Introduction
Chronic subdural hematomas (CSDHs) are neurological
events characterized by the gradual accumulation of blood and its
breakdown products in the subdural space between the dura mater and
the cerebral cortex. CSDHs are most commonly associated with minor
head trauma, which can rupture the bridging veins spanning the
subdural space that then drain into the dural venous sinuses
(1). CSDHs primarily affect the
elderly population and have become one of the most frequently
encountered neurosurgical disorders in clinical settings. Recent
epidemiological data indicate that the annual incidence of CSDH
ranges from 1 to 13.5 per 100,000 individuals in the general
population, with a marked increase observed with advancing age. In
individuals aged 65 years and older, the incidence rises to ~58.1
per 100,000 individuals per year (2). The incidence of CSDHs has steadily
increased over recent decades; for example, a temporal analysis
conducted in Finland from 1990 to 2015 reported a doubling in
annual incidence, from 8.2 to 17.6 per 100,000 individuals per
year, likely driven by population aging and the widespread use of
antithrombotic agents (3). These
epidemiological trends highlight the growing clinical burden of
CSDHs and the need for improved therapeutic strategies.
The pathophysiology of a CSDH is multifactorial,
involving a complex interplay of biological processes, among which
inflammation has a central role. Following the initial hemorrhagic
event, a persistent inflammatory cascade is triggered, which
facilitates immune cell infiltration, activation of the
fibrinolytic system and the formation of a vascularized outer
membrane encasing the hematoma. The neovasculature within this
membrane is typically immature and highly permeable, resulting in
recurrent microbleeding and the extravasation of plasma components.
This self-perpetuating cycle, often referred to as the ‘CSDH
cycle’, drives progressive hematoma expansion over time (2,3).
Considering the key roles of inflammation and
aberrant angiogenesis in CSDH progression, recent pharmacological
efforts have increasingly focused on targeting these pathways using
established anti-inflammatory and anti-angiogenic therapies
(4,5). Atorvastatin, a widely prescribed
3-hydroxy-3-methylglutaryl-CoA reductase inhibitor primarily used
to treat hyperlipidemia and atherosclerosis, has demonstrated a
variety of biological effects beyond lipid-lowering (6). Notably, the anti-inflammatory and
angiogenesis-modulating properties of atorvastatin render it a
promising candidate for the management of CSDHs (7,8).
Preliminary clinical investigations have indicated that
atorvastatin may facilitate hematoma resolution and improve
neurological outcomes in patients with CSDHs (9-11).
These observations are further supported by animal studies showing
similar therapeutic benefits (12,13);
however, the precise molecular mechanisms by which atorvastatin
mediates its effects on CSDHs have not yet been fully
elucidated.
Network pharmacology offers an integrative framework
for investigating the complex pharmacodynamic profiles of
therapeutic agents by identifying their interactions with a number
of molecular targets (14). This
approach, which integrates computational modeling with systems
biology, enables the construction of comprehensive drug-target
networks and facilitates the discovery of novel mechanisms of
action as well as drug repurposing opportunities. In the present
study, network pharmacology analysis was combined with experimental
in vitro validation to investigate the potential molecular
mechanisms by which atorvastatin exerts therapeutic effects in the
treatment of CSDHs. Specifically, this study aimed to identify the
key molecular targets and signaling pathways through which
atorvastatin modulates inflammation in CSDHs, and to provide
mechanistic insights supporting its potential as a therapeutic
agent for this condition.
Materials and methods
Prediction of the therapeutic targets
of atorvastatin in CSDHs
Potential molecular targets of atorvastatin in CSDHs
were systematically retrieved from the ChEMBL (https://www.ebi.ac.uk/chembl/), NCBI PubChem Compound
(https://www.ncbi.nlm.nih.gov/pccompound) and
SwissTargetPrediction (http://www.swisstargetprediction.ch/) databases using
the keyword ‘atorvastatin’. All identified targets were
standardized to the official gene symbols via the UniProt database
(https://www.uniprot.org/) and duplicate entries
were removed to generate a non-redundant gene list. Concurrently,
CSDH-related genes were obtained from the GeneCards database
(https://www.genecards.org/) using the
keyword ‘CSDH’ and from the DisGeNET database (https://www.disgenet.org/) using the term ‘subdural
hematoma’ and duplicates were similarly excluded to ensure
uniqueness. The intersection of the atorvastatin-associated and
CSDH-associated genes was determined using Venny 2.1 (https://bioinfogp.cnb.csic.es/tools/venny/), and the
overlapping genes were defined as the putative therapeutic targets
of atorvastatin in CSDHs.
Enrichment analysis
Enrichment analysis of the identified target genes
was performed using R software (version 4.2.1; RStudio, Inc.). The
gene identifiers were converted to Entrez IDs using the
‘org.Hs.eg.db’ package (15). Gene
Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG)
pathway analyses were conducted using the ‘clusterProfiler,’
(16) ‘enrichplot,’ (17) ‘org.Hs.eg.db’ and ‘pathview’
(18) packages. P<0.05 and
Q<0.05 was considered to indicate a statistically significant
difference. Visualization of the enrichment results was performed
using the ‘ggplot2’ (19)
package.
Protein-protein interaction (PPI)
network construction
To investigate the functional interactions among the
candidate target proteins, a PPI network was constructed using the
STRING database (https://string-db.org/). The resulting network was
subsequently visualized using Cytoscape software (version 3.9.1;
Cytoscape Consortium) (20).
Molecular docking analysis
The 2D chemical structure of atorvastatin was
retrieved from the ChEMBL database (https://www.ebi.ac.uk/chembl/), whereas the 3D
structures of the target proteins were obtained from the RCSB
Protein Data Bank (https://www.rcsb.org/; accession numbers: 8QY5, 6WZM,
8H78, 6ESM and 4G8O). Molecular docking simulations were conducted
using AutoDockTools (version 1.5.7; Molecular Graphics Laboratory;
The Scripps Research Institute). The ligand-receptor binding
conformations and interactions were visualized using PyMOL software
(version 2.5.4; Schrödinger, Inc.).
Cell culture
Human umbilical vein endothelial cells (HUVECs; cat.
no. QS-H002; Keycell Biotechnology Co., Ltd.), authenticated via
short tandem repeat profiling by the manufacturer, were purchased
at passage 3 and cultured in endothelial cell-specific medium (cat.
no. QS-H002A; Keycell Biotechnology Co., Ltd.) supplemented with
10% fetal bovine serum (HyClone™; Cytiva). The cells were
maintained in a humidified incubator at 37˚C with 5% CO2
and all experiments were performed using cells between passages 5
and 8.
Induction of inflammation and
atorvastatin treatment
HUVECs were seeded into 6-well culture plates at a
density of 5x105 cells per well. The cells were treated
with 10 µg/l tumor necrosis factor-α (TNF-α; cat. no. 300-01A-10UG;
PeproTech Inc.; Thermo Fisher Scientific, Inc.) at 37˚C for 24 h to
induce inflammation. To assess the effects of atorvastatin
dose-dependently, cells were assigned to five groups: Control group
(untreated), inflammation group (TNF-α treated only) and three
intervention groups co-treated with TNF-α and 2.5, 10.0 or 20.0
µmol/l atorvastatin (cat. no. S5715: Selleck Chemicals) at 37˚C for
24 h; both agents were added to the culture medium at the same time
to evaluate the direct protective effect of atorvastatin against
TNF-α-induced inflammation. Based on the results of the
dose-response experiments and previous studies (21,22),
10 µmol/l was selected to investigate the effect of atorvastatin on
the expression of inflammation-related genes in endothelial cell
models.
Quantification of inflammatory
cytokine levels by enzyme-linked immunosorbent assay (ELISA)
After 24 h of treatment, cell culture supernatants
were collected and centrifuged at 560 x g for 5 min at 4˚C. The
concentrations of interleukin (IL)-6, C-X-C motif chemokine ligand
8 (CXCL-8)/IL-8, intercellular adhesion molecule 1 (ICAM-1) and
vascular cell adhesion molecule 1 (VCAM-1) were measured using
ELISA kits (IL-6, cat. no. EH0201; CXCL-8, cat. no. EH0205; ICAM-1,
cat. no. EH0161; VCAM-1, cat. no. EH0326; Wuhan Fine Biotech Co.,
Ltd), in accordance with the manufacturers' instructions.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from the HUVECs in each
group after 24 h of treatment using TRIzol™ reagent (cat. no.
15596-026; Ambion; Thermo Fisher Scientific, Inc.), then RT was
performed with the HiScript® II Q Select RT SuperMix kit (cat. no.
R233; Vazyme Biotech Co., Ltd.) under the following conditions:
50˚C for 15 min, 85˚C for 5 sec and 4˚C for 10 min. qPCR was
conducted using the AceQ qPCR SYBR Green Master Mix (cat. no. Q111;
Vazyme Biotech Co., Ltd.) on a ViiA™ 7 Real-Time PCR System
(Applied Biosystems; Thermo Fisher Scientific, Inc.). The qPCR
cycling conditions were: 95˚C for 10 min, followed by 40 cycles of
95˚C for 10 sec and 60˚C for 60 sec. A melting curve was obtained
by 95˚C for 15 sec, 60˚C for 60 sec, and a gradual increase to 95˚C
with continuous fluorescence acquisition to verify amplification
specificity. Data were analyzed using QuantStudio™ Real-Time PCR
software (v1.6.1; Thermo Fisher Scientific, Inc.). All primers were
synthesized by Beijing Tsingke Biotech Co., Ltd. (Table I). Gene expression was normalized
to GAPDH and the relative changes in gene expression were
quantified using the 2-ΔΔCq method (23).
 | Table IPrimers used for reverse
transcription-quantitative PCR. |
Table I
Primers used for reverse
transcription-quantitative PCR.
| Gene | Forward sequence
(5' to 3') | Reverse sequence
(5' to 3') | Size, bp |
|---|
| MMP-2 |
GTGTGAAGTATGGGAACGCC |
CCTGGAAGCGGAATGGAAAC | 231 |
| MMP-9 |
GCTACCACCTCGAACTTTGAC |
TCAGTGAAGCGGTACATAGGG | 161 |
| SERPINE-1 |
TGCCCTCACCAACATTCT |
TGCCACTCTCGTTCACCT | 54 |
| GAPDH |
TCAAGAAGGTGGTGAAGCAGG |
TCAAAGGTGGAGGAGTGGGT | 115 |
FITC-dextran permeability assay for
detecting endothelial cell barrier function
HUVECs were seeded into the upper chambers of
Transwell inserts (0.4 µm microporous PET membrane; BD Biosciences)
at a density of 5x104 cells per well. The cells were
cultured for 3-5 days until confluency was achieved, and a
continuous monolayer was formed. The same experimental grouping and
treatments as aforementioned were applied. After 24 h of
intervention, the medium in the upper chamber was removed and
replaced with 200 µl serum-free medium (cat. no. QS-H002A; Keycell
Biotechnology Co., Ltd.) containing 1 g/l FITC-dextran (10 kDa;
cat. no. HY-128868; MedChemExpress). Simultaneously, 800 µl
serum-free medium was added to the lower chamber. The plates were
then incubated at 37˚C with 5% CO2 for 3 h. Following
incubation, 100 µl of the medium was collected from the lower
chamber and transferred to a 96-well plate for fluorescence
measurement. As a reference, 100 µl of the initial FITC-dextran
solution was used to determine the fluorescence intensity of the
upper chamber. The permeability rate (%) was calculated as follows:
(Fluorescence intensity of the lower chamber/initial fluorescence
intensity of the upper chamber) x100.
Matrigel tube formation assay
To evaluate the effect of atorvastatin on the
angiogenesis of endothelial cell models, a Matrigel tube formation
assay was performed. Experimental procedures were conducted as
previously described (24).
Briefly, HUVECs (5x105 cells/well) were seeded on
Matrigel, which had been thawed overnight at 4˚C and used to
precoat the culture plates at 37˚C for 1 h before cell seeding, and
were treated as aforementioned. Tube formation was visualized and
images were collected under an inverted light microscope (IX51;
Olympus Corporation).
Statistical analysis
The data are presented as the mean ± standard
deviation. Statistical analysis was conducted using SPSS 25.0
software (IBM Corp.). Data were first assessed for normality and
homogeneity of variances using the Shapiro-Wilk test and Levene's
test, respectively. Then, one-way analysis of variance (ANOVA) was
employed to compare the differences among groups, followed by the
least significant difference or Tukey's honestly significant
difference post hoc test for pairwise comparisons. The overall
P-values from ANOVA are reported in the text, whereas the
significance levels from pairwise comparisons are indicated in the
figures. P<0.05 was considered to indicate a statistically
significant difference.
Results
Potential targets of atorvastatin in
the treatment of CSDHs
An initial list of 324 potential molecular targets
of atorvastatin was obtained without applying a specific confidence
score threshold, retaining only those with annotated gene symbols
and functional annotations. After standardization and
deduplication, 271 unique targets were retained. To identify the
genes associated with CSDHs, 125 genes were retrieved from
databases, resulting in 121 non-redundant CSDH-related genes.
Cross-referencing these genes with the atorvastatin targets
identified 19 overlapping genes through strict symbol matching via
Venny, which were defined as the candidate therapeutic targets of
atorvastatin in the context of CSDHs (Table II).
 | Table IIPotential target genes of
atorvastatin in the treatment of chronic subdural hematoma. |
Table II
Potential target genes of
atorvastatin in the treatment of chronic subdural hematoma.
| Gene symbol | Gene name |
|---|
| MMP-9 | Matrix
metalloproteinase 9 |
| SERPINE-1 | Serpin family e
member 1 |
| MMP-2 | Matrix
metalloproteinase 2 |
| FGF-2 | Fibroblast growth
factor 2 |
| CXCL-10 | C-X-C motif
chemokine 10 |
| PLAT | Tissue-type
plasminogen activator |
| AKT-1 | AKT
serine/threonine kinase 1 |
| ANGPT-2 | Angiopoietin 2 |
| CCL-2 | C-C motif chemokine
ligand 2 |
| CD-36 | CD36 molecule |
| CRP | C-reactive
protein |
| CXCL-8/IL-8 | C-X-C motif
chemokine ligand 8/Interleukin 8 |
| EDN-1 | Endothelin 1 |
| F2 | Coagulation factor
II |
| F5 | Coagulation factor
V |
| IL-6 | Interleukin 6 |
| INS | Insulin |
| VWF | Von Willebrand
factor |
| F7 | Coagulation factor
VII |
Target gene enrichment analysis in the
treatment of CSDHs with atorvastatin
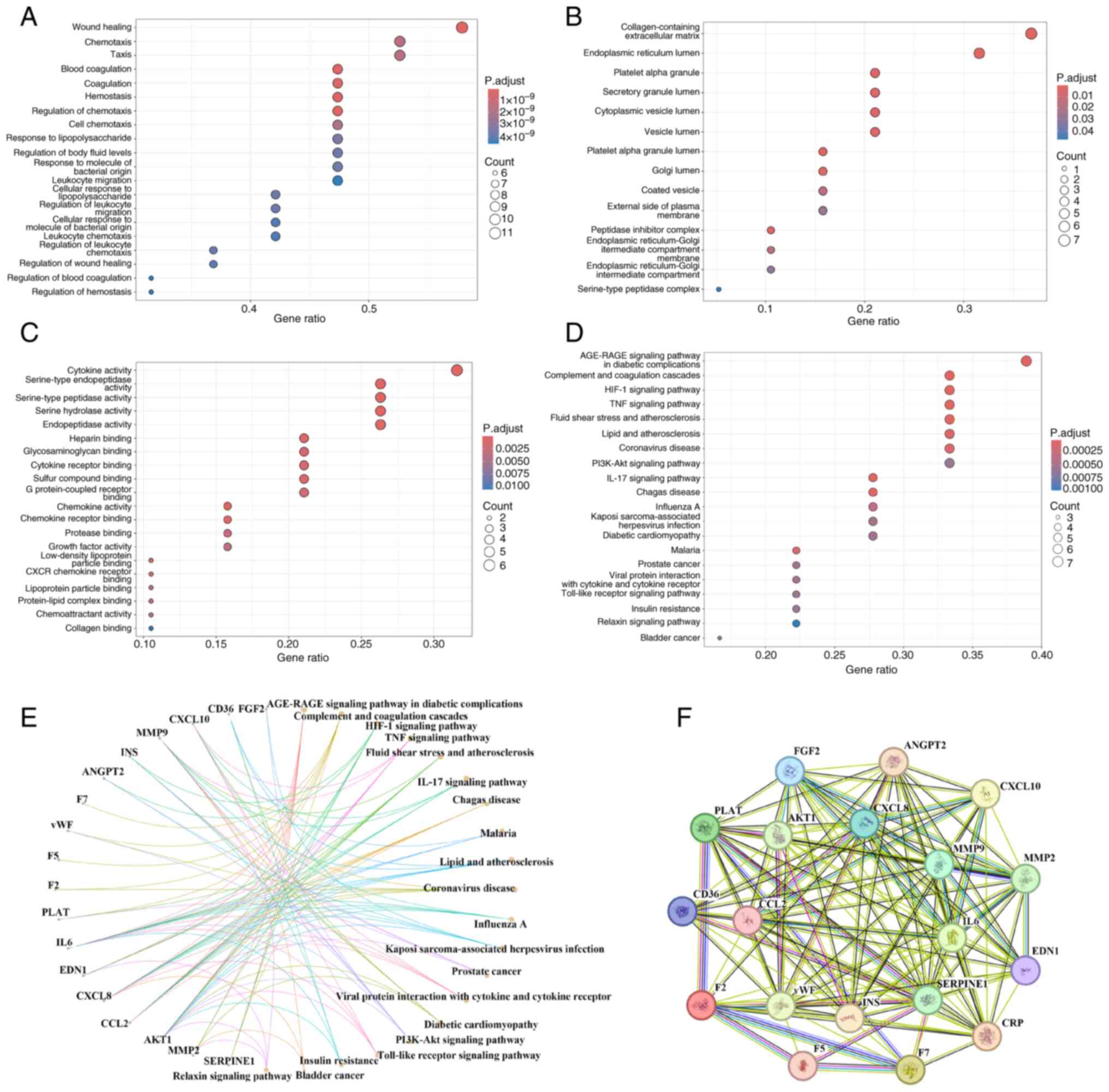
GO enrichment analysis of the 19 candidate genes
yielded 948 enriched terms, comprising 897 biological processes
(BPs), 15 cellular components (CCs) and 36 molecular functions
(MFs). The top 20 terms in each category are presented in Fig. 1A-C. These results suggest that
atorvastatin may exert therapeutic effects in CSDH management by
modulating BPs such as ‘Wound healing’, ‘Coagulation’,
‘Chemotaxis’, ‘Hemostasis’ and ‘Leukocyte migration’. The enriched
MFs included ‘Cytokine receptor binding’, ‘Cytokine activity’ and
‘Serine hydrolase activity’. The associated CCs included
‘Collagen-containing extracellular matrix’, ‘Peptidase inhibitor
complex’, ‘Secretory granule lumen’ and ‘cytoplasmic vesicle
lumen’.
KEGG pathway enrichment analysis revealed 62
pathways associated with the putative targets of atorvastatin in
treating CSDHs, with the top 20 pathways shown in Fig. 1D and E. These pathways are primarily involved
in the regulation of inflammation, coagulation and angiogenesis,
including the ‘HIF-1 signaling pathway’, ‘TNF signaling pathway’,
‘Complement and coagulation cascades’, ‘Toll-like receptor
signaling pathway’, ‘PI3K-Akt signaling pathway’ and ‘IL-17
signaling pathway’.
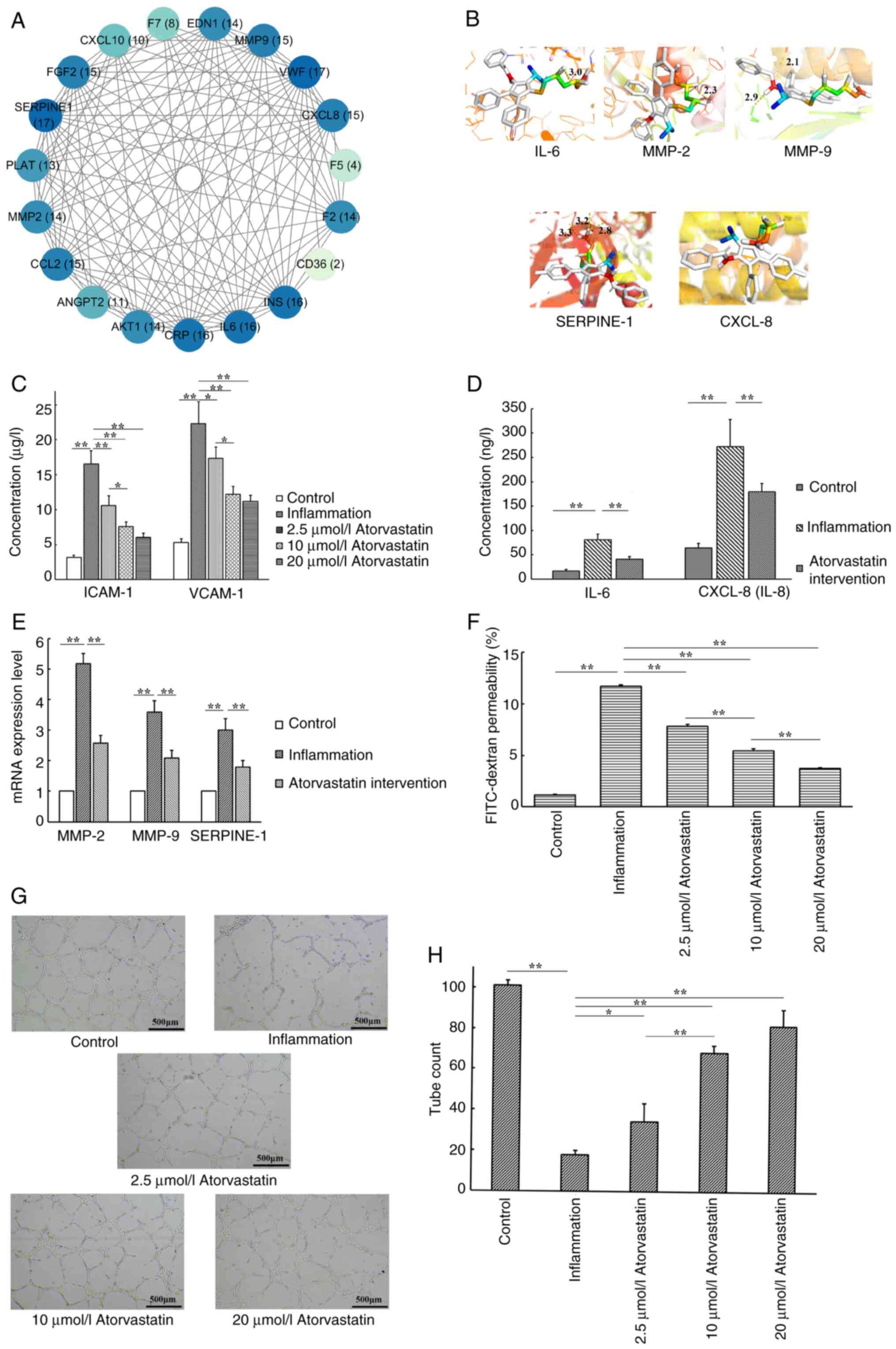
PPI network analysis
A PPI network was constructed using the STRING
database (Fig. 1F). Node degree
centrality was assessed to identify the highly connected hub genes
(Fig. 2A), with darker colors
representing nodes of higher degree (greater connectivity). Based
on both the topological importance and functional relevance, five
core targets [matrix metalloproteinase (MMP)-2, MMP-9, IL-6,
CXCL-8/IL-8 and serpin family E member 1 (SERPINE-1)] were selected
for further validation through molecular docking and in
vitro experiments. Other network metrics such as betweenness
and closeness centrality were also calculated, although they were
not used as primary selection criteria. These parameters reflect
the extent to which a node acts as a bridge or is close to other
nodes in the network, respectively, and were included to provide a
more comprehensive topological characterization. However, node
degree centrality was prioritized since it more directly represents
the number of connections a protein has, which is commonly regarded
as a reliable indicator of functional importance in biological
networks.
Molecular docking
Results of the molecular docking simulations
indicated that atorvastatin had strong binding affinities with all
five core target proteins, with binding energies ≤-5.0 kcal/mol.
This threshold value indicates strong ligand-receptor interactions
(25), with binding energies
<-7.0 kcal/mol suggesting a particularly high affinity (26). Furthermore, compared with the
binding energies of these target proteins with other known ligands
(27-31),
atorvastatin also exhibited a moderate to strong binding affinity
(Table III). Visualization of
the molecular docking results (Fig.
2B) revealed stable binding conformations characterized by key
hydrogen bonds and hydrophobic interactions (Table SI). These results provide
mechanistic insights into the potential modulation of these targets
by atorvastatin.
 | Table IIIBinding energies of potential target
gene proteins with atorvastatin and other known ligands. |
Table III
Binding energies of potential target
gene proteins with atorvastatin and other known ligands.
| Target | Binding energy with
Atorvastatin, kcal/mol | Binding energy with
other ligands, kcal/mol |
|---|
| IL-6 | -8.1 | -5.9-5.8 |
| MMP-2 | -6.0 | -8.2-5.7 |
| MMP-9 | -8.0 | -8.1-5.9 |
| SERPINE-1 | -6.7 | -8.2-5.3 |
| CXCL-8/IL-8 | -6.1 | -8.7-4.4 |
Atorvastatin reduces the inflammatory
response in an endothelial cell inflammation model
As shown in Fig.
2C, stimulation with TNF-α significantly increased the
secretion of the inflammatory biomarkers, ICAM-1 and VCAM-1, in the
HUVEC culture supernatants compared with the control group.
Treatment with atorvastatin at 2.5, 10.0 and 20.0 µmol/l
significantly reduced the ICAM-1 (F=60.544; P<0.001) and VCAM-1
(F=41.848; P<0.001) levels in a dose-dependent manner, compared
with the inflammation group. However, no statistically significant
difference was observed between the 10 and 20 µmol/l atorvastatin
treatment groups (P>0.05).
Inhibitory effects of atorvastatin on
TNF-α-induced inflammatory cytokine secretion and gene
expression
As shown in Fig. 2D
and E, TNF-α significantly
increased the secretion of IL-6 and CXCL-8 in the HUVEC culture
supernatants and significantly upregulated the mRNA expression of
MMP-2, MMP-9 and SERPINE-1 in HUVECs. Treatment with 10 µmol/l
atorvastatin significantly reduced the TNF-α-induced IL-6
(F=64.526; P<0.001) and CXCL-8 (F=37.779; P<0.001) secretion
levels and significantly downregulated the mRNA expression of MMP-2
(F=264.413; P<0.001), MMP-9 (F=86.675; P<0.001) and SERPINE-1
(F=71.180; P<0.001).
Atorvastatin attenuates
inflammation-induced endothelial barrier dysfunction
As shown in Fig.
2F, TNF-α stimulation markedly impaired the integrity of the
endothelial barrier, resulting in a significant increase in
FITC-dextran permeability from 1.14±0.08% in the control group to
11.68±0.13% in the inflammation group. Atorvastatin treatment
effectively restored barrier function in a dose-dependent manner,
reducing permeability to 7.83±0.17, 5.44±0.20 and 3.71±0.06% at
2.5, 10.0 and 20.0 µmol/l, respectively (F=2490.788; P<0.001).
These results further support the hypothesis that atorvastatin has
a protective effect on endothelial barrier integrity under
inflammatory conditions.
Atorvastatin protects endothelial
angiogenesis from inflammation-induced inhibition
As shown in Fig. 2G
and H, TNF-α stimulation
significantly impaired tube formation in HUVECs compared with the
control group. Co-treatment with atorvastatin at 2.5, 10.0 and 20.0
µmol/l significantly preserved angiogenic capacity in a
dose-dependent manner, attenuating the inflammation-induced
inhibition (F=99.859; P<0.001). However, no statistically
significant difference was observed between the 10 and 20 µmol/l
atorvastatin treatment groups (P>0.05).
Discussion
The pathophysiological mechanisms underlying the
onset and progression of CSDHs have been increasingly clarified in
recent decades. It is now accepted that the initial hemorrhage
originates from ruptured bridging veins, often triggered by minor
head trauma (particularly in elderly individuals with cerebral
atrophy) or by low intracranial pressure and neurosurgical
interventions (2,32). In the early phase following
hemorrhage, inflammatory cells (including neutrophils, lymphocytes,
macrophages and eosinophils) are recruited to the subdural space to
initiate tissue repair (2,3). However, these immune cells also
release inflammatory mediators and pro-angiogenic factors that
contribute to the formation of the hematoma capsule and to the
development of the neovascularization within it. The resulting
vasculature is typically immature and highly permeable, allowing
plasma leakage into the hematoma cavity and sustaining a cycle of
inflammation and hematoma enlargement, a process often referred to
as the ‘CSDH cycle’ (2,3). In addition to inflammation and
angiogenesis, the dysregulation of coagulation and fibrinolysis is
also involved in the pathogenesis of CSDHs. Enhanced fibrinolytic
activity may lead to persistent microhemorrhage and progressive
hematoma enlargement (33,34). Accordingly, fibrinolytic biomarkers
such as tissue-type plasminogen activator (tPA) have been proposed
as potential predictors of CSDH recurrence (35).
In the present study, the overlapping genes
identified in the network pharmacology analysis of the treatment of
CSDHs with atorvastatin were notably enriched in BPs related to
inflammation such as ‘Hemostasis’ and ‘Leukocyte migration’, as
well as key pathways such as the ‘TNF signaling pathway’, ‘IL-17
signaling pathway’ and ‘HIF-1 signaling pathway’. These results
provide support for the hypothesis that atorvastatin may interrupt
multiple pathogenic associations in the CSDH cycle. In the present
study, GO enrichment analysis revealed that the
atorvastatin-associated genes were significantly enriched in
biological processes related to these key mechanisms. Among the top
10 enriched processes, ‘Wound healing’ reflected an association to
angiogenesis and inflammation (36), ‘Blood coagulation’ and ‘Hemostasis’
were associated with coagulation-fibrinolysis balance (37) and terms such as ‘Leukocyte
chemotaxis,’ ‘Leukocyte migration’ and ‘Cell chemotaxis’ were
indicative of inflammatory regulation (38). Based on the subsequent PPI network
analysis performed in the present study and supporting literature
(1-3),
five key genes (IL-6, CXCL-8, MMP-2, MMP-9 and SERPINE-1) were
identified as central regulators of inflammation, angiogenesis and
coagulation/fibrinolysis in the treatment of CSDHs with
atorvastatin. These genes were selected for further mechanistic
investigation to elucidate the molecular basis of the therapeutic
effects of atorvastatin. IL-6 and CXCL-8 (also known as IL-8) are
pro-inflammatory cytokines predominantly secreted by fibroblasts,
endothelial cells and various immune cells such as monocytes,
macrophages, neutrophils and T lymphocytes (39). IL-6 mediates acute-phase
inflammatory responses, immune cell differentiation, thrombopoiesis
and leukocyte recruitment (40),
whereas CXCL-8 has a key role in immune cell chemotaxis and
promotes angiogenesis by stimulating endothelial cell proliferation
and tube formation (41). Elevated
levels of both cytokines have been detected in the hematoma fluid
from patients with CSDHs, particularly those with recurrent
hematomas (42). MMP-2 and MMP-9
are involved in the degradation of extracellular matrix (ECM)
components and are functionally linked to both inflammation and
angiogenesis, and aberrant upregulation of these enzymes
contributes to neovascular instability and increases the risk of
recurrent hemorrhage in CSDHs (43). Additionally, MMP-2 and MMP-9
participate in inflammatory signaling cascades by modulating the
activity of cytokines and their receptors (44). Elevated levels of both enzymes have
been observed in the hematoma fluid from patients with CSDHs
(45). SERPINE-1, also known as
plasminogen activator inhibitor-1, is a serine protease inhibitor
predominantly expressed by endothelial cells; it regulates
fibrinolysis by inhibiting tissue-type and urokinase-type
plasminogen activators and has been implicated in multiple
pathological processes, including inflammation, oxidative stress,
fibrosis and macrophage migration (46).
In the present study, molecular docking simulations
demonstrated that atorvastatin stably bound to the protein
structures encoded by the five identified target genes. Consistent
with these predictions, in vitro experiments revealed that
atorvastatin treatment significantly reduced the secretion of
ICAM-1 and VCAM-1 in TNF-α-stimulated HUVECs. These adhesion
molecules are well-established biomarkers of endothelial
inflammation (47), and their
downregulation indicates effective suppression of TNF-α-mediated
inflammatory responses. Additionally, atorvastatin significantly
decreased the secretion of IL-6 and CXCL-8 and the mRNA expression
of MMP-2 and MMP-9, supporting the mechanistic insights derived
from both the network pharmacology and docking analyses. Therefore,
the reduction in IL-6/CXCL-8 secretion and MMP-2/MMP-9 expression
in HUVECs, coupled with the improvement in endothelial barrier
integrity, may help limit microvascular leakage and inflammatory
cell infiltration in hematoma membranes. These effects could, in
turn, contribute to stabilization of neovessels and reduced
re-bleeding, ultimately facilitating hematoma resolution in
vivo; these mechanisms will be validated in animal models of
CSDH in future studies. It is noteworthy that fibrinolysis may
exert bidirectional effects in the pathogenesis and progression of
CSDHs; in the pathophysiological setting of CSDHs, excessive
fibrinolysis may accelerate ECM breakdown, potentially triggering
pro-MMP activation and compromising vascular stability, thereby
increasing the risk of microvascular leakage or re-bleeding
(33,34). Paradoxically, hyperfibrinolysis
could also promote hematoma resolution by facilitating clot
degradation and reducing postoperative hematoma recurrence
(48). Therefore, as an important
regulator of fibrinolysis, it is necessary to avoid overly
simplistic conclusions regarding the regulation of SERPINE-1 in
CSDHs. Future work should employ time-course analyses of
fibrinolytic markers (such as tPA, plasmin activity and D-dimer),
ECM degradation products, MMP/tissue inhibitors of
metalloproteinase activity profiles and fibrosis markers (such as
collagen I/III, α-smooth muscle actin and fibronectin) in both
in vitro co-culture systems and in vivo CSDH models.
Such integrative approaches will enable a more precise
understanding of whether the effects of atorvastatin on SERPINE-1
confer a benefit or risk in the context of CSDH management.
Notably, in the present study, atorvastatin was
demonstrated to significantly improve endothelial barrier integrity
under inflammatory conditions as evidenced by the reduced
FITC-dextran permeability, a key pathological feature implicated in
CSDH progression (2,3). This improvement was accompanied by a
concentration-dependent decrease in both cytokine secretion and
permeability, underscoring the therapeutic window and potency of
atorvastatin in mitigating endothelial dysfunction. Moreover, the
present study demonstrated that TNF-α suppressed tube formation in
HUVECs, supporting the notion that inflammatory cytokines impair
normal angiogenesis and contribute to the formation of fragile,
leaky vessels in CSDH (2,3). Atorvastatin alleviated this
inhibitory effect, which may be attributed to its anti-inflammatory
and endothelial-protective properties. A previous study has
demonstrated that atorvastatin reduces CSDH in animal models by
promoting angiogenesis (49).
Collectively, these in vitro findings support the hypothesis
that atorvastatin may exert therapeutic benefits in CSDH not only
through inflammation suppression but also by improving the quality
of angiogenesis within the hematoma capsule.
Taken together, the findings of the present study
suggest that atorvastatin may interrupt the self-sustaining CSDH
cycle by inhibiting inflammatory signaling, modulating fibrinolytic
and angiogenic processes, suppressing the formation of immature
leaky vessels, limiting immune cell infiltration and preserving
endothelial function. These combined effects may facilitate
hematoma liquefaction and enhance spontaneous resorption. Despite
these promising results, it is important to recognize that the
pathogenesis of CSDHs is multifaceted and that the regulatory
effects of atorvastatin likely extend beyond the five core targets
identified in the present study. Therefore, the proposed mechanism
should be viewed as a partial representation of the therapeutic
actions of atorvastatin. In the present study, KEGG pathway
enrichment analysis further indicated that multiple classical
inflammatory and angiogenic signaling pathways may be implicated in
the mode of action of atorvastatin, including the ‘HIF-1 signaling
pathway’, ‘TNF signaling pathway’, ‘IL-17 signaling pathway’,
‘Toll-like receptor signaling pathway’ and the ‘PI3K-Akt signaling
pathway’. Furthermore, clinical evidence has identified diabetes
mellitus and atherosclerosis as notable risk factors for both the
onset and recurrence of CSDH (50,51).
Consistently, the KEGG analysis in the present study revealed the
enrichment of pathways related to these metabolic and vascular
disorders, such as ‘AGE-RAGE signaling pathway in diabetic
complications’, ‘Fluid shear stress and atherosclerosis’ and
‘Diabetic cardiomyopathy’, providing potential avenues for
exploring the broader mechanistic effects of atorvastatin in CSDHs.
Additionally, several pathways associated with infectious diseases
and malignancies were also enriched. Prior studies have reported
associations between CSDH recurrence and underlying malignancy
(52) or chronic infection
(53). These findings suggest that
the systemic health status of patients may influence disease
progression and that statins may offer a benefit in patient
subgroups with these comorbidities. The roles of these
comorbidities in CSDH pathophysiology, as well as their potential
modulation by atorvastatin, warrant further investigation. Although
atorvastatin is primarily used to regulate lipid metabolism
(6), the results of the enrichment
analysis in the present study did not clearly indicate a potential
role of lipid metabolism regulation in its treatment of CSDH.
Therefore, the in vitro endothelial inflammation model
established in the present study does not involve lipid metabolism,
and the observed molecular effects are more likely attributable to
the pleiotropic effects of atorvastatin rather than its
lipid-lowering properties. Nonetheless, it must be acknowledged
that in vivo, lipid regulation may synergistically
contribute to endothelial protection (54). In future studies, control compounds
with lipid-lowering effects but minimal pleiotropic activity will
be used to help delineate the relative contributions of each
mechanism.
Despite the promising findings of the present study,
several limitations should be acknowledged. First, the study relied
solely on in vitro experiments using HUVECs as a model for
endothelial inflammation; although this approach provides
mechanistic insights, it may not fully replicate the
cerebrovascular endothelial environment of CSDHs, nor simulate
interactions with macrophages and fibroblasts. Second, the key
inflammatory regulatory signaling pathways of statins, such as the
NF-κB pathway, have not been directly evaluated. Third, the network
pharmacology analysis was based on existing public databases, which
may not cover all relevant molecular interactions or reflect
tissue-specific gene expression profiles. In terms of target gene
selection, while the method employed in the present study provides
clear traceability, it may underestimate the extent of functional
overlap. The selection of five core target genes, while supported
by the established PPI network and functional relevance, may
overlook other key targets involved in CSDH progression. Moreover,
the present study did not examine the long-term treatment outcomes
of atorvastatin. Finally, the lack of in vivo validation,
such as animal model experiments, limits the translational
applicability of the current findings. Future studies should
incorporate in vivo CSDH models for validation of the core
molecular targets, integrate proteomic and transcriptomic analyses
to delineate the downstream signaling networks and assess the
dose-response relationships and pharmacokinetic profiles of
atorvastatin in animal models. In addition, broader target
selection and verification such as scoring-based or
relevance-weighted intersections, along with the incorporation of
clinical data, will be essential to more comprehensively elucidate
the therapeutic mechanisms of atorvastatin on CSDHs. The next phase
of research will also focus on p65 nuclear translocation assays and
phosphorylated p65 expression by western blotting to investigate
whether the anti-inflammatory effects of atorvastatin are mediated,
at least in part, through NF-κB inhibition. Meanwhile,
pathway-specific verifications, including HIF-1α and IL-17 protein
levels, will also be prioritized in future mechanistic studies.
Furthermore, future studies will incorporate direct assessments of
ECM remodeling, such as collagen degradation assays, gelatin
zymography and immunofluorescent staining for ECM components, to
confirm whether the downregulation of MMP-2 and MMP-9 contributes
to the preservation of ECM integrity.
In conclusion, the present study identified key
molecular targets and signaling pathways potentially involved in
the therapeutic effects of atorvastatin on CSDHs. The results from
the integrated network pharmacology analysis and in vitro
experimental validation suggest that atorvastatin may exert its
effects by modulating inflammation, angiogenesis and fibrinolysis.
These findings provide a mechanistic foundation and valuable
reference for future preclinical and clinical studies aimed at
elucidating the therapeutic potential of atorvastatin in managing
CSDHs.
Supplementary Material
Key hydrogen bonds and hydrophobic
interactions between atorvastatin and potential target gene
proteins predicted by molecular docking.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by grant from Key
Research and Development Plan of Shaanxi Province, China (grant no.
2024SF-YBXM-047).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
CL conceived and designed the study. JY performed
the in vitro experiments. XW conducted the bioinformatics
analyses. CL and JY contributed primarily to manuscript drafting
and confirm the authenticity of all the raw data. All authors
reviewed and approved the final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Use of artificial intelligence tools
During the preparation of this work, AI tools were
used to improve the readability and language of the manuscript, and
subsequently, the authors revised and edited the content produced
by the AI tools as necessary, taking full responsibility for the
ultimate content of the present manuscript.
References
|
1
|
Nouri A, Gondar R, Schaller K and Meling
T: Chronic subdural hematoma (cSDH): A review of the current state
of the art. Brain Spine. 1(100300)2021.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Zhong D, Cheng H, Xian Z, Ren Y, Li H, Ou
X and Liu P: Advances in pathogenic mechanisms, diagnostic methods,
surgical and non-surgical treatment, and potential recurrence
factors of Chronic Subdural Hematoma: A review. Clin Neurol
Neurosurg. 242(108323)2024.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Feghali J, Yang W and Huang J: Updates in
chronic subdural hematoma: Epidemiology, etiology, pathogenesis,
treatment, and outcome. World Neurosurg. 141:339–345.
2020.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Liu T, Zhao Z, Liu M, An S, Nie M, Liu X,
Qian Y, Tian Y, Zhang J and Jiang R: The pharmacological landscape
of chronic subdural hematoma: A systematic review and network
meta-analysis of randomized and non-randomized controlled studies.
Burns Trauma. 12(tkae034)2024.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Verma Y, Abdelghaffar M, Verma O, Gajjar
A, Ghozy S and Kallmes DF: Bevacizumab: The future of chronic
subdural hematoma. Interv Neuroradiol: November 21, 2024 (Epub
ahead of print).
|
|
6
|
Morofuji Y, Nakagawa S, Ujifuku K,
Fujimoto T, Otsuka K, Niwa M and Tsutsumi K: Beyond lipid-lowering:
Effects of statins on cardiovascular and cerebrovascular diseases
and cancer. Pharmaceuticals (Basel). 15(151)2022.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Araujo FA, Rocha MA, Mendes JB and Andrade
SP: Atorvastatin inhibits inflammatory angiogenesis in mice through
down regulation of VEGF, TNF-alpha and TGF-beta1. Biomed
Pharmacother. 64:29–34. 2010.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Kim SK, Choe JY, Kim JW, Park KY and Kim
B: Anti-Inflammatory effect of atorvastatin and rosuvastatin on
monosodium urate-induced inflammation through IL-37/Smad3-complex
activation in an in vitro study using THP-1 macrophages.
Pharmaceuticals (Basel). 17(883)2024.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Chan DY, Chan DT, Sun TF, Ng SC, Wong GK
and Poon WS: The use of atorvastatin for chronic subdural
haematoma: A retrospective cohort comparison study. Br J Neurosurg.
31:72–77. 2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Jiang R, Zhao S, Wang R, Feng H, Zhang J,
Li X, Mao Y, Yuan X, Fei Z, Zhao Y, et al: Safety and efficacy of
atorvastatin for chronic subdural hematoma in chinese patients: A
randomized ClinicalTrial. JAMA Neurol. 75:1338–1346.
2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Monteiro GA, Queiroz TS, Goncalves OR,
Cavalcante-Neto JF, Batista S, Rabelo NN, Welling LC, Figueiredo
EG, Leal PRL and Solla DJF: Efficacy and safety of atorvastatin for
chronic subdural hematoma: An updated systematic review and
meta-analysis. World Neurosurg. 188:177–184. 2024.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Quan W, Zhang Z, Li P, Tian Q, Huang J,
Qian Y, Gao C, Su W, Wang Z, Zhang J, et al: Role of regulatory T
cells in atorvastatin induced absorption of chronic subdural
hematoma in rats. Aging Dis. 10:992–1002. 2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Yang L, Li N, Yang L, Wang D, Qiang S and
Zhao Z: Atorvastatin-induced absorption of chronic subdural
hematoma is partially attributed to the polarization of
macrophages. J Mol Neurosci. 72:565–573. 2022.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Liang C, Zhang B, Li R, Guo S and Fan X:
Network pharmacology -based study on the mechanism of traditional
Chinese medicine in the treatment of glioblastoma multiforme. BMC
Complement Med Ther. 23(342)2023.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Carlson M: Genome wide annotation for
Human. Bioconductor, 2022. https://bioconductor.org/packages/org.Hs.eg.db.
|
|
16
|
Yu G, Wang LG, Han Y and He QY:
ClusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Yu G: enrichplot: Visualization of
Functional Enrichment Result. Bioconductor, 2022. https://bioconductor.org/packages/enrichplot.
|
|
18
|
Luo W and Brouwer C: Pathview: An
R/Bioconductor package for pathway-based data integration and
visualization. Bioinformatics. 29:1830–1831. 2013.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Wickham H: ggplot2: Elegant Graphics for
Data Analysis. 1st Edition. Springer, New York, NY, 2009.
|
|
20
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Bao XM, Wu CF and Lu GP: Atorvastatin
attenuates homocysteine-induced apoptosis in human umbilical vein
endothelial cells via inhibiting NADPH oxidase-related oxidative
stress-triggered p38MAPK signaling. Acta Pharmacol Sin.
30:1392–1398. 2009.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Li D, Chen H, Romeo F, Sawamura T, Saldeen
T and Mehta JL: Statins modulate oxidized low-density
lipoprotein-mediated adhesion molecule expression in human coronary
artery endothelial cells: Role of LOX-1. J Pharmacol Exp Ther.
302:601–605. 2002.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Livak KJ and Schmittgen TD: . Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Liang C, Wei T, Zhang T and Niu C:
Adipose-derived stem cell-mediated alphastatin targeting delivery
system inhibits angiogenesis and tumor growth in glioma. Mol Med
Rep. 28(215)2023.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Shityakov S and Förster C: In silico
predictive model to determine vector-mediated transport properties
for the blood-brain barrier choline transporter. Adv Appl Bioinform
Chem. 7:23–36. 2014.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Hsin KY, Ghosh S and Kitano H: Combining
machine learning systems and multiple docking simulation packages
to improve docking prediction reliability for network pharmacology.
PLoS One. 8(e83922)2013.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Xiong L, Chen Q and Liu H: Network
pharmacology and molecular docking identified IL-6 as a critical
target of Qing Yan He Ji against COVID-19. Medicine (Baltimore).
103(e40720)2024.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Ahmad A, Sayed A, Ginnebaugh KR, Sharma V,
Suri A, Saraph A, Padhye S and Sarkar FH: Molecular docking and
inhibition of matrix metalloproteinase-2 by novel
difluorinatedbenzylidene curcumin analog. Am J Transl Res.
7:298–308. 2015.PubMed/NCBI
|
|
29
|
Kumar NK, Geervani VS, Kumar RSM, Singh S,
Abhishek M and Manimozhi M: Data-driven dentistry: Computational
revelations redefining pulp capping. J Conserv Dent Endod.
27:649–653. 2024.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Dos Santos RV, Barrionuevo MVF, Vieira
MRF, Mazoni I and Tasic L: Plasminogen activator inhibitors in
thrombosis: Structural analysis and potential natural inhibitors.
ACS Omega. 10:27348–27362. 2025.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Madej M, Halama A, Chrobak E and Gola JM:
Time-dependent impact of betulin and its derivatives on IL-8
expression in colorectal cancer cells with molecular docking
studies. Int J Mol Sci. 26(6186)2025.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Weigel R, Schilling L and Krauss JK: The
pathophysiology of chronic subdural hematoma revisited: Emphasis on
aging processes as key factor. Geroscience. 44:1353–1371.
2022.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Jensen TSR, Olsen MH, Lelkaitis G, Kjaer
A, Binderup T and Fugleholm K: Urokinase plasminogen activator
receptor: An important focal player in chronic subdural hematoma?
Inflammation. 47:1015–1027. 2024.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Tamura R, Sato M, Yoshida K and Toda M:
History and current progress of chronic subdural hematoma. J Neurol
Sci. 429(118066)2021.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Katano H, Kamiya K, Mase M, Tanikawa M and
Yamada K: Tissue plasminogen activator in chronic subdural
hematomas as a predictor of recurrence. J Neurosurg. 104:79–84.
2006.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Shi Z, Yao C, Shui Y, Li S and Yan H:
Research progress on the mechanism of angiogenesis in wound repair
and regeneration. Front Physiol. 14(1284981)2023.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Costantini TW, Kornblith LZ, Pritts T and
Coimbra R: The intersection of coagulation activation and
inflammation after injury: What you need to know. J Trauma Acute
Care Surg. 96:347–356. 2024.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Cambier S, Gouwy M and Proost P: The
chemokines CXCL8 and CXCL12: molecular and functional properties,
role in disease and efforts towards pharmacological intervention.
Cell Mol Immunol. 20:217–251. 2023.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Adumitrachioaiei H, Sasaran MO and
Marginean CO: The diagnostic and prognostic role of interleukin 6
and interleukin 8 in childhood acute gastroenteritis-a review of
the literature. Int J Mol Sci. 25(7655)2024.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Schumertl T, Lokau J, Rose-John S and
Garbers C: Function and proteolytic generation of the soluble
interleukin-6 receptor in health and disease. Biochim Biophys Acta
Mol Cell Res. 1869(119143)2022.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Matsushima K, Yang D and Oppenheim JJ:
Interleukin-8: An evolving chemokine. Cytokine.
153(155828)2022.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Hong HJ, Kim YJ, Yi HJ, Ko Y, Oh SJ and
Kim JM: Role of angiogenic growth factors and inflammatory cytokine
on recurrence of chronic subdural hematoma. Surg Neurol.
71:161–165; discussion 165-166. 2009.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Mustafa S, Koran S and AlOmair L: Insights
into the role of matrix metalloproteinases in cancer and its
various therapeutic aspects: A review. Front Mol Biosci.
9(896099)2022.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Muneer PMA, Pfister BJ, Haorah J and
Chandra N: Role of matrix metalloproteinases in the pathogenesis of
traumatic brain injury. Mol Neurobiol. 53:6106–6123.
2016.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Hua C, Zhao G, Feng Y, Yuan H, Song H and
Bie L: Role of matrix metalloproteinase-2, matrix
metalloproteinase-9, and vascular endothelial growth factor in the
development of chronic subdural hematoma. J Neurotrauma. 33:65–70.
2016.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Kwak SY, Park S, Kim H, Lee SJ, Jang WS,
Kim MJ, Lee S, Jang WI, Kim AR, Kim EH, et al: Atorvastatin
inhibits endothelial PAI-1-mediated monocyte migration and
alleviates radiation-induced enteropathy. Int J Mol Sci.
22(1828)2021.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Hou X and Pei F: Estradiol inhibits
cytokine-induced expression of VCAM-1 and ICAM-1 in cultured human
endothelial cells via AMPK/PPARalpha activation. Cell Biochem
Biophys. 72:709–717. 2015.PubMed/NCBI View Article : Google Scholar
|
|
48
|
O YM, Tsang SL and Leung GK:
Fibrinolytic-facilitated chronic subdural hematoma drainage-A
systematic review. World Neurosurg. 150:e408–e419. 2021.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Wang D, Li T, Wei H, Wang Y, Yang G, Tian
Y, Zhao Z, Wang L, Yu S, Zhang Y, et al: Atorvastatin enhances
angiogenesis to reduce subdural hematoma in a rat model. J Neurol
Sci. 362:91–99. 2016.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Chen M, Da L, Zhang Q, Liu J, Tang J and
Zha Z: Development of a predictive model for assessing the risk
factors associated with recurrence following surgical treatment of
chronic subdural hematoma. Front Surg. 11(1429128)2024.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Jensen TSR, Thiesson EM, Fugleholm K,
Wohlfahrt J and Munch TN: Inflammatory risk factors for chronic
subdural hematoma in a nationwide cohort. J Inflamm Res.
17:8261–8270. 2024.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Hori YS, Aoi M, Oda K and Fukuhara T:
Presence of a malignant tumor as a novel predictive factor for
repeated recurrences of chronic subdural hematoma. World Neurosurg.
105:714–719. 2017.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Dubinski D, Won SY, Trnovec S, Gounko K,
Baumgarten P, Warnke P, Cantre D, Behmanesh B, Bernstock JD,
Freiman TM, et al: Recurrence of chronic subdural hematoma due to
low-grade infection. Front Neurol. 13(1012255)2022.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Hermida N and Balligand JL: Low-density
lipoprotein-cholesterol-induced endothelial dysfunction and
oxidative stress: The role of statins. Antioxid Redox Signal.
20:1216–1237. 2014.PubMed/NCBI View Article : Google Scholar
|