Introduction
The coronavirus disease 2019 (COVID-19) pandemic is
characterized by high-level infection by severe acute respiratory
syndrome coronavirus 2 (SARS-CoV-2). The most severe of its
symptoms is caused by a hyperinflammatory condition due to the
excessive production of cytokines and chemokines, known as
‘cytokine storm’ (1,2). COVID-19 cytokine storm primarily
originates from T cells, macrophages, monocytes, dendritic cells
and endothelial cells (3-5),
where it is induced by a molecular interaction between the
SARS-CoV-2 S-protein and angiotensin-converting enzyme 2 (ACE2).
This is followed by complex intracellular changes that include
hyperactivation of the transcription factor NF-κB by the IL-6/STATs
axis (6). These cellular changes
are eventually associated with the life-threatening acute
respiratory distress syndrome (ARDS), which mainly affects the lung
and is a typical feature of patients with COVID-19(7) exhibiting a severe form of the
pathology (8,9). Accordingly, pharmacological
anti-inflammatory strategies for anti-SARS-CoV-2 treatment based on
targeting of IL-6 and IL-8 are highly impactful (10,11).
Despite these important developments, further novel pharmacological
approaches for treating hyperinflammatory ARDS are required,
because different patients with COVID-19 can respond differently to
the available treatments (12).
In this respect, various biomolecules derived from
herbal medicinal extracts have been proposed for anti-inflammatory
strategies to reduce COVID-19 ‘cytokine storm’ and associated ARDS
(13,14). Among the herbal medicinal extracts
hypothesized to confer anti-inflammatory activities, aged garlic
extract (AGE) is of particular interest (15). The preparation of AGE is performed
by the immersion (which can be performed at room temperature) of
fresh garlic in an aqueous ethanol solution over a prolonged period
(≤20 months) (16). Experimental
evidence exists demonstrating that this natural product possesses
immunomodulatory and anticancer properties (15,16).
Among the bioactive compounds that can be isolated from AGE,
S-1-propenyl-l-cysteine (S1PC) has previously been studied, where
it has been found to retain in vitro and in vivo
biological (including anti-inflammatory) activities of interest in
biomedicine (17-20).
The main aim of the present study was to assess the
effects of S1PC on the expression of genes involved in the COVID-19
‘cytokine storm’. The effects were studied using an experimental
in vitro model system based on the human bronchial
epithelial cell line IB3-1(21)
exposed to the COVID-19 BNT162b2 vaccine, according to previously
validated protocols (22). After
exposure to the BNT162b2 vaccine, the cells were cultured for 48 h
in the presence of increasing concentrations of S1PC, before they
were harvested for reverse transcription-quantitative PCR (RT-qPCR)
analysis.
Materials and methods
Materials
All chemicals and reagents were analytical grade.
S1PC™ (S-1-propenyl-l-cysteine) were obtained from Wakunaga
Pharmaceutical Co., Ltd., Japan. SARS-COV-2 Spike recombinant
glycoprotein (cat. no. ab49046) was purchased by Abcam. The purity
was >90% as determined by SDS-PAGE.
Gas chromatography (GC)-mass
spectrometry (MS) analysis
S1PC was analyzed by GC-MS as TBDMS derivatives
according to Jiménez-Martín et al (23).
S1PC was dissolved in 0.1 N HCl to a final
concentration of 4 mg/ml. In total, 5 µl these solutions were
spiked with 10 µl internal standard (3,4-dimethoxybenzoic acid, 0.1
mg/ml) and dried under N2. Subsequently, 30 µl pure
N-tert-butyldimethylsilyl-N-methyltrifluoroacetamide, followed by
30 µl pyridine, was added. The mixture was then heated at 80˚C for
1 h. The sample was neutralized afterwards with sodium bicarbonate
and subjected to GC-MS analysis. The same derivatization protocol
was used for the AGE powder (4 mg/ml 0.1 M HCl).
For all GC-MS analyses, an Agilent 7890B gas
chromatograph coupled to a 5977B quadrupole mass selective detector
(Agilent Technologies, Inc.) was employed. For chromatographic
separations, an Agilent HP5ms fused-silica capillary column (30 m x
0.25 mm i.d.) (Agilent Technologies, Inc.) was used, coated with
5%-phenyl-95%-dimethylpolysiloxane (film thickness, 0.25 µm) as
stationary phase. Splitless injection was performed at 280˚C. The
column temperature program was set to 70˚C (1 min), then to 300˚C
at a rate of 20˚C/min and held for 10 min. The carrier gas was
helium at a constant flow of 1.0 ml/min. The spectra were obtained
in the electron impact mode at 70 eV ionization energy, ion source
of 280˚C and ion source vacuum of 10-5 Torr. MS analysis was
performed simultaneously in total ion current (mass range scan in
the range of m/z 50-600 at a rate of 0.42 scans per sec) and
selected ion chromatogram mode. GC-SIM-MS analysis was performed by
selecting the following ions: m/z 332 for S1PC and m/z 239 for
3,4-dimethoxybenzoic acid (internal standard).
Cell culture conditions and treatment
with the BNT162b2 vaccine
The human bronchial epithelial IB3-1 cell line
(Thermo Fischer Scientific, Inc.) (21) was cultured in LHC-8 medium (Gibco;
Thermo Fischer Scientific, Inc.), supplemented with 5% FBS
(Biowest) without antibiotics at a temperature of 37˚C and 5%
CO2 (21). The BNT162b2
vaccine (COMIRNATY™; lot. No. FP8191) was obtained from the
Hospital Pharmacy of the University of Padova. The S1PC powder was
freshly dissolved in culture medium, normally up to 50 mM, before
each experiment. The solution was kept in the dark and used only
once (15). For treatment with the
BNT162b2 vaccine, IB3-1 cells were seeded at 200,000 cells/ml
concentration. After 24 h at 37˚C, 0.5 µg/ml vaccine (22) was added just before the indicated
concentrations of S1PC were added for an additional 48 h at 37˚C of
treatment, for determining the effects on S1PC on BNT162b2-induced
gene expression. Following incubation, cells were detached from the
plate by trypsinization, counted using a Beckman
Coulter® Z2 cell counter (Beckman Coulter, Inc.)
viability assay was performed using the Muse Annexin V & Dead
Cell reagent (Merck Millipore), and RNA was extracted for RT-qPCR
analysis using TRIzol reagent (Thermo Fischer Scientific,
Inc.).
Quantitative analyses of mRNAs
For quantification of the relative mRNA content, 500
ng of total cellular RNA was reverse transcribed to cDNA with the
Taq-Man Reverse Transcription Kit (cat no. N8080234; Applied
Biosystems; Thermo Fisher Scientific, Inc.), as described by
Gasparello et al (24).
qPCR experiments were performed using an assay consisting of a PCR
primer pair and a fluorescently labeled 5' nuclease probe or SYBR
Green. Assays IDs: i) Hs.PT.58.40226675 (HEX) for IL-6; ii)
Hs.PT.58.38869678.g (Cy5) for IL-8; iii) Hs.PT.58.20610757 (FAM)
for granulocyte-colony stimulation factor (G-CSF); iv)
Hs.PT.58.38905484 (FAM) for NF-κB p50; and v) Hs.PT.58.22880470
(FAM) for NF-κB p65. The primers and probes for ribosomal protein
L13a (RPL13A) and b-actin and were as follows: β-actin forward,
5'-ACGATGGAGGGGAAGACG-3' and reverse, 5'-ACAGAGCCTCGCCTTTG-3';
β-actin probe, 5'-/5Cy5/CCTTGCACATGCCGGAGCC/3IAbRQSp/-3'; RPL13A
forward, 5'-GGCAATTTCTACAGAAACAAGTTG-3' and reverse,
5'-GTTTTGTGGGGCAGCATACC-3'; RPL13A probe,
5'-/5HEX/CGCACGGTC/ZEN/CGCCAGAAGAT/3IABkFQ/-3' (Applied Biosystems;
Thermo Fisher Scientific, Inc.). The primers used for the
amplification of BNT162b2 Spike sequences using a Master Mix with
the SYBR Green intercalating Dye were forward
5'-CGAGGTGGCCAAGAATCTGA-3' and reverse,
5'-TAGGCTAAGCGTTTTGAGCTG-3', and β-actin forward
5'-CCTCGCCTTTGCCGATCC-3' and reverse, 5'-GGATCTTCATGAGGTAGTCAGTC-3'
(Integrated DNA Technologies), according to Aldén et al
(25). qPCR amplification of cDNA
was performed at 95˚C for 1 min, then 50 cycles of 95˚C for 15 sec
and 60˚C for 1 min, using the CFX96 Touch Real-Time PCR Detection
System (Bio-Rad Laboratories, Inc.). Relative expression was
calculated using the comparative quantification cycle (Cq) method
(2-ΔΔCq method) (26)
and the endogenous controls human β-actin and RPL13A, used as
normalizer. RT-qPCR reactions were performed in duplicate for both
target and normalizer genes (24).
Computational studies
All the computational methodologies were performed
on a 32 Core AMD Ryzen 93,905x, 3.5 GHz Linux Workstation (O.S.
Ubuntu 20.04) equipped with GPU (Nvidia Quadro RTX 4000, 8 GB;
Nvidia Corporation). The Toll-like receptor 4 (TLR4) dimer
structure was derived from available information (27). The structure of S1PC was drawn and
minimized using the Avogadro software (ver. 1.2.0; Avogadro
Chemistry) (28). A blind docking
simulation was performed on the entire TLR4 dimer surface using the
AutoDock Vina software (ver. 1.2.3; Center for Computational
Structural Biology) (29). The top
scoring complex was submitted to all-atom unbiased molecular
dynamics (MDs) simulation, as described by Zurlo et al
(22), using the GROMACS ver.
2025.1 software, open source (30), patched with Plumed ver.
2.6.5(31) under the Charmm36
force field (32). The complex was
included in a rectangular box of 8x10x7 nm length, solvated and
neutralized using 0.15 M sodium chloride. The full system was
submitted to energy minimization and equilibrated under constant
temperature and volume and constant temperature and pressure (NPT)
conditions. Long-range electrostatic interactions were modelled
using the Particle Mesh Ewald algorithm (33). The LINCS (34), Nosé-Hoover (35) and Parrinello and Rahman (36) algorithms were used in the
simulations for restraints and as thermostat and barostat,
respectively. MDs were conducted under the NPT conditions for 50
nsec with 2 fsec time steps. Root-mean-squared deviation (RMSD),
root-mean-squared fluctuation, number of hydrogen bonds and
interaction energy were obtained through the ‘rms’, ‘rmsf’, ‘hbond’
and ‘energy’ routines implemented in GROMACS.
Cell viability assay
Effects on cellular viability and apoptosis Annexin
V and Dead Cell assay were performed using the flow cytometry-based
Muse Cell Analyzer (Merck Millipore) instrument, according to the
protocols supplied by the manufacturer. Cells were washed with
sterile DPBS 1X, trypsinized, and 150,000 cells were suspended in
LHC-8 medium and diluted (1:2) with Muse Annexin V & Dead Cell
reagent (Annexin V-PE and 7-AAD) (Merck Millipore) and analyzed.
After an incubation of 15 min at room temperature in the dark,
samples were acquired and data were analysed using the Muse 1.5
Analysis Software with the Annexin V and Dead Cell Software Module
(Merck Millipore) (37).
Western blotting
For NF-κB (p105/p50 and p65) protein quantification,
20 µg total protein extract in RIPA Buffer (Thermo Fisher
Scientific, Inc.) quantified with BCA kit (Pierce; Thermo Fisher
Scientific, Inc.) were denatured for 5 min at 98˚C and loaded onto
a SDS polyacrylamide (8%) gel in Tris-glycine buffer (25 mM Tris,
192 mM glycine and 0.1% SDS). The electrotransfer to 0.2-µm
nitrocellulose membranes was performed overnight at 360 mA and 4˚C
in electrotransfer with CAPS buffer (25 mM Tris, 192 mM glycine,
CAPS 10 mM and 10% methanol). Obtained membranes were stained in
Ponceau S solution (Sigma-Aldrich; Merck KGaA) to verify proteins
transfer and incubated in 25 ml blocking buffer (TBS-T with 5%
nonfat dry milk (Cell Signalling Technology, Inc.) for 1 h at room
temperature. After three washes in TBS-T 1X (containing Tween-20 at
0.1%), membranes were incubated overnight at 4˚C in primary
antibodies (NF-κB p105/p50 Ab; cat. no. GTX133711; 1:5,000
dilution; GeneTex, Inc.). The day after, membranes were washed in
TBST 1X and incubated for 1 h at room temperature, with an
appropriate HRP-conjugated secondary antibody (anti-rabbit IgG
HRP-conjugated; cat. no. 7074P3; 1:2,000 dilution; Cell Signalling
Technology, Inc.). β-actin (primary antibody: Cat. no. 4970S;
1:1,000; Cell Signalling Technology, Inc.) was used as a
normalization control. After incubation with the ECL Solution
(Claity™ ECL Substrate; Bio-Rad Laboratories, Inc.) the gel images
were acquired with the ChemiDoc (Bio-Rad Laboratories, Inc.) with
the software Image Lab version 6.1.0, used also for densitometric
analysis (Bio-Rad Laboratories, Inc.).
Analysis of cytokines, chemokines and
growth factors
Proteins released into culture supernatants were
measured using Bio-Plex Human Cytokine 27-plex Assay (cat. no.
M500KCAF0Y; Bio-Rad Laboratories, Inc.), as suggested by the
manufacturer. The assay allows the multiplexed quantitative
measurement of 27 cytokines/chemokines [including FGF basic,
Eotaxin, G-CSF, granulocyte macrophage-colony stimulating factor,
IFN-γ, IL-1β, IL-1ra, IL-2, IL-4, IL-5, IL-6, IL-7, IL-8, IL-9,
IL-10, IL-12 (p70), IL-13, IL-15, IL-17A, C-X-C motif chemokine
ligand 10, chemokine ligand (CCL)2, CCL3, CCLe4, PDGFBB, CCL5,
TNF-α and VEGF] in a single well (21).
Briefly, an amount of 50 µl cytokine standards or
samples (diluted supernatants recovered from IB3-1 cells) was
incubated with 50 µl anti-cytokine conjugated beads in a 96-well
filter plate for 30 min at room temperature with shaking. The plate
was washed by vacuum filtration three times with 100 µl Bio-Plex
Wash Buffer, 25 µl diluted detection antibody was added to each
well and the plate was incubated for 30 min at room temperature
with shaking. After three filter washes, 50 µl
streptavidin-phycoerythrin was added and the plate was incubated
for 10 min at room temperature with shaking. Finally, the plate was
washed by vacuum filtration three times, the beads were suspended
in Bio-Plex Assay Buffer and the plate was read by a Bio-Rad
96-well plate reader. Collected data were analyzed by the Bio-Plex
Manager Software (version 6.2; Bio-Rad Laboratories, Inc.)
(21).
Statistical analysis
The data are presented as the mean ± standard
deviation of at least three independent experiments. Statistical
differences between/among groups were analyzed using one-way ANOVA.
Prism (v. 9.02) by GraphPad software (Dotmatics) was used (followed
by Bonferroni's test). P#x003C;0.05 was considered to indicate a
significant difference.
Results
Preliminary characterization of the
bronchial epithelial IB3-1 cellular system after exposure to the
COVID-19 BNT162b2 vaccine
The effects of exposure of IB3-1 cells to the
COVID-19 BNT162b2 vaccine (22)
were first analyzed. After the treatment, total cellular RNA was
extracted for RT-qPCR analysis, whereas culture supernatants were
isolated for the analysis of secreted proteins, to characterize
BNT162b2-induced alteration of the secretome profile. IB3-1 cells
treated with BNT162b2 vaccine were found to produce large amounts
of SARS-CoV-2 Spike protein. To obtain this information, western
blotting was performed using protein extracts from IB3-1 cells
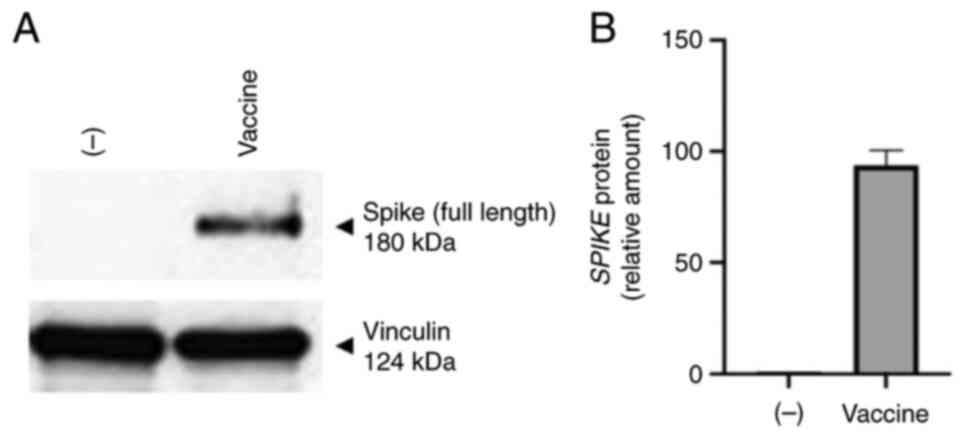
treated with the BNT162b2 vaccine (Fig. 1). A concentration of 0.5 µg/ml of
BNT162b2 was found to be sufficient to induce S-protein (Figs. 1, S1 and S2) and cytokine and chemokine production
(Fig. S3), with lower inhibitory
effects on cell viability compared with 1 and 2 µg/ml of
BNT162b2(22). The western
blotting data shown in Fig. 1
confirm that S-protein is produced by BNT162b2 treated cells, which
was reported elsewhere in other cellular model systems (22,38,39).
In addition, IB3-1 cells treated with the BNT162b2 vaccine were
found to accumulate large amounts of SARS-CoV-2 Spike mRNA
(Fig. S2) as discussed elsewhere
(40). This suggests that the
Spike protein was expressed by IB3-1 cells treated with the
BNT162b2 vaccine. When RT-qPCR and Bio-plex analyses were performed
using RNA or secreted materials from BNT162b2-treated IB3-1 cells,
it was found that they exhibited the increased expression and
production of pro-inflammatory factors, including IL-6, IL-8,
G-CSF, GM-CSF and IP-10 (Figs. S3
and S4).
GC-MS analysis of S1PC
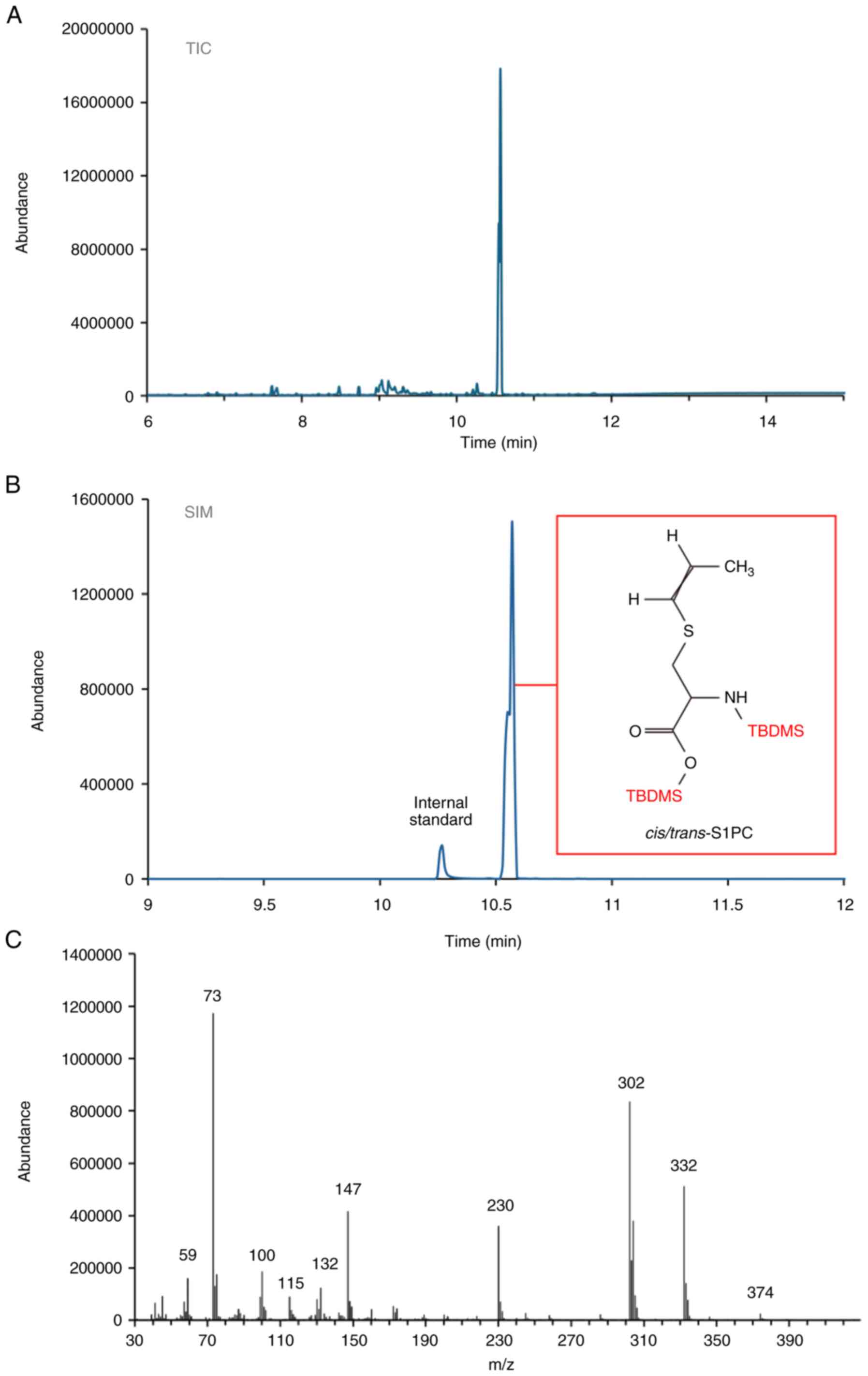
Fig. 2 shows the
GC-MS analysis of S1PC as a di-TBDMS derivative. Both the total ion
current (TIC) chromatogram and the selected ion chromatogram (SIM)
are shown. Fig. 2A and B revealed the presence of two partially
co-eluting peaks at 10.55 and 10.57 min (Fig. 2B), exhibiting identical electron
impact mass spectra (Fig. 2C) and
the corresponding cis- and trans-isomers of S1PC. Both cis- and
trans-S1PC have previously been identified in AGE, with the
cis-form believed to be generated through isomerization of its
trans form (41). Furthermore, the
GC-MS analyses presented in Fig. 2
confirmed the purity levels of the S1PC preparation procedure,
which was found to be >95%.
S1PC inhibits NF-κB expression in
IB3-1 cells treated with the BNT162b2 vaccine
Considering the effects of the spike protein on the
NF-κB pathway (42-44),
RT-qPCR was next performed to measure p50 and p65 mRNA expression
in BNT162b2-treated IB3-1 cells (Fig.
3), following on from a previous study on the same system using
SARS-CoV-2 spike protein (45).
Cells were therefore exposed for 24 h to BNT162b2 and then cultured
for an additional 48 h in the presence of increasing concentrations
of S1PC.
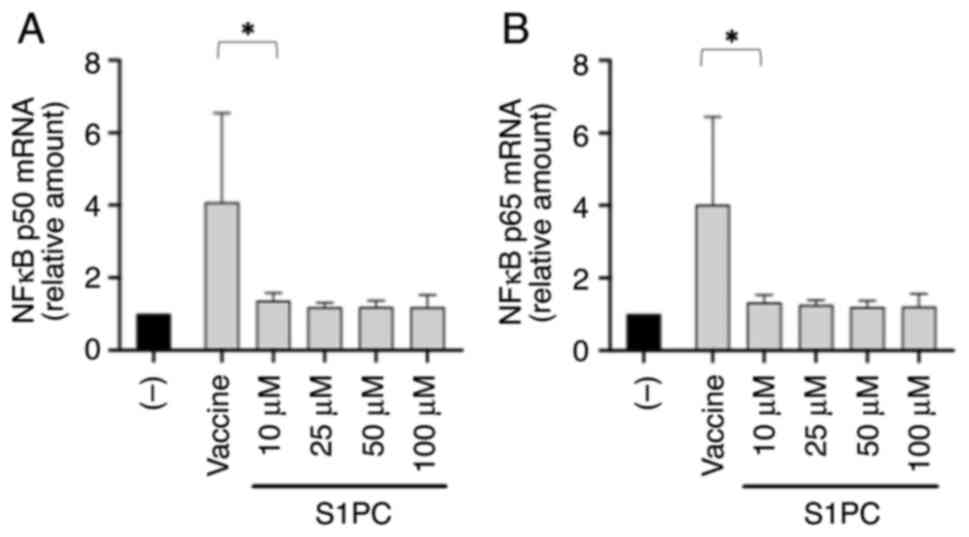
In total, two important conclusions could be
gathered from the results shown in Fig. 3. BNT162b2 stimulated the
accumulation of NF-κB p50 (Fig.
3A) and NF-κB p65 (Fig. 3B)
mRNA expression, even though NF-κB was already present at high
concentrations in the cytoplasm of IB3-1 cells before BNT162b2
treatment (21,45). In addition, after the
BNT162b2-treated IB3-1 cells were cultured with S1PC, reversal of
the accumulation of NF-κB p50 (Fig.
3A) and NF-κB p65 (Fig. 3B)
mRNA expression was observed. The effect of S1PC resembled that
observed using the NF-κB inhibitor sulforaphane (46) on the IB3-1 cellular model system
(45). In agreement with the
results shown in Fig. 3, the
effects of 50 mM S1PC on NF-κB were also evident on protein level
according to the western blot analysis using an antibody
recognizing the p50/p105 NF-κB subunits (Fig. S5).
S1PC inhibits the accumulation of
proinflammatory mRNAs in IB3-1 cells treated with the BNT162b2
vaccine
To verify if the BNT162b2-induced production of
proinflammatory mRNA expression could be affected in IB3-1 cells
treated with the BNT162b2 vaccine in the presence of S1PC, RT-qPCR
analysis was performed (Fig. 4).
The BNT162b2 vaccine was used at 0.5 µg/ml to minimize its
anti-proliferative effects (22).
Analysis of mRNA accumulation was performed 48 h after treatment.
IL-1β, IL-6 and IL-8(47), G-CSF
(48) and GM-CSF (49) were first considered, before
experimentally focusing on IL-6, IL-8 and G-CSF, which are much
more expressed in the IB3-1 experimental model system employed as
reported by Gasparello et al (21,45).
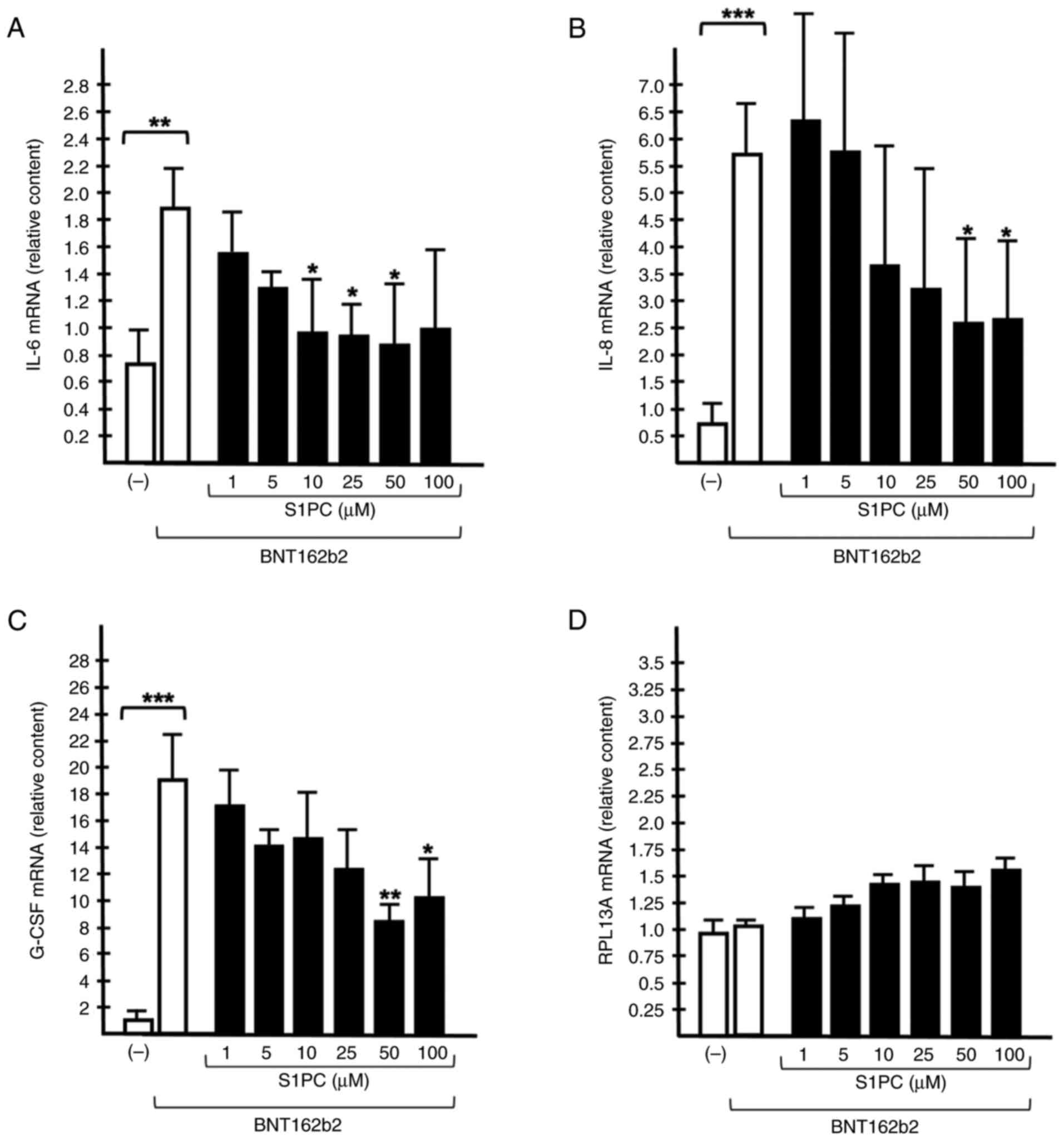
The results shown in Fig. 4 demonstrate that the expression of
IL-6 (Fig. 4A), IL-8 (Fig. 4B) and G-CSF (Fig. 4C) mRNA expression was significantly
upregulated in IB3-1 cells after exposure to the BNT162b2 vaccine.
The BNT162b2-mediated induction of expression was more efficient
compared with that of SARS-CoV-2 Spike protein (Fig. S4 and data not shown). No major
changes in the accumulation of mRNA expression of RPL13A were
observed. The RPL13A mRNA was studied, due to its role in
mitochondrial metabolism and that its expression is highly stable
in several cellular systems (50,51).
The effects of different concentrations (range 1-100
µM) of S1PC on BNT162b2-stimulated IB3-1 cells were next studied by
RT-qPCR, using β-actin as internal control (Fig. 4). A concentration-dependent
inhibition of the accumulation of IL-6 (Fig. 4A), IL-8 (Fig. 4B) and G-CSF (Fig. 4C) mRNAs was detectable, suggesting
that 10 µM S1PC is sufficient to inhibit to some extent the
expression of IL-6, IL-8 and G-CSF induced in these cells by the
BNT162b2 vaccine. However, higher S1PC concentrations were found to
be more effective. By contrast, no inhibitory effects were observed
in the RPL13A mRNA expression levels (Fig. 4D).
The results shown in Fig. 4 indicate that S1PC can be proposed
as an anti-inflammatory component of AGE to be considered in
pre-clinical studies (52,53). However, further studies on the
biological effects of S1PC are required to assess possible
anti-proliferative and/or cytotoxic effects associated with the
inhibition of proinflammatory gene expression.
Effects of S-1-propenyl-1-cysteine on
BNT162b2 treated IB3-1 cells: analysis of cell proliferation
efficiency and cell viability
To analyze in depth the effects of S1PC on cell
viability, the proportion of live/dead cells was analyzed using the
MUSE® Annexin V & Dead Cell Kit, which was used to
discriminate among live cells, apoptotic cells and dead cells
(24,37).
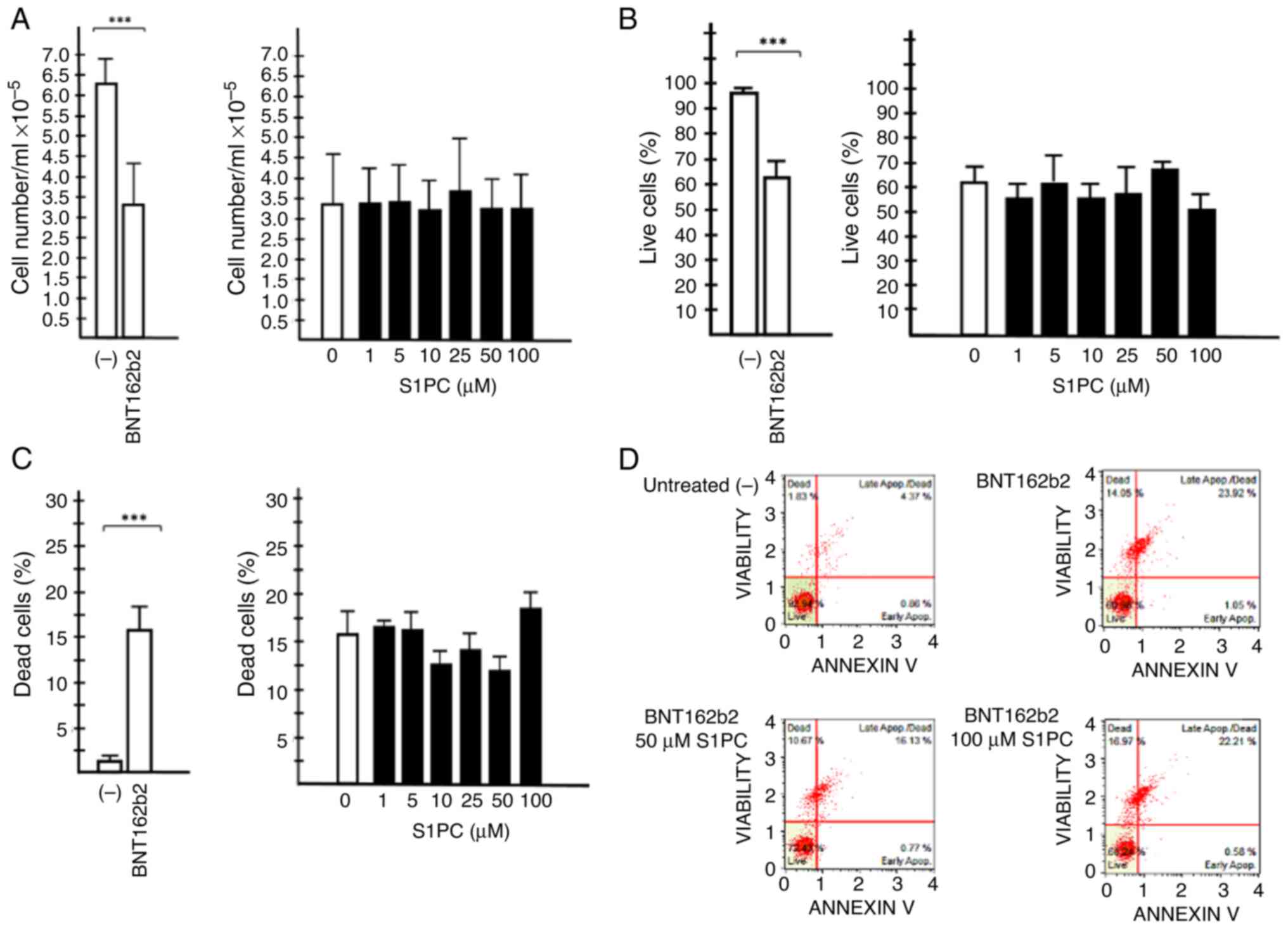
The results obtained are shown in Fig. 5. Fig.
5A shows that treatment of IB3-1 cells with the BNT162b2
vaccine is associated with the reduction of cell viability, whereas
S1PC did not induce anti-proliferative effects in IB3-1 cells
treated with 0.5 µg/ml of the BNT162b2 vaccine. In addition,
Fig. 5B-D indicates that treatment
of IB3-1 cells with the BNT162b2 vaccine is associated with a
decrease of the % live cells (Fig.
5B) and an increase of the % of dead cells (Fig. 5C). No major effects of S1PC on %
live cells or % dead cells could be observed (Fig. 5B and C).
Molecular docking and molecular
dynamics support the hypothesis that S-1-propenyl-1-cysteine
efficiently interacts with TLR4
To propose the possible mechanism of action of S1PC,
a molecular docking analysis was performed. Preliminary analyses
demonstrated a lack of binding of S1PC to NF-κB. The binding of
low-molecular-weight drugs to this protein has however been
previously reported, such as trimethylangelicin and analogues
(54), corilagin (55) and sulforaphane (45,47).
In these cases, efficient interactions with NF-κB were found.
However, no evidence of molecular interaction between S1PC and
NF-κB could be found in in the present docking analysis (data not
shown). Since TLR are upstream regulators of the NF-κB signaling
(56-58),
the possible interaction between S1PC and TLR4 was assessed using
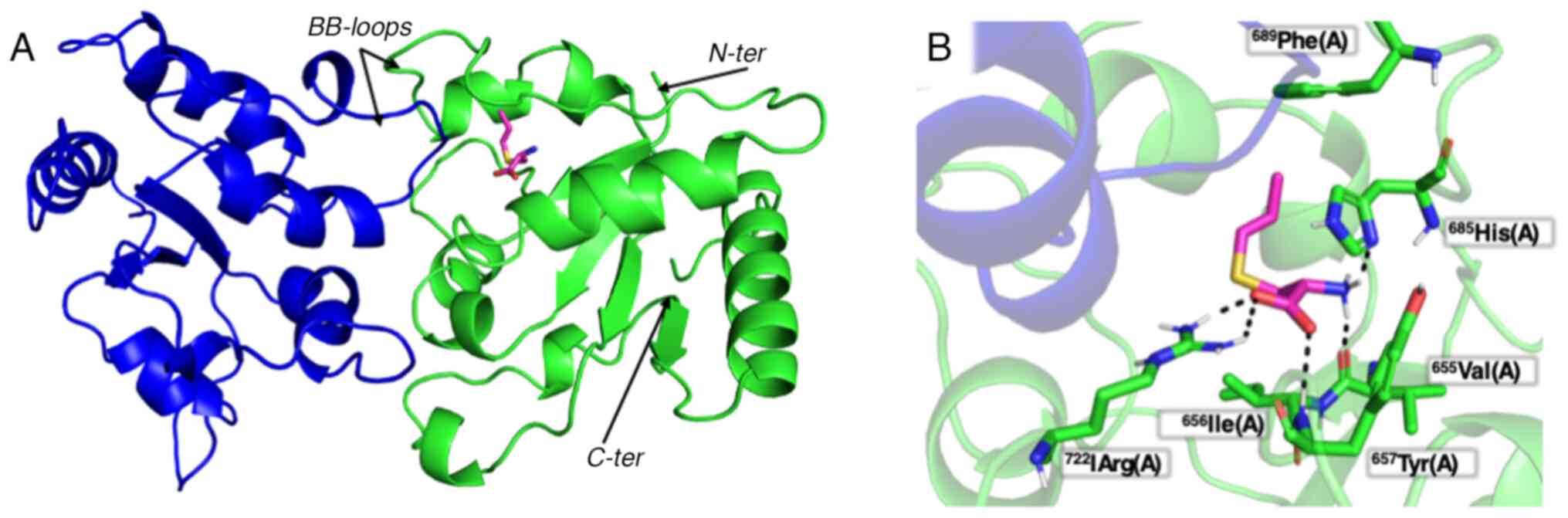
the docking AutoDock Vina software (Fig. 6) (29). The results obtained indicate that
S1PC is able to bind in silico that to the Toll-IL-1
receptor domain of TLR4. The amino acids involved in the S1PC-TLR4
interactions are His685, Val655, Arg722 and Tyr657.
To further sustain the reliability of the molecular
interaction reported in Fig. 6,
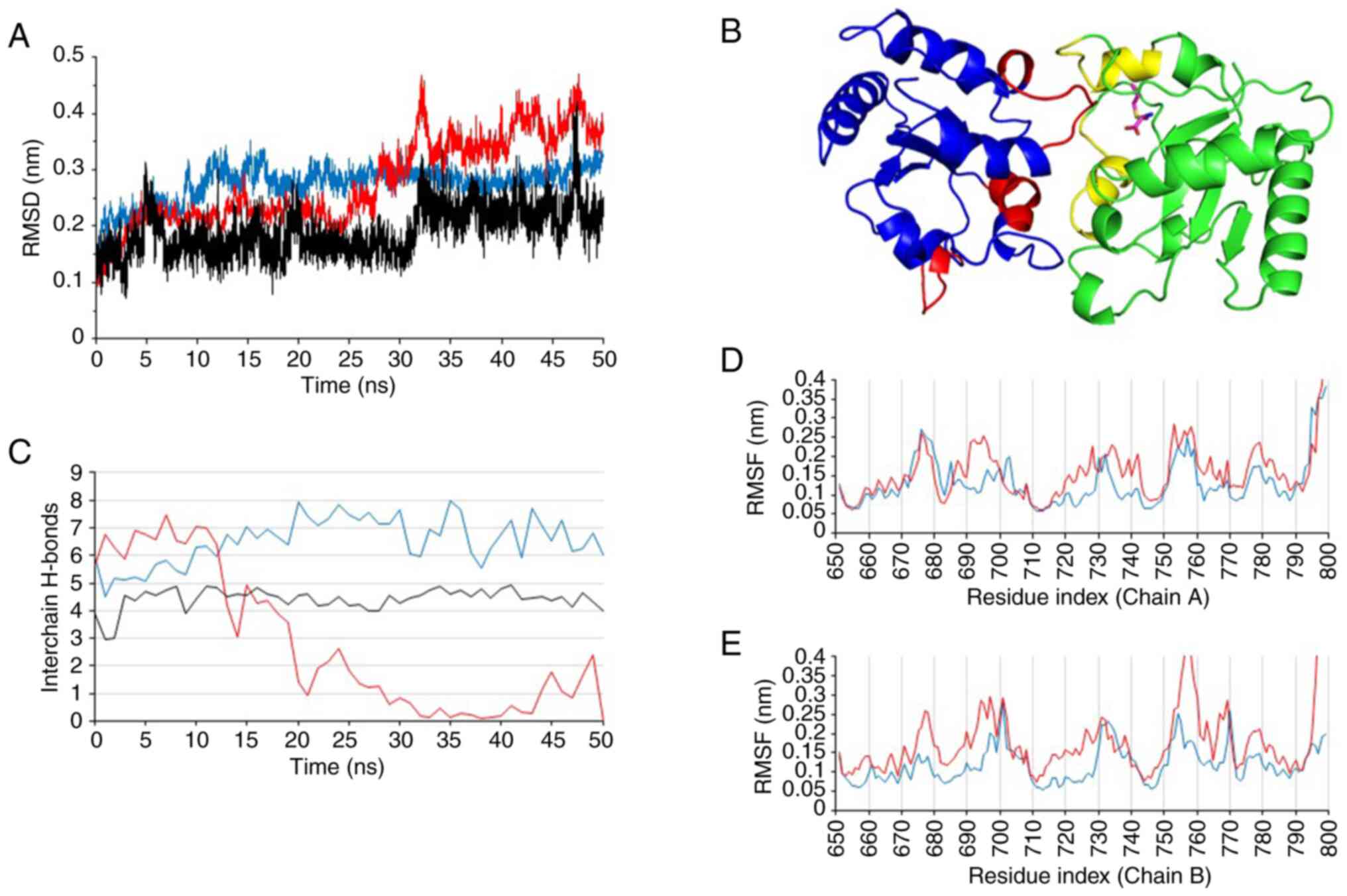
the computed model was submitted to 50 nsec of all-atom unbiased
molecular dynamics simulation. The results obtained demonstrated
that the complex remained stable, as can be seen from the Cα-RMSD
values calculated over the simulation time (Fig. 7A). In particular, the hydrogen
bonds reported in Fig. 7B were
retained during the entire molecular dynamic's simulation, yielding
an estimated interaction energy of -65.2±7.3 Kcal/mol (computed as
the sum of short-range Lennard-Jones and short-range Coulomb
contributions over the 50 nsec of simulation). In addition, binding
with S1PC was found to reduce the intermolecular interaction
between the TLR4 domains, as revealed by both the reduction in
H-bond numbers (Fig. 7B) and the
increased per-residue RMSF values in comparison to the apo complex
(Fig. 7C-E).
Discussion
One of the most important and clinically relevant
characteristics of COVID-19 is the high expression of IL-6, IL-8
and several other cytokines, chemokines and growth factors
(2). This is frequently associated
with a hyperinflammatory state and severe forms of
COVID-19(59). Del Valle et
al (60) previously reported
that high serum IL-6, IL-8 and TNF-α levels at the time of
hospitalization are associated with poor prognosis. In another
study, Burke et al (61)
also found that increased IL-6 and IL-8 levels can be used to
predict clinical outcomes in patients with COVID-19. Therefore,
anti-inflammatory molecules and novel anti-inflammatory strategies
are highly needed.
In the present study, the human bronchial epithelial
IB3-1 cell line was used, where it was stimulated by the COVID-19
BNT-162b2 vaccine to express proinflammatory factors. The IB3-1
cell line has been previously used to study the inflammatory
response (62-64)
and effects of anti-inflammatory agents on the expression of
proinflammatory genes known to be involved in COVID-19 cytokine
Storm (21,45,54,55,65).
The main aim of the present study was to determine
the possible effects of a major component of AGE, S1PC, on the
expression of proinflammatory factors and hypothesize the possible
mechanism of action. The beneficial effects of garlic have been
previously reported, where they were proposed to be due to the
presence of several bioactive molecules within its preparations,
including lipid-soluble allyl sulfur compounds and water-soluble
derivatives, such as SAC and S1PC (66). S1PC is a stereoisomer of SAC
(17). This sulfur-containing
amino acid has important properties for the beneficial
pharmacological roles of AGE (20). S1PC is present only in trace
amounts in raw garlic, but its concentration will increase,
approaching that of SAC levels, during the aging process of AGE
(41). S1PC has been observed to
show immunomodulatory functions both in vitro and in
vivo, in addition to reduce blood pressure in a hypertensive
animal model (67,68). In addition, a previous
pharmacokinetic investigation showed that S1PC is rapidly absorbed
after oral administration in rats and dogs, with high
bioavailability (~100%) (17). In
addition, S1PC exhibited a low inhibitory effect on human
cytochrome P450 activities, even when it was used at a
concentration of 1 mM (67,68).
Considering all these findings regarding the potential medicinal
value of S1PC, this molecule was suggested to be another
pharmacologically active and safe derivative of AGE similar to SAC,
consistent with the proposal made by Kodera et al (17).
The present GC/MS analysis confirmed that the S1PC
preparation contains both cis- and trans-isomers of S1PC.
Concerning the biological activity of the applied S1PC preparation,
the results presented in the present study suggest that the release
of key proteins of the COVID-19 ‘cytokine storm’ (69) can be inhibited by S1PC. Since
control of this ‘cytokine storm’ is a major issue in the management
of patients with COVID-19(70),
results from the present study may stimulate the development of
protocols for controlling the hyperinflammatory state associated
with SARS-CoV-2 infection. In addition, experimental activity on
plant extracts and food supplements containing S1PC for supporting
the possibility of using ‘phyto-preparations’ in combination with
‘conventional’ medicine focusing on COVID-19 treatment is
encouraged as a result of the present study. The present study also
extended recent observations by Gasparello et al (71) on the effects of AGE and its
component SAC on expression of pro-inflammatory genes. In the
present study, S1PC was considered for the first time and G-CSF was
included in the analysis, sustaining the conclusions of the
previous study on AGE and SAC (71), suggesting an inhibitory effect of
these agents on the expression of pro-inflammatory genes. To the
best of our knowledge, the present study was the first to consider
potential cytotoxicity and anti-proliferative effects of an AGE
component on bronchial epithelial cells exposed to the BNT162b2
vaccine. In addition, in the present study low concentrations (1
and 5 µM) of the AGE component S1PC were considered.
One of the limitations of the present study is the
lack of a full explanation on the mechanism of action of S1PC. This
should be considered in future research plans, since it can be used
to identify novel targets for therapeutic strategies. Among the
several possibilities, S1PC can exert its anti-inflammatory
activity by inhibiting the JNK/activator protein-1 (AP-1)/NF-κB
pathway. This may be associated with the activity of TLRs, such as
TLR4 (72-74).
The present study supports the hypothesis that S1PC interacts with
and possibly inhibits TLR4, by destabilizing the dimer
interactions. TLRs (including TLR4) are involved in SARS-CoV-2
entry into infected cells (75,76)
and in NF-κB expression (77,78).
Therefore, it can be hypothesized that inhibitors of TLR4 can
inhibit the early phases of SARS-CoV-2 infection, including
activation of the NF-κB pathway. The inhibition of NF-κB by S1PC
may be relevant in the context of possible inhibition by S1PC of
proinflammatory genes, which contain NF-κB binding sites in their
promoter sequence. The effect of S1PC resembled that observed using
the NF-κB inhibitor sulforaphane (46) on the IB3-1 cellular model system
(45,47). The present study supports the
hypothesis that TLR-4 should be considered as a target of
anti-SARS-CoV-2 therapeutic strategies (78,79).
In particular, Yang et al (80) previously showed that an aptamer
blocking the Spike-TLR4 interaction is able to selectively inhibit
SARS-CoV-2-induced inflammation. Docking data from the present
study sustained this hypothesis, which showed in silico that
S1PC is possibly able to bind to the Toll-IL-1 receptor domain of
TLR4. The amino acids involved in the S1PC-TLR4 interactions are
His685, Val655, Arg722 and Tyr657, belonging to a TLR4 region,
serving a role in the molecular interaction with the SARS-CoV-2
Spike protein (81). Therefore,
S1PC should be considered in further studies aimed at verifying its
possible anti-SARS-CoV-2 activity.
In conclusion, a simple experimental model system
was developed and validated for the identification and
characterization of molecules able to inhibit the expression of
genes involved in the COVID-19 ‘cytokine storm’ in the present
study. Specifically, exposure of epithelial IB3-1 cells to the
COVID-19 BNT162b2 vaccine is associated with a potent increase in
the expression of the transcription factor NF-κB and
NF-κB-regulated genes, including IL-6, IL-8 and G-CSF. However, the
present study did not explain the mechanism of action of BNT162b2
in inducing the upregulation of proinflammatory gene expression.
The activity of BNT162b2 may be due to the liposomal vaccine
vector, to the Spike mRNA or both of these BNT162b2 components.
Lipid-based RNA nanoparticles can stimulate inflammatory responses
(82-84),
whereas the purified SARS-CoV-2 spike protein (encoded by the
BNT162b2 mRNA vaccine) can also induce the upregulation of
proinflammatory gene expression, despite to an extent lower
compared with BNT162b2. Therefore, both lipid formulation and spike
RNAs can contribute to the activity of BNT162b2 on the expression
of proinflammatory genes.
Treatment with S1PC was not found to be toxic but it
reversed the proinflammatory cytokine IL-6, IL-8 and G-CSF
upregulation induced by the BNT162b2 vaccine in IB3-1 cells.
Considering the TLR4-NF-κB interplay, molecular dynamic results
obtained in the present study suggest that the anti-inflammatory
effects of S1PC may be due to an inhibition of the JNK/AP-1/NF-κB
interaction. Therefore, S1PC should be further evaluated as a
potential inhibitor of proinflammatory factors involved in the
COVID-19 ‘cytokine storm’, moving the study from in vitro
experimental model systems to in vivo treatments (80-85)
and clinical trials. Examples of clinical trials based on AGE and
reporting anti-inflammatory effects are those published by
Wlosinska et al (86) and
by Xu et al (87). Although
in vitro experimental systems are viable for the analysis of
short-term effects, studies using in vivo systems and
clinical trials are needed to study long-term effects, including
potential negative effects.
The present study has several limits that should be
considered in future studies. Only one cell line has been studied
(the bronchial epithelial IB3-1 cell line). Exposure of IB3-1 cells
to the BNT162b2 vaccine should be just considered as a simple
method to induce the increased expression of proinflammatory genes
to an extent higher than that exhibited by SARS-CoV2 Spike-treated
IB3-1 cells (45,47). The possible inhibitory effects of
S1PC on other experimental model systems closely resembling the
cell types that are involved in the COVID-19 cytokine storm, such
as T cells, macrophages, monocytes, dendritic cells and endothelial
cells, should also be investigated (3-5).
In addition, other well validated experimental systems mimicking
the induced proinflammatory state can be employed, such as the same
IB3-1 exposed to Pseudomonas aeruginosa (88), to TNF-α (54,55)
or to lipopolysaccharide (89). In
all of these aforementioned experimental model systems, cytokines
and chemokines involved in COVID-19 cytokine storm were found to be
upregulated, including Il-6 and IL-8 (54,55,88,89).
Furthermore, other model systems have been proposed, based on the
Pseudomonas aeruginosa infection of other cell lines, such
as NuLi (88), CuFi (88-90)
and A549(91). Furthermore,
experiments performed on skeletal muscle cells exposed to the
BNT162b2 vaccine and on SARS-CoV-2-infected lung epithelial Calu-3
cells (47) can be considered to
closely mimic the vaccination procedure by intramuscular
administration (92) and COVID-19
lung infection, respectively. A possible inhibitory effect of S1PC
on NF-κB signaling in SARS-CoV-2-infected lung epithelial cells
should be considered, as it was previously reported using the NF-κB
inhibitor sulforaphane (45,47).
Another limitation of the present study is that
possible negative effects on gene expression of S1PC have not been
considered. Global transcriptomic and proteomic analyses will be
necessary to clarify this point. In terms of the docking and
molecular dynamics experiments, a proposed model of possible
interactions between S1PC and TLR4, along with their effects on
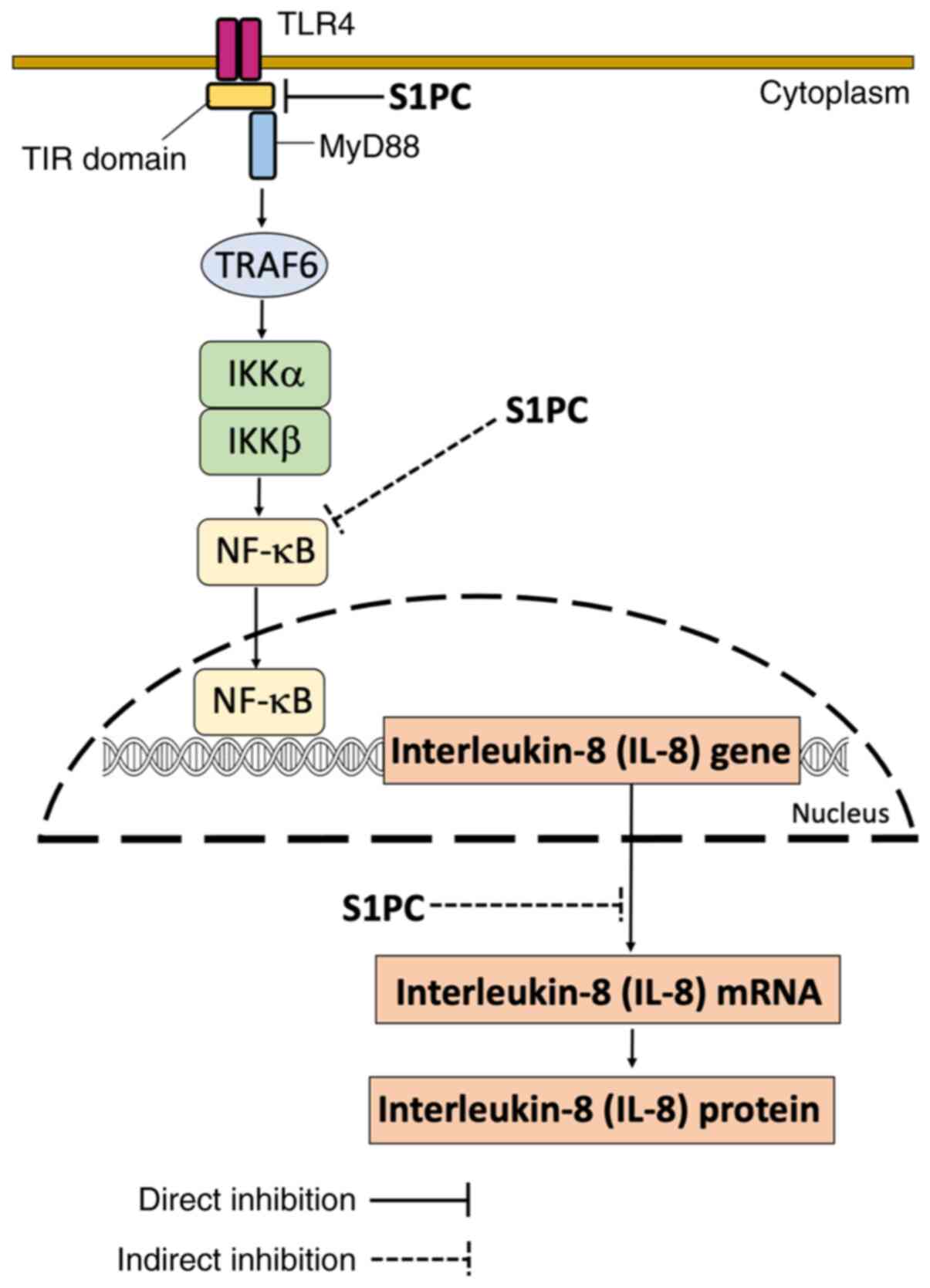
TLR4 functions, was constructed (Fig.
8). It must be emphasized that it is a speculative model at
this stage, where further experimental work based on biochemical
evidence validating the predicted S1PC-TLR4 interaction and its
effects on downstream TLR4 signaling is encouraged. The effects of
the S1PC homologue SAC on TLR4 have been also reported previously
(93,94). An extensive study of AGE
constituents (SAC and S1PC) on other members of the TLR family are
highly warranted. In addition, further experimental effort is
needed to identify other agents within the list of AGE components
that can modify gene expression induced by the COVID-19 BNT162b2
vaccine. This is to verify whether combined treatment with S1PC
could be possible to obtain the highest inhibitory effects, using
also SARS-CoV-2 infected cells.
In conclusion, the present study should encourage
further experiments based on western blotting and Bio-plex analyses
to verify whether the inhibitory effects of S1PC on the
accumulation of NF-κB and proinflammatory mRNAs are associated with
a decrease of proinflammatory protein production and release. This
will clarify the clinical impact of the present study. In addition,
possible validation in clinical trials should be considered, given
the potential relevance of the present study for COVID-19
management. Furthermore, the present study has clinical relevance,
considering the role of inflammation in lung pathologies, such as
cystic fibrosis, asthma and COPD (95-97),
in neurological diseases (98), in
osteoarthritis (53) and in
skeletal muscular atrophy (94-99),
cardiovascular diseases (100),
diabetes (101) and cancer
(102,103).
Supplementary Material
Uncropped original gel images of the
western blot experiment from Fig.
1A. Bronchial epithelial IB3-1 cells have been either untreated
(#1) or treated with 0.5 μg/ml BNT182b2 vaccine (#2). After
48 h, cell lysates were analyzed by western blotting.
(A) Representative RT-qPCR analysis of
SARS-CoV-2 S-protein mRNA expression. (B) Expression of the
SARS-CoV-2 mRNA (fold increase with respect to trace amount of
hybridizable material found in control cells, set as 1). SARS-CoV-2
S-protein mRNA expression was measured by RT-qPCR using protocols
used by Aldén et al (25). Results represent the means ±
standard deviation from three independent experiments (P<0.001).
Endogenous β-actin was used as internal control. The threshold line
and the amplification curves (two representative vaccine-treated
samples) are reported in green and pink, respectively. SARS-CoV-2,
Severe Acute Respiratory Syndrome Coronavirus 2; RT-qPCR, reverse
transcription-quantitative PCR; RFU, relative fluorescence unit;
(-) untreated samples.
Bio-plex analysis showing the increase
in the release of (A) IL-6, (B) IL-8, (C) G-CSF, (D) GM-CSF and (E)
IP-10 following 48 h treatment of IB3-1 cells with 0.5 μg/ml
BNT182b2 vaccine. Results represent the means ± standard deviation
from four independent experiments. *P<0.05 and
**P<0.01. CSF, colony stimulating factor; G,
granulocyte; GM, granulocyte macrophage; IP-10, C-X-C motif
chemokine ligand 10; (-) untreated samples.
Effects of exposure to the SARS-CoV-2
S-protein and BNT162b2 vaccine on the expression levels of IL-8
mRNA. The (A) SARS-CoV-2 S-protein and the (B) BNT162b2 vaccine
were used at 5 nM and 0.5 μg/ml concentrations, respectively
(22,40). IB3-1 cells were seeded at 200,000
cells/ml and treatments were conducted for 48 h before RNA
isolation. Results represent the means ± standard deviation from
three independent experiments. *P<0.05 and
**P<0.01. SARS-CoV-2, Severe Acute Respiratory
Syndrome Coronavirus 2; (-) untreated samples.
Effects of 50 μM S1PC on NF-κB
p105/p50 accumulation in BNT162b2-stimulated IB3-1 cells. (A)
Original version of the western blotting images relative to the
effects of S1PC (lane #2) on NF-κB p105/p50 accumulation (arrows)
on BNT162b2-stimulated IB3-1 cells. Lane #1 represents control
IB3-1 cells treated with BNT162b2 in the absence of S1PC. (B)
Original version of the western blotting images relative to the
effects of S1PC (lane #2) on β-actin accumulation (arrow) in
BNT162b2-stimulated IB3-1 cells. Lane #1 represents control IB3-1
cells treated with BNT162b2 in the absence of S1PC. (C) Relative
protein/β-actin ratios obtained after densitometry analysis.
ChemiDoc instrument and Image Lab software (both from Bio-Rad
Laboratories, Inc.) were used for densitometry analysis of the
obtained bands. *P<0.05 from three independent experiments.
S1PC, S-1-propenyl-l-cysteine.
Acknowledgements
The authors would like to thank Dr Takahiro Ogawa
and Dr Toshiaki Matsutomo (Wakunaga Pharmaceutical Co., Ltd.) for
organizing the shipment of S1PC.
Funding
Funding: The present study was funded by the MUR-FISR
COVID-miRNAPNA Project (grant no. FISR2020IP_04128), by FIRD 2024
funds from the University of Ferrara (grant no. FIRD-Finotti 2024),
by the Interuniversity Consortium for Biotechnologies, Italy (grant
no. CIB-Unife-2020) and by Wakunaga Pharmaceutical Co. Ltd. (grant
no. IPFO2024).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
AF, EA and RG designed the experimental plan. CP,
JG, MZ, FDP and AF were involved in the methodology development.
CP, JG, GM, AM, MZ, FDP and PF performed the experiments. CP and
FDP performed the treatments, the western blotting and the reverse
transcription-quantitative PCR analyses. JG and MZ developed and
characterized the experimental model system based on exposure of
IB3-1 cells to the BNT162b2 vaccine. FDP and AF performed the
cytotoxicity tests. EA, AM and PF performed the gas
chromatography-mass spectrometry analyses. GM performed the
molecular docking and molecular dynamics experiments. AF, CP, JG
and RG curated the data and analyzed the results. RG wrote the
original draft. AF, RG and EA reviewed and edited the draft. All
authors read and approved the final version of the manuscript. RG
and AF confirm the authenticity of all the raw data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Pascarella G, Strumia A, Piliego C, Bruno
F, Del Buono R, Costa F, Scarlata S and Agrò FE: COVID-19 diagnosis
and management: A comprehensive review. J Intern Med. 288:192–206.
2020.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Pelaia C, Tinello C, Vatrella A, De Sarro
G and Pelaia G: Lung under attack by COVID-19-induced cytokine
storm: Pathogenic mechanisms and therapeutic implications. Ther Adv
Respir Dis. 14(1753466620933508)2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Fajgenbaum DC and June CH: Cytokine storm.
N Engl J Med. 383:2255–2273. 2020.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Mangalmurti N and Hunter CA: Cytokine
storms: Understanding COVID-19. Immunity. 53:19–25. 2020.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Fara A, Mitrev Z, Rosalia RA and Assas BM:
Cytokine storm and COVID-19: A chronicle of pro-inflammatory
cytokines. Open Biol. 10(200160)2020.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Ratajczak MZ, Bujko K, Ciechanowicz A,
Sielatycka K, Cymer M, Marlicz W and Kucia M: SARS-CoV-2 entry
receptor ACE2 is expressed on very small CD45-
precursors of hematopoietic and endothelial cells and in response
to virus spike protein activates the Nlrp3 inflammasome. Stem Cell
Rev Rep. 17:266–277. 2021.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Soumagne T, Winiszewski H, Besch G, Mahr
N, Senot T, Costa P, Grillet F, Behr J, Mouhat B, Mourey G, et al:
Pulmonary embolism among critically ill patients with ARDS due to
COVID-19. Respir Med Res. 78(100789)2020.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Grasselli G, Tonetti T, Protti A, Langer
T, Girardis M, Bellani G, Laffey J, Carrafiello G, Carsana L,
Rizzuto C, et al: Pathophysiology of COVID-19-associated acute
respiratory distress syndrome: A multicentre prospective
observational study. Lancet Respir Med. 8:1201–1208.
2020.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Matthay MA, Leligdowicz A and Liu KD:
Biological mechanisms of COVID-19 acute respiratory distress
syndrome. Am J Respir Crit Care Med. 202:1489–1491. 2020.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Nasonov E and Samsonov M: The role of
interleukin 6 inhibitors in therapy of severe COVID-19. Biomed
Pharmacother. 131(110698)2020.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Andreakos E, Papadaki M and Serhan CN:
Dexamethasone, pro-resolving lipid mediators and resolution of
inflammation in COVID-19. Allergy. 76:626–628. 2021.PubMed/NCBI View Article : Google Scholar
|
|
12
|
de Simone G and Mancusi C: Finding the
right time for anti-inflammatory therapy in COVID-19. Int J Infect
Dis. 101:247–248. 2020.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Khalifa SAM, Yosri N, El-Mallah MF,
Ghonaim R, Guo Z, Musharraf SG, Du M, Khatib A, Xiao J, Saeed A, et
al: Screening for natural and derived bio-active compounds in
preclinical and clinical studies: One of the frontlines of fighting
the coronaviruses pandemic. Phytomedicine.
85(153311)2021.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Matveeva T, Khafizova G and Sokornova S:
In search of herbal anti-SARS-Cov2 compounds. Front Plant Sci.
11(589998)2020.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Ohkubo S, Dalla Via L, Grancara S,
Kanamori Y, García-Argáez AN, Canettieri G, Arcari P, Toninello A
and Agostinelli E: The antioxidant, aged garlic extract, exerts
cytotoxic effects on wild-type and multidrug-resistant human cancer
cells by altering mitochondrial permeability. Int J Oncol.
53:1257–1268. 2018.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Lee J, Zhao N, Fu Z, Choi J, Lee HJ and
Chung M: Effects of garlic intake on cancer: A systematic review of
randomized clinical trials and cohort studies. Nutr Res Pract.
15:773–788. 2021.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Kodera Y, Ushijima M, Amano H, Suzuki JI
and Matsutomo T: Chemical and biological properties of
S-1-propenyl-l-cysteine in aged garlic extract. Molecules.
22(570)2017.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Yeh YY and Liu L: Cholesterol-lowering
effect of garlic extracts and organosulfur compounds: Human and
animal studies. J Nutr. 131 (3S):989S–993S. 2001.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Nango H and Ohtani M:
S-1-propenyl-L-cysteine suppresses lipopolysaccharide-induced
expression of matrix metalloproteinase-1 through inhibition of
tumor necrosis factor-α converting enzyme-epidermal growth factor
receptor axis in human gingival fibroblasts. PLoS One.
18(e0284713)2023.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Ushijima M, Takashima M, Kunimura K,
Kodera Y, Morihara N and Tamura K: Effects of S-1-propenylcysteine,
a sulfur compound in aged garlic extract, on blood pressure and
peripheral circulation in spontaneously hypertensive rats. J Pharm
Pharmacol. 70:559–565. 2018.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Gasparello J, d'Aversa E, Breveglieri G,
Borgatti M, Finotti A and Gambari R: In vitro induction of
interleukin-8 by SARS-CoV-2 spike protein is inhibited in bronchial
epithelial IB3-1 cells by a miR-93-5p agomiR. Int Immunopharmacol.
101(108201)2021.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Zurlo M, Gasparello J, Verona M, Papi C,
Cosenza LC, Finotti A, Marzaro G and Gambari R: The anti-SARS-CoV-2
BNT162b2 vaccine suppresses mithramycin-induced erythroid
differentiation and expression of embryo-fetal globin genes in
human erythroleukemia K562 cells. Exp Cell Res.
433(113853)2023.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Jiménez-Martín E, Ruiz J, Pérez-Palacios
T, Silva A and Antequera T: Gas chromatography-mass spectrometry
method for the determination of free amino acids as their
dimethyl-tert-butylsilyl (TBDMS) derivatives in animal source food.
J Agric Food Chem. 60:2456–2463. 2012.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Gasparello J, Lomazzi M, Papi C, D'Aversa
E, Sansone F, Casnati A, Donofrio G, Gambari R and Finotti A:
Efficient delivery of MicroRNA and AntimiRNA molecules using an
argininocalix[4]arene macrocycle. Mol Ther Nucleic Acids.
18:748–763. 2019.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Aldén M, Olofsson Falla F, Yang D,
Barghouth M, Luan C, Rasmussen M and De Marinis Y: Intracellular
reverse transcription of pfizer BioNTech COVID-19 mRNA vaccine
bnt162b2 in vitro in human liver cell line. Curr Issues Mol Biol.
44:1115–1126. 2022.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Patra MC, Kwon HK, Batool M and Choi S:
Computational insight into the structural organization of
full-length Toll-like receptor 4 dimer in a model phospholipid
bilayer. Front Immunol. 9(489)2018.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Hanwell MD, Curtis DE, Lonie DC,
Vandermeersch T, Zurek E and Hutchison GR: Avogadro: An advanced
semantic chemical editor, visualization, and analysis platform. J
Cheminform. 4(17)2012.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Eberhardt J, Santos-Martins D, Tillack AF
and Forli S: AutoDock Vina 1.2.0: New docking methods, expanded
force field, and python bindings. J Chem Inf Model. 61:3891–3898.
2021.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Abraham MJ, Murtola T, Schulz R, Páll S,
Smith JC, Hess B and Lindahl E: GROMACS: High performance molecular
simulations through multi-level parallelism from laptops to
supercomputers. SoftwareX. 1-2:19–25. 2015.
|
|
31
|
Bonomi M, Branduardi D, Bussi G, Camilloni
C, Provasi D, Raiteri P, Donadio D, Marinelli F, Pietrucci F,
Broglia RA and Parrinello M: PLUMED: A portable plugin for
free-energy calculations with molecular dynamics. Comput Phys
Commun. 180:1961–1972. 2009.
|
|
32
|
Huang J and MacKerell AD Jr: CHARMM36
all-atom additive protein force field: Validation based on
comparison to NMR data. J Comput Chem. 34:2135–2145.
2013.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Darden T, York D and Pedersen L: Particle
mesh Ewald: An N·log(N) method for Ewald sums in large systems. J
Chem Phys. 98:10089–10092. 1993.
|
|
34
|
Hess B: P-LINCS: A parallel linear
constraint solver for molecular simulation. J Chem Theory Comput.
4:116–122. 2008.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Evans D and Holian B: The nose-hoover
thermostat. J Chem Phys. 83:4069–4074. 1985.
|
|
36
|
Parrinello M and Rahman A: Polymorphic
transitions in single crystals: A new molecular dynamics method. J
Appl Phys. 52:7182–7190. 1981.
|
|
37
|
Tupini C, Zurlo M, Gasparello J, Lodi I,
Finotti A, Scattolin T, Visentin F, Gambari R and Lampronti I:
Combined treatment of cancer cells using allyl palladium complexes
bearing purine-based NHC ligands and molecules targeting MicroRNAs
miR-221-3p and miR-222-3p: Synergistic effects on apoptosis.
Pharmaceutics. 15(1332)2023.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Cari L, Naghavi Alhosseini M, Mencacci A,
Migliorati G and Nocentini G: Differences in the expression levels
of SARS-CoV-2 spike protein in cells treated with mRNA-based
COVID-19 vaccines: A study on vaccines from the real world.
Vaccines (Basel). 11(879)2023.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Bansal S, Perincheri S, Fleming T, Poulson
C, Tiffany B, Bremner RM and Mohanakumar T: Cutting edge:
Circulating exosomes with COVID spike protein are induced by
BNT162b2 (Pfizer-BioNTech) Vaccination prior to Development of
Antibodies: A novel mechanism for immune activation by mRNA
vaccines. J Immunol. 207:2405–2410. 2021.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Gambari R, Papi C, Gasparello J,
Agostinelli E and Finotti A: Preliminary results and a theoretical
perspective of co-treatment using a miR-93-5p mimic and aged garlic
extract to inhibit the expression of the pro-inflammatory
interleukin-8 gene. Exp Ther Med. 29(85)2025.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Kodera Y, Matsutomo T and Itoh K: The
evidence for the production mechanism of cis-S-1-propenylcysteine
in aged garlic extract based on a model reaction approach using its
isomers and deuterated solvents. Planta Med Lett. 2:e69–e72.
2015.
|
|
42
|
Kircheis R and Planz O: Could a lower
Toll-like receptor (TLR) and NF-κB activation due to a changed
charge distribution in the spike protein be the reason for the
lower pathogenicity of omicron? Int J Mol Sci.
23(5966)2022.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Robles JP, Zamora M, Adan-Castro E,
Siqueiros-Marquez L, Martinez de la Escalera G and Clapp C: The
spike protein of SARS-CoV-2 induces endothelial inflammation
through integrin α5β1 and NF-κB signaling. J Biol Chem.
298(101695)2022.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Forsyth CB, Zhang L, Bhushan A, Swanson B,
Zhang L, Mamede JI, Voigt RM, Shaikh M, Engen PA and Keshavarzian
A: The SARS-CoV-2 S1 spike protein promotes MAPK and NF-κB
activation in human lung cells and inflammatory cytokine production
in human lung and intestinal epithelial cells. Microorganisms.
10(1996)2022.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Gasparello J, D'Aversa E, Papi C, Gambari
L, Grigolo B, Borgatti M, Finotti A and Gambari R: Sulforaphane
inhibits the expression of interleukin-6 and interleukin-8 induced
in bronchial epithelial IB3-1 cells by exposure to the SARS-CoV-2
spike protein. Phytomedicine. 87(153583)2021.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Múnera-Rodríguez AM, Leiva-Castro C,
Sobrino F, López-Enríquez S and Palomares F: Sulforaphane-mediated
immune regulation through inhibition of NF-κB and MAPK signaling
pathways in human dendritic cells. Biomed Pharmacother.
177(117056)2024.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Gasparello J, Marzaro G, Papi C, Gentili
V, Rizzo R, Zurlo M, Scapoli C, Finotti A and Gambari R: Effects of
sulforaphane on SARS-CoV-2 infection and NF-κB dependent expression
of genes involved in the COVID-19 ‘cytokine storm’. Int J Mol Med.
52(76)2023.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Dharra R, Kumar Sharma A and Datta S:
Emerging aspects of cytokine storm in COVID-19: The role of
proinflammatory cytokines and therapeutic prospects. Cytokine.
169(156287)2023.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Leavis HL, van de Veerdonk FL and Murthy
S: Stimulating severe COVID-19: The potential role of GM-CSF
antagonism. Lancet Respir Med. 10:223–224. 2022.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Cherubini A, Rusconi F and Lazzari L:
Identification of the best housekeeping gene for RT-qPCR analysis
of human pancreatic organoids. PLoS One.
16(e0260902)2021.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Gentile AM, Lhamyani S, Coín-Aragüez L,
Oliva-Olivera W, Zayed H, Vega-Rioja A, Monteseirin J, Romero-Zerbo
SY, Tinahones FJ, Bermúdez-Silva FJ and El Bekay R: RPL13A and
EEF1A1 are suitable reference genes for qPCR during adipocyte
differentiation of vascular stromal cells from patients with
different BMI and HOMA-IR. PLoS One. 11(e0157002)2016.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Pease JE and Sabroe I: The role of
interleukin-8 and its receptors in inflammatory lung disease:
Implications for therapy. Am J Respir Med. 1:19–25. 2002.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Molnar V, Matišić V, Kodvanj I, Bjelica R,
Jeleč Ž, Hudetz D, Rod E, Čukelj F, Vrdoljak T, Vidović D, et al:
Cytokines and chemokines involved in osteoarthritis pathogenesis.
Int J Mol Sci. 22(9208)2021.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Marzaro G, Lampronti I, D'Aversa E,
Sacchetti G, Miolo G, Vaccarin C, Cabrini G, Dechecchi MC, Gambari
R and Chilin A: Design, synthesis and biological evaluation of
novel trimethylangelicin analogues targeting nuclear factor kB
(NF-κB). Eur J Med Chem. 151:285–293. 2018.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Gambari R, Borgatti M, Lampronti I, Fabbri
E, Brognara E, Bianchi N, Piccagli L, Yuen MCW, Kan CW, Hau DKP, et
al: Corilagin is a potent inhibitor of NF-kappaB activity and
downregulates TNF-alpha induced expression of IL-8 gene in cystic
fibrosis IB3-1 cells. Int Immunopharmacol. 13:308–315.
2012.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Doyle SL and O'Neill LAJ: Toll-like
receptors: From the discovery of NFkappaB to new insights into
transcriptional regulations in innate immunity. Biochem Pharmacol.
72:1102–1113. 2006.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Carmody RJ and Chen YH: Nuclear
factor-kappaB: Activation and regulation during Toll-like receptor
signaling. Cell Mol Immunol. 4:31–41. 2007.PubMed/NCBI
|
|
58
|
Verstrepen L, Bekaert T, Chau TL,
Tavernier J, Chariot A and Beyaert R: TLR-4, IL-1R and TNF-R
signaling to NF-kappaB: Variations on a common theme. Cell Mol Life
Sci. 65:2964–2978. 2008.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Zeng Z, Yu H, Chen H, Qi W, Chen L, Chen
G, Yan W, Chen T, Ning Q, Han M and Wu D: Longitudinal changes of
inflammatory parameters and their correlation with disease severity
and outcomes in patients with COVID-19 from Wuhan, China. Crit
Care. 24(525)2020.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Del Valle DM, Kim-Schulze S, Huang HH,
Beckmann ND, Nirenberg S, Wang B, Lavin Y, Swartz TH, Madduri D,
Stock A, et al: An inflammatory cytokine signature predicts
COVID-19 severity and survival. Nat Med. 26:1636–1643.
2020.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Burke H, Freeman A, Cellura DC, Stuart BL,
Brendish NJ, Poole S, Borca F, Phan HTT, Sheard N, Williams S, et
al: Inflammatory phenotyping predicts clinical outcome in COVID-19.
Respir Res. 21(245)2020.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Aldallal N, McNaughton EE, Manzel LJ,
Richards AM, Zabner J, Ferkol TW and Look DC: Inflammatory response
in airway epithelial cells isolated from patients with cystic
fibrosis. Am J Respir Crit Care Med. 166:1248–1256. 2002.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Maiuri L, Luciani A, Giardino I, Raia V,
Villella VR, D'Apolito M, Pettoello-Mantovani M, Guido S, Ciacci C,
Cimmino M, et al: Tissue transglutaminase activation modulates
inflammation in cystic fibrosis via PPARgamma down-regulation. J
Immunol. 180:7697–7705. 2008.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Bhattacharyya S, Balakathiresan NS,
Dalgard C, Gutti U, Armistead D, Jozwik C, Srivastava M, Pollard HB
and Biswas R: Elevated miR-155 promotes inflammation in cystic
fibrosis by driving hyperexpression of interleukin-8. J Biol Chem.
286:11604–11615. 2011.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Balakathiresan NS, Bhattacharyya S, Gutti
U, Long RP, Jozwik C, Huang W, Srivastava M, Pollard HB and Biswas
R: Tristetraprolin regulates IL-8 mRNA stability in cystic fibrosis
lung epithelial cells. Am J Physiol Lung Cell Mol Physiol.
296:L1012–L1018. 2009.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Kanamori Y, Via LD, Macone A, Canettieri
G, Greco A, Toninello A and Agostinelli E: Aged garlic extract and
its constituent, S-allyl-L-cysteine, induce the apoptosis of
neuroblastoma cancer cells due to mitochondrial membrane
depolarization. Exp Ther Med. 19:1511–1521. 2020.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Amano H, Kazamori D and Itoh K:
Pharmacokinetics and N-acetylation metabolism of
S-methyl-l-cysteine and trans-S-1-propenyl-l-cysteine in rats and
dogs. Xenobiotica. 46:1017–1025. 2016.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Amano H, Kazamori D and Itoh K: Evaluation
of the effects of S-allyl-L-cysteine, S-methyl-L-cysteine,
trans-S-1-propenyl-L-cysteine, and their N-acetylated and
S-oxidized metabolites on human CYP activities. Biol Pharm Bull.
39:1701–1707. 2016.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Soy M, Keser G, Atagündüz P, Tabak F,
Atagündüz I and Kayhan S: Cytokine storm in COVID-19: Pathogenesis
and overview of anti-inflammatory agents used in treatment. Clin
Rheumatol. 39:2085–2094. 2020.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Mustafa MI, Abdelmoneim AH, Mahmoud EM and
Makhawi AM: Cytokine storm in COVID-19 patients, its impact on
organs and potential treatment by QTY code-designed detergent-free
chemokine receptors. Mediators Inflamm.
2020(8198963)2020.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Gasparello J, Papi C, Marzaro G, Macone A,
Zurlo M, Finotti A, Agostinelli E and Gambari R: Aged garlic
extract (AGE) and its constituent S-allyl-cysteine (SAC) inhibit
the expression of pro-inflammatory genes induced in bronchial
epithelial IB3-1 cells by exposure to the SARS-CoV-2 spike protein
and the BNT162b2 vaccine. Molecules. 29(5938)2024.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Choudhury A and Mukherjee S: In silico
studies on the comparative characterization of the interactions of
SARS-CoV-2 spike glycoprotein with ACE-2 receptor homologs and
human TLRs. J Med Virol. 92:2105–2113. 2020.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Shirato K and Kizaki T: SARS-CoV-2 spike
protein S1 subunit induces pro-inflammatory responses via Toll-like
receptor 4 signaling in murine and human macrophages. Heliyon.
7(e06187)2021.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Zhao Y, Kuang M, Li J, Zhu L, Jia Z, Guo
X, Hu Y, Kong J, Yin H, Wang X and You F: SARS-CoV-2 spike protein
interacts with and activates TLR41. Cell Res. 31:818–820.
2021.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Chakraborty C, Mallick B, Bhattacharya M
and Byrareddy SN: SARS-CoV-2 omicron spike shows strong binding
affinity and favourable interaction landscape with the TLR4/MD2
compared to other variants. J Genet Eng Biotechnol.
22(100347)2024.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Aboudounya MM and Heads RJ: COVID-19 and
Toll-Like Receptor 4 (TLR4): SARS-CoV-2 may bind and activate TLR4
to increase ACE2 expression, facilitating entry and causing
hyperinflammation. Mediators Inflamm. 2021(8874339)2021.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Kawai T and Akira S: Signaling to
NF-kappaB by Toll-like receptors. Trends Mol Med. 13:460–469.
2007.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Tang S, Liang Y, Wang M, Lei J, Peng Y,
Tao Q, Ming T, Yang W, Zhang C, Guo J and Xu H: Qinhuo Shanggan
oral solution resolves acute lung injury by down-regulating
TLR4/NF-κB signaling cascade and inhibiting NLRP3 inflammasome
activation. Front Immunol. 14(1285550)2023.PubMed/NCBI View Article : Google Scholar
|
|
79
|
Kaushik D, Bhandari R and Kuhad A: TLR4 as
a therapeutic target for respiratory and neurological complications
of SARS-CoV-2. Expert Opin Ther Targets. 25:491–508.
2021.PubMed/NCBI View Article : Google Scholar
|
|
80
|
Yang G, Zhang S, Wang Y, Li L, Li Y, Yuan
D, Luo F, Zhao J, Song X and Zhao Y: Aptamer blocking S-TLR4
interaction selectively inhibits SARS-CoV-2 induced inflammation.
Signal Transduct Target Ther. 7(120)2022.PubMed/NCBI View Article : Google Scholar
|
|
81
|
Asaba CN, Ekabe CJ, Ayuk HS, Gwanyama BN,
Bitazar R and Bukong TN: Interplay of TLR4 and SARS-CoV-2:
Unveiling the complex mechanisms of inflammation and severity in
COVID-19 infections. J Inflamm Res. 17:5077–5091. 2024.PubMed/NCBI View Article : Google Scholar
|
|
82
|
Verbeke R, Hogan MJ, Loré K and Pardi N:
Innate immune mechanisms of mRNA vaccines. Immunity. 55:1993–2005.
2022.PubMed/NCBI View Article : Google Scholar
|
|
83
|
Sartorius R, Trovato M, Manco R, D'Apice L
and De Berardinis P: Exploiting viral sensing mediated by Toll-like
receptors to design innovative vaccines. NPJ Vaccines.
6(127)2021.PubMed/NCBI View Article : Google Scholar
|
|
84
|
Delehedde C, Even L, Midoux P, Pichon C
and Perche F: Intracellular routing and recognition of lipid-based
mRNA nanoparticles. Pharmaceutics. 13(945)2021.PubMed/NCBI View Article : Google Scholar
|
|
85
|
Da Costa CBP, Cruz ACM, Penha JCQ, Castro
HC, Da Cunha LER, Ratcliffe NA, Cisne R and Martins FJ: Using in
vivo animal models for studying SARS-CoV-2. Expert Opin Drug
Discov. 17:121–137. 2022.PubMed/NCBI View Article : Google Scholar
|
|
86
|
Wlosinska M, Nilsson AC, Hlebowicz J,
Fakhro M, Malmsjö M and Lindstedt S: Aged garlic extract reduces
IL-6: A double-blind placebo-controlled trial in females with a low
risk of cardiovascular disease. Evid Based Complement Alternat Med.
2021(6636875)2021.PubMed/NCBI View Article : Google Scholar
|
|
87
|
Xu C, Mathews AE, Rodrigues C, Eudy BJ,
Rowe CA, O'Donoughue A and Percival SS: Aged garlic extract
supplementation modifies inflammation and immunity of adults with
obesity: A randomized, double-blind, placebo-controlled clinical
trial. Clin Nutr ESPEN. 24:148–155. 2018.PubMed/NCBI View Article : Google Scholar
|
|
88
|
Fabbri E, Borgatti M, Montagner G, Bianchi
N, Finotti A, Lampronti I, Bezzerri V, Dechecchi MC, Cabrini G and
Gambari R: Expression of microRNA-93 and interleukin-8 during
Pseudomonas aeruginosa-mediated induction of proinflammatory
responses. Am J Respir Cell Mol Biol. 50:1144–1155. 2014.PubMed/NCBI View Article : Google Scholar
|
|
89
|
De Stefano D, Ungaro F, Giovino C,
Polimeno A, Quaglia F and Carnuccio R: Sustained inhibition of IL-6
and IL-8 expression by decoy ODN to NF-κB delivered through
respirable large porous particles in LPS-stimulated cystic fibrosis
bronchial cells. J Gene Med. 13:200–208. 2011.PubMed/NCBI View Article : Google Scholar
|
|
90
|
Lampronti I, Dechecchi MC, Rimessi A,
Bezzerri V, Nicolis E, Guerrini A, Tacchini M, Tamanini A, Munari
S, D'Aversa E, et al: β-Sitosterol reduces the expression of
chemotactic cytokine genes in cystic fibrosis bronchial epithelial
cells. Front Pharmacol. 8(236)2017.PubMed/NCBI View Article : Google Scholar
|
|
91
|
Hawdon NA, Aval PS, Barnes RJ, Gravelle
SK, Rosengren J, Khan S, Ciofu O, Johansen HK, Høiby N and Ulanova
M: Cellular responses of A549 alveolar epithelial cells to serially
collected Pseudomonas aeruginosa from cystic fibrosis
patients at different stages of pulmonary infection. FEMS Immunol
Med Microbiol. 59:207–220. 2010.PubMed/NCBI View Article : Google Scholar
|
|
92
|
Demonbreun AR, Velez MP, Saber R, Ryan DT,
Sancilio A, McDade TW and McNally EM: mRNA intramuscular
vaccination produces a robust IgG antibody response in advanced
neuromuscular disease. Neuromuscul Disord. 32:33–35.
2022.PubMed/NCBI View Article : Google Scholar
|
|
93
|
Elmazoglu Z, Aydın Bek Z, Sarıbaş SG,
Özoğul C, Goker B, Bitik B, Aktekin CN and Karasu Ç:
S-allylcysteine inhibits chondrocyte inflammation to reduce human
osteoarthritis via targeting RAGE, TLR4, JNK, and Nrf2 signaling:
Comparison with colchicine. Biochem Cell Biol. 99:645–654.
2021.PubMed/NCBI View Article : Google Scholar
|
|
94
|
Shao Z, Pan Z, Lin J, Zhao Q, Wang Y, Ni
L, Feng S, Tian N, Wu Y, Sun L, et al: S-allyl cysteine reduces
osteoarthritis pathology in the tert-butyl hydroperoxide-treated
chondrocytes and the destabilization of the medial meniscus model
mice via the Nrf2 signaling pathway. Aging (Albany NY).
12:19254–19272. 2020.PubMed/NCBI View Article : Google Scholar
|
|
95
|
Cantin AM, Hartl D, Konstan MW and Chmiel
JF: Inflammation in cystic fibrosis lung disease: Pathogenesis and
therapy. J Cyst Fibros. 14:419–430. 2015.PubMed/NCBI View Article : Google Scholar
|
|
96
|
Lemanske RF Jr: Inflammatory events in
asthma: An expanding equation. J Allergy Clin Immunol.
105:S633–S636. 2000.PubMed/NCBI View Article : Google Scholar
|
|
97
|
Xu J, Zeng Q, Li S, Su Q and Fan H:
Inflammation mechanism and research progress of COPD. Front
Immunol. 15(1404615)2024.PubMed/NCBI View Article : Google Scholar
|
|
98
|
Degan D, Ornello R, Tiseo C, Carolei A,
Sacco S and Pistoia F: The role of inflammation in neurological
disorders. Curr Pharm Des. 24:1485–1501. 2018.PubMed/NCBI View Article : Google Scholar
|
|
99
|
Ji Y, Li M, Chang M, Liu R, Qiu J, Wang K,
Deng C, Shen Y, Zhu J, Wang W, et al: Inflammation: Roles in
skeletal muscle atrophy. Antioxidants (Basel).
11(1686)2022.PubMed/NCBI View Article : Google Scholar
|
|
100
|
Henein MY, Vancheri S, Longo G and
Vancheri F: The role of inflammation in cardiovascular disease. Int
J Mol Sci. 23(12906)2022.PubMed/NCBI View Article : Google Scholar
|
|
101
|
Tsalamandris S, Antonopoulos AS, Oikonomou
E, Papamikroulis GA, Vogiatzi G, Papaioannou S, Deftereos S and
Tousoulis D: The role of inflammation in diabetes: Current concepts
and future perspectives. Eur Cardiol. 14:50–59. 2019.PubMed/NCBI View Article : Google Scholar
|
|
102
|
Coussens LM and Werb Z: Inflammation and
cancer. Nature. 420:860–867. 2002.PubMed/NCBI View Article : Google Scholar
|
|
103
|
Zhao H, Wu L, Yan G, Chen Y, Zhou M, Wu Y
and Li Y: Inflammation and tumor progression: Signaling pathways
and targeted intervention. Signal Transduct Target Ther.
6(263)2021.PubMed/NCBI View Article : Google Scholar
|