Introduction
SRY-related high-mobility group box protein B5
(SOX5), a member of the SRY-related HMG-box gene family, has gained
interest for its functions in developmental biology and disease
pathogenesis (1-5).
In tumor biology, abnormal SOX5 expression and activity is
associated with the advancement of a number of human malignancies,
including hepatocellular carcinoma [promoting epithelial-mesencymal
transition (EMT) and invasion], bladder cancer (modulating
migration and therapy resistance), prostate cancer (driving
metastasis via EMT), ovarian cancer (regulating glycolysis and
proliferation), gastric cancer (enhancing proliferation and
migration), tongue carcinoma (facilitating tumorigenesis) and
esophageal squamous cell carcinoma (where its downregulation is
associated with a poor prognosis) (6-8).
While SOX5 may have tumor-suppressive characteristics in some
tissues, its upregulation in numerous malignancies is acknowledged
as a notable contributor to tumor growth, rendering it a key
subject within cancer research.
The protein encoded by SOX5 is part of the high
mobility group of proteins, which modulate transcription through
DNA binding and are key in cell differentiation. In typical
physiological conditions, SOX5 regulates collagen synthesis, neural
development and cartilage creation. In cancer, the function of SOX5
is altered, potentially facilitating tumor initiation and
progression by regulating the cell cycle and promoting malignant
behaviors (such as inducing EMT, enhancing cell migration and
invasion, maintaining cancer stem cell properties) or enabling
immune evasion (9-12).
Altered SOX5 expression has been documented in a
number of malignancies, including lung, breast, gastric cancers and
melanoma (13,14). In lung adenocarcinoma (LUAD),
increased SOX5 expression is associated with adverse prognosis,
including reduced survival and enhanced recurrence rates. This
modified expression indicates that SOX5 may enhance tumor cell
proliferation and survival by activating downstream signaling
pathways, such as Wnt/β-catenin and PI3K/AKT. In addition to its
intrinsic effects on tumor cells, SOX5 also affects the tumor
microenvironment (TME). It can regulate the activity of
tumor-associated fibroblasts, thereby influencing immunological
responses and facilitating immune evasion (15-17).
Additionally, SOX5 has been associated with the regulation of tumor
angiogenesis, highlighting its complex involvement in cancer
advancement.
Understanding the varied expression patterns and
roles of SOX5 across different tumor types is key for the
advancement of novel cancer therapeutics. Consequently, the aims of
the present study are threefold: i) To delineate the molecular
regulatory network of SOX5; ii) to characterize its distinct
involvement in carcinogenesis; and iii) to explore methodologies
for improving therapeutic efficacy through the targeting of
SOX5-associated pathways. Pan-cancer analysis provides a thorough
method to elucidate the role of SOX5 role in tumorigenesis,
establishing a theoretical basis and prospective targets for
forthcoming clinical therapies.
Materials and methods
Data processing
Within the present study, data from The Cancer
Genome Atlas (TCGA; https://portal.gdc.cancer.gov/) was utilized. TCGA is
a large-scale cancer genome project jointly funded and managed by
the National Cancer Institute and the National Human Genome
Research Institute to advance the scientific understanding of
cancer. Specifically, RNA-sequencing data (fragments per kilobase
format) and clinical annotations from TCGA Pan-Cancer Atlas
(version 2018) were obtained through the University of California,
Santa Cruz Xena browser (https://xena.ucsc.edu/). Normalization and batch
correction were performed using the ComBat algorithm from the ‘sva’
R package (version 4.3.1) (18) to
mitigate technical variations across sequencing centers. Samples
with incomplete clinical metadata or low sequencing quality (read
count <10 million) were excluded.
For validation, Gene Expression Omnibus (GEO;
https://www.ncbi.nlm.nih.gov/geo/)
datasets (GSE30219, GSE31210 and GSE50081) (19-21)
were retrieved using the ‘GEOquery’ R package (22). Probes were annotated to gene
symbols based on platform-specific annotation files (such as the
platform name ‘GPL570’ for the array name ‘Affymetrix Human
Genome-U133 Plus 2.0’). For genes mapped by multiple probes, the
duplicates were collapsed by selecting the single probe with the
maximum mean expression value across all samples for that gene.
Additionally, the ‘TCGAbiolinks’ R package (23) was employed to download and access
TCGA data, facilitating the integration and bioinformatics analysis
of numerous data types.
Expression of SOX5 gene in a number of
cancers
Differential SOX5 expression across tumor and
corresponding normal tissues in TCGA was examined through use of
the ‘TCGAplot’ R package (version 1.0) (24). For cancers lacking normal tissue
data in TCGA [including diffuse large B cell lymphoma (DLBC) and
mesothelioma (MESO)], GEO datasets (GSE12453 and GSE51024)
(25,26) were supplemented. Normalized
log2-transformed expression values from both sources
were compared using Wilcoxon rank-sum tests. This analysis provided
SOX5 expression profiles in normal and tumor tissues for cancers
including DLBC, low-grade glioma, MESO, ovarian serous
cystadenocarcinoma, testicular germ cell tumor (TGCT), uterine
carcinosarcoma and uveal melanoma.
Diagnosis and prognosis analysis
Using TCGA data, the diagnostic potential of SOX5
was assessed by generating receiver-operating characteristic (ROC)
curves with the ‘pROC’ R package (27). An area under the curve (AUC) value
of 0.5-0.7 indicated a low diagnostic value, 0.7-0.9 indicated a
moderate value and >0.9 indicated a high diagnostic value.
Prognostic associations of SOX5 expression with overall survival
(OS), disease-specific survival (DSS), disease-free interval (DFI)
and progression-free interval (PFI) were evaluated using the
SangerBox database (version 3.0; http://sangerbox.com/). Statistical significance was
determined using multiple-testing correction (the
Benjamini-Hochberg method) (28).
Patients were stratified by median SOX5 expression and multivariate
Cox regression adjusted for age, stage and sex was applied where
appropriate.
Correlation analysis of immune
infiltration
Immune infiltration in the TME was assessed by
calculating Stromal Score, Immune Score and Estimation of Stromal
and Immune Cells in Malignant Tumor Tissues Using Expression Data
(ESTIMATE) Score using the ‘ESTIMATE’ R package (29). The cancer immunity cycle was
analyzed using the Tumor Immune Estimation Resource 2.0 web tool
(http://timer.cistrome.org/) to evaluate
correlations between SOX5 expression and immune activation steps
such as T cell recruitment and antigen presentation. Immune cell
fractions were estimated using Cell-Type Identification by
Estimating Relative Subsets of RNA Transcripts (CIBERSORT version
1.04) (30) deconvolution and
single-sample Gene Set Enrichment Analysis (ssGSEA) through the
Gene Set Variation (‘GSVA’) R package (31) quantified immune cell enrichment.
Associations between SOX5 expression and microsatellite instability
(MSI), tumor mutational burden (TMB) and immune checkpoint proteins
across tumors were analyzed in SangerBox (http://sangerbox.com/) using Pearson correlation
tests.
Drug sensitivity analysis
Drug sensitivity data from the Genomics of Drug
Sensitivity in Cancer 2 database (version Release 8.5) (https://www.cancerrxgene.org/) were processed using
the ‘oncoPredict’ R package (version 0.2) (32). IC50 values were
estimated through ridge regression and normalized to Z-scores.
Drugs with a Z-score >2 were considered to be significantly
associated with SOX5 expression.
Analysis of tumor mutation
patterns
The cBioPortal database (https://www.cbioportal.org/) was used to explore
associations between SOX5 expression and tumor mutation patterns.
Somatic mutations and copy-number alterations were retrieved from
cBioPortal (TCGA Pan-Cancer Atlas; version 2018). Patients were
divided into high and low SOX5 expression groups (top/bottom 25%;
high SOX5, n=250; low SOX5, n=261). Mutational signatures were
visualized using the ‘Maftools’ package (version 2.6.05) (33), enabling comprehensive analysis of
somatic mutation patterns in TCGA-LUAD cohort.
Functional enrichment analysis
Co-expression networks were constructed using
LinkedOmics (www.linkedomics.org; TCGA_LUAD cohort; HiSeq platform)
with Pearson correlation [r>0.3; false discovery rate (FDR)
<0.05] to identify genes co-expressed with SOX5. GSEA of
Molecular Signatures Database Hallmark and Kyoto Encyclopedia of
Genes and Genomes (KEGG) gene sets, as well as GSVA of Gene
Ontology terms were performed to explore associated biological
functions and pathways. Analyses were performed with 1,000
permutations and results with FDR-adjusted P<0.05 were
considered significant.
Cell culture
Human non-small cell lung cancer (NSCLC) cell lines
A549 and H1299, as well as the normal human bronchial epithelial
cell line BEAS-2B, were obtained from the Cell Bank of the Chinese
Academy of Sciences (Shanghai Institute of Biochemistry and Cell
Biology). H1299, A549 and BEAS-2B normal lung epithelial cells were
cultured in DMEM supplemented with 10% FBS (Gibco; Thermo Fisher
Scientific, Inc.) at 37˚C in a humidified incubator with 5%
CO2. Cells were digested every 48 h with 0.25%
trypsin-EDTA (Gibco; Thermo Fisher Scientific, Inc.) and passaged
at a 1:3 ratio. Experiments utilized cells between passages
5-20.
Cell transfection
For SOX5 knockdown, A549 cells were transfected with
50 nM of a SOX5-targeting siRNA (sense, 5'-GGGUCGUGUUCAAUCUUAUU-3'
and antisense, 5'-UUAAGAUUGAACACGACCCUU-3'; Shanghai GenePharma
Co., Ltd.) or a scrambled control siRNA (sense,
5'-GGCUGGUAGUUGGCAUUUAUU-3' and antisense,
5'-UAAAUGCCAACUACCAGCCUU-3'; Shanghai GenePharma Co., Ltd.) using
Lipofectamine™ RNAiMAX (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions. Following
transfection, cells were incubated at 37˚C in a 5% CO2
atmosphere for 24 h to allow for efficient knockdown. Experimental
groups consisted of the untreated control, scrambled siRNA control
and SOX5 knockdown. SOX5 expression was measured after the 24-h
incubation period using quantitative PCR (qPCR).
Cell Counting Kit-8 (CCK-8) assay
At 24 h post-transfection, cells were seeded in
96-well plates (5x103 cells/well) and incubated for 24
h. Furthermore, 10 µl of Cell Counting Kit-8 (CCK-8) solution
(Dojindo Molecular Technologies, Inc.) was added to each well and
the plates were incubated for 4 h. A total of 150 µl DMSO was added
to each well to dissolve the formazan crystals. The absorbance at
490 nm was then measured using a microplate reader (BioTek Synergy
HTX; BioTek; Agilent Technologies, Inc.). Cytotoxic activity (%)
using optical density (OD) values was calculated as:
Wound healing assay
Cell migration was evaluated by monitoring wound
closure over 24 h. Cells were grown to ~80% confluence in 6-well
plates,, scratched with a sterile 200 µl pipette tip, washed with
PBS and cultured in serum-free DMEM. Images were captured at 0 and
24 h using the Axiovert 200 microscope (Zeiss GmbH) and analyzed
with ImagePro Plus software (version 6.0). Experiments were
performed in triplicate.
Transwell invasion assay
After 24 h of transfection, cells
(5x104/well) were seeded into Transwell chambers. The
chambers were pre-coated with Matrigel by incubating with a diluted
Matrigel solution at 37˚C for 1 h to allow for polymerization. The
cells were then plated in serum-free medium. Medium containing 10%
FBS was added to the lower chamber. After a 24-h incubation period
at 37˚C, non-migrated cells on the upper surface of the membrane
were carefully removed with a cotton swab. The invaded cells on the
lower surface were fixed with 95% ethanol for 15 min at room
temperature and subsequently stained with 0.1% crystal violet for
20 min at room temperature. After washing and air-drying, the cells
were imaged in three random fields per chamber at x100
magnification using an inverted light microscope (Olympus
Corporation).
Reverse Transcription Quantitative PCR
(RT-qPCR)
Total RNA was isolated from cultured A549 cells
using the TRIzol™ reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol. RNA concentration
and purity were determined spectrophotometrically. Subsequently, 1
µg of total RNA was reverse-transcribed into cDNA using the
PrimeScript™ RT Reagent Kit (Takara Bio Inc.) in strict accordance
with the manufacturer's instructions. qPCR was performed using
PowerUp™ SYBR™ Green Master Mix (cat. no. A25742; Applied
Biosystems) on an Applied Biosystems 7500 Real-Time PCR System. The
thermal cycling conditions were as follows: Initial denaturation at
95˚C for 2 min; followed by 40 cycles of denaturation at 95˚C for
15 sec and annealing/extension at 60˚C for 1 min. A melting curve
analysis was performed post-amplification to verify reaction
specificity. Relative mRNA expression levels were calculated using
the 2-ΔΔCq method (34), with GAPDH serving as the internal
reference gene for normalization using the following formulae:
∆Cq=Cqtarget-CqGAPDH;
∆∆Cq=∆Cqreference-∆Cqtest. The primer
sequences were as follows: GAPDH forward,
5'-CATCATCCCTGCCTCTACTGG-3' and reverse,
5'-GTGGGTGTCGCTGTTGAAGTC-3'; and SOX5 forward,
5'-CAGATGGAGAGGTAGCCATGG-3 and reverse,
5'-CCATTGTATTGTGCTGAGAAGTG-3'.
Statistical analysis
Statistical analyses were performed using SPSS
(version 26.0; IBM Corp.) and R (version 4.3.1). For comparisons
between two groups of continuous variables, the unpaired Student's
t-test was used for normally distributed data and the unpaired
Mann-Whitney U test for non-normally distributed data. For
comparisons among three or more groups, one-way ANOVA (parametric)
was applied, followed by Tukey's post hoc test for multiple
comparisons. The prognostic value of SOX5 expression was evaluated
by univariate Cox regression analysis and Kaplan-Meier survival
analysis with the log-rank test. Associations between SOX5
expression and immune-related regulatory factors were assessed
using Spearman's rank correlation analysis. A two-sided P-value of
<0.05 was considered to indicate a statistically significant
difference.
Results
Differential expression of SOX5 in
pan-cancer between tumor and normal tissue
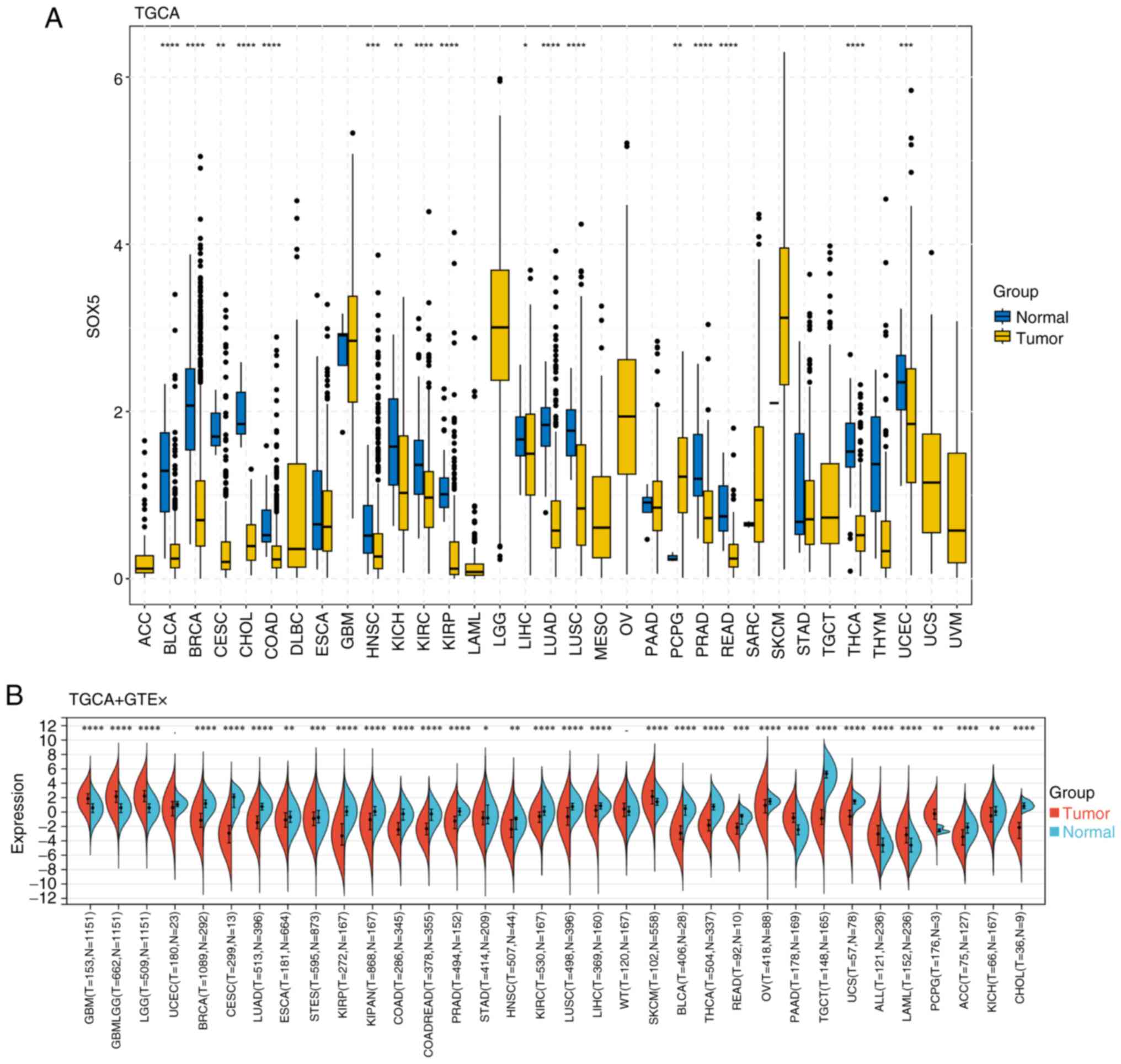
Using box plots generated from TCGA database, the
expression levels of SOX5 mRNA in tumor and normal tissues were
analyzed. As shown in Fig. 1A,
SOX5 expression was significantly higher in tumor tissues compared
with normal tissues in certain cancer types, such as PCPG
(P<0.01). By contrast, in numerous cancers, including LUAD, lung
squamous cell carcinoma (LUSC) and breast invasive carcinoma
(BRCA), SOX5 mRNA expression was markedly lower in tumor tissues
compared with adjacent normal tissues (P<0.05). Fig. 1B further demonstrated this pattern,
showing lower SOX5 expression was observed in tumor tissues across
the majority of cancers analyzed (P<0.05). Collectively, these
results indicate that SOX5 expression is generally reduced in
numerous cancer tissues
Predictive value of SOX5 expression
for diagnosis and prognosis
Fig. 2 illustrates
the diagnostic and prognostic significance of SOX5 across cancers.
ROC curve analysis (Fig. 2A)
demonstrated good diagnostic performance for head-neck squamous
cell carcinoma (AUC=0.953; 95% CI, 0.928-0.978) and LUAD
(AUC=0.916; 95% CI, 0.894-0.940), with notable discrimination in 15
additional cancer types (AUC range, 0.508-0.893), all exceeding
random chance. Differential expression analysis (Fig. 2B) showed that SOX5 plays an
environment-dependent role in tumorigenesis and is progressively
dysregulated at various tumor stages. We observed significant
differences in six types of tumors, such as STES (Stage I=76;
II=201; III=230; IV=57) (P<0.05), STAD (Stage I=58; II=121;
III=169; IV=41) (P<0.05), THYM (Stage I=36;II=61;III=14;IV=6)
(P<0.05), PAAD (Stage I=21;II=147; III=3;IV=4) (P<0.05), BLCA
(Stage II=130;III=140;IV=133) (P<0.05), and KICH (Stage
I=21;II=25;III=14;IV=6) (P<0.05). Notably, 78% (18/23) of
carcinomas showed marked downregulation of SOX5.
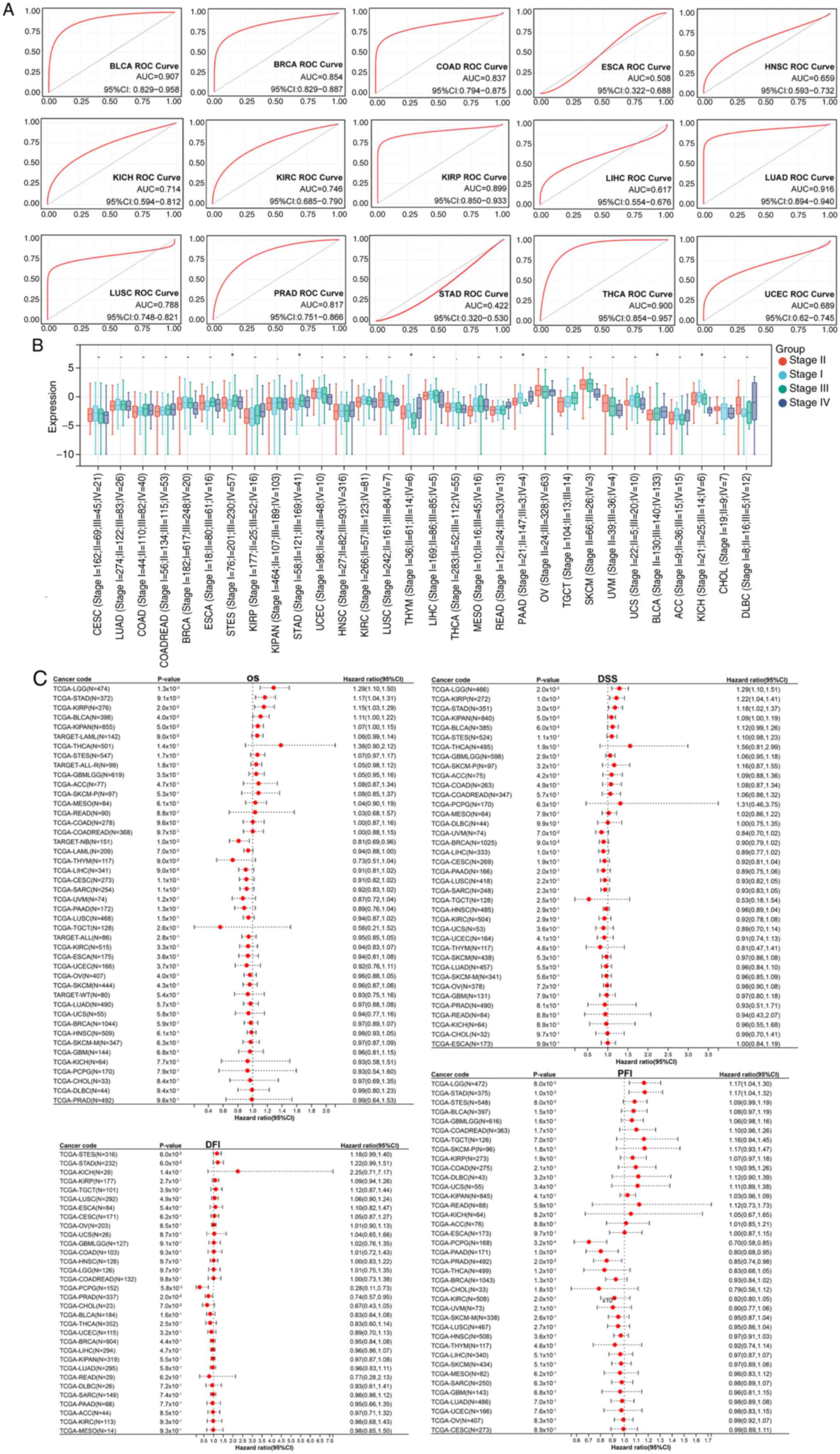 | Figure 2Diagnostic and prognostic value of
SOX5 in pan-cancer. (A) ROC curves for diagnosing 15 cancers based
on SOX5 expression. (B) Boxplots showing the association between
SOX5 expression levels and seven clinical characteristics in 30
cancer types. (C) Circular histogram of OS, DSS, DFI and PFI of
SOX5 in 44 types of cancer using Kaplan-Meier analysis.
*P<0.05; **P<0.01;
***P<0.001. ns, not significant; ROC, receiver
operator characteristic; SOX5, SRY-related high-mobility group box
protein B5; OS, overall survival; DSS, disease-specific survival;
DFI, disease-free interval; PFI, progression-free interval; BLCA,
bladder urothelial carcinoma; BRCA, breast invasive carcinoma;
COAD, colon adenocarcinoma; ESCA, esophageal carcinoma; KICH,
kidney chromophobe; KIRC, kidney renal clear cell carcinoma; KIRP,
kidney renal papillary cell carcinoma; LIHC, liver hepatocellular
carcinoma; HNSC, head and neck squamous cell carcinoma; LUAD, lung
adenocarcinoma; LUSC, lung squamous cell carcinoma; PRAD, prostate
adenocarcinoma; STAD, stomach adenocarcinoma; THCA, thyroid
carcinoma; UCEC, uterine corpus endometrial carcinoma. |
Multivariate survival analysis (Fig. 2C) demonstrated clinically relevant
prognostic stratification. Forest plots for OS, DSS, DFI and PFI
revealed that elevated SOX5 expression was associated with worse
outcomes [hazard ratio (HR) >1; 95% CI, excluding 1] in a number
of cancers, while low expression was associated with protective
effects (HR <1). These integrated findings suggest that SOX5 is
a key dual-purpose biomarker for early cancer detection and precise
prognostication.
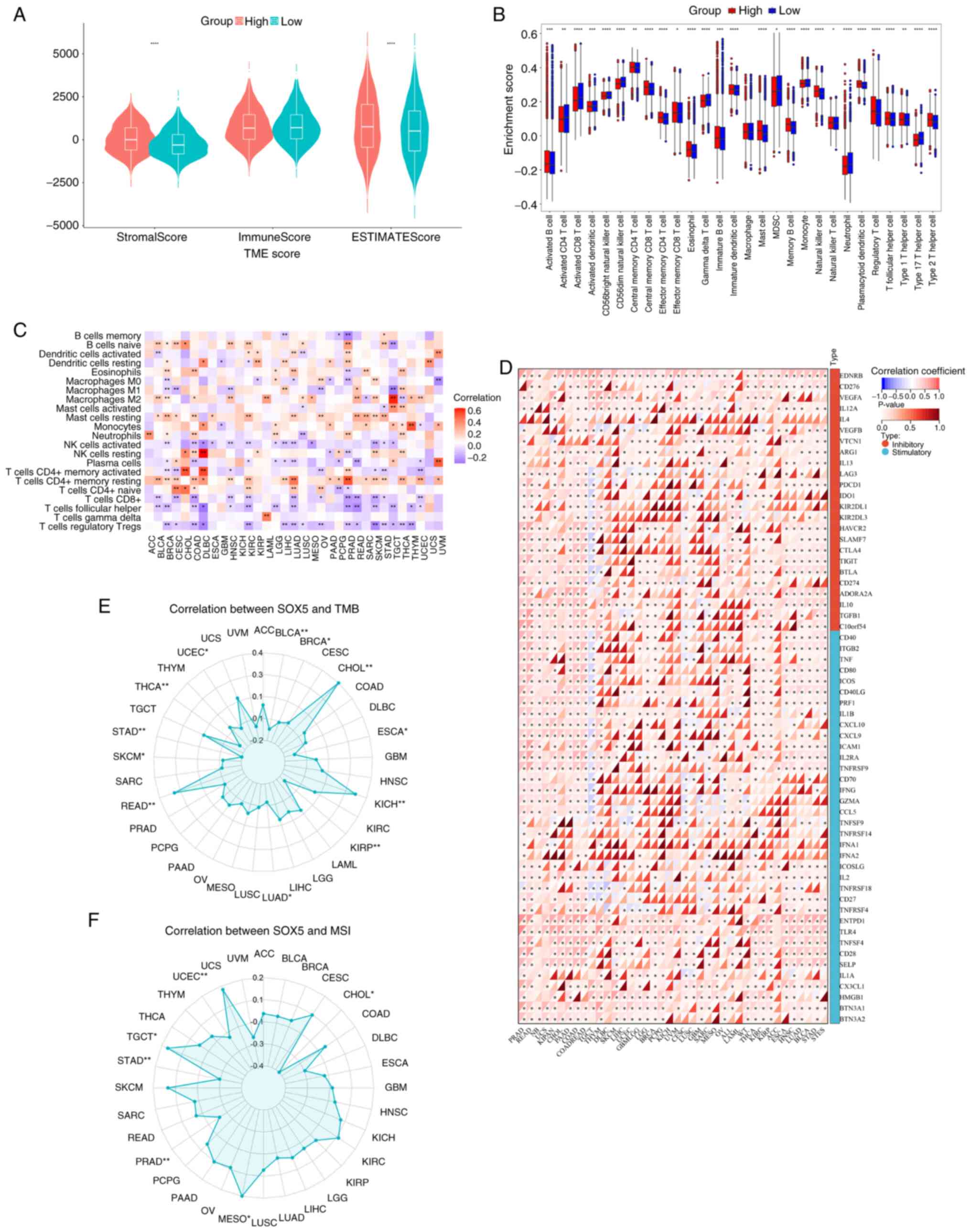
SOX5 in pan-cancer immune
infiltration
The present analysis identified SOX5 as a notable
modulator of tumor immune dynamics. Violin plots (Fig. 3A) showed significantly higher SOX5
expression in tumor tissues compared with normal tissues
(P<1x10-15), while box plots (Fig. 3B) revealed notable inter-cohort
heterogeneity. ESTIMATE, CIBERSORT and ssGSEA analyses demonstrated
that high SOX5 expression is associated with an activated TME and
increased immune scores (P<0.05), particularly in LUAD and
UCEC.
Heatmap analysis (Fig.
3C) revealed a negative correlation between SOX5 and cytotoxic
T cell infiltration (Pearson r=-0.68; P<2x10-16) and
a positive correlation between SOX5 and regulatory T cell abundance
(r=0.59; P<5x10-9), indicating immunosuppressive
reprogramming. Radar charts (Fig.
3E and F) further showed that
SOX5 expression was inversely correlated with TMB (r=-0.43;
P<0.001) and microsatellite instability (r=-0.51; P<0.0001),
although subtype-specific variations existed. Correlation networks
(Fig. 3D) revealed significant
co-expression of SOX5 with immune checkpoint molecules [programmed
death-ligand 1 (PD-L1), r=0.62; cytotoxic T-lymphocyte associated
protein 4 (CTLA-4), r=0.57; P<0.05], positioning SOX5 as a key
regulator of immune evasion and DNA damage response in cancer.
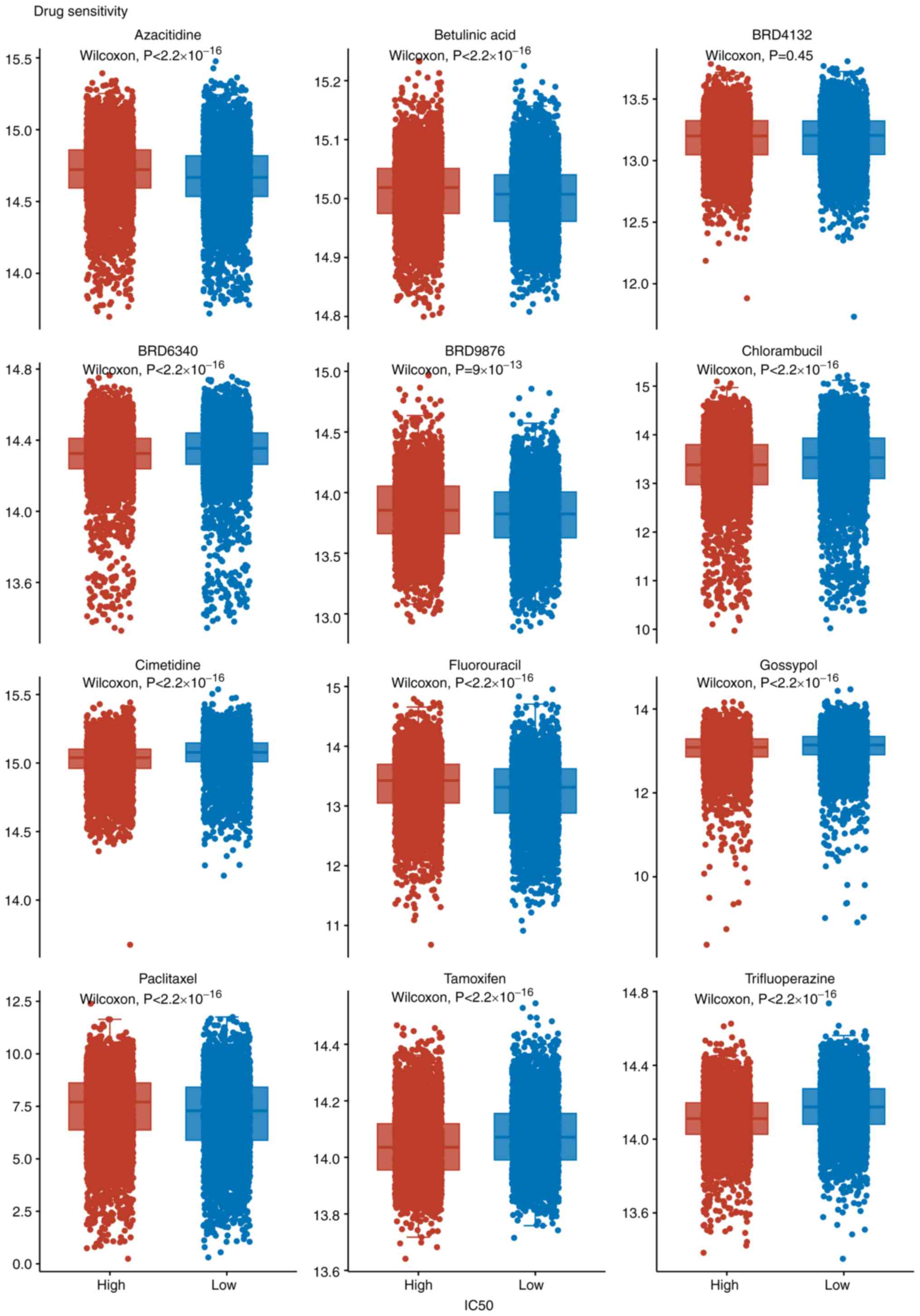
Drug sensitivity analysis
The drug sensitivity screening revealed
statistically significant differences in inhibitory potency between
the high and low groups for the majority of tested compounds. With
the exception of BRD4132, all agents, including azacitidine,
betulinic acid, dorsomorphin, etoposide, GW843682X, mitotane,
PX-478, S-trityl-L-cysteine, sanguinarine, and selenium,
demonstrated markedly enhanced efficacy against the high group (all
P<0.001). By contrast, BRD4132 exhibited no significant
difference in activity between the two groups (P=0.45). These
findings successfully identify a panel of candidate compounds with
high potential for selective efficacy against the defined cellular
model, providing a substantive basis for further investigation into
targeted therapeutic strategies (Fig.
4). Pharmacogenomic profiling of 12 drugs revealed notably
distinct IC50 distributions between ‘SOX5_High’ and
‘SOX5_Low’ cohorts. Wilcoxon rank-sum tests showed significant
differences (P<2.2x10-16) for 11 compounds, including
azacitidine and betulinic acid, with BRD4132 as the only exception
(P=0.45). These results highlight SOX5 expression as a potential
predictor of therapeutic response, underscoring its value as a
precision oncology biomarker.
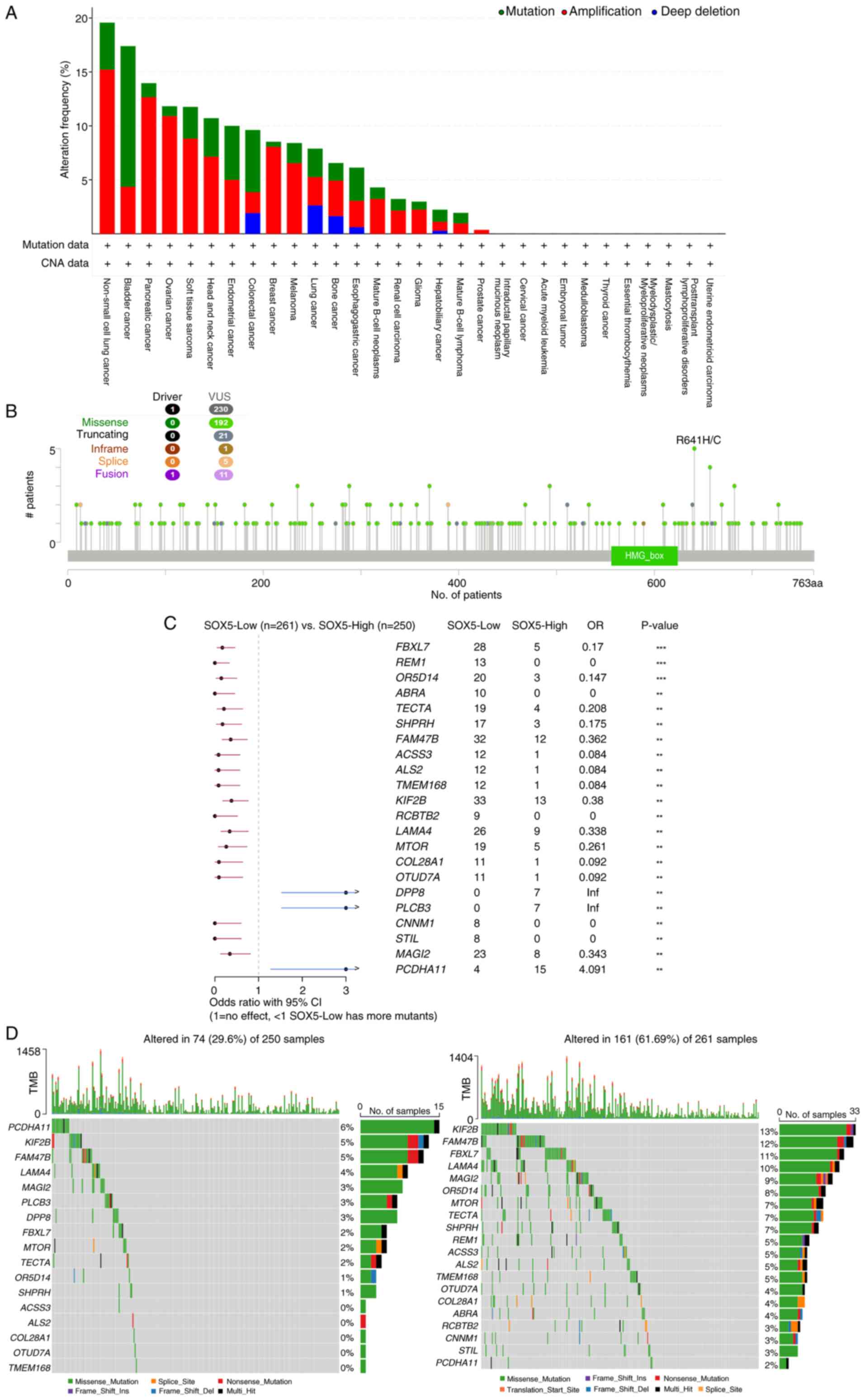
Tumor mutation pattern analysis
Pan-cancer genomic profiling revealed that
SOX5-altered tumors exhibit distinct molecular signatures.
Alteration frequency analysis (Fig.
5A) demonstrated prevalent SOX5 mutations and copy-number
changes with lineage-specific prevalence, notably in gastric and
pancreatic cancers. Detailed characterization (Fig. 5B) showed truncating mutations as
the predominant alteration subtype.
Expression stratification (Fig. 5C) revealed that ‘SOX5_Low’ tumors
were associated with transcriptional suppression of cardiac
morphogenesis pathways [odds ratio (OR)=0.32;
P<1x10-15] and upregulation of cell-cycle regulators
(OR=3.41; P<8x10-10). Waterfall plots (Fig. 5D) demonstrated mutually exclusive
alterations between ‘SOX5_High’ and ‘SOX5_Low’ groups, with
truncating mutations concentrated in ‘SOX5_Low_tumors’ (P=0.0028)
and the analysis demonstrated significant associations between SOX5
expression status and mutations in specific signaling pathways,
primarily the PI3K-AKT-mTOR pathway and PD-L1 associated immune
regulation (P<0.05). (P<0.05). These findings establish SOX5
loss-of-function as a driver of gene expression changes, especially
in LUAD and other cancers with frequent SOX5 variation and
amplification.
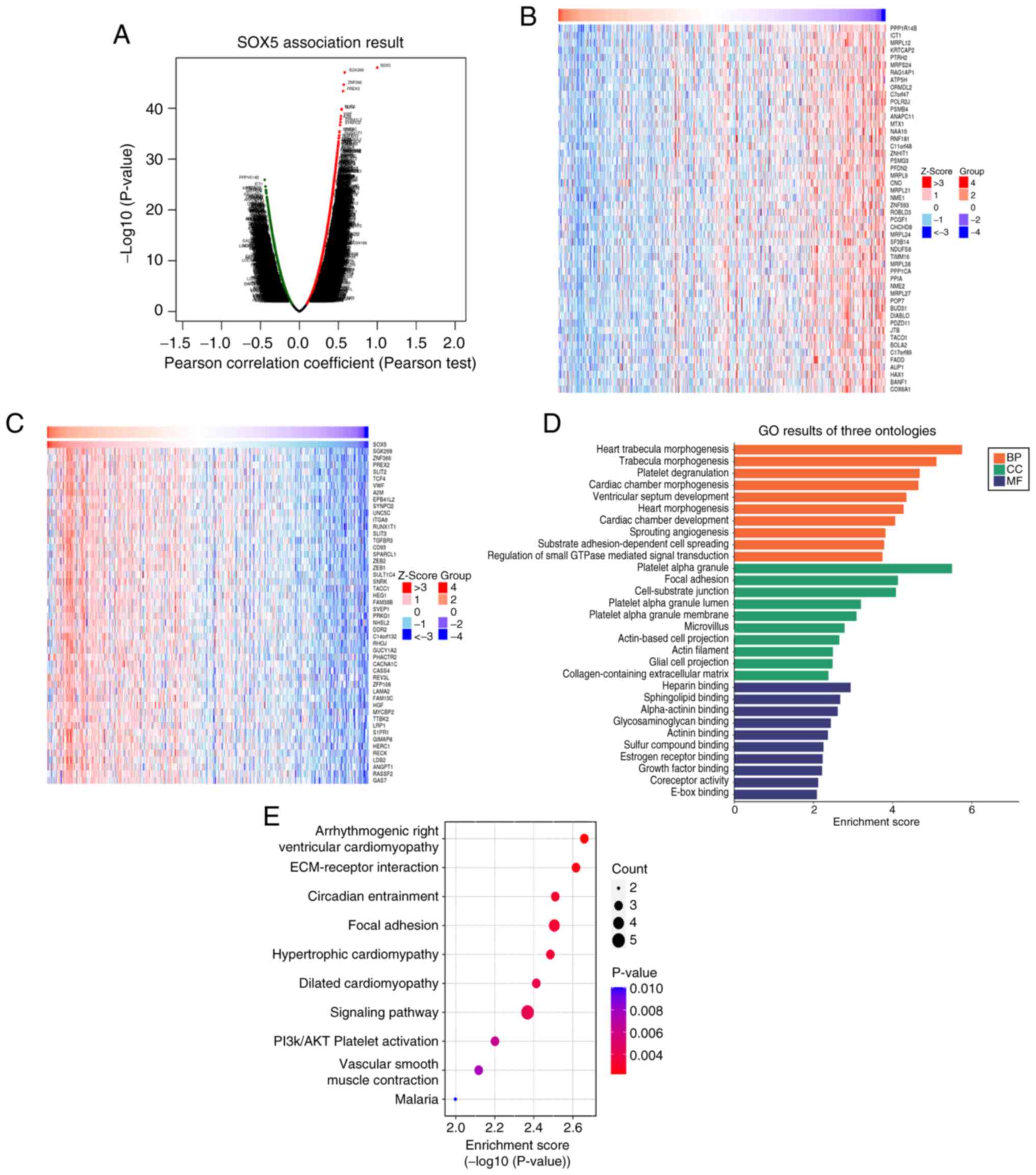
Functional enrichment analysis
Multi-omics integration revealed SOX5 as a central
regulator of cardiac morphogenesis and tumor biology. Pearson
correlation analysis (Fig. 6A)
showed strong associations (r=1; P<1x10-30) with
select targets. Z-score clustering (Fig. 6B and C) demonstrated SOX5 co-activation of
morphogenetic pathways, such as heart trabecula morphogenesis
(Z>3) and cardiac chamber development, alongside suppression of
proliferative pathways (cell cycle, Z<-3; small GTPase
signaling, Z<-2). KEGG and Hallmark enrichment analyses revealed
SOX5 involvement in biological processes including cardiac
morphogenesis and angiogenesis, cellular components such as the
actin cytoskeleton and molecular functions including actin binding.
KEGG analysis (Fig. 6E) implicated
SOX5 dysregulation in ‘Arrhythmogenic right ventricular
cardiomyopathy’ pathogenesis (-log10P>20), mediated
by altered adhesion molecules, extracellular matrix receptors and
metalloproteinases (P<0.05). These findings highlight SOX5 as a
key regulator of structural integrity and tumor biology in
LUAD.
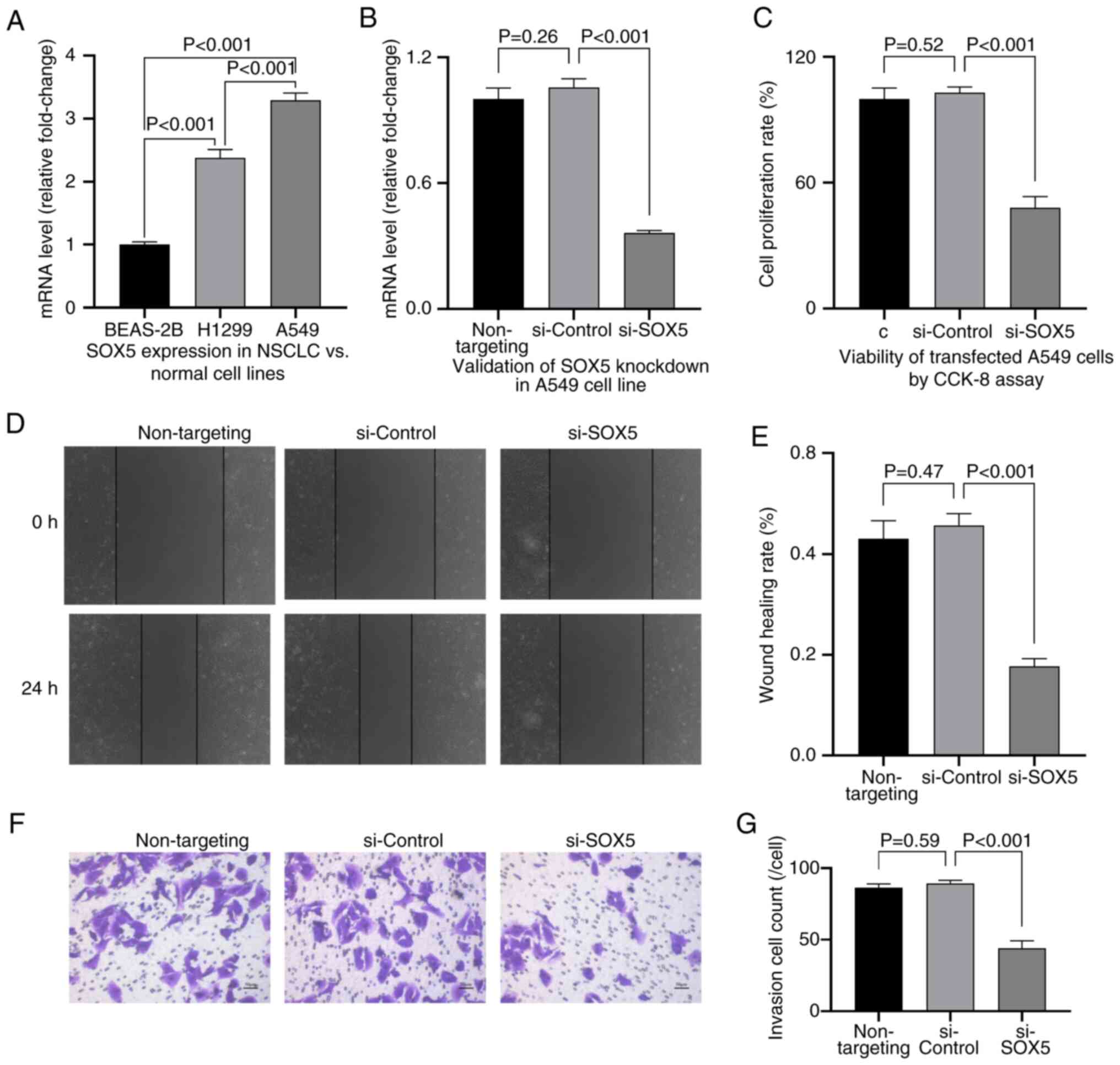
SOX5 silencing inhibits NSCLC
progression in vitro
Functional studies revealed that SOX5 is upregulated
in NSCLC cell lines (H1299 and A549) compared with normal bronchial
epithelial cells (BEAS-2B; P<0.001; Fig. 7A). SOX5 knockdown in A549 cells,
confirmed using qPCR (Fig. 7B),
significantly inhibited proliferation (P<0.01; Fig. 7C), migration (scratch assay;
P<0.001; Fig. 7D and E) and invasion (Transwell assay;
P<0.001; Fig. 7F and G).
The study findings are consistent with the
established oncogenic role of SOX5 in other cancers. In
triple-negative breast cancer, SOX5 drives stemness, EMT and immune
evasion (35). In melanoma, SOX5
promotes migration and invasion via transcriptional regulation by
the oncogenic long non-coding RNA (lncRNA) SLNCR1(36). Mechanistically, SOX5 interacts with
co-activators, such as transcriptional co-activator with
PDZ-binding motif, stabilizing effectors, such as collagen type X
α1 chain, and enhancing extracellular matrix remodeling (37,38).
In gliomas, SOX5:anaplastic lymphoma kinase (ALK) fusion drives
tumor progression, with ALK inhibitors suppressing growth in
vivo (39). Collectively,
these data validate SOX5 as a driver of NSCLC aggressiveness and a
potential therapeutic target.
Discussion
Research into the mechanisms driving malignant tumor
development and progression has advanced beyond initial
understandings. The rapid development of molecular biological
technologies, particularly the application of omics approaches and
bioinformatics, has been key in elucidating the functional roles of
tumor-associated genes and evaluating their potential as diagnostic
biomarkers and therapeutic targets (40-43).
SOX5 displays diverse expression patterns and
biological roles across human cancers. In the present study, the
role of SOX5 in tumor development was investigated, with a focus on
the impact of SOX5 on the TME and its potential mechanisms
underlying tumor progression, diagnosis and prognosis, employing a
range of bioinformatics tools and statistical approaches.
Results indicate that the functional role of SOX5 in
tumorigenesis is highly context-dependent. Dysregulation of SOX5
(upregulation or downregulation) varies notably among cancer types
and is strongly influenced by cell-of-origin, driver mutations and
the surrounding signaling microenvironment (44-46).
Specifically, SOX5 acts predominantly as a tumor suppressor in
epithelial carcinomas driven by somatic mutations, where it is
frequently downregulated or inactivated (47-49),
however exhibits oncogenic properties in specific malignancies such
as germ cell tumors, where it is maintained or upregulated
(45). This dual role underscores
the importance of evaluating SOX5 as a diagnostic or prognostic
biomarker and therapeutic target within the context of specific
cancer types (45,48,50).
The differential expression of SOX5 across cancers
not only reflects its complexity in tumor development but also
highlights distinct underlying biological mechanisms (51-53).
Reduced SOX5 expression in LUAD, LUSC and BRCA is consistent with a
tumor-suppressive role, potentially through regulation of the cell
cycle, induction of senescence, or promotion of apoptosis.
Conversely, elevated SOX5 expression in germ cell tumors suggests
an oncogenic role, possibly by activating pro-proliferative
pathways, enhancing migration and invasion or inhibiting
apoptosis.
Previous studies further illustrate these divergent
roles (3,54,55).
In Kaposi's sarcoma, SOX5 overexpression inhibits
epithelial-to-mesenchymal transition in Kaposi's sarcoma-associated
herpesvirus-infected cells, suggesting a tumor-suppressive function
and therapeutic potential. In germ cell tumors, where germ
cell-specific expression patterns prevail, increased SOX5 levels
may reflect the physiological role it has in reproductive system
development, repurposed within the TME to drive uncontrolled
growth. This variability in expression patterns underscores the
necessity for detailed, cancer type-specific research into SOX5
function. For example, the role of SOX5 in promoting germ cell
tumors may involve interactions with specific transcription factors
or growth factors that are lost in other types of cancer. In TGCTs,
amplification of the 12p11.1-p12.1 region (which contains the SOX5
gene) has been identified as a key event (56), and this 12p gain [including i(12p)
and 12p11.2-12.1 amplification] is associated with aggressive tumor
progression (55). Although SOX5
is located within this critical region, its expression is lost in
some 12p amplification-positive TGCTs, suggesting that its role may
be mediated through complex regulatory networks. This mechanism may
involve interactions with germ cell tumor-specific signaling
pathways such as the KIT/KITLG pathway, or exert effects through
regulating cell proliferation, migration and EMT (50).
Beyond its tissue-specific functions, SOX5 shows
notable potential as a biomarker. In cancer diagnosis and
prognosis, biomarker identification is key to improving patient
management and therapeutic efficacy (48,57).
The present findings position SOX5 as a promising biomarker with
clinical relevance. In LUAD, a high AUC value (0.916) highlights
its diagnostic accuracy, suggesting potential utility for early
detection. This supports the notion that SOX5 serves a role in
early tumor formation and progression, making it a good candidate
for early diagnosis, particularly in LUAD, through its expression
profiling.
Notable associations between SOX5 expression and
survival outcomes, including DSS, DFI and PFI, further reinforce
its prognostic value. Variations in SOX5 expression may reflect
differences in tumor invasiveness and therapeutic response, thereby
impacting patient survival and recurrence risk. In LUAD, low SOX5
expression was associated with worse survival rates, potentially
due to its involvement in regulating cell proliferation and
apoptosis pathways. Thus, SOX5 expression represents a key
prognostic indicator to guide clinical decision-making.
Notably, SOX5 appears to influence the TME,
particularly immune evasion mechanisms. The complexity of the TME
markedly affects therapeutic outcomes. SOX5 may modulate the immune
landscape by regulating immune cell infiltration and related
factors, thereby influencing tumor immune surveillance and
treatment response (58). The
observed positive correlation between elevated SOX5 expression and
immune checkpoint levels (PD-L1 and CTLA-4) suggests that SOX5
serves a key immunomodulatory role, potentially facilitating immune
evasion within the TME. ‘SOX5_High’ tumors exhibited activated yet
exhausted immune features (including increased immune infiltration,
immune scores and TMB, especially in LUAD and UCEC), suggesting
SOX5 coordinates both immune recognition and immunosuppressive
checkpoint expression.
These findings position SOX5 as a promising
predictive biomarker for immune checkpoint blockade (ICB)
responsiveness. Patients with high SOX5 expression may benefit more
from ICB due to pre-existing checkpoint enrichment. Moreover,
targeting SOX5 could enhance ICB efficacy by reducing intrinsic
immunosuppression. Future work should aim to validate the
mechanistic regulation of immune checkpoints SOX5 may exhibit and
associate its expression with clinical ICB outcomes.
Analysis using ESTIMATE and CIBERSORT demonstrated
that elevated SOX5 expression is associated with an activated TME
and high immune scores, particularly in LUAD and UCEC and is
associated with high TMB. SOX5 may regulate the TME through
multiple mechanisms, influencing immune cell recruitment,
activation and immune escape. Elevated SOX5 could enhance secretion
of cytokines and chemokines, recruiting immune cells such as T
cells and macrophages. While such infiltration could support
antitumor immunity, it may also promote immune escape by recruiting
immunosuppressive cells. SOX5 may also regulate immune cell
activity by modulating the expression of cell surface molecules
such as MHC proteins or immune checkpoint ligands (such as PD-L1),
directly affecting T cell activation and inhibition.
The correlation between SOX5 expression and TMB
suggests an association with tumor genetic complexity and
neoantigen production. As high TMB often predicts improved
immunotherapy responses, SOX5 may influence sensitivity to
immunotherapy through TMB modulation.
The present study also revealed therapeutic
implications of SOX5 expression. Pharmacogenomic analyses showed
high SOX5 expression correlates with increased sensitivity to
various chemotherapeutics, including azacitidine and betulinic
acid. This aligns with the regulation of DNA methyltransferase 1
(DNMT1)/p21 signaling in bladder cancer progression exhibited by
SOX5(48) and suggests that
SOX5-overexpressing tumors may be particularly susceptible to DNMT1
inhibitor therapies. Furthermore, targeting the HuR-lncRNA-SOX5
axis, which stabilizes SOX5 transcripts in carcinomas, could
exploit this therapeutic vulnerability (59). In addition, SOX5 may sensitize
tumor cells to drug-induced apoptosis by regulating cell death
pathways, enhancing cell cycle arrest through p53 activation,
altering cancer cell metabolism and influencing drug transport and
metabolism gene expression (60).
Pan-cancer analysis of SOX5 mutation patterns
indicates that alterations in SOX5, particularly in gastric and
pancreatic cancers, may drive invasion and therapy resistance. Such
mutations could disrupt normal regulatory networks and activate
oncogenic pathways (such as Wnt and Notch), contributing to tumor
progression. While this may confer resistance to conventional
therapies, it offers opportunities for targeted interventions
against specific SOX5 alterations.
Although this study provides a comprehensive
pan-cancer analysis of SOX5, several important limitations should
be acknowledged. First, the research primarily relies on
bioinformatic analyses of public databases such as TCGA, and lacks
validation in large-scale independent cohorts, which may introduce
potential biases or overfitting. For instance, while citation3
developed a prognostic model, it was based solely on TCGA data
without involving multi-center samples, thereby limiting its
clinical applicability. Second, functional experiments were
confined to in vitro cell lines (e.g., A549 and H1299), and
lacked in vivo animal model validation, which is essential
for fully recapitulating the complexity of the tumor
microenvironment. Third, the mechanistic insights remain
insufficient; for example, the specific pathways through which SOX5
regulates immune infiltration or drug sensitivity were not fully
elucidated. Fourth, while the mutation pattern analysis revealed
associations between SOX5 alterations and specific signaling
pathways, it did not account for tumor heterogeneity and subtype
variations, which may affect the generalizability of the findings.
Finally, the drug sensitivity data were based on computational
predictions and lacked pre-clinical experimental support,
necessitating further validation of actual therapeutic
efficacy.
In conclusion, the present pan-cancer analysis
established SOX5 as a context-dependent regulator with dual roles
in tumorigenesis, acting as a tumor suppressor in numerous
epithelial cancers while driving oncogenesis in select
malignancies. The dysregulation of SOX5 is associated with immune
evasion, TME remodeling, genomic instability and patient prognosis.
Functional validation demonstrates the role of SOX5 in enhancing
proliferation, migration and invasion in NSCLC. These findings
position SOX5 as a robust biomarker for cancer diagnosis and
prognosis and a promising therapeutic target, offering a strategic
avenue for precision oncology across diverse tumor types.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the Hunan Natural
Science Foundation-Regional Joint Fund (grant no. 2024JJ7601) and
Basic Research Guiding Program of Yueyang City Science and
Technology Bureau 2024(20).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
QY, LO and YT contributed to the conception and
design of the study. Data collection was performed by YT, HD, KT,
SL, QZ, LY, SB and LL. Formal analysis was conducted by YT, HD, KT,
SL, QZ, LY, SB, LL and QY. YT, HD and QY wrote the original draft
of the manuscript. The manuscript was reviewed and edited by YT,
HD, KT, QZ, LO, QY and SB. LL, SB, LO and QY supervised the study.
YT, HD, KT, QZ, SL, LL and QY confirm the authenticity of all the
raw data. All authors have read and approved the final version of
the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Li Q, Wang W, Yang T, Li D, Huang Y, Bai G
and Li Q: LINC00520 up-regulates SOX5 to promote cell proliferation
and invasion by miR-4516 in human hepatocellular carcinoma. Biol
Chem. 403:665–678. 2022.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Wu B, Wang X, Yu R and Xue X: CircWHSC1
serves as a prognostic biomarker and promotes malignant progression
of non-small-cell lung cancer via miR-590-5p/SOX5 axis. Environ
Toxicol. 38:2440–2449. 2023.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Yuan WM, Fan YG, Cui M, Luo T, Wang YE,
Shu ZJ, Zhao J, Zheng J and Zeng Y: SOX5 regulates cell
proliferation, apoptosis, migration and invasion in KSHV-infected
cells. Virol Sin. 36:449–457. 2021.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Hu C, Li Q, Xiang L, Luo Y, Li S, An J, Yu
X, Zhang G, Chen Y, Wang Y and Wang D: Comprehensive pan-cancer
analysis unveils the significant prognostic value and potential
role in immune microenvironment modulation of TRIB3. Comput Struct
Biotechnol J. 23:234–250. 2023.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Yu D, Sun R, Shen D, Ge L, Xue T and Cao
Y: Nuclear heme oxygenase-1 improved the hypoxia-mediated
dysfunction of blood-spinal cord barrier via the miR-181c-5p/SOX5
signaling pathway. Neuroreport. 32:112–120. 2021.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Liang Q, Chu F, Zhang L, Jiang Y, Li L and
Wu H: circ-LDLRAD3 knockdown reduces cisplatin chemoresistance and
inhibits the development of gastric cancer with cisplatin
resistance through miR-588 enrichment-mediated SOX5 inhibition. Gut
Liver. 17:389–403. 2023.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Sadeghi Z, Dodangeh F and Raheb J: SOX2
overlapping transcript (SOX2-OT) enhances the lung cancer
malignancy through interaction with miR-194-5p/SOX5 axis. Iran J
Biotechnol. 21(e3530)2023.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Tenorio-Castano J, Gómez ÁS, Coronado M,
Rodríguez-Martín P, Parra A, Pascual P, Cazalla M, Gallego N, Arias
P, Morales AV, et al: Lamb-Shaffer syndrome: 20 Spanish patients
and literature review expands the view of neurodevelopmental
disorders caused by SOX5 haploinsufficiency. Clin Genet.
104:637–647. 2023.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Jing Y, Jiang X, Ji Q, Wu Z, Wang W, Liu
Z, Guillen-Garcia P, Esteban CR, Reddy P, Horvath S, et al:
Genome-wide CRISPR activation screening in senescent cells reveals
SOX5 as a driver and therapeutic target of rejuvenation. Cell Stem
Cell. 30:1452–1471.e10. 2023.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Shi L, Zhang H, Sun J, Gao X and Liu C:
CircSEC24A promotes IL-1β-induced apoptosis and inflammation in
chondrocytes by regulating miR-142-5p/SOX5 axis. Biotechnol Appl
Biochem. 69:701–713. 2022.PubMed/NCBI View
Article : Google Scholar
|
|
11
|
Qiu M, Lu Y, Li J, Gu J, Ji Y, Shao Y,
Kong X and Sun W: Interaction of SOX5 with SOX9 promotes
warfarin-induced aortic valve interstitial cell calcification by
repressing transcriptional activation of LRP6. J Mol Cell Cardiol.
162:81–96. 2022.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Chen Z, Shang Y, Zhang X, Duan W, Li J,
Zhu L, Ma L, Xiang X, Jia J, Ji X and Gong S: METTL3 mediates SOX5
m6A methylation in bronchial epithelial cells to attenuate Th2 cell
differentiation in T2 asthma. Heliyon. 10(e28884)2024.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Gu Y, Tang S, Wang Z, Cai L, Lian H, Shen
Y and Zhou Y: A pan-cancer analysis of the prognostic and
immunological role of β-actin (ACTB) in human cancers.
Bioengineered. 12:6166–6185. 2021.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Chen R, Zhang C, Cheng Y, Wang S, Lin H
and Zhang H: LncRNA UCC promotes epithelial-mesenchymal transition
via the miR-143-3p/SOX5 axis in non-small-cell lung cancer. Lab
Invest. 101:1153–1165. 2021.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Li C, Zhang J and Bi Y: Unveiling the
prognostic significance of SOX5 in esophageal squamous cell
carcinoma: A comprehensive bioinformatic and experimental analysis.
Aging (Albany NY). 15:7565–7582. 2023.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Innella G, Greco D, Carli D, Magini P,
Giorgio E, Galesi O, Ferrero GB, Romano C, Brusco A and Graziano C:
Clinical spectrum and follow-up in six individuals with
Lamb-Shaffer syndrome (SOX5). Am J Med Genet A. 185:608–613.
2021.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Edgerley K, Bryson L, Hanington L, Irving
R, Joss S, Lampe A, Maystadt I, Osio D, Richardson R, Split M, et
al: SOX5: Lamb-Shaffer syndrome-A case series further expanding the
phenotypic spectrum. Am J Med Genet A. 191:1447–1458.
2023.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Leek JT, Johnson WE, Parker HS, Jaffe AE
and Storey JD: The sva package for removing batch effects and other
unwanted variation in high-throughput experiments. Bioinformatics.
28:882–883. 2012.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Rousseaux S, Debernardi A, Jacquiau B,
Vitte AL, Vesin A, Nagy-Mignotte H, Moro-Sibilot D, Brichon PY,
Lantuejoul S, Hainaut P, et al: Ectopic activation of germline and
placental genes identifies aggressive metastasis-prone lung
cancers. Sci Transl Med. 5(186ra66)2013.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Okayama H, Kohno T, Ishii Y, Shimada Y,
Shiraishi K, Iwakawa R, Furuta K, Tsuta K, Shibata T, Yamamoto S,
et al: Identification of genes upregulated in ALK-positive and
EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res.
72:100–111. 2012.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Der SD, Sykes J, Pintilie M, Zhu CQ,
Strumpf D, Liu N, Jurisica I, Shepherd FA and Tsao MS: Validation
of a histology-independent prognostic gene signature for
early-stage, non-small-cell lung cancer including stage IA
patients. J Thorac Oncol. 9:59–64. 2014.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Davis S and Meltzer PS: GEOquery: A bridge
between the gene expression omnibus (GEO) and BioConductor.
Bioinformatics. 23:1846–1847. 2007.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Colaprico A, Silva TC, Olsen C, Garofano
L, Cava C, Garolini D, Sabedot TS, Malta TM, Pagnotta SM,
Castiglioni I, et al: TCGAbiolinks: An R/Bioconductor package for
integrative analysis of TCGA data. Nucleic Acids Res.
44(e71)2016.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Liao C and Wang X: TCGAplot: An R package
for integrative pan-cancer analysis and visualization of TCGA
multi-omics data. BMC Bioinformatics. 24(483)2023.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Brune V, Tiacci E, Pfeil I, Döring C,
Eckerle S, van Noesel CJ, Klapper W, Falini B, von Heydebreck A,
Metzler D, et al: Origin and pathogenesis of nodular
lymphocyte-predominant Hodgkin lymphoma as revealed by global gene
expression analysis. J Exp Med. 205:2251–2268. 2008.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Suraokar MB, Nunez MI, Diao L, Chow CW,
Kim D, Behrens C, Lin H, Lee S, Raso G, Moran C, et al: Expression
profiling stratifies mesothelioma tumors and signifies deregulation
of spindle checkpoint pathway and microtubule network with
therapeutic implications. Ann Oncol. 25:1184–1192. 2014.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Robin X, Turck N, Hainard A, Tiberti N,
Lisacek F, Sanchez JC and Müller M: pROC: An open-source package
for R and S+ to analyze and compare ROC curves. BMC Bioinformatics.
12(77)2011.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Benjamini Y and Hochberg Y: Controlling
the False Discovery Rate: A Practical and Powerful Approach to
Multiple Testing. J R Statist Soc В. 57:289–300. 1995.
|
|
29
|
Yoshihara K, Shahmoradgoli M, Martínez E,
Vegesna R, Kim H, Torres-Garcia W, Treviño V, Shen H, Laird PW,
Levine DA, et al: Inferring tumour purity and stromal and immune
cell admixture from expression data. Nat Commun.
4(2612)2013.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Chen B, Khodadoust MS, Liu CL, Newman AM
and Alizadeh AA: Profiling tumor infiltrating immune cells with
CIBERSORT. Methods Mol Biol. 1711:243–259. 2018.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Hänzelmann S, Castelo R and Guinney J:
GSVA: Gene set variation analysis for microarray and RNA-seq data.
BMC Bioinformatics. 14(7)2013.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Maeser D, Gruener RF and Huang RS:
oncoPredict: An R package for predicting in vivo or cancer patient
drug response and biomarkers from cell line screening data. Brief
Bioinform. 22(bbab260)2021.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Mayakonda A, Lin DC, Assenov Y, Plass C
and Koeffler HP: Maftools: Efficient and comprehensive analysis of
somatic variants in cancer. Genome Res. 28:1747–1756.
2018.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Dong G, Wang X, Wang X, Jia Y, Jia Y, Zhao
W and Tong Z: Circ_0084653 promotes the tumor progression and
immune escape in triple-negative breast cancer via the
deubiquitination of MYC and upregulation of SOX5. Int J Biol
Macromol. 280 (Pt 1)(135655)2024.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Cong L, Zhao Q, Sun H, Zhou Z, Hu Y, Li C,
Hao M and Cong X: A novel long non-coding RNA SLNCR1 promotes
proliferation, migration, and invasion of melanoma via
transcriptionally regulating SOX5. Cell Death Discov.
10(160)2024.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Li Y and Yang S, Qin L and Yang S: TAZ is
required for chondrogenesis and skeletal development. Cell Discov.
7(26)2021.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Medina-Menéndez C, Tirado-Melendro P, Li
L, Rodríguez-Martín P, Melgarejo-de la Peña E, Díaz-García M,
Valdés-Bescós M, López-Sansegundo R and Morales AV: Sox5 controls
the establishment of quiescence in neural stem cells during
postnatal development. PLoS Biol. 23(e3002654)2025.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Tsai CC, Huang MH, Fang CL, Hsieh KL,
Hsieh TH, Ho WL, Chang H, Tsai ML, Kao YC, Miser JS, et al: An
infant-type hemispheric glioma with SOX5::ALK: A novel fusion. J
Natl Compr Canc Netw. 22(e237102)2024.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Sara H, Kallioniemi O and Nees M: A decade
of cancer gene profiling: From molecular portraits to molecular
function. Methods Mol Biol. 576:61–87. 2010.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Legge F, Ferrandina G and Scambia G: From
bio-molecular and technology innovations to clinical practice:
Focus on ovarian cancer. Ann Oncol. 17 (Suppl 7):vii46–vii48.
2006.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Lu DY, Qu RX, Lu TR and Wu HY: Cancer
bioinformatics for updating anticancer drug developments and
personalized therapeutics. Rev Recent Clin Trials. 12:101–110.
2017.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Shi Y, Wang Y, Zhang W, Niu K, Mao X, Feng
K and Zhang Y: N6-methyladenosine with immune infiltration and
PD-L1 in hepatocellular carcinoma: Novel perspective to
personalized diagnosis and treatment. Front Endocrinol (Lausanne).
14(1153802)2023.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Chen X, Fu Y, Xu H, Teng P, Xie Q, Zhang
Y, Yan C, Xu Y, Li C, Zhou J, et al: SOX5 predicts poor prognosis
in lung adenocarcinoma and promotes tumor metastasis through
epithelial-mesenchymal transition. Oncotarget. 9:10891–10904.
2017.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Ueda R, Yoshida K, Kawase T, Kawakami Y
and Toda M: Preferential expression and frequent IgG responses of a
tumor antigen, SOX5, in glioma patients. Int J Cancer.
120:1704–1711. 2007.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Chen M, Zou S, He C, Zhou J, Li S, Shen M,
Cheng R, Wang D, Zou T, Yan X, et al: Transactivation of SOX5 by
Brachyury promotes breast cancer bone metastasis. Carcinogenesis.
41:551–560. 2020.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Yang B, Zhang W, Sun D, Wei X, Ding Y, Ma
Y and Wang Z: Downregulation of miR-139-5p promotes prostate cancer
progression through regulation of SOX5. Biomed Pharmacother.
109:2128–2135. 2019.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Wu L, Yang Z, Dai G, Fan B, Yuan J, Liu Y,
Liu P and Ou Z: SOX5 promotes cell growth and migration through
modulating the DNMT1/p21 pathway in bladder cancer. Acta Biochim
Biophys Sin (Shanghai). 54:987–998. 2022.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Pei XH, Lv XQ and Li HX: SOX5 induces
epithelial to mesenchymal transition by transactivation of Twist1.
Biochem Biophys Res Commun. 46:322–327. 2014.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Xue JD, Xiang WF, Cai MQ and Lv XY:
Biological functions and therapeutic potential of SRY related high
mobility group box 5 in human cancer. Front Oncol.
14(1332148)2024.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Docherty AR, Mullins N, Ashley-Koch AE,
Qin X, Coleman JRI, Shabalin A, Kang J, Murnyak B, Wendt F, Adams
M, et al: GWAS meta-analysis of suicide attempt: identification of
12 genome-wide significant loci and implication of genetic risks
for specific health factors. Am J Psychiatry. 180:723–738.
2023.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Vinyals A, Ferreres JR, Calbet-Llopart N,
Ramos R, Tell-Martí G, Carrera C, Marcoval J, Puig S, Malvehy J,
Puig-Butillé JA and Fabra À: Oncogenic properties via MAPK
signaling of the SOX5-RAF1 fusion gene identified in a wild-type
NRAS/BRAF giant congenital nevus. Pigment Cell Melanoma Res.
35:450–460. 2022.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Collins FL, Roelofs AJ, Symons RA, Kania
K, Campbell E, Collie-Duguid ESR, Riemen AHK, Clark SM and De Bari
C: Taxonomy of fibroblasts and progenitors in the synovial joint at
single-cell resolution. Ann Rheum Dis. 82:428–437. 2023.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Sridharan S and Basu A: Distinct roles of
mTOR targets S6K1 and S6K2 in breast cancer. Int J Mol Sc.
21(1199)2020.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Blanco L and Tirado CA: Testicular germ
cell tumors: A cytogenomic update. J Assoc Genet Technol.
44:128–133. 2018.PubMed/NCBI
|
|
56
|
Mostert MC, Verkerk AJ, van de Pol M,
Heighway J, Marynen P, Rosenberg C, van Kessel AG, van Echten J, de
Jong B, Oosterhuis JW and Looijenga LH: Identification of the
critical region of 12p over-representation in testicular germ cell
tumors of adolescents and adults. Oncogene. 16:2617–2627.
1998.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Cai Y and Jia Y: Circular RNA SOX5
promotes the proliferation and inhibits the apoptosis of the
hepatocellular carcinoma cells by targeting miR-502-5p/synoviolin 1
axis. Bioengineered. 13:3362–3370. 2022.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Knights AJ, Farrell EC, Ellis OM, Lammlin
L, Junginger LM, Rzeczycki PM, Bergman RF, Pervez R, Cruz M, Knight
E, et al: Synovial fibroblasts assume distinct functional
identities and secrete R-spondin 2 in osteoarthritis. Ann Rheum
Dis. 82:272–282. 2023.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Wang L, Ye S, Wang J, Gu Z, Zhang Y, Zhang
C and Ma X: HuR stabilizes lnc-SOX5 mRNA to promote tongue
carcinogenesis. Biochemistry (Mosc). 82:438–445. 2017.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Li QS, Shabalin AA, DiBlasi E, Gopal S and
Canuso CM: FinnGen, International Suicide Genetics Consortium.
Palotie A, Drevets WC, Docherty AR and Coon H: Genome-wide
association study meta-analysis of suicide death and suicidal
behavior. Mol Psychiatry. 28:891–900. 2023.PubMed/NCBI View Article : Google Scholar
|