Introduction
Wasabi [Wasabia japonica (Miq.) Matsumura],
called Japanese horseradish, is a member of the Brassicaceae family
of vegetables. Its rhizome is a very popular pungent spice in
Japan. Several studies have shown that wasabi has multiple
biological activities, such as anti-microbial activity (1), inhibition of platelet aggregation
(2) and the suppression of
N-methyl-N'-nitro-N-nitrosoguanidine (MNNG)-induced rat
gastric carcinogenesis (3).
Accumulated data reveal that a series of allyl isothiocyanates are
present in its rhizome as active compounds (4) of which 6-(methylsulfinyl)hexyl
isothiocyanate (6-MSITC or 6-MITC in previous reports) is a major
allyl isothiocyanate (5). Our
group recently reported that 6-MSITC inhibits lipopolysaccharide
(LPS)-induced expression of cyclooxygenase-2 (COX-2) (6) and inducible nitric oxide synthase
(iNOS) (7). These data suggest
that 6-MSITC may have potential as an anti-inflammatory agent.
Inflammation is essential for defense against
bacterial infection. Expression of inflammatory cytokines,
chemokines and other mediators are involved in inflammatory
processes (8). Although
recognition of invading pathogens by host cells initiates the
binding of specific cellular receptors to pathogen molecules with
distinct patterns (9), toll-like
receptors (TLRs) are the major pattern-recognition receptors. In
particular, inflammation caused by LPS is one of the most
extensively studied cases (10).
LPS liberated from gram-negative bacteria associates with LPS
binding protein, an acute phase protein present in the bloodstream
and subsequently binds to CD14 antigen (CD14), a
glycosylphosphatidylinositol-linked protein expressed on the cell
surface of phagocytes. LPS is then transferred to MD-2, which
associates with the extracellular portion of toll-like receptor 4
(TLR4), followed by oligomerization of TLR4, a key molecule of LPS
signaling (11,12). TLR4 signaling promptly induces
potent innate immune responses through adaptor molecules, such as
myeloid differentiation factor 88 (MyD88), toll/interleukin (IL)-1
receptor (TIR) domain containing adaptor protein, TRIF-related
adaptor molecule (TRAM) and TIR domain containing adaptor inducing
interferon-β (TRIF) to activate intracellular signaling pathways
involving interleukin-1 receptor-associated kinase (IRAK), Tnf
receptor-associated factor 6 (TRAF6), inhibitor of κB kinase (IKK)
and mitogen-activated protein kinase (MAPK) (13,14).
These signaling pathways ultimately stimulate transcription
factors, nuclear factor κB (NF-κB), activator protein 1 (AP-1) and
interferon regulatory factors to induce antibacterial and antiviral
responses including the induction of tumor necrosis factor (TNF),
interleukin-1 (IL1) and interleukin-6 (IL6) (15).
To evaluate the anti-inflammatory function and
underlying genes targeted by wasabi 6-MSITC, we used
oligonucleotide DNA microarray to investigate the effects of
6-MSITC on genome-wide gene expression in an inflammatory cell
model, murine macrophage-like RAW264. Furthermore, we explored the
molecular interaction networks from gene expression data, using
Ingenuity pathway analysis (IPA) software. These data revealed a
molecular basis for the anti-inflammatory activity of 6-MSITC.
Materials and methods
Materials and cell culture
6-MSITC was purified by reversephase HPLC to
>99%, and dissolved in dimethyl sulfoxide for cell culture (DMSO
final concentration, 0.2%). LPS (Escherichia coli serotype
055:B5) and other reagents used in the chemical analysis were
purchased from Sigma Chemical. Murine macrophage-like RAW264 cells
were obtained from Riken BioResource Center Cell Bank, Japan (cell
no. RCB0535) and cultured at 37°C in a 5% CO2 atmosphere
in Dulbecco's modified Eagle's medium containing 10% fetal bovine
serum.
RNA preparation and microarray
hybridization
RAW264.7 cells were pretreated with or without
6-MSITC for 30 min and then exposed to 40 ng/ml LPS for 6 h. Total
RNA was extracted using Qiagen™ RNeasy mini kit (Valencia, CA)
following the manufacturer's protocol. The RNA quantity was
assessed using an Agilent 2100 bioanalyzer (Palo Alto, CA). Total
RNA (500 ng) was amplified at 40°C for 2 h by the Agilent low RNA
input fluorescent linear amplification kit following the
manufacturer's protocol. cRNAs were labeled at 40°C for 2 h with
cyanine 5 (Cy5) for samples and with cyanine 3 (Cy3) for the
universal mouse reference RNA (Agilent Technologies). After the
amplification and labeling, the yields and dye incorporation
efficiencies were determined using a spectrophotometer. Agilent
mouse 22,050 oligonucleotide microarrays were used for this study
following Agilent microarray processing protocol. Briefly,
Cy3-labeled samples and Cy5-labeled references were mixed and
incubated with an Agilent microarray slide for 17 h using an
Agilent in situ hybridization kit. After washing with
stabilization and drying solution (Agilent Technologies),
microarray signals were scanned in an Agilent model G2505A
microarray scanner.
Data analysis
Images were processed using Agilent Feature
Extraction software, which provides normalized Cy3 and Cy5 channel
intensity values for each spot on an array (for a detailed
description of the Agilent Feature Extraction software and the
algorithms, see the Agilent Feature Extraction User's Manual). The
selected genes were further classified into the biological process,
molecular function and signaling pathway by the Gene Ontology
software (http://www.geneontology.org/).
Reverse transcription and real-time
PCR
The primers used in the present study have been
described in previous studies (16). The primers for IL1α, IL1β, IL6,
IFI1 and IFI47 were designed according to the NCBI sequence
database using the software Primer3 (16). The primers for TNF (17), COX-2 (20), CCL22 (18) and PTGS2 (19) have been described previously.
Reverse transcription and real-time PCR were performed with DyNAmo™
SYBR® Green 2-Step qRT-PCR Kit (Finnzymes Oy., Espoo,
Finland) according to the manufacturer's manual. Briefly, RNA (200
ng) was reversed to cRNA using Oligo dT and M-MuLV RNase at 37°C
for 30 min, and the reaction was then terminated at 85°C for 5 min.
Quantitative PCR was performed with the Roter-Gene-3000AKAA
(Corbett Research) in triplicates using the standard curve. The
Tm-value of PCR was determined according to each primer sequence
(https://www.finnzymes.fi/tm.determination.html). Each
PCR reaction contained 250 ng of reverse transcripts, 75 ng of each
primer and 10 μl Master mix (Finnzymes Oy.). The thermal cycling
condition was held at 95°C for 15 min followed by 55 cycles of 30
sec at 94°C, 30 sec at Tm-value (16) and 30 sec at 72°C. The result was
represented by the relative expression level normalized with
control cells.
Pathway analysis and network
generation
Gene accession numbers, the fold change upon 6-MSTIC
treatment vs. the control cells, and the t-test P-value were
imported into the IPA software. IPA was carried out with P<0.002
as the cutoff point. The genes were categorized according to the
molecular functions using the software. The identified genes were
also mapped to genetic networks in the IPA database and ranked by a
score that denotes the probability that a collection of genes equal
to or greater than the number in a network could be achieved by
chance alone. The genetic network analysis focuses on the
functional relationships that are present in the literature to
create a network of genes with similar functions and recorded
interactions. A network pathway is a graphical representation of
the molecular relationships between genes or gene products. Genes
or gene products are represented as nodes, and the biological
relationship between two nodes is represented as an edge (line).
All edges are supported by at least one reference from the
literature, from a textbook, or from canonical information stored
in the Ingenuity Pathways Knowledge Base. The intensity of the node
color indicates the degree of up- (red) or down-regulation (green).
Nodes are displayed using various shapes that represent the
functional class of the gene product. Edges are displayed with
various labels that describe the nature of the relationship between
the nodes (e.g., P for phosphorylation, T for transcription).
Statistical analysis
Differences between treated and control cells were
analyzed by the Student's t-test. A probability of P<0.05 was
considered significant.
Results
Gene expression profiles
Based on the results of our preexperiments, RAW264.7
cells were treated with or without 8 μM 6-MSITC for 30 min and then
exposed to 40 ng/ml lipopolysaccharide (LPS) for another 6 h. Under
this condition, RAW264.7 cells had a favorable response to 6-MSITC
without cytotoxicity. Cell mRNA was prepared and processed for
hybridization to the mouse oligonucleotide DNA microarray, as
described in Materials and methods. Comparing the signals of
LPS-treatment with those of the control revealed that a total of
1123 gene signals were changed by ≥3-fold, of which, 406 gene
signals were increased and 717 gene signals were decreased by
≥3-fold (Table I). Among the 1123
genes responding to LPS, treatment with 6-MSITC reduced 238 gene
signals by ≥2-fold and enhanced 336 gene signals, respectively
(Table I). Therefore, 6-MSITC
affected ∼51% of the LPS-responsive genes by ≥2-fold. This result
suggests that 6-MSITC might be effective in attenuating LPS-induced
responses. To validate the accuracy of the microarray data, 6
representative genes with changes in expression of several fold,
several ten fold and several hundred fold by LPS were chosen, and
their expression levels were detected by real-time PCR with the
same RNA. The real-time PCR results exhibited a similar expression
pattern with that of the DNA microarray, suggesting the DNA
microarray data obtained in the present study is valid (data not
shown).
 | Table I.Number of genes that are regulated by
6-MSITC. |
Table I.
Number of genes that are regulated by
6-MSITC.
| Fold change | LPS/CTL |
(6-MSITC+LPS)/LPS |
|---|
| ≥9 | 90 | 74 |
| ≥6 to <9 | 85 | 59 |
| ≥3 to <6 | 231 | 105 |
| Subtotal | 406 | 238 |
| > −6 to ≤ −3 | 664 | 321 |
| > −9 to ≤ −6 | 26 | 12 |
| ≤ −9 | 7 | 3 |
| Subtotal | 717 | 336 |
| Total | 1123 | 574 |
Grouping of genes targeted by
6-MSITC
The genes that showed response to LPS only and LPS
plus 6-MITC were classified into biological process, molecular
function and signaling pathway by the Gene Ontology software. As
shown in Table II, 332 genes with
≥3-fold change by LPS were classified into 35 groups of which 206
genes were regulated by 6-MSITC with ≥2-fold and classified into 30
groups hitting for biological process (81), molecular function
(108) and signaling pathway (17)
categories. The remaining 11,480 genes were unclassified. The gene
groups highly affected by wasabi 6-MSITC were associated with
‘inflammatory response, signal transduction, cytokine activities,
hydrolase activity, kinase activity, receptor activities,
transferase activity, nucleic acid binding and apoptosis’
categories.
 | Table II.Classification of genes targeted by
6-MSITC in LPS-activated macrophages. |
Table II.
Classification of genes targeted by
6-MSITC in LPS-activated macrophages.
| Category | CTL | LPS | (M+LPS)/LPS |
|---|
| Biological
process | | | |
| Apoptosis | 192 | 17 | 9 |
| Carbohydrate
metabolism | 107 | 4 | 2 |
| Cell adhesion | 289 | 5 | 2 |
| Cell cycle | 247 | 3 | 2 |
| Cell
proliferation | 69 | 5 | 2 |
| Defense | 121 | 4 | 4 |
| Signal
transduction | 546 | 29 | 21 |
| Transport | 1001 | 10 | 7 |
| Lipid metabolic
process | 125 | 1 | 1 |
| Inflammatory
response | 88 | 16 | 13 |
| Cytokine
activity | 139 | 23 | 18 |
| Molecular
function | | | |
| Cell adhesion
molecule activity | 289 | 5 | 2 |
| Cytoskeletal
protein binding | 25 | 1 | 0 |
| Defense
response | 121 | 4 | 4 |
| Hydrolase
activity | 977 | 27 | 17 |
| Ion channel
activity | 233 | 1 | 0 |
| Kinase
activity | 621 | 20 | 9 |
| Lyase
activity | 78 | 2 | 1 |
| Protein
ubiquitination | 120 | 2 | 1 |
| Nucleic acid
binding | 394 | 7 | 5 |
| Oxidoreductase
activity | 384 | 4 | 3 |
| Receptor
activity | 1513 | 38 | 21 |
| Signal
transduction | 546 | 29 | 21 |
| Transcription
factor activity | 500 | 14 | 10 |
| Transferase
activity | 1015 | 25 | 13 |
| Transporter
activity | 142 | 2 | 1 |
| Pathway | | | |
| Angiogenesis | 63 | 2 | 2 |
| Apoptosis | 192 | 17 | 9 |
| B-cell
activation | 8 | 3 | 3 |
| Blood
coagulation | 42 | 1 | 0 |
| Cell
adhesion | 289 | 5 | 2 |
| Cytokine and
chemokine-mediated signaling pathway | 21 | 3 | 1 |
| Integrin-mediated
signaling pathway | 62 | 2 | 0 |
| JAK-STAT
cascade | 11 | 1 | 0 |
Profiling of anti-inflammatory effects by
6-MSITC
To investigate the anti-inflammatory effects by
6-MSITC, we next profiled the genes that are related to
inflammatory response, defense, cytokine activity and receptor
activity. The changes in gene expression by 6-MSITC (≥2-fold) are
partially listed in Table III. The
induction of pro-inflammatory genes, including TNF, IL1β, IL6,
prostaglandin-endoperoxide synthase 2 (PTGS2) and COX-2 by LPS were
reduced by 6-MSITC. In addition, the inductions of various CC and
CXC chemokines including chemokine (C-C motif) ligand 17 (CCL17),
22 (CCL22), chemokine (C-X3-C motif) ligand 1 (CX3CL1), chemokine
(C-X-C motif) ligand 11 (CXCL11) and 16 (CXCL16) by LPS were
suppressed by 6-MSITC. LPS-induced interferon-inducible protein 1
(IFI1) and 47 (IFI47), and interleukin receptor (IL10ra, IL23r and
IL4ra) levels were also attenuated by 6-MSITC. On the other hand,
LPS decreased the expression levels of a few CC chemokines CCL11
and CCL25), interleukins (IL3) and receptors (IL1ra12, IL8ra,
TNFRSF23 and TNFSF4) while 6-MSITC restored them to normal
levels.
 | Table III.List of genes targeted by 6-MSITC in
categories of ‘defense, inflammatory response, cytokine activity
and receptor activity’. |
Table III.
List of genes targeted by 6-MSITC in
categories of ‘defense, inflammatory response, cytokine activity
and receptor activity’.
| Gene | Accession no. | LPS/CTL | (M+LPS)/CTL |
|---|
| CCL12 | NM_011331 | 8.48 | 2.83 |
| CCL17 | NM_011332 | 17.64 | 3.63 |
| CCL2 | NM_011333 | 22.01 | 1.79 |
| CCL22 | NM_009137 | 6.79 | 0.51 |
| CCL4 | NM_013652 | 91.36 | 40.87 |
| CCL5 | NM_013653 | 29.18 | 2.14 |
| CCL7 | NM_013654 | 302.69 | 1.34 |
| CCL9 | NM_011338 | 4.51 | 1.34 |
| CCR1 | NM_009912 | 7.94 | 2.51 |
| CCRL2 | NM_017466 | 28.93 | 2.04 |
| CD300le | NM_172050 | 8.24 | 0.64 |
| CD86 | NM_019388 | 3.70 | 1.22 |
| COX-2 | AF344876 | 5.85 | 1.78 |
| CSF2 | NM_009969 | 124.48 | 15.85 |
| CX3CL1 | NM_009142 | 3.56 | 1.37 |
| CCCL10 | NM_021274 | 63.04 | 1.13 |
| ICOSL | NM_015790 | 3.68 | 1.59 |
| IFI1 | NM_008326 | 3.97 | 0.64 |
| IFI47 | NM_008330 | 2.71 | 1.27 |
| IFNB1 | NM_010510 | 35.30 | 2.41 |
| IL10 | NM_010548 | 10.80 | 1.08 |
| IL1f6 | NM_019450 | 11.63 | 1.10 |
| IL1rn | NM_031167 | 7.88 | 1.14 |
| IL6 | NM_031168 | 718.90 | 53.26 |
| IRAK2 | NM_172161 | 5.41 | 2.06 |
| NFKBIZ | NM_030612 | 57.08 | 16.83 |
| PTGS2 | NM_011198 | 416.00 | 142.40 |
| PTGER2 | NM_008964 | 28.78 | 3.82 |
| TLR1 | NM_030682 | 3.68 | 1.41 |
| TNF | NM_013693 | 2.49 | 1.00 |
| TNFRSF5 | NM_011611 | 6.81 | 1.57 |
| CCL11 | NM_011330 | 0.24 | 0.75 |
| CCL25 | NM_009138 | 0.28 | 0.65 |
| IL3 | NM_010556 | 0.22 | 0.57 |
| IL6ra | NM_010559 | 0.28 | 0.63 |
| TNFSF4 | NM_009452 | 0.22 | 0.54 |
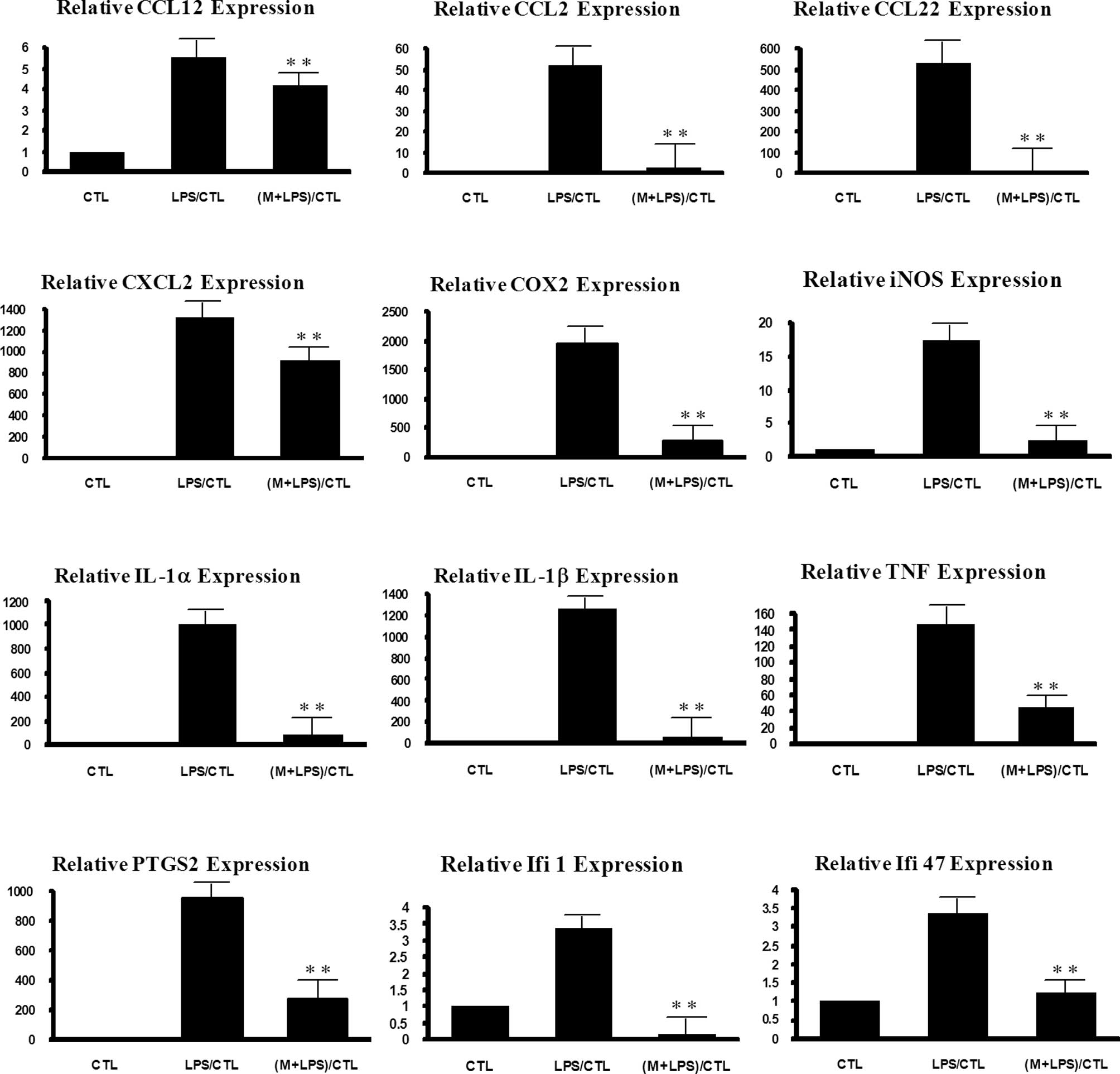
To confirm the results, the expression levels of
IL1β, IL6, TNF, COX-2, PTGS2, CCL22, IFI47 and IFI1 were further
detected by real-time PCR (Fig.
1). Most showed a similar expression pattern between the
microarray and real-time PCR data. For example, the level of
LPS-stimulated expression of TNF was reduced by 85% by 6-MSITC in
the real-time PCR experiment, while the extent of reduction was 68%
in the microarray experiment. The inhibitory effect of 6-MSITC on
IFI1 was 60% in the real-time PCR and 57% in the microarray
experiments. IL1β showed a somewhat lower extent of 6-MSITC effect
in the microarray (40%) than the extent in the real-time PCR
experiment (93%).
Biologically relevant networks and
pathways
To determine the biologically relevant networks and
pathways of the differentially expressed genes, pathway analysis
was conduced on genes with 2-fold changes upon 6-MSITC treatment,
using the Ingenuity Pathway Knowledge Base. The most significant
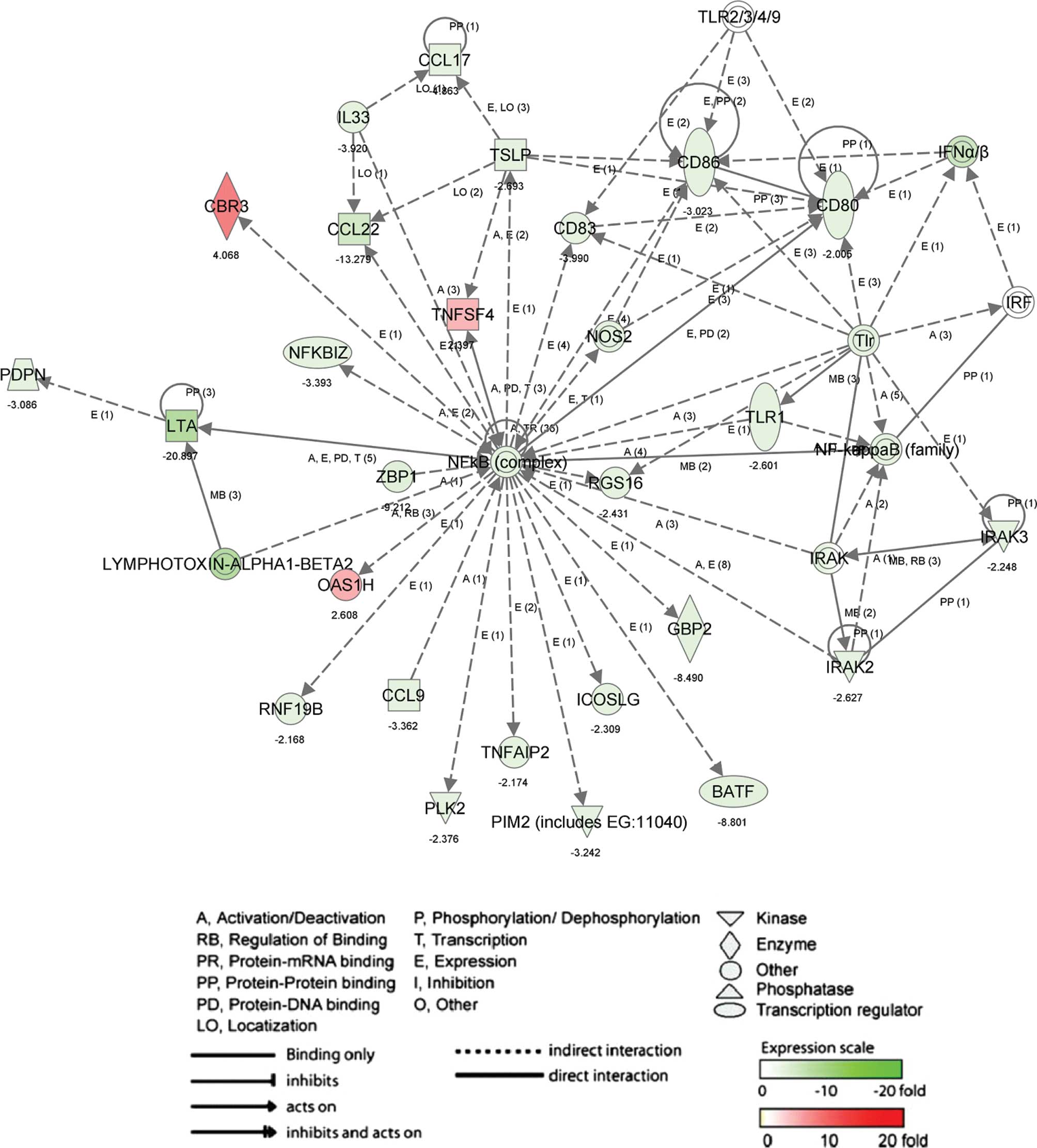
network in up- and down-regulated genes by 6-MSITC (Fig. 2; score, 42 and 26 genes) including
inducible T-cell co-stimulator ligand (ICOSLG), tumor necrosis
factor (ligand) superfamily member 4 (TNFSF4), CD86 antigen (CD86),
nuclear factor of κ light polypeptide gene enhancer in B-cell
inhibitor ζ (NFKBIZ), CCL17 and CCL22 was associated with
inflammatory response (P-value, 2.46E-4 to 4.47E-2) and immune
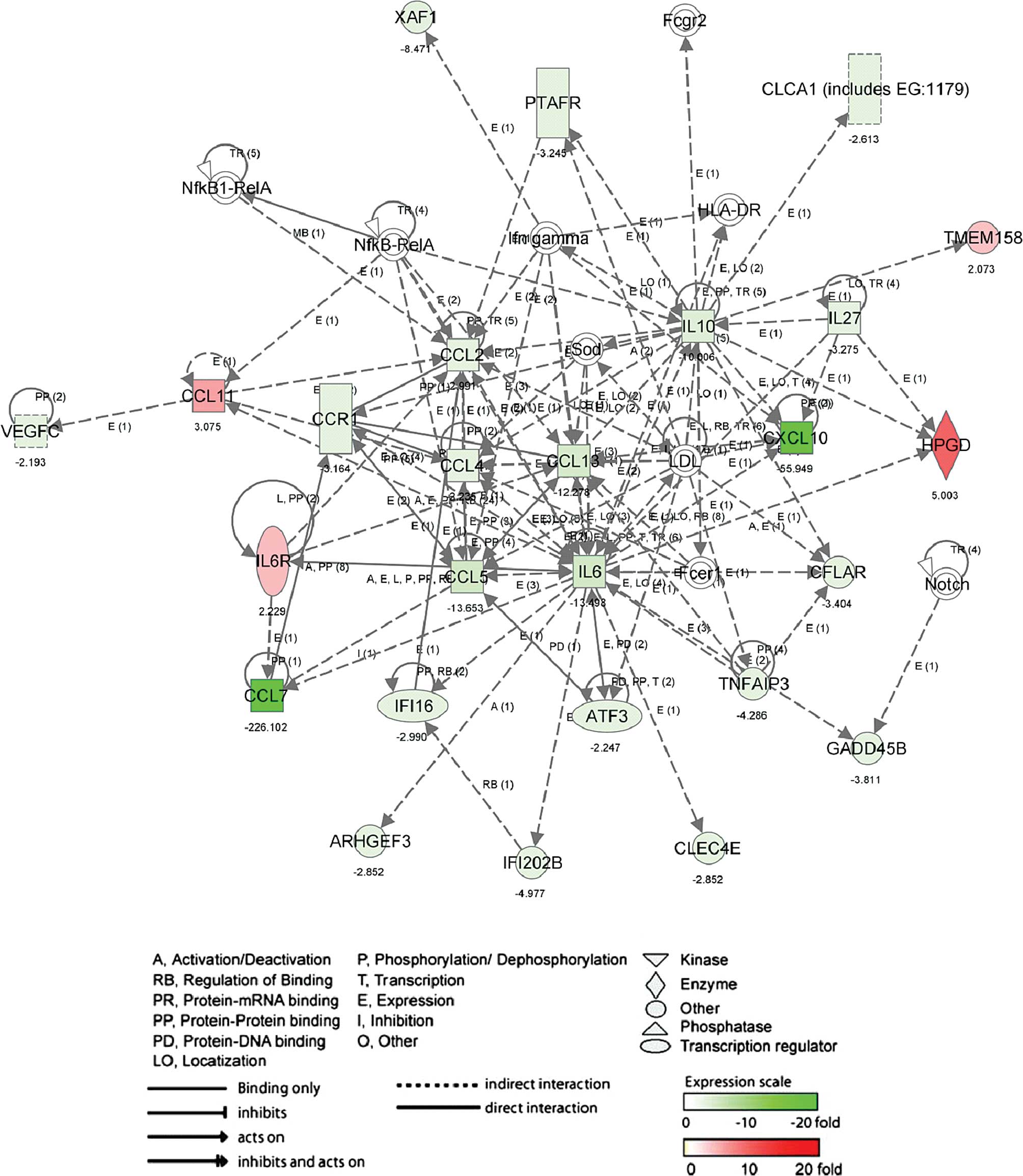
response (P-value, 3.06E-5 to 7.16E-3). The other network (Fig. 3; score, 42 and 26 genes) in up- and
down-regulated genes by 6-MSITC, including CCL11, CCL13, CCL2,
CCL4, CCL5, CCL7, CCR1, CXCL10, IL6 and IL10 was associated with
inflammatory (P-value, 2.27E-9 to 1.15E-3) and immune response
(P-value, 6.31E-8 to 6.31E-8).
Discussion
6-(Methylsulfinyl)hexyl isothiocyanate has been
reported to attenuate expression of COX-2 (6) and iNOS (7), and production of prostaglandin E
synthase 2 (PGE2) (6) and NO
(7), suggesting 6-MSITC may have
anti-inflammatory activity. In the present study, we, for the first
time, showed the gene expression profiles upon 6-MSITC treaatment
in RAW264.7 cells, a cell model system to study inflammation, by
genome-wide DNA microarray.
Among a total of 22,050 gene probes, LPS treatment
at 40 ng/ml for 6 h up-regulated signals of 406 gene probes (1.8%
of the total gene probes) and down-regulated signals of 717 gene
probes (3.2% of the total gene probes) by ≥3-fold. These genes were
categorized into 35 groups and hit for biological processes,
molecular functions and signaling pathways. The number of genes
affected by LPS was fewer than that upon treatment with 1 μg/ml for
6 h as reported previously (18),
suggesting that RAW264 cells responded to LPS in a dose-dependent
manner. In our preliminary treatment, we found that a high dose of
LPS appeared cytotoxic to RAW246.7 cells; we thus treated the cells
with 40 ng/ml LPS in the present study to mimic an inflammatory
response under normal conditions.
The number of genes affected by 6-MSITC treatment
consisted of 58% of genes down-regulated by LPS and 47% of genes
up-regulated by LPS. Analysis of the genes targeted by 6-MSITC
revealed that a number of inflammation and defense-related genes
were affected. The up-regulation of pro-inflammatory genes, such as
TNF, IL1β, IL6, PTGS and COX-2 by LPS has been suggested to be a
major factor for the inflammatory effects observed in LPS-activated
macrophages (20,21), and 6-MSITC appeared to attenuate
their expression. In addition, 6-MSITC reduced the expression of
interferon-inducible genes (IFI1 and IFI47). These factors are
documented to be involved in interferon-mediated immunity and cell
proliferation and differentiation (22). The inductions of interleukin
receptors (IL10ra, IL23r and IL4ra) by LPS were also suppressed by
6-MSITC. These results suggest that 6-MSITC inhibition of the
expression of various pro-inflammatory genes may explain its
anti-inflammatory effects. On the other hand, 6-MSITC also restored
the expression levels of LPS-reduced CC chemokines (CCL11 and
CCL25), interleukins (IL3) and receptors (IL1ra12, IL8ra, TNFRSF23
and TNFRSF4) to control levels. The data suggest that wasabi
6-MSITC might not only attenuate the expression of certain
pro-inflammatory genes induced by LPS but also restore the
expression level of anti-inflammatory genes reduced by LPS although
detailed information must be obtained through further studies.
In summary, our DNA microarray data, for the first
time, revealed gene expression profiling of wasabi 6-MSITC in an
inflammatory cell model, RAW264.7. 6-MSITC may target immune and
inflammation-related genes including chemokines, interleukins and
interferons to exert its anti-inflammatory function. These data
provide a basis for understanding the molecular mechanisms of the
anti-inflammatory effects of wasabi.
Abbreviations:
|
6-MSITC
|
6-(methylsulfinyl)hexyl
isothiocyanate;
|
|
CCL
|
chemokine (C-C motif) ligand;
|
|
COX-2
|
cyclooxygenase-2;
|
|
CX3CL
|
chemokine (C-X3-C motif) ligand;
|
|
IKK
|
inhibitor of κB kinase;
|
|
iNOS
|
inducible nitric oxide synthase;
|
|
IPA
|
Ingenuity pathways analysis;
|
|
IRAK
|
interleukin-1 receptor-associated
kinase;
|
|
MyD88
|
myeloid differentiation factor 88;
|
|
PGE2
|
prostaglandin E synthase 2;
|
|
PTGS2
|
prostaglandin-endoperoxide synthase
2;
|
|
TIR
|
toll/interleukin (IL)-1 receptor;
|
|
TLR
|
toll-like receptor;
|
|
TRAF6
|
Tnf receptor-associated factor 6;
|
|
TRAM
|
TRIF-related adaptor molecule;
|
|
TRIF
|
TIR domain containing adaptor inducing
interferon (IFN)-β
|
Acknowledgements
This study was supported, in part,
through a fund of the Frontier Science Research Center of Kagoshima
University to De-Xing Hou.
References
|
1.
|
Isshiki K and Tokuoka K: Allyl
isothiocyanate and wholesomeness of food. Jpn J Food Microbiol.
12:1–6. 1993.
|
|
2.
|
Kumagai H, Kashima N, Seki T, Sakurai H,
Ishii K and Ariga T: Analysis of components in essential oil of
upland wasabi and their inhibitory effects on platelet aggregation.
Biosci Biotech Biochem. 58:2131–2135. 1994. View Article : Google Scholar
|
|
3.
|
Tanida N, Kawaura A, Takahashi A, Sawada K
and Shimoyama T: Suppressive effect of wasabi (pungent Japanese
spice) on gastric carcinogenesis induced by MNNG in rats. Nutr
Cancer. 16:53–58. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Ono H, Adach K, Fuke Y and Shinohara K:
Purification and structural analysis of substances in wasabi
(Eutrema wasabi Maxim.) that suppress the growth of MKN-28
human stomach cancer cells. J Jpn Food Sci Technol. 43:1092–1097.
1996.
|
|
5.
|
Morimitsu Y, Nakagawa Y, Hayashi K, Fujii
H, Kumagai T and Nakamura Y: A sulforaphane analogue that potently
activates the Nrf2-dependent detoxification pathway. J Biol Chem.
277:3456–3463. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Uto T, Fujii M and Hou DX: Inhibition of
lipopolysaccharideinduced cyclooxygenase-2 transcription by
6-(methylsulfinyl) hexyl isothiocyanate, a chemopreventive compound
from Wasabia japonica (Miq.) Matsumura, in mouse
macrophages. Biochem Pharmacol. 70:1772–1784. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Uto T, Fujii M and Hou DX:
6-(Methylsulfinyl)hexyl isothiocyanate suppresses inducible nitric
oxide synthase expression through the inhibition of Janus kinase
2-mediated JNK pathway in lipopolysaccharide-activated murine
macrophages. Biochem Pharmacol. 70:1211–1221. 2005. View Article : Google Scholar
|
|
8.
|
Baggiolini M: Chemokines and leukocyte
traffic. Nature. 392:565–568. 1998. View
Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Akira S, Takeda K and Kaisho T: Toll-like
receptors: critical proteins linking innate and acquired immunity.
Nat Immunol. 2:675–680. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Erridge C, Bennett-Guerrero E and Poxton
IR: Structure and function of lipopolysaccharides. Microbes Infect.
4:837–851. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Shimazu R, Akashi S, Ogata H, Nagai Y,
Fukudome K, Miyake K and Kimoto M: MD-2, a molecule that confers
lipopolysaccharide responsiveness on Toll-like receptor 4. J Exp
Med. 189:1777–1782. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Poltorak A, He X, Smirnova I, Liu MY, van
Huffel C, Du X, Birdwell D, Alejos E, Silva M, Galanos C,
Freudenberg M, Ricciardi-Castagnoli P, Layton B and Beutler B:
Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations
in Tlr4 gene. Science. 282:2085–2088. 1998. View Article : Google Scholar
|
|
13.
|
Beutler B, Jiang Z, Georgel P, Crozat K,
Croker B and Rutschmann S: Genetic analysis of host resistance:
toll-like receptor signaling and immunity at large. Annu Rev
Immunol. 24:353–389. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
O'Neill LA and Bowie AG: The family of
five: TIR-domaincontaining adaptors in Toll-like receptor
signalling. Nat Rev Immunol. 7:353–364. 2007.
|
|
15.
|
Akira S, Uematsu S and Takeuchi O:
Pathogen recognition and innate immunity. Cell. 124:783–801. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Chen JH, Uto T, Tanigawa S, Kumamoto T,
Fujii M and Hou DX: Expression profiling of genes targeted by
bilberry (Vaccinium myrtillus) in macrophages through DNA
microarray. Nutr Cancer. 60:43–50. 2008. View Article : Google Scholar
|
|
17.
|
Tao JY, Zhao L, Huang ZJ, Zhang XY, Zhang
SL, Zhang QG, Fei-Xiao, Zhang BH, Feng QL and Zheng GH:
Anti-inflammatory effects of ethanol extract from Kummerowia
striata (Thunb.) Schindl on LPS-stimulated RAW 2647 cell.
Inflammation. 31:154–166. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Beaty SR, Rose CE Jr and Sung SS: Diverse
and potent chemokine production by lung CD11b high dendritic cells
in homeostasis and in allergic lung inflammation. J Immunol.
178:1882–1895. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Huang H, Chang EJ, Lee Y, Kim JS, Kang SS
and Kim HH: A genome-wide microarray analysis reveals
anti-inflammatory target genes of paeonol in macrophages. Inflamm
Res. 26:189–198. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Alcorn JF and Wright JR: Surfactant
protein A inhibits alveolar macrophage cytokine production by
CD14-independent pathway. Am J Physiol Lung Cell Mol Physiol.
286:129–136. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Fassbender K, Mielke O, Bertsch T,
Muehlhauser F, Hennnerici M, Kurimoto M and Rossol S:
Interferon-gamma-inducing factor (IL-18) and interferon-gamma in
inflammatory CNS diseases. Neurology. 53:1104–1106. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Asefa B, Klarmann KD, Copeland NG, Gilbert
DJ, Jenkins NA and Keller JR: The interferon-inducible p200 family
of proteins: a perspective on their roles in cell cycle regulation
and differentiation. Blood Cells Mol Dis. 32:155–167. 2004.
View Article : Google Scholar : PubMed/NCBI
|