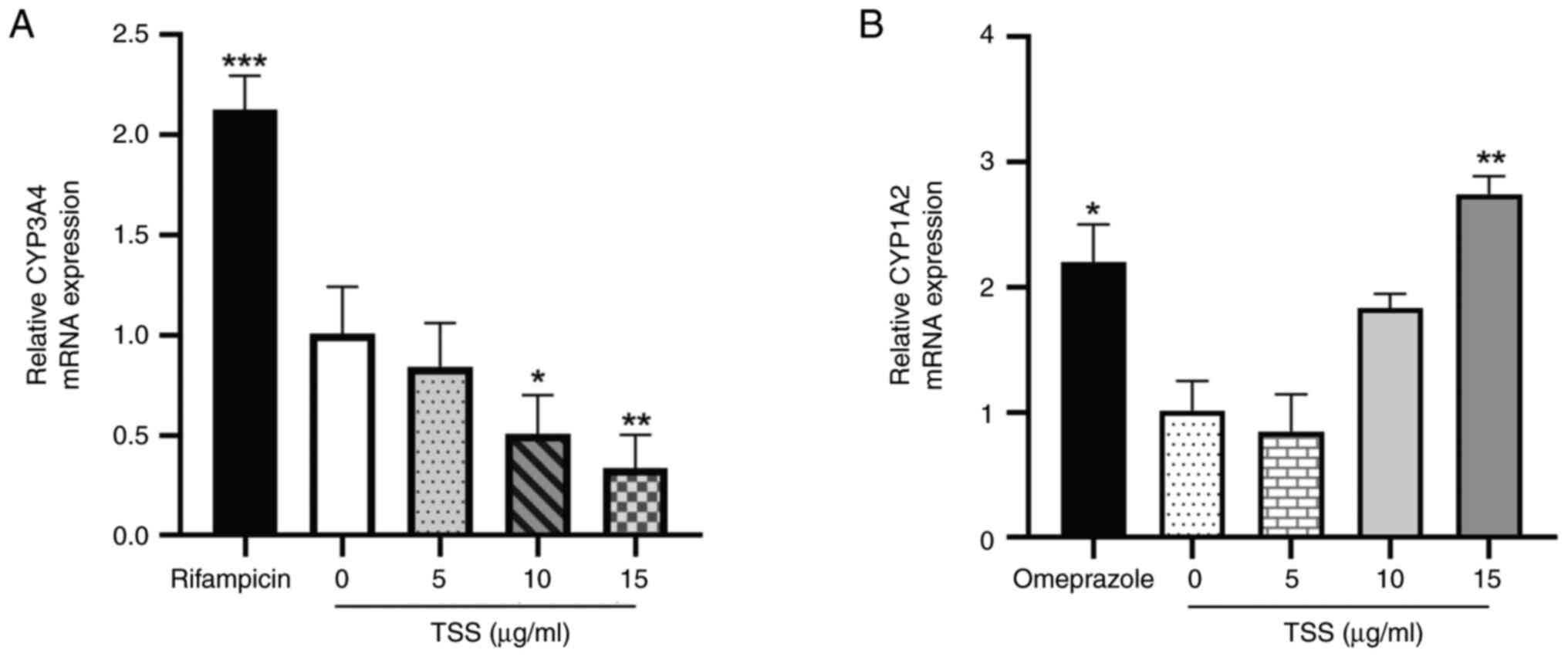
Effects of TSS on the expression
levels of CYP3A4 and CYP1A2 mRNA
As presented in Fig.
3, compared with those in the control (0 µg/ml TSS), 50 µM
rifampicin and 50 µM omeprazole significantly increased CYP3A4 and
CYP1A2 mRNA expression, respectively. However, 10 and 15 µg/ml TSS
significantly reduced the mRNA expression levels of CYP3A4 in a
dose-dependent manner compared with those in the control group
(Fig. 3A). By contrast, CYP1A2
expression was observed to be significantly increased by 15 µg/ml
TSS compared with that in the control group (Fig. 3B).
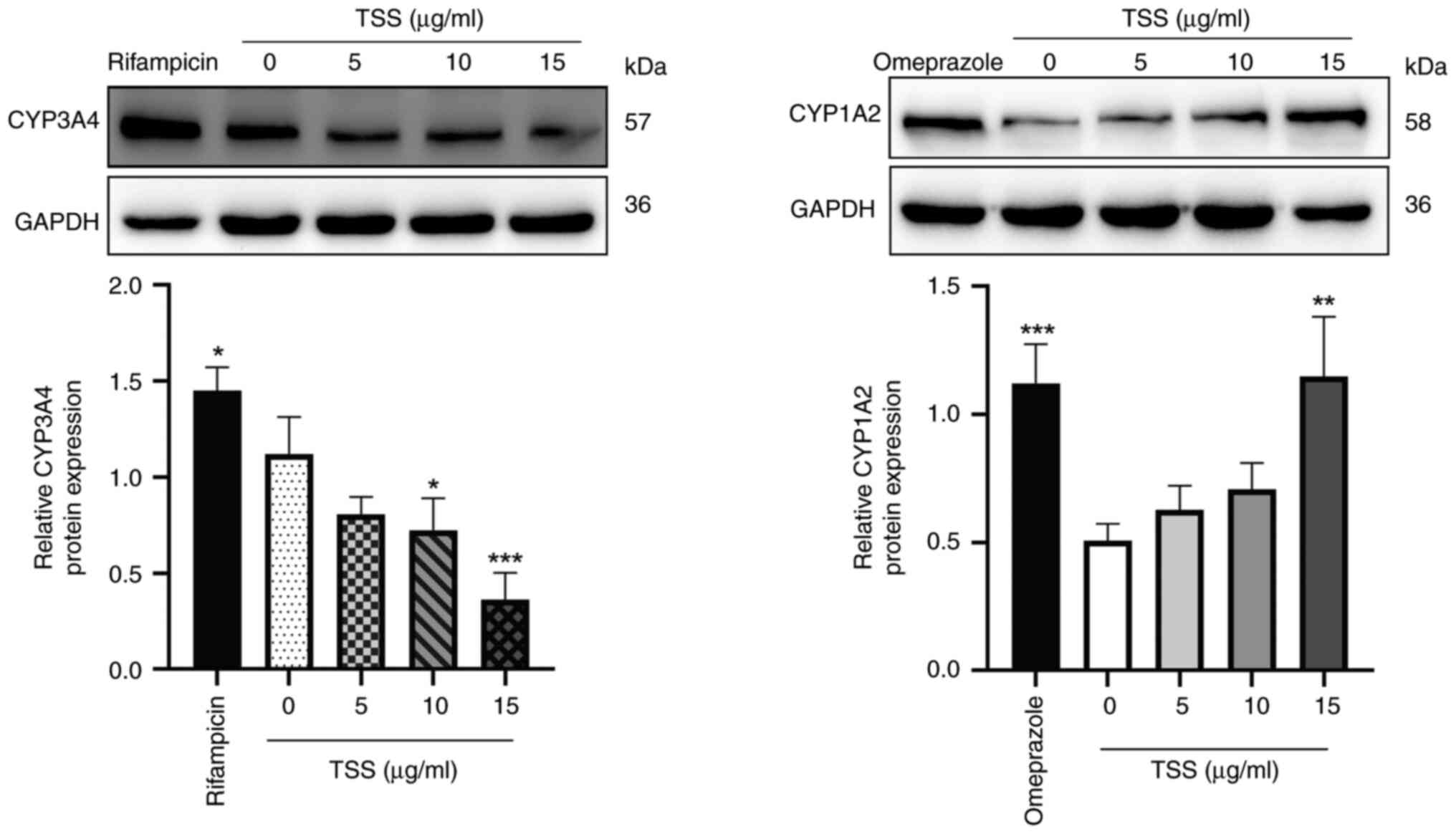
Effects of TSS on the expression
levels of CYP3A4 and CYP1A2 protein
Protein expression levels of CYP3A4 and CYP1A2 in
HepaRG cells treated with TSS for 72 h were next measured (Fig. 4). Compared with those in the
control group (0 µg/ml TSS), 50 µM rifampicin and 50 µM omeprazole
significantly increased the CYP3A4 and CYP1A2 protein expression
levels, respectively. In addition, 10 and 15 µg/ml TSS
significantly reduced the protein expression levels of CYP3A4 in a
dose-dependent manner compared with those in the control group
(Fig. 4A). However, CYP1A2
expression levels were observed to be significantly increased by 15
µg/ml TSS compared with those in the control group (Fig. 4B).
Discussion
Pharmacokinetic drug-drug interactions, resulting
from the induction or inhibition of CYP enzymes, are commonly known
concerns in clinical practice as these interactions can potentially
lead to changes in the drug concentration in the blood, which may
result in adverse reactions or therapeutic failure (27,28).
Screening and metabolism studies of novel drug candidates in Europe
and the United States mandate CYP assays, which are also mandatory
for new drug applications (29).
Similarly, China's Guiding Principles for Non-Clinical
Pharmacokinetic Studies of Chemical Drug stipulates that novel
prospective drugs should be evaluated for their effects on CYPs
induction or inhibition (30-32).
Therefore, drug interactions involving CYPs have become a topic of
interest for research in the field of clinical pharmacy.
Traditionally, rodents and primates have been used
as experimental models for studying drug metabolism and drug
interactions. However, species differences can affect the relevance
of these models for human metabolism (33-35).
The liver is the main organ for drug metabolism (36). Primary human hepatocytes (PHH) have
been considered to be the gold standard for in vitro drug
metabolism research. However, the practicality and reliability of
PHH as a research model remain limited due to difficulties in cell
culture, early phenotypic changes, short maintenance time of
metabolic enzyme activity and donor-to-donor variations (13,37).
The human liver cancer cell line HepG2 has been proposed to be an
alternative in vitro drug metabolism model. However, HepG2
cells express relatively low levels of human liver-specific
functions such as drug-metabolizing enzymes (CYP1A2, CYP2B6,
CYP2C9, CYP2C19, CYP2D6 and CYP3A4) despite being readily
available, limiting their application in drug metabolism and
interaction research (37).
Another cell line, HuH7(38),
suffer from similar limitations (12,39-41).
By contrast, the HepaRG cell line, which was originally isolated
from a patient with a chronic hepatitis C viral infection (11), has shown promising characteristics
for in vitro drug metabolism research. When cultured with
DMSO, HepaRG cells can differentiate into hepatocyte-like cells
that express various enzymes, including CYPs. These differentiated
cells maintain liver-specific functional properties and have been
proposed to be an appropriate model for drug metabolism research
(12,42,43).
They have been used in studies associated with drug metabolism,
drug interactions and toxicology (44-47).
Furthermore, HepaRG cells can be induced to increase the expression
levels of CYP1A2, CYP2B6 and CYP3A4 enzymes by specific inducers,
such as omeprazole, phenobarbital and rifampicin (48).
In the present study, TSS, the main active component
of Bupleurum, was investigated. TSS has been used in clinical
practice due to its various reported biological activities,
including antidepressive, anti-inflammatory, antiviral,
antiendotoxin, antitumor, anti-pulmonary fibrosis and anti-gastric
ulcer effects (49-51).
TSS has also gained attention over the past decade for its
neuroprotective and nephroprotective effects (52,53).
Given its extensive use and potential interactions with drugs
metabolized by CYPs, TSS is an active ingredient worthy of
attention in drug development. However, despite TSS being the main
bioactive component of Bupleurum, its effects on CYPs have, to the
best of our knowledge, only been investigated twice (31,54).
A previous study reported the downregulation of CYP3A4 protein
expression by TSS in mice, supporting the potential inhibitory
effects of TSS on CYP3A4(54).
Additionally, TSS has been found to exert inhibitory effects on
CYP3A11 while inducing CYP1A2 in the liver and intestines of mice
(31). In addition, the effects of
Xiaochaihu decoction extract (containing TSS) on the mRNA and
protein expression levels of CYP1A2, CYP3A1, CYP2D6 and CYP1B1 in
rats have been examined (55).
However, the effects of TSS alone on CYPs still requires further
investigation.
A previous study has reported an increase in the
blood concentration of CsA when combined with Xiaochaihu decoction,
but the specific substance and pathway involved in this mechanism
remains unknown (8). In our
previous studies, it was revealed that one of the saponins isolated
from Bupleurum, SSD, can inhibit the activity of the CYP3A4 enzyme
in HepaRG cells whilst increasing CYP1A2 enzymes (18,19).
In the present study, it was observed that compared with those in
the control group, TSS downregulated the mRNA and protein
expression levels of the CYP3A4 enzyme in HepaRG cells but
upregulated the mRNA and protein expression levels of CYP1A2. CsA
is a substrate of the drug metabolism enzyme CYP3A4(56). Therefore, it could be hypothesized
that the increase in the blood concentration of CsA caused by the
Xiaochaihu decoction may be due to the inhibition of CYP3A4 by TSS,
leading to an impaired CsA metabolism.
Consistent with the findings of the present study,
another previous study has also reported the downregulation of
CYP3A4 protein expression by TSS in mice, supporting the potential
inhibitory effect of TSS on CYP3A4(54). Additionally, TSS has been found to
exert an inhibitory effect on CYP3A11 (which corresponds to human
CYP3A4) whilst inducing CYP1A2 in the liver and intestines of mice
(31). Our previous study on the
effect of the Xiaochaihu decoction in mouse found that it induced
the mRNA and protein expression levels of both CYP1A2 and CYP3A1
(the mouse homolog of human CYP3A4) (55). This discrepancy may be attributed
to species differences and the interactions of other components in
the Xiaochaihu decoction. At present, it is challenging to
determine the concentrations and the active forms of all components
in TSS. Due to the metabolism of TSS, it can be transformed into
specific metabolites in vivo by various transformation
factors, such as the intestinal microflora, gastrointestinal
enzymes and gastric acids, making it difficult to determine the
actual pharmacological substances after TSS enters the body. CYPs
can be influenced by both endogenous and exogenous substances such
as anticoagulants, antidiabetics and antimicrobials (36), leading to modifications in enzyme
quantity and activity (57). In
the present study, the effects of TSS on CYP3A4 and CYP1A2 protein
and mRNA levels were investigated, without assessing enzyme
activity. The lack of experiments investigating enzyme activity was
a limitation of the present study.
The results of the present study suggest that TSS
may inhibit CYP3A4 whilst inducing CYP1A2 expression. Further
research is warranted to investigate the bioavailability and
gastrointestinal absorption of each SS, whilst also exploring the
impact of TSS on the enzyme activity of CYP3A4 and CYP1A2. In
addition, the effects of other substances in the Xiaochaihu
decoction on CYP enzymes and the clinical implications of
TSS-associated interactions with drugs metabolized by CYPs are
worthy of further investigation. These aspects are important for a
more comprehensive understanding of the implications of the present
findings.
In the present study, established protocols and
guidelines for using rifampicin as a positive control inducer for
CYP3A4 and omeprazole as a positive control inducer for CYP1A2 were
followed. Rifampicin is recommended by the U.S. Food and Drug
Administration (FDA) as a reference compound for evaluating the
CYP3A4 induction potential and, as a well-known potent inducer of
CYP3A4, it is used as a positive control inducer for studying the
expression of CYP3A4 in vitro (58,59).
Similarly, omeprazole is a known inducer of CYP1A2, and it is used
as a positive control inducer for studying the expression of CYP1A2
in vitro, which is also recommended by the FDA as a standard
reference compound for assessing the CYP1A2 induction potential
(58,59). By using these compounds, the
present study aimed to ensure the reliability and comparability of
the present results with previous studies.
In conclusion, utilizing HepaRG cells as a model for
drug metabolism enzymes revealed that compared with those in the
control group, TSS reduced the mRNA and protein expression levels
of CYP3A4 whilst increasing those of CYP1A2. These findings
highlight the potential for drug interactions when TSS or Bupleurum
associated preparations are used in combination with drugs
metabolized by CYP3A4 and CYP1A2 enzymes in clinical practice.
Particular attention should be given to drug interactions to
optimize therapeutic effects and minimize potential adverse
reactions.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by grants from Zunyi
Science and Technology Bureau Zunyi Medical University Joint Fund
Project 2021 (Zunyi Science and Technology Cooperation HZ; grant
no. [2021]-289); Zunyi Medical University Academic New Seed
Cultivation and Innovation Exploration Special Project 2020 (Qianke
Platform Talent; grant no. [2020]-015); and National Natural
Science Foundation of China (grant no. 32160225).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
FT, YT and HL conceived and designed the
experiments. HL, LH and YL performed the experiments. HL, YT, FM
and JT analyzed the data. FT, HL and YT confirm the authenticity of
all the raw data. FT, HL and YT prepared the manuscript. All
authors read and approved the final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Yuan B, Yang R, Ma YS, Zhou S, Zhang XD
and Liu Y: A systematic review of the active saikosaponins and
extracts isolated from Radix Bupleuri and their applications. Pharm
Biol. 55:620–635. 2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Zhang X, Liu Z, Chen S, Li H, Dong L and
Fu X: A new discovery: Total Bupleurum saponin extracts can inhibit
the proliferation and induce apoptosis of colon cancer cells by
regulating the PI3K/Akt/mTOR pathway. J Ethnopharmacol.
283(114742)2022.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Wang H and Liu X: Research progress on
Bupleurum marginatum var. stenophyllum. World Latest Med Info.
19:110–113. 2019.(In Chinese).
|
|
4
|
Ashour ML and Wink M: Genus Bupleurum: A
review of its phytochemistry, pharmacology and modes of action. J
Pharm Pharmacol. 63:305–321. 2011.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Liu YF, Luo WT, Tang TT, Tang C and Xiao
YP: A meta-analysis of clinical efficacy of the Dachaihu decoction
plus hypoglycemic medicine on type 2 diabetes mellitus. Clin J Chin
Med. 14(5)2022.(In Chinese).
|
|
6
|
Li HX, Yu Q, Wang S, Li HQ, Wu Q, Chen Al
and Diao LM: Meta-analysis of efficacy and safety of Chaihushugan
decoction combined with antiepileptic drugs in treatment of
epilepsy. Chin Arch Tradit Chin Med. 8:1673–7717. 2020.(In
Chinese).
|
|
7
|
Zhu MJ, Cai MQ, Yang HP, Wang L and Zhao
WX: Meta-analysis of Xiaochaihu decoction combined with entecavir
in the treatment of chronic viral hepatitis B. Western J Trad Chin
Med. 34:62–66. 2021.(In Chinese).
|
|
8
|
Zhou Y and Chu XM: Effects of oral
administration of minor Radix Buplenri granule on whole blood
cyclosporin A concentration in kidney transplantation patients.
Chin J Hosp Pharm. 19(713)1999.(In Chinese).
|
|
9
|
Lewis DF: P450 structures and oxidative
metabolism of xenobiotics. Pharmacogenomics. 4:387–395.
2003.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Faber MS, Jetter A and Fuhr U: Assessment
of CYP1A2 activity in clinical practice: Why, how, and when? Basic
Clin Pharmacol Toxicol. 97:125–134. 2005.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Goutelle S, Bourguignon L, Bleyzac N,
Berry J, Clavel-Grabit F and Tod M: In vivo quantitative prediction
of the effect of gene polymorphisms and drug interactions on drug
exposure for CYP2C19 substrates. AAPS J. 15:415–426.
2013.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Zanger UM and Schwab M: Cytochrome P450
enzymes in drug metabolism: regulation of gene expression, enzyme
activities, and impact of genetic variation. Pharmacol Ther.
138:103–141. 2013.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Chen Y, Zeng L, Wang Y, Tolleson WH, Knox
B, Chen S, Ren Z, Guo L, Mei N, Qian F, et al: The expression,
induction and pharmacological activity of CYP1A2 are
post-transcriptionally regulated by microRNA hsa-miR-132-5p.
Biochem Pharmacol. 145:178–191. 2017.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Gripon P, Rumin S, Urban S, Le Seyec J,
Glaise D, Cannie I, Guyomard C, Lucas J, Trepo C and
Guguen-Guillouzo C: Infection of a human hepatoma cell line by
hepatitis B virus. Proc Natl Acad Sci USA. 99:15655–15660.
2002.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Wang Z, Luo X, Anene-Nzelu C, Yu Y, Hong
X, Singh NH, Xia L, Liu S and Yu H: HepaRG culture in tethered
spheroids as an in vitro three-dimensional model for drug safety
screening. J Appl Toxicol. 35:909–917. 2015.PubMed/NCBI View
Article : Google Scholar
|
|
16
|
Andersson TB, Kanebratt KP and Kenna JG:
The HepaRG cell line: A unique in vitro tool for understanding drug
metabolism and toxicology in human. Expert Opin Drug Metab Toxicol.
8:909–920. 2012.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Li HF, Sun SS, Zhang T and Tang FS:
Research progress on the application of HepaRG cells in drug
metabolism. Herald Med. 38:1038–1043. 2019.(In Chinese).
|
|
18
|
Li H, Tang Y, Wang Y, Wei W, Yin C and
Tang F: Effects of saikosaponin D on CYP1A2 and CYP2D6 in HepaRG
cells. Drug Des Devel Ther. 14:5251–5258. 2020.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Li H, Tang Y, Wei W, Yin C and Tang F:
Effects of saikosaponin-D on CYP3A4 in HepaRG cell and
protein-ligand docking study. Basic Clin Pharmacol Toxicol.
128:661–668. 2021.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Liu QX, Tan L, Bai YJ, Liang H and Zhao
YY: A survey of the studies on saponins from Bupleurum in past 10
years. Zhongguo Zhong Yao Za Zhi. 27(7-11.45)2002.PubMed/NCBI(In Chinese).
|
|
21
|
Huang W, Lv Z and Sun R: Research
development on chemical compositions in Bupleurum chinense related
with efficacy and toxicity. Chin J Pharmacovigil. 10:545–548.
2013.(In Chinese).
|
|
22
|
Wang YZ, Zhang XN, Zhang Y, Duan CC and
Zhang JY: Study on metabolic profiling of total saponins of
Bupleurum chinense DC in Rats. J Zunyi Med Uni. 46:21–29. 2023.(In
Chinese).
|
|
23
|
Yu P, Qiu H, Wang M, Tian Y, Zhang Z and
Song R: In vitro metabolism study of saikosaponin D and its
derivatives in rat liver microsomes. Xenobiotica. 47:11–19.
2017.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Liu G, Tian Y, Li G, Xu L, Song R and
Zhang ZJ: Metabolism of saikosaponin A in rats: Diverse oxidations
on the aglycone moiety in liver and intestine in addition to
hydrolysis of glycosidic bonds. Drug Metab Dispos. 41:622–633.
2013.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Aninat C, Piton A, Glaise D, Le
Charpentier T, Langouët S, Morel F, Guguen-Guillouzo C and
Guillouzo A: Expression of cytochromes P450, conjugating enzymes
and nuclear receptors in human hepatoma HepaRG cells. Drug Metab
Dispos. 34:75–83. 2006.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta C (T)) method. Methods. 25:402–408. 2001.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Mar PL, Gopinathannair R, Gengler BE,
Chung MK, Perez A, Dukes J, Ezekowitz MD, Lakkireddy D, Lip GYH,
Miletello M, et al: Drug interactions affecting oral anticoagulant
use. Circ Arrhythm Electrophysiol. 15(e007956)2022.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Hakkola J, Hukkanen J, Turpeinen M and
Pelkonen O: Inhibition and induction of CYP enzymes in humans: An
update. Arch Toxicol. 94:3671–3722. 2020.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Huang SM, Temple R, Throckmorton DC and
Lesko LJ: Drug interaction studies: Study design, data analysis,
and implications for dosing and labeling. Clin Pharmacol Ther.
81:298–304. 2007.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Song FX, Wang R and Yuan YF: Application
progress in cytochrome P450 enzymes and research methods of drug
metabolism. Med Recapitulate. 23:665–669. 2017.(In Chinese).
|
|
31
|
Wang YH, Qi JF and Lin M: Effects of total
saikosaponins on intestinal first-pass effect and CYP3A, CYP2E1.
Chin J Clin Pharm Ther. 16:740–748. 2011.(In Chinese).
|
|
32
|
Tang YY, Lan X, Shang DC and Tang FS:
Research overview of Xiaochuhu decoction combined with chemical
drugs and their interaction. China Pharm. 30:846–850. 2019.(In
Chinese).
|
|
33
|
Xue ZK and Wei H: Comparison of
pharmacokinetics between recombinant liver microsomes with CYP3A4
and CYP3A29. Herald Med. 34:15–21. 2015.
|
|
34
|
Kang KJ, Min BH, Lee JH, Kim ER, Sung CO,
Cho JY, Seo SW and Kim JJ: Alginate hydrogel as a potential
alternative to hyaluronic acid as submucosal injection material.
Dig Dis Sci. 58:1491–1496. 2013.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Michaut A, Le Guillou D, Moreau C, Bucher
S, McGill MR, Martinais S, Gicquel T, Morel I, Robin MA, Jaeschke H
and Fromenty B: A cellular model to study drug-induced liver injury
in nonalcoholic fatty liver disease: Application to acetaminophen.
Toxicol Appl Pharmacol. 292:40–55. 2016.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Bozina N, Bradamante V and Lovri M:
Genetic polymorphism of metabolic enzymes P450 (CYP) as a
susceptibility factor for drug response, toxicity, and cancer risk.
Arh Hig Rada Toksikol. 60:217–242. 2009.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Liu ZJ, Zhang Y, Yin PH and Zhao L:
Toxicity evaluation of landfill leachate before and after SND/UF/RO
treatment on HepG2 cells. Acta Scientiae Circumstantiae.
41:660–669. 2021.(In Chinese).
|
|
38
|
Godoy P, Hewitt NJ, Albrecht U, Andersen
ME, Ansari N, Bhattacharya S, Bode JG, Bolleyn J, Borner C, Böttger
J, et al: Recent advances in 2d and 3d in vitro systems using
primary hepatocytes, alternative hepatocyte sources and
non-parenchymal liver cells and their use in investigating
mechanisms of hepatotoxicity, cell signaling and ADME. Arch
Toxicol. 87:1315–1530. 2013.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Lübberstedt M, Müller-Vieira U, Mayer M,
Biemel KM, Knöspel F, Knobeloch D, Nüssler AK, Gerlach JC and
Zeilinger K: HepaRG human hepatic cell line utility as a surrogate
for primary human hepatocytes in drug metabolism assessment in
vitro. J Pharmacol Toxicol Methods. 63:59–68. 2011.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Pernelle K, Le Guevel R, Glaise D, Stasio
CG, Le Charpentier T, Bouaita B, Corlu A and Guguen-Guillouzo C:
Automated detection of hepatotoxic compounds in human hepatocytes
using HepaRG cells and image-based analysis of mitochondrial
dysfunction with Jc-1 dye. Toxicol Appl Pharmacol. 254:256–266.
2011.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Sugiyama I, Murayama N, Kuroki A, Kota J,
Iwano S, Yamazaki H and Hirota T: Evaluation of cytochrome P450
inductions by anti-epileptic drug oxcarbazepine,
10-hydroxyoxcarbazepine, and carbamazepine using human hepatocytes
and HepaRG cells. Xenobiotica. 46:765–774. 2006.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Leite SB, Wilk-Zasadna I, Zaldivar JM,
Airola E, Reis-Fernandes MA, Mennecozzi M, Guguen-Guillouzo C,
Chesne C, Guillou C, Alves PM and Coecke S: Three-dimensional
HepaRG model as an attractive tool for toxicity testing. Toxicol
Sci. 130:106–116. 2012.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Jiang L and Wang YM: Biological
characteristics and application of HepaRG cells. Chin Hepatol.
15:380–383. 2010.(In Chinese).
|
|
44
|
Wuerger LTD, Hammer HS, Hofmann U,
Kudiabor F, Sieg H and Braeuning A: Okadaic acid influences
xenobiotic metabolism in HepaRG cells. EXCLI J. 21:1053–1065.
2022.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Ashraf MN, Asghar MW, Rong Y, Doschak MR
and Kiang TKL: Advanced in vitro HepaRG culture systems for
xenobiotic metabolism and toxicity characterization. Eur J Drug
Metab Pharmacokinet. 44:437–458. 2019.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Hammour MM, Othman A, Aspera-Werz R, Braun
B, Weis-Klemm M, Wagner S, Nadalin S, Histing T, Ruoß M and Nüssler
AK: Optimisation of the HepaRG cell line model for drug toxicity
studies using two different cultivation conditions: Advantages and
limitations. Arch Toxicol. 96:2511–2521. 2022.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Meirinho S, Rodrigues M, Fortuna A, Falcão
A and Alves G: Study of the metabolic stability profiles of
perampanel, rufinamide and stiripentol and prediction of drug
interactions using HepaRG cells as an in vitro human model. Toxicol
In Vitro. 82(105389)2022.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Anthérieu S, Chesné C, Li R, Camus S,
Lahoz A, Picazo L, Turpeinen M, Tolonen A, Uusitalo J,
Guguen-Guillouzo C and Guillouzo A: Stable expression, activity,
and inducibility of cytochromes P450 in differentiated HepaRG
cells. Drug Metab Dispos. 38:516–525. 2010.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Xiao LX, Zhou HN and Jiao ZY: Present and
future prospects of the anti-cancer activities of saikosaponins.
Curr Cancer Drug Targets. 23:2–14. 2022.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Li J, Zou B, Cheng XY, Yang XH, Li J, Zhao
CH, Ma RX, Tian JX and Yao Y: Therapeutic effects of total
saikosaponins from Radix Bupleuri against Alzheimer’s disease.
Front Pharmacol. 13(940999)2022.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Zhou Z, Chen H, Tang X, He B, Gu L and
Feng H: Total saikosaponins attenuates depression-like behaviors
induced by chronic unpredictable mild stress in rats by regulating
the PI3K/AKT/NF-ΚappaB signaling axis. Evid Based Complement
Alternat Med. 2022(4950414)2022.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Sun X, Li X, Pan R, Xu Y, Wang Q and Song
M: Total saikosaponins of Bupleurum yinchowense reduces depressive,
anxiety-like behavior and increases synaptic proteins expression in
chronic corticosterine-treated mice. BMC Complement Altern Med.
18(117)2018.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Li X, Li X, Huang N, Liu R and Sun R: A
comprehensive review and perspectives on pharmacology and
toxicology of saikosaponins. Phytomedicine. 50:73–87.
2018.PubMed/NCBI View Article : Google Scholar
|
|
54
|
He ZH, Fan R, Zhang CH, Tang T, Cui HJ,
Liu X, Liang QH and Wang Y: Research on total saponins of Radix
Bupleuri regulating cytochrome P450 similar to Chaihushugan powder
exerting antidepressant effects. J Hunan Univ Tradit Chin Med.
39:693–698. 2019.(In Chinese).
|
|
55
|
Li H, Tang Y, Wei W, Yin C and Tang F:
Effects of Xiaochaihu decoction on the expression of cytochrome
P450s in rats. Exp Ther Med. 21(588)2021.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Kong Q, Gao N, Wang Y, Hu G, Qian J and
Chen B: Functional evaluation of cyclosporine metabolism by CYP3A4
variants and potential drug interactions. Front Pharmacol.
13(1044817)2023.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Wu B, Liu P, Gao Y and Wang Y: Effect of
water extract from traditional Chinese medicines Rehmannia
glutinosa, Scrophularia ningpoensis, Asparagus cochinchinensis and
Ophiopogon japonicas on contents of CYP450 and activities of CYP3A,
CYP2E1 and CYP1A2 in rat. Zhongguo Zhong Yao Za Zhi. 36:2710–2714.
2011.PubMed/NCBI(In Chinese).
|
|
58
|
Liu H, Narayanan R, Hoffmann M and
Surapaneni S: The uremic toxin indoxyl-3-sulfate induces CYP1A2 in
primary human hepatocytes. Drug Metab Lett. 10:195–199.
2016.PubMed/NCBI View Article : Google Scholar
|
|
59
|
FDA, (2023). Drug development and drug
interactions | table of substrates, inhibitors and inducers.
https://www.fda.gov/drugs/drug-interactions-labeling/drug-development-and-drug-interactions-table-substrates-inhibitors-and-inducers.
|