Introduction
O6-methylguanine-DNA methyltransferase (MGMT) is a
DNA repair protein responsible for combating adducts from
O6-guanine (1,2). Alkylation at the O6 position of
guanine renders the DNA vulnerable to point mutation and the
formation of DNA crosslinks; all of which can predispose an
individual to cancer (3). MGMT
protein is expressed in lower amounts in some cancers,
predominantly due to the hypermethylation of the MGMT promoter CpG
(4,5). Abnormal methylation of gene promoters
leads to a loss of gene function, which can confer a selective
advantage to neoplastic cells, similar to the effects seen with
genetic mutations (6). MGMT serves
a pivotal role in repairing DNA damage caused by environmental
carcinogens and alkylating chemotherapy drugs such as temozolomide
(3); therefore, loss of MGMT
function due to promoter methylation compromises DNA repair
mechanisms, rendering tumor cells more vulnerable to alkylating
agents and increasing DNA damage and apoptosis (7-9).
MGMT methylation has been extensively studied in
glioblastoma, where numerous clinical studies have demonstrated
that methylation of the MGMT promoter is an independent predictor
of improved survival. For instance, a meta-analysis of 11 studies
exclusively involving patients with glioblastoma revealed that
patients with a methylated MGMT promoter experienced improved
outcomes, with improvements in overall survival (OS) (10). MGMT promoter methylation also
serves as a predictive marker for chemotherapy response in patients
with glioblastoma (11,12).
Similar to high-grade gliomas (13), emerging data suggests MGMT
methylation is also a prognostic factor for response to treatment
in advanced neuroendocrine tumors (NETs). Studies have documented
an association between MGMT status and clinical outcomes with
alkylating agents (temozolomide, dacarbazine and
streptozotocin-based therapies) in NET (14-16).
A hypermethylated MGMT phenotype has been identified in multiple
premalignant and malignant solid tumors, including colorectal
(17), urothelial (18), pheochromocytomas and paragangliomas
(19) and oral (20) tumors. However, the prognostic role
of MGMT methylation on OS in solid tumors remains to be
determined.
In the present systematic review and meta-analysis,
the prognostic importance of MGMT methylation on OS in solid tumors
was investigated. By pooling data from diverse studies, the
analysis sought to evaluate the prognostic significance of MGMT
methylation across different tumor types and identify cancers where
MGMT methylation may serve as a predictive biomarker.
Materials and methods
The present meta-analysis and systematic review were
performed in accordance with the Preferred Reporting Items for
Systematic Reviews and Meta-Analyses (PRISMA) guidelines (21).
Eligibility criteria
Studies were included in the meta-analysis if they
met the following inclusion criteria: i) Population: Only studies
involving human subjects were included, with research based solely
on cell lines without accompanying clinical data, as well as
studies focused on pediatric populations, being excluded.
Additionally, studies concerning gliomas, lymphomas, leukemias or
benign/premalignant conditions were not considered. However,
investigations of brain metastases originating from solid tumors
were included; ii) Methodology: MGMT methylation status had to be
assessed using a method that was stated in detail, and the research
needed to focus on malignant solid tumors; iii) Outcome: OS had to
be reported as a study outcome and presented numerically, through
hazard ratios (HRs) and 95% confidence intervals (95% CIs). HR
estimates derived from either univariate or multivariate analyses
comparing MGMT-methylated vs. non-methylated patients were
accepted. When HR or 95% CI values were not provided in a paper,
the paper was excluded; and iv) Language: Finally, only
English-language articles published in peer-reviewed international
journals were included, whereas review papers, meta-analyses and
conference abstracts were excluded.
Search strategy
A systematic search of the literature was conducted
in the PubMed database (https://pubmed.ncbi.nlm.nih.gov/) up to October 26,
2024. The search strategy in PubMed was structured as follows:
[‘MGMT’ (title/abstract) OR ‘O6-methylguanine-DNA
methyltransferase’ (title/abstract)] AND [‘methylation’
(title/abstract) OR ‘promoter methylation’ (title/abstract) OR
‘epigenetic’ (title/abstract)] AND [‘prognosis’ (title/abstract) OR
‘survival’ (title/abstract) OR ‘outcome’ (title/abstract)] OR
‘mortality’ (title/abstract) OR ‘prognostic’ (title/abstract) OR
‘predictive’ (title/abstract)] AND [‘solid tumor’ (title/abstract)
OR ‘solid cancer’ (title/abstract) OR ‘carcinoma’ (title/abstract)
OR ‘sarcoma’ (title/abstract) OR ‘neoplasms’ (Medical Subject
Headings terms) NOT [‘review’ (publication type) OR ‘meta-analysis’
(publication type) OR ‘systematic review’ (publication type)] NOT
[‘glioma’ (title/abstract) OR ‘glioblastoma’ (title/abstract) OR
‘astrocytoma’ (title/abstract) OR ‘brain tumor’ (title/abstract) OR
‘brain cancer’ (title/abstract)].
Study selection and data
extraction
The titles and abstracts were independently screened
two by reviewers, followed by a full-text assessment of studies
that potentially satisfied the eligibility criteria. Any
disagreements were resolved by consulting a third reviewer. Data
extraction was performed using a standard form, capturing the
title, first author, publication year, country, study design,
cancer type, total sample size, number of cases with MGMT
methylation, age, metastatic status, follow-up period, sample type,
detection method, analysis type [multivariate (MV); yes or no], OS
HR and upper and lower CIs.
Meta-analysis
Statistical analyses were performed in R (version
4.4.2; RStudio, Inc.), using the ‘meta’ (22) and ‘metafor’ packages (23). HR and corresponding 95% CI were
utilized to assess the association between MGMT methylation status
and OS. The overall effect estimate was derived using a
fixed-effects model (Mantel-Haenszel method) and a random-effects
model (DerSimonian-Laird method). Heterogeneity was assessed via
Cochran's Q test and the I2 statistic. Heterogeneity was
considered significant when the χ2 test P<0.05 and
the I2 statistic >50% (24); in the presence of significant
heterogeneity, a random-effects model was adopted. Subgroup
analyses stratified by cancer type were performed to explore
heterogeneity sources. Publication bias was evaluated through
funnel plot inspection and Egger's linear regression test. A
sensitivity analysis, excluding individual studies iteratively, was
conducted to assess the robustness of the results. P<0.05 was
considered to indicate a statistically significant difference.
Results
Included studies
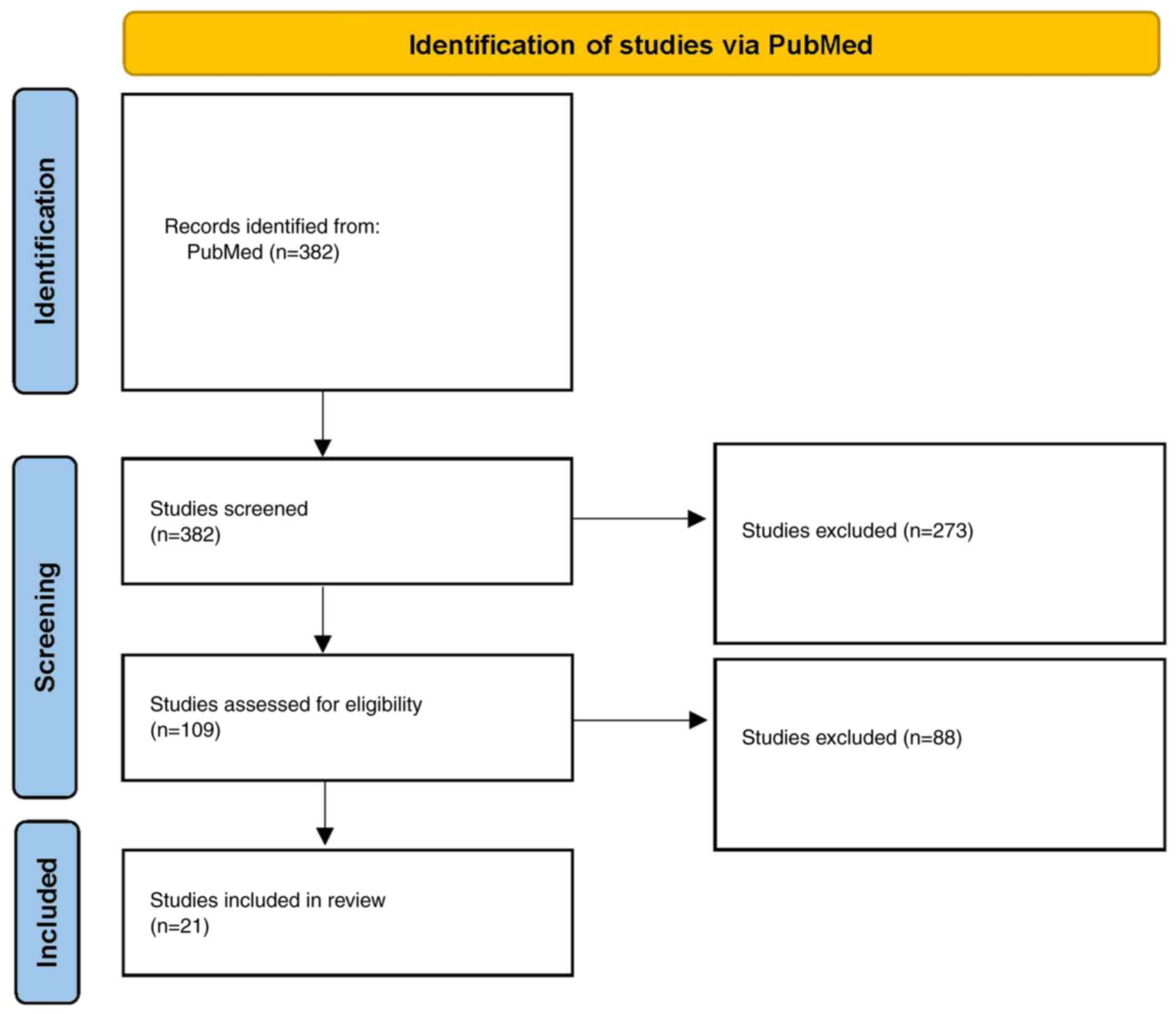
A total of 21 studies met the inclusion criteria,
comprising 2,946 patients (Fig. 1;
Table I). The most frequently
represented cancer types were colorectal cancer (n=6) and head and
neck cancer (n=6), followed by non-small cell lung cancer (NSCLC)
(n=2). The remaining cancer types were each represented by a single
study: Pancreatic NET, biliary tract cancer, cervical cancer,
duodenal cancer, esophageal cancer, leiomyosarcoma and
melanoma.
 | Table ICharacteristics of included
studies. |
Table I
Characteristics of included
studies.
| Author, year | Country | Study design | Cancer type | Age (median or
mean) | Patients, n | Patients with MGMT
methylation status | Metastatic patients
included | Metastatic
patients, % | Sampling
method | Methylation
detection method | Multi-variable
analysis | OS, HR (95%
CI) | PFS, HR (95%
CI) | (Refs.) |
|---|
| Gupta et al,
2023 | India | Retrospective | Cervical | 48.44 | 220 | 48 | Yes | 7.30 | Serum | Nested MSP | Yes | 2.50
(1.20-5.19) | Not reported | (25) |
| Cannella et
al, 2023 | Italy | Retrospective |
Leiomyo-sarcoma | 58 | 32 | 32 | Yes | 100 | Tissue | MSP | No | 1.70
(0.66-4.20) | 2.2 (1-4.8) | (26) |
| Jamai et al,
2023 | Tunisia | Retrospective | Colorectal | 63 | 111 | 111 | Yes | 50 | Tissue | MSP | Yes | 1.75
(1.30-2.35) | Not reported | (37) |
| Shima et al,
2011 | USA | Retrospective | Colorectal | 66.2 | 855 | 855 | Yes | NA | Tissue | MethyLight
(Q-MSP) | Yes | 1.08
(0.88-1.32) | Not reported | (39) |
| Fu et al,
2016 | China | Retrospective | Duodenal | 64.2 | 64 | 64 | No | 0 | Tissue | MethyLight
(MSP) | Yes | 4.25
(2.00-9.05) | 2.80
(1.43-5.48) | (33) |
| Niger et al,
2022(29) | Italy | Retrospective | Biliary tract
cancer | 66 | 164 | 164 | Yes | NA | Tissue | Pyrosequencing for
methylation | Yes | 2.31
(1.44-3.71) | 1.24
(0.78-1.98) | (40) |
| Guadagni et
al, 2017 | Italy | Retrospective | Melanoma | 64.1 | 27 | 27 | No | 0 | Tissue | MS-MLPA | Yes | 0.32
(0.12-0.89) | Not reported | (41) |
| Viúdez et
al, 2016(31) | USA | Retrospective | Pancreatic
neuroendocrine cancer | 57 | 92 | 92 | No | 0 | Tissue | MSP | Yes | 2.06
(0.44-9.57) | 1.09
(0.52-2.29) | (42) |
| Supic et al,
2011 | Serbia | Retrospective | Head and neck | 58 | 47 | 47 | No | 0 | Tissue | MSP | No | 1.23
(0.41-3.87) | Not reported | (43) |
| Taioli et
al, 2009 | USA | Retrospective | Head and neck | 62.2 | 88 | 88 | Yes | 39.80 | Tissue | MSP; Q-MS using
pyrosequencing | Yes | 2.17
(1.11-4.23) | 3.49
(1.62-7.52) | (44) |
| Chen et al,
2009(42) | Taiwan | Retrospective | Colorectal | 65.5 | 117 | 117 | Yes | 21.40 | Tissue | MSP | Yes | 1.33
(0.61-2.92) | Not reported | (34) |
| Morano et
al, 2018 | Italy | Prospective | Colorectal | 62 | 25 | 25 | Yes | 100 | Tissue | MSP | No | 0.75
(0.24-2.11) | 0.46
(0.13-0.95) | (45) |
| Safar et al,
2005 | USA | Retrospective | NSCLC | 67 | 105 | 105 | Yes | 22 | Tissue | MSP | Yes | 0.59
(0.21-1.63) | Not reported | (27) |
| Nilsson et
al, 2013(36) | Sweden | Retrospective | Colorectal | NA | 111 | 111 | No | 0 | Tissue | Pyrosequencing for
methylation | Yes | 0.36
(0.15-0.87) | Not reported | (28) |
| Lu et al,
2011 | China | Retrospective | Esophageal | 61.8 | 125 | 120 | Yes | 1 | Tissue | MSP | No | 1.00
(0.55-1.79) | Not reported | (35) |
| Brabender et
al, 2003(37) | Multinational | Retrospective | NSCLC | 63.3 | 90 | 90 | No | 0 | Tissue | Q-MSP (TaqMan) | Yes | 1.89
(1.06-3.37) | 2.60
(1.60-3.60) | (29) |
| Dikshit et
al, 2007 | Multinational | Retrospective | Head and neck | 60.4 | 235 | 212 | No | 0 | Tissue | MSP | Yes | 1.02
(0.66-1.58) | Not reported | (30) |
| Šupić et al,
2009 | Serbia | Retrospective | Head and neck | 58 | 77 | 77 | No | 0 | Tissue | MSP | Yes | 0.67
(0.34-1.33) | Not reported | (31) |
| Li et al,
2014(45) | China | Retrospective | Colorectal | 58.8 | 282 | 282 | Yes | NA | Tissue | MS-HRM | Yes | 1.05
(0.69-1.61) | Not reported | (38) |
| Zuo et al,
2004 | USA | Retrospective | Head and neck | 63.5 | 94 | 94 | Yes | 38 | Tissue | MS-HRM | No | 1.66
(1.11-5.18) | 2.38
(1.14-7.26) | (32) |
| Sun et al,
2012 | USA | Prospective | Head and neck | 58 | 197 | 185 | Yes | 66 | Saliva | Q-MSP | Yes | 0.91
(0.47-1.75) | 0.96
(0.53-1.73) | (36) |
In the included studies, a range of methylation
detection techniques were used, including methylation-specific
polymerase chain reaction (MSP), real-time quantitative MSP and
pyrosequencing. A summary of the study characteristics is provided
in Table I (25-45).
Pooled analysis
A total of two meta-analytic models were computed,
fixed-effects and random-effects. Using the fixed-effects model
(Mantel-Haenszel method), MGMT promoter methylation was
significantly associated with worse OS, with a pooled HR of 1.27
(95% CI, 1.13-1.42; P<0.0001). However, due to significant
heterogeneity across studies [Q=56.68; degrees of freedom (df)=20;
P<0.0001; I2=64.7%], a random-effects model using the
DerSimonian-Laird estimator with Hartung-Knapp adjustment was
employed. Under this model, the association between MGMT
methylation and OS was not statistically significant, with a pooled
HR of 1.26 (95% CI, 0.97-1.65; P=0.084).
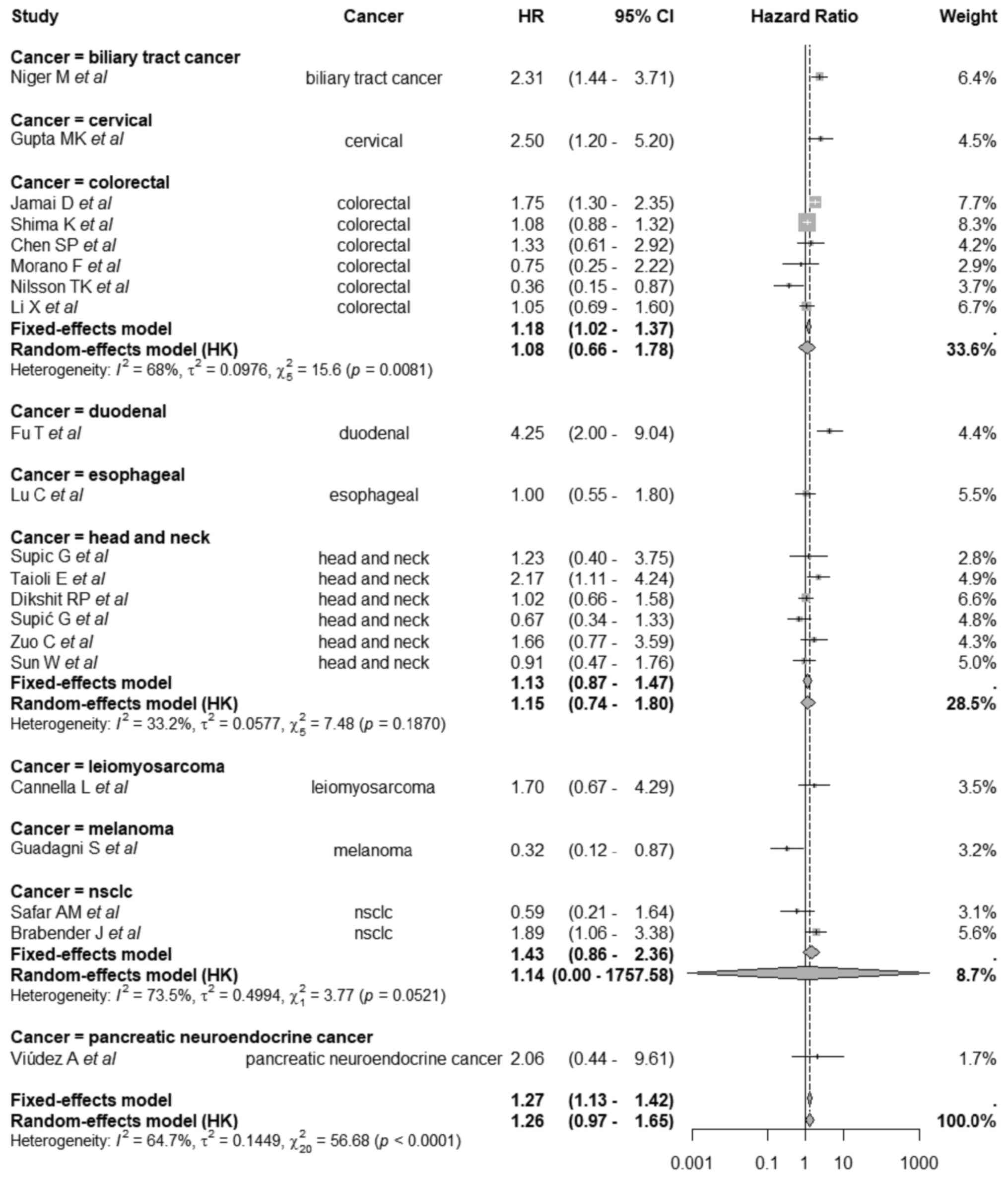
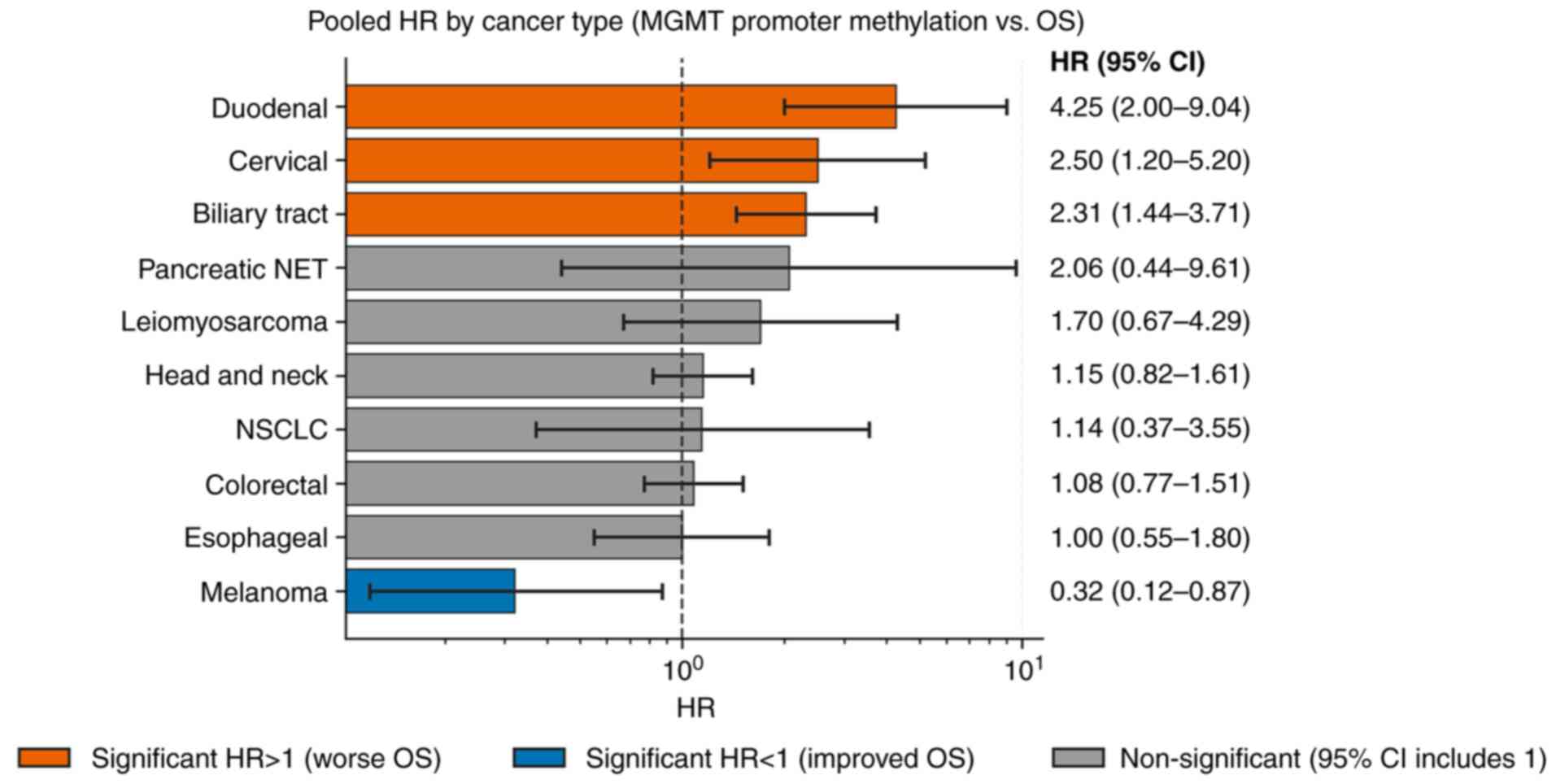
Subgroup analysis
Subgroup analyses stratified by cancer type
demonstrated a significant difference in the effect of MGMT
methylation on OS across tumor types (Q=28.60; df=9; P=0.0008). The
corresponding pooled subgroup HRs and CIs are shown in Figs. 2 and 3. In biliary tract cancer, MGMT
methylation was associated with a significantly worse OS (HR=2.31;
95% CI, 1.44-3.71). A similar detrimental effect was observed in
cervical cancer (HR=2.50; 95% CI, 1.20-5.20) and duodenal cancer,
which showed the highest HR among all subgroups (HR=4.25; 95% CI,
2.00-9.04). By contrast, melanoma demonstrated a significant
association with improved survival (HR=0.32; 95% CI, 0.12-0.87).
For colorectal cancer, the pooled estimate was HR=1.08 (95% CI,
0.77-1.51), indicating a non-significant relationship; similarly,
head and neck cancer showed a non-significant association (HR=1.15;
95% CI, 0.82-1.61). Other cancer types, including esophageal cancer
(HR=1.00; 95% CI, 0.55-1.80), leiomyosarcoma (HR=1.70; 95% CI,
0.67-4.29), NSCLC (HR=1.14, 95% CI, 0.37-3.55) and pancreatic NET
(HR=2.06; 95% CI, 0.44-9.61), were also not significantly
associated with OS.
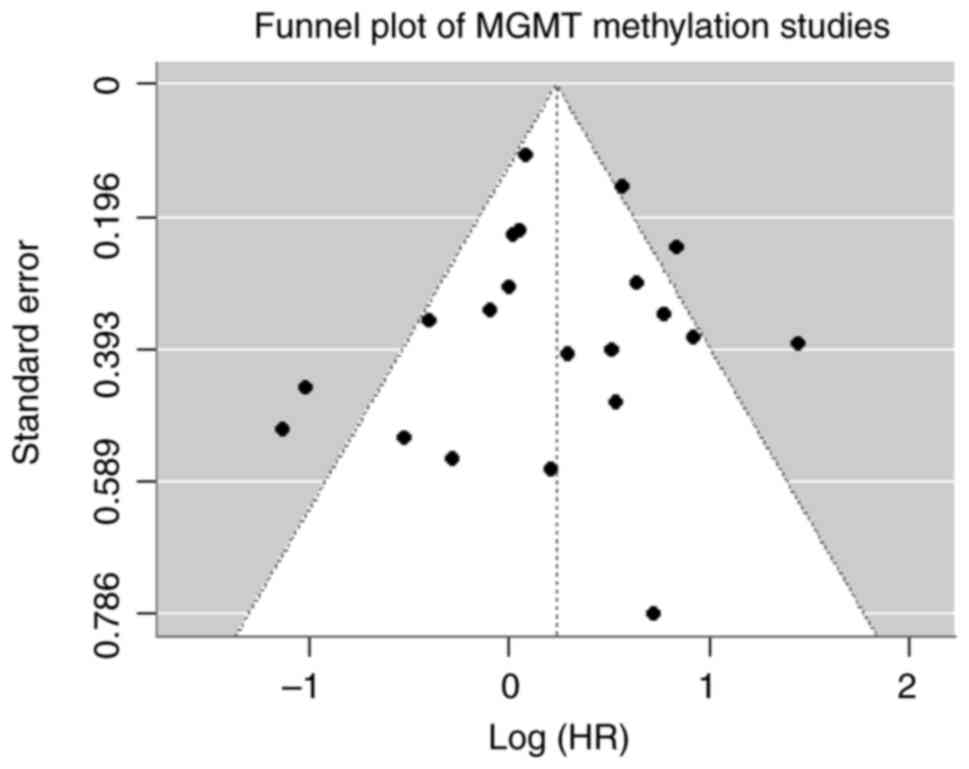
Publication bias
The Egger's regression test was not significant
(t=-0.13; P=0.895), suggesting no substantial evidence of
publication bias among the included studies (Fig. 4).
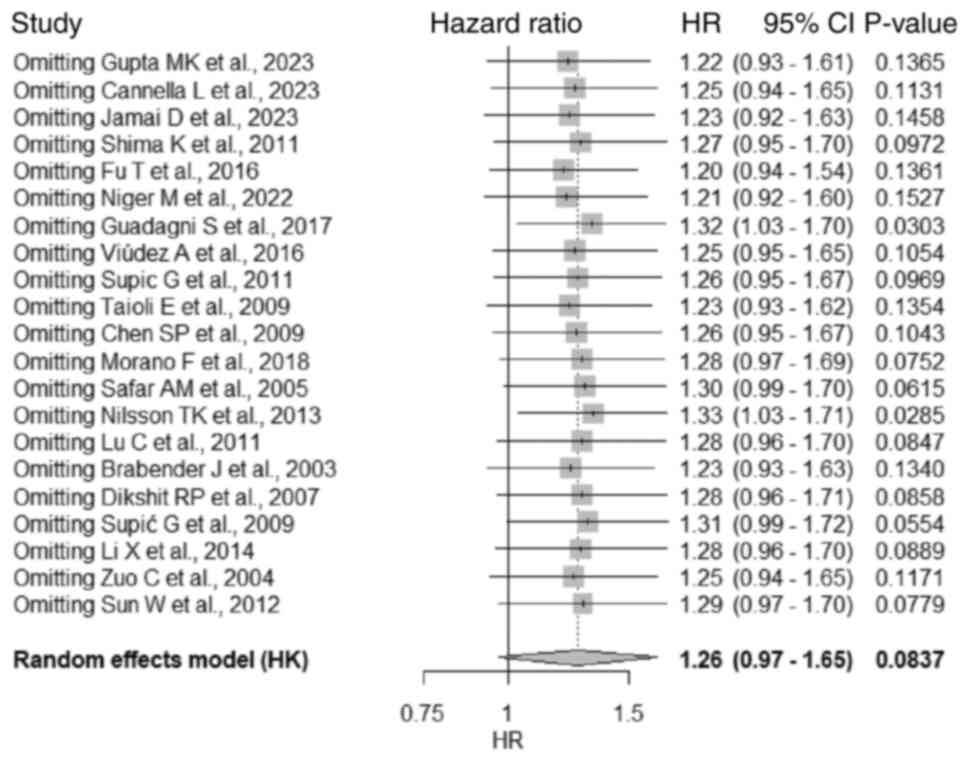
Sensitivity analysis
A leave-one-out sensitivity analysis was performed
for the overall pooled meta-analysis using a random-effects model
with Hartung-Knapp adjustment. The analysis demonstrated that no
single study exerted a disproportionate influence on the overall
pooled estimate. Across all iterations, the HR remained relatively
stable (ranging from 1.20 to 1.33), and heterogeneity
(I2) was within a moderate range (59-67%). Notably, the
removal of certain studies [such as that by Guadagni et al,
2017(41) or that by Nilsson et
al, 2013(28)] led to a
significant association (P<0.05), but the overall findings
remained generally consistent (Fig.
5).
Discussion
Against the backdrop of the need for prognostic
biomarkers for cancer, to the best of our knowledge, the present
meta-analysis investigated for the first time the prognostic
relevance of MGMT promoter methylation on OS across a mix of
non-glioma solid cancers in adult patients. A total of 21 studies
involving 2,946 patients were included, encompassing diverse solid
cancer types. The findings provide a nuanced understanding of the
potential role of MGMT methylation as a prognostic biomarker.
MGMT promoter methylation is a relatively common
epigenetic finding in numerous solid tumors, which are found in
~33% of colorectal cancers (46),
frequently arising early in the adenoma-carcinoma sequence
(47). Similarly, the frequency of
MGMT promoter methylation appears on the order of 20-30% in
patients with duodenal cancer, independent of microsatellite
instability or KRAS status (33).
In head and neck cancers, MGMT promoter methylation occurs in ~47%
of cases (48), and is
consistently observed in a notable portion of oral, pharyngeal and
laryngeal tumors. Such alteration often co-occurs with tobacco and
alcohol exposure and human papillomavirus (HPV) infection,
suggesting a field defect in carcinogenesis (48). Similarly, MGMT promoter methylation
is often part of the HPV-driven CpG island hypermethylation
phenotype in cervical carcinogenesis (49). It occurs in a minority of cervical
carcinomas but rises with disease progression, with 20-30% of
invasive cervical cancers harboring MGMT methylation, increasing
from <5% in low-grade lesions to >25% in invasive cancer
(49).
In NSCLC, frequency of this alteration ranges
between 20-35% of tumors, with MGMT methylation being slightly more
common in advanced-stage NSCLC compared with early-stage disease
(50). Smoking appears to affect
MGMT promoter methylation status in tumors; for example, in
patients with esophageal cancer with a history of heavy tobacco or
betel use, MGMT promoter methylation has been detected in up to 70%
of cases using serum DNA assays (51). Notably, tissue-based studies report
lower frequencies; MGMT methylation is noted in 30-40% of
esophageal tumors (51,52).
Moreover, one-third of pancreatic neuroendocrine
tumors exhibit MGMT hypermethylation and loss of MGMT expression
(53), with MGMT protein loss
occurring more frequently in higher-grade tumors (54). MGMT promoter methylation is
observed in ~38% of biliary tract cancers, with intrahepatic
cholangiocarcinomas showing 38% methylation, extrahepatic
cholangiocarcinomas showing 26% and gallbladder cancers showing up
to 62% (55). In leiomyosarcoma,
MGMT methylation is less studied, but it was reported in a study
that it was present in 5 of 9 uterine leiomyosarcoma cases
(56). By contrast, among
soft-tissue sarcomas overall, MGMT methylation is less common,
occurring in ~13% of cases (57),
and in melanoma, MGMT promoter methylation occurs in 15-30% of
cases (58).
In the present study the pooled analysis under the
fixed-effects model revealed a significant association between MGMT
promoter methylation and worse OS. However, the presence of
substantial heterogeneity (I2=64.7%) warranted the use
of a random-effects model with Hartung-Knapp adjustment, which
yielded a non-significant association.
In the subgroup analysis performed in the present
study, MGMT promoter methylation had variable impact on OS across
cancer types, occasionally reaching significance. The divergent
prognostic effects of MGMT methylation noted on subgroup analysis
may be explained by the role of the MGMT gene in DNA repair.
Encoded by the gene, MGMT protein is a key DNA repair enzyme that
counteracts adduct formation at the O6 position of guanine
(1,2). When alkylated, DNA becomes prone to
point mutations and the formation of crosslinks (3). The resultant compromise in DNA repair
capacity has two major consequences; this includes increased
chemo-sensitivity and genomic instability and progression. Tumor
cells lacking MGMT cannot easily repair the lethal O6 guanine
lesions induced by alkylating chemotherapeutic agents (including
temozolomide and dacarbazine). As a result, MGMT-methylated cancers
are more sensitive to these drugs, manifesting as improved
treatment response and survival. This mechanism underlies the use
of MGMT status to predict benefit from temozolomide in patients
with glioblastoma. In a prospective study, MGMT-promoter
methylation was associated with improved progression-free survival
and OS (although 95% CI were not mentioned) with temozolomide-based
treatment in patients with advanced NETs (53); this was also reported in melanomas
(58). Notably, in the present
analysis, the only cancer showing a significant increase in OS with
MGMT methylation was melanoma, with a HR of 0.32.
On the other hand, MGMT loss permits accumulation of
O6-methylguanine lesions from spontaneous alkylation via
environmental carcinogens or cellular processes (51). These unrepaired lesions lead to a
characteristic G:C to A:T mutation pattern due to misrepair
(51); consequently,
MGMT-methylated cells can acquire additional driver mutations
(46), therefore contributing to
cancer development and progression. This may be associated with the
aforementioned relationship between the exposure to smoking and
betel nut and MGMT promoter methylation status. Moreover, the
mutator phenotype from MGMT loss might produce more aggressive
subclones, associating with advanced stage or recurrence; for
example, MGMT-methylated head and neck squamous cell carcinoma have
been associated with more frequent lymph node metastases and tumor
recurrences (48). In cervical
cancer, methylation-induced MGMT downregulation is associated with
poor survival (49), likely as it
occurs as part of a broader CpG island hypermethylation phenotype
leading to concurrent epigenetic inactivation of multiple tumor
suppressor genes (such as CDKN2A, DAPK, RARB and HIC1) (59,60).
Therefore, in the absence of effective alkylator therapy, MGMT
methylation often portends a worse prognosis due to increased
genomic instability and tumor aggressiveness. According to the
present analysis, cancers demonstrating a significant decrease in
survival were duodenal, biliary tract and cervical cancers.
Finally, the remaining cancer types, colorectal,
esophageal, head and neck, leiomyosarcoma, NSCLC and pancreatic
NET, demonstrated no significant relationship with OS in the
present analysis. Lack of association in the present results does
not exclude the presence of a clinically relevant relationship, as
a subset of patients may still derive an advantage or disadvantage
from harboring MGMT promoter methylation, but further studies are
needed to confirm this.
The present study has several limitations. First,
the evidence is drawn mostly from observational studies, a number
of which studied MGMT promoter methylation status as part of a
larger methylation panel, making results of such studies prone to
bias and confounding. Additionally, such studies are also
predisposed to residual confounding; MGMT promoter methylation
might coincide with other genetic or epigenetic alterations that
drive prognosis, making it challenging to isolate the effect of
MGMT promoter methylation per se. Second, a number of tumor
types were represented by a single study with modest sample sizes;
this limited evidence base within each subgroup reduces statistical
power and confidence in the subgroup-specific estimates. Also, the
inclusion of different cancer subtypes in a single analysis makes
it hard to draw insights on such a heterogeneous group of patients.
Third, there was considerable heterogeneity in the methodologies
for determining MGMT methylation status; for example, there was no
centralized standard for what threshold defines the ‘methylated’
status; this can result in misclassification and variability in
results across studies. Fourth, the high between-study
heterogeneity in the pooled analysis limits the strength of
conclusions from the summary estimate; thus, the lack of a
significant overall effect in the random-effects model may be due
to true differences in effect rather than absence of any biological
effect. Publication bias is another concern, as studies finding a
significant prognostic effect of a biomarker are more likely to be
published. While the present study attempted to assess this (via
funnel plot symmetry and Egger's test), the interpretability was
limited by the small number of studies in numerous subgroups.
Overall, these limitations suggest that the findings should be
interpreted with caution.
There is a need for well-designed prospective
studies that uniformly evaluate MGMT promoter methylation and
follow patients for survival outcomes, especially OS, as
progression-free survival or other potentially surrogate survival
outcomes may not be truly significant for the clinical journey for
the patient. Studies should also work on harmonizing the MGMT
promoter methylation evaluation procedures, which includes agreeing
on an optimal laboratory method and defining clinically relevant
cut-off values.
In summary, MGMT promoter methylation may be
associated with worse OS in select cancer types, but the evidence
remains inconsistent when accounting for heterogeneity. The
findings highlight the importance of tumor-specific context in
biomarker research and reinforce the need for further high-quality
studies to validate the prognostic utility of MGMT methylation.
Moreover, laboratory studies are needed to understand the
mechanisms behind MGMT promoter methylation impact on tumor
behavior, which could inform targeted therapeutic strategies.
Acknowledgements
This abstract was presented at the American Society
of Clinical Oncology annual meeting, June 2025, which was held in
Chicago, Illinois, and was published as Abstract no. 3146.
Funding
Funding: No funding was received.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
AA and FA designed the overall concept and outline
of the manuscript. AA, AK, MAH, RA, IM, HAR and FA contributed to
data collection and review of literature; in addition these authors
contributed to the writing and editing of the manuscript. AA
performed the statistical analysis. All authors have read and
approved the final version of the manuscript. AA and FA confirm the
authenticity of all the raw data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Montesano R, Becker R, Hall J, Likhachev
A, Lu SH, Umbenhauer D and Wild CP: Repair of DNA alkylation
adducts in mammalian cells. Biochimie. 67:919–928. 1985.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Pegg AE, Dolan ME and Moschel RC:
Structure, function, and inhibition of O6-alkylguanine-DNA
alkyltransferase. Prog Nucleic Acid Res Mol Biol. 51:167–223.
1995.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Jacinto FV and Esteller M: MGMT
hypermethylation: A prognostic foe, a predictive friend. DNA Repair
(Amst). 6:1155–1160. 2007.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Esteller M, Hamilton SR, Burger PC, Baylin
SB and Herman JG: Inactivation of the DNA repair gene
O6-methylguanine-DNA methyltransferase by promoter hypermethylation
is a common event in primary human neoplasia. Cancer Res.
59:793–797. 1999.PubMed/NCBI
|
|
5
|
Qian XC and Brent TP: Methylation hot
spots in the 5'flanking region denote silencing of the
O6-methylguanine-DNA methyltransferase gene. Cancer Res.
57:3672–3677. 1997.PubMed/NCBI
|
|
6
|
Jones PA and Baylin SB: The fundamental
role of epigenetic events in cancer. Nat Rev Genet. 3:415–428.
2002.PubMed/NCBI View
Article : Google Scholar
|
|
7
|
Bernd K, Fritz G, Mitra S and Coquerelle
T: Transfection and expression of human O6-methylguanine-DNA
methyltransferase (MGMT) cDNA in Chinese hamster cells: The role of
MGMT in protection against the genotoxic effects of alkylating
agents. Carcinogenesis. 12:1857–1867. 1991.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Glassner BJ, Weeda G, Allan JM, Broekhof
JLM, Carls NHE, Donker I, Engelward BP, Hampson RJ, Hersmus R,
Hickman MJ, et al: DNA repair methyltransferase (Mgmt) knockout
mice are sensitive to the lethal effects of chemotherapeutic
alkylating agents. Mutagenesis. 14:339–347. 1999.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Hansen RJ, Nagasubramanian R, Delaney SM,
Samson LD and Dolan ME: Role of O6-methylguanine-DNA
methyltransferase in protecting from alkylating agent-induced
toxicity and mutations in mice. Carcinogenesis. 28:1111–1116.
2007.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Wang S, Song C, Zha Y and Li L: The
prognostic value of MGMT promoter status by pyrosequencing assay
for glioblastoma patients' survival: A meta-analysis. World J Surg
Oncol. 14(261)2016.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Hegi ME, Diserens AC, Gorlia T, Hamou MF,
de Tribolet N, Weller M, Kros JM, Hainfellner JA, Mason W, Mariani
L, et al: MGMT gene silencing and benefit from temozolomide in
glioblastoma. N Engl J Med. 352:997–1003. 2005.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Hegi ME, Liu L, Herman JG, Stupp R, Wick
W, Weller M, Mehta MP and Gilbert MR: Correlation of
O6-Methylguanine Methyltransferase (MGMT) promoter methylation with
clinical outcomes in glioblastoma and clinical strategies to
modulate MGMT activity. J Clin Oncol. 26:4189–4199. 2008.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Esteller M, Garcia-Foncillas J, Andion E,
Goodman SN, Hidalgo OF, Vanaclocha V, Baylin SB and Herman JG:
Inactivation of the DNA-repair gene MGMT and the clinical response
of gliomas to alkylating agents. N Engl J Med. 343:1350–1354.
2000.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Kulke MH, Hornick JL, Frauenhoffer C,
Hooshmand S, Ryan DP, Enzinger PC, Meyerhardt JA, Clark JW, Stuart
K, Fuchs CS and Redston MS: O6-methylguanine DNA methyltransferase
deficiency and response to Temozolomide-based therapy in patients
with neuroendocrine tumors. Clin Cancer Res. 15:338–345.
2000.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Walter T, Van Brakel B, Vercherat C,
Hervieu V, Forestier J, Chayvialle JA, Molin Y, Lombard-Bohas C,
Joly MO and Scoazec JY: O6-Methylguanine-DNA methyltransferase
status in neuroendocrine tumours: Prognostic relevance and
association with response to alkylating agents. Br J Cancer.
112:523–531. 2015.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Kunz PL, Graham NT, Catalano PJ, Nimeiri
HS, Fisher GA, Longacre TA, Suarez CJ, Martin BA, Yao JC, Kulke MH,
et al: Randomized study of temozolomide or temozolomide and
capecitabine in patients with advanced pancreatic neuroendocrine
tumors (ECOG-ACRIN E2211). J Clin Oncol. 41:1359–1369.
2023.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Svrcek M, Buhard O, Colas C, Coulet F,
Dumont S, Massaoudi I, Lamri A, Hamelin R, Cosnes J, Oliveira C, et
al: Methylation tolerance due to an O6-methylguanine DNA
methyltransferase (MGMT) field defect in the colonic mucosa: An
initiating step in the development of mismatch repair-deficient
colorectal cancers. Gut. 59:1516–1526. 2010.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Millis SZ, Bryant D, Basu G, Bender R,
Vranic S, Gatalica Z and Vogelzang NJ: Molecular profiling of
infiltrating urothelial carcinoma of bladder and nonbladder origin.
Clin Genitourin Cancer. 13:e37–e49. 2015.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Hadoux J, Favier J, Scoazec JY, Leboulleux
S, Ghuzlan AAl, Caramella C, Déandreis D, Borget I, Loriot C,
Chougnet C, et al: SDHB mutations are associated with response to
temozolomide in patients with metastatic pheochromocytoma or
paraganglioma. Int J Cancer. 135:2711–2720. 2014.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Mithani SK, Mydlarz WK, Grumbine FL, Smith
IM and Califano JA: Molecular genetics of premalignant oral
lesions. Oral Dis. 13:126–133. 2007.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Page MJ, McKenzie JE, Bossuyt PM, Boutron
I, Hoffmann TC, Mulrow CD, Shamseer L, Tetzlaff JM, Akl EA, Brennan
SE, et al: The PRISMA 2020 statement: An updated guideline for
reporting systematic reviews. PLoS Med. 18(e1003583)2021.PubMed/NCBI View
Article : Google Scholar
|
|
22
|
Balduzzi S, Rücker G and Schwarzer G: How
to perform a Meta-analysis with R: A practical tutorial. Evid Based
Ment Health. 22:153–160. 2019.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Viechtbauer W: Conducting Meta-analyses in
R with the metafor package. J Stat Softw. 36:1–48. 2010.
|
|
24
|
Higgins JPT, Thompson SG, Deeks JJ and
Altman DG: Measuring inconsistency in meta-analyses. BMJ.
327:557–560. 2003.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Gupta MK, Kushwah AS, Singh R, Srivastava
K and Banerjee M: Genetic and epigenetic alterations in MGMT gene
and correlation with concomitant chemoradiotherapy (CRT) in
cervical cancer. J Cancer Res Clin Oncol. 149:15159–15170.
2023.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Cannella L, Della Monica R, Marretta AL,
Iervolino D, Vincenzi B, De Chiara AR, Clemente O, Buonaiuto M,
Barretta ML, Di Mauro A, et al: The impact of O6-Methylguanine-DNA
methyltransferase (MGMT) promoter methylation on the outcomes of
patients with leiomyosarcoma treated with dacarbazine. Cells.
12(1635)2023.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Safar AM, Spencer H III, Su X, Coffey M,
Cooney CA, Ratnasinghe LD, Hutchins LF and Fan CY: Methylation
profiling of archived non-small cell lung cancer: A promising
prognostic system. Clin Cancer Res. 11:4400–4405. 2005.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Nilsson TK, Löf-Öhlin ZM and Sun XF: DNA
methylation of the p14ARF, RASSF1A and APC1A genes as an
independent prognostic factor in colorectal cancer patients. Int J
Oncol. 42:127–133. 2013.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Brabender J, Usadel H, Metzger R,
Schneider PM, Park J, Salonga D, Tsao-Wei DD, Groshen S, Lord RV,
Takebe N, et al: Quantitative O(6)-methylguanine DNA
methyltransferase methylation analysis in curatively resected
non-small cell lung cancer: Associations with clinical outcome.
Clin Cancer Res. 9:223–227. 2003.PubMed/NCBI
|
|
30
|
Dikshit RP, Gillio-Tos A, Brennan P, De
Marco L, Fiano V, Martinez-Peñuela JM, Boffetta P and Merletti F:
Hypermethylation, risk factors, clinical characteristics, and
survival in 235 patients with laryngeal and hypopharyngeal cancers.
Cancer. 110:1745–1751. 2007.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Šupić G, Kozomara R, Branković-Magić M,
Jović N and Magić Z: Gene hypermethylation in tumor tissue of
advanced oral squamous cell carcinoma patients. Oral Oncol.
45:1051–1057. 2009.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Zuo C, Ai L, Ratliff P, Suen JY, Hanna E,
Brent TP and Fan CY: O6-Methylguanine-DNA methyltransferase gene:
Epigenetic silencing and prognostic value in head and neck squamous
cell carcinoma. Cancer Epidemiol Biomarkers Prev. 13:967–975.
2009.PubMed/NCBI
|
|
33
|
Fu T, Sharmab A, Xie F, Liu Y, Li K, Wan
W, Baylin SB, Wolfgang CL and Ahuja N: Methylation of MGMT is
associated with poor prognosis in patients with stage III duodenal
adenocarcinoma. PLoS One. 11(e0162929)2016.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Chen SP, Chiu SC, Wu CC, Lin SZ, Kang JC,
Chen YL, Lin PC, Pang CY and Harn HJ: The association of
methylation in the promoter of APC and MGMT and the prognosis of
Taiwanese CRC patients. Genet Test Mol Biomarkers. 13:67–71.
2009.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Lu C, Xie H, Wang F, Shen H and Wang J:
Diet folate, DNA methylation and genetic polymorphisms of MTHFR
C677T in association with the prognosis of esophageal squamous cell
carcinoma. BMC Cancer. 11(91)2025.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Sun W, Zaboli D, Wang H, Liu Y,
Arnaoutakis D, Khan T, Khan Z, Koch WM and Califano JA: Detection
of TIMP3 promoter hypermethylation in salivary rinse as an
independent predictor of local recurrence-free survival in head and
neck cancer. Clin Cancer Res. 18:1082–1091. 2012.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Jamai D, Gargouri R, Selmi B and Khabir A:
ERCC1 and MGMT methylation as a predictive marker of relapse and
FOLFOX response in colorectal cancer patients from south tunisia.
Genes (Basel). 14(1467)2023.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Li X, Hu F, Wang Y, Yao X, Zhang Z, Wang
F, Sun G, Cui BB, Dong X and Zhao Y: CpG island methylator
phenotype and prognosis of colorectal cancer in Northeast China.
Biomed Res Int. 2014(236361)2014.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Shima K, Morikawa T, Baba Y, Nosho K,
Suzuki M, Yamauchi M, Hayashi M, Giovannucci E, Fuchs CS and Ogino
S: MGMT promoter methylation, loss of expression and prognosis in
855 colorectal cancers. Cancer Causes Control. 22:301–309.
2011.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Niger M, Nichetti F, Casadei-Gardini A,
Morano F, Pircher C, Tamborini E, Perrone F, Canale M, Lipka DB,
Vingiani A, et al: MGMT inactivation as a new biomarker in patients
with advanced biliary tract cancers. Mol Oncol. 16:2733–2746.
2022.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Guadagni S, Fiorentini G, Clementi M,
Palumbo G, Masedu F, Deraco M, De Manzoni G, Chiominto A, Valenti M
and Pellegrini C: MGMT methylation correlates with melphalan pelvic
perfusion survival in stage III melanoma patients: A pilot study.
Melanoma Res. 27:439–447. 2017.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Viúdez A, Carvalho FLF, Maleki Z, Zahurak
M, Laheru D, Stark A, Azad NS, Wolfgang CL, Baylin S, Herman JG, et
al: A new immunohistochemistry prognostic score (IPS) for
recurrence and survival in resected pancreatic neuroendocrine
tumors (PanNET). Oncotarget. 7:24950–24961. 2016.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Supic G, Kozomara R, Jovic N, Zeljic K and
Magic Z: Prognostic significance of tumor-related genes
hypermethylation detected in cancer-free surgical margins of oral
squamous cell carcinomas. Oral Oncol. 47:702–708. 2011.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Taioli E, Ragin C, Wang X hong, Chen J,
Langevin SM, Brown AR, Gollin SM, Garte S and Sobol RW: Recurrence
in oral and pharyngeal cancer is associated with quantitative MGMT
promoter methylation. BMC Cancer. 9(354)2009.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Morano F, Corallo S, Niger M, Barault L,
Milione M, Berenato R, Moretto R, Randon G, Antista M, Belfiore A,
et al: Temozolomide and irinotecan (TEMIRI regimen) as salvage
treatment of irinotecan-sensitive advanced colorectal cancer
patients bearing MGMT methylation. Ann Oncol. 29:1800–1806.
2018.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Ogino S, Kawasaki T, Kirkner GJ, Suemoto
Y, Meyerhardt JA and Fuchs CS: Molecular correlates with MGMT
promoter methylation and silencing support CpG island methylator
phenotype-Low (CIMP-Low) in colorectal cancer. Gut. 56:1564–1571.
2017.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Li Y, Lyu Z, Zhao L, Cheng H, Zhu D, Gao
Y, Shang X and Shi H: Prognostic value of MGMT methylation in
colorectal cancer: A meta-analysis and literature review. Tumour
Biol. 36:1595–1601. 2015.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Cai F, Xiao X, Niu X, Shi H and Zhong Y:
Aberrant Methylation of MGMT Promoter in HNSCC: A Meta-Analysis.
PLoS One. 11(e0163534)2016.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Choi B, Na Y, Whang MY, Ho JY, Han MR,
Park SW, Song H, Hur SY and Choi YJ: MGMT Methylation is associated
with human papillomavirus infection in cervical dysplasia: A
longitudinal study. J Clin Med. 12(6188)2023.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Chen L, Wang Y, Liu F, Xu L, Peng F, Zhao
N, Fu B, Zhu Z, Shi Y, Liu J, et al: A systematic review and
meta-analysis: Association between MGMT hypermethylation and the
clinicopathological characteristics of non-small-cell lung
carcinoma. Sci Reports. 8(1439)2018.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Das M, Sharma SK, Sekhon GS, Saikia BJ,
Mahanta J and Phukan RK: Promoter methylation of MGMT gene in serum
of patients with esophageal squamous cell carcinoma in North East
India. Asian Pac J Cancer Prev. 15:9955–9960. 2014.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Zhang L, Lu W, Miao X, Xing D, Tan W and
Lin D: Inactivation of DNA repair gene O6-methylguanine-DNA
methyltransferase by promoter hypermethylation and its relation to
p53 mutations in esophageal squamous cell carcinoma.
Carcinogenesis. 24:1039–1044. 2003.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Brighi N, Lamberti G, Andrini E, Mosconi
C, Manuzzi L, Donati G, Lisotti A and Campana D: Prospective
evaluation of MGMT-Promoter methylation status and correlations
with outcomes to Temozolomide-based chemotherapy in
Well-differentiated neuroendocrine tumors. Curr Oncol.
30:1381–1394. 2023.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Yagi K, Ono H, Kudo A, Kinowaki Y, Asano
D, Watanabe S, Ishikawa Y, Ueda H, Akahoshi K, Tanaka S and Tanabe
M: MGMT is frequently inactivated in pancreatic NET-G2 and is
associated with the therapeutic activity of STZ-based regimens. Sci
Reports. 13(7535)2023.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Niger M, Morano F, Manglaviti S, Nichetti
F, Tamborini E, Perrone F, Marcuzzo M, Peverelli G, Brambilla M,
Pagani F, et al: 732P-Is MGMT methylation a new therapeutic target
for biliary tract cancer? Ann Oncol. 30(v281)2019.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Bujko M, Kowalewska M, Danska-Bidzinska A,
Bakula-Zalewska E, Siedecki JA and Bidzinski M: The promoter
methylation and expression of the O6-methylguanine-DNA
methyltransferase gene in uterine sarcoma and carcinosarcoma. Oncol
Lett. 4:551–555. 2012.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Jakob J, Hille M, Sauer C, Ströbel P, Wenz
F and Hohenberger P: O6-methylguanine-DNA methyltransferase (MGMT)
Promoter methylation is a rare event in soft tissue sarcoma. Radiat
Oncol. 7(180)2012.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Schraml P, Von Teichman A, Mihic-Probst D,
Simcock M, Ochsenbein A, Dummer R, Michielin O, Seifert B, Schläppi
M, Moch H and von Moos R: Predictive value of the MGMT promoter
methylation status in metastatic melanoma patients receiving
First-line temozolomide plus bevacizumab in the trial SAKK 50/07.
Oncol Rep. 28:654–668. 2012.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Kazanets A, Shorstova T, Hilmi K, Marques
M and Witcher M: Epigenetic silencing of tumor suppressor genes:
Paradigms, puzzles, and potential. Biochim Biophys Acta.
1865:275–288. 2025.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Esteller M: CpG island hypermethylation
and tumor suppressor genes: A booming present, a brighter future.
Oncogene. 21:5427–5440. 2002.PubMed/NCBI View Article : Google Scholar
|