1. Introduction
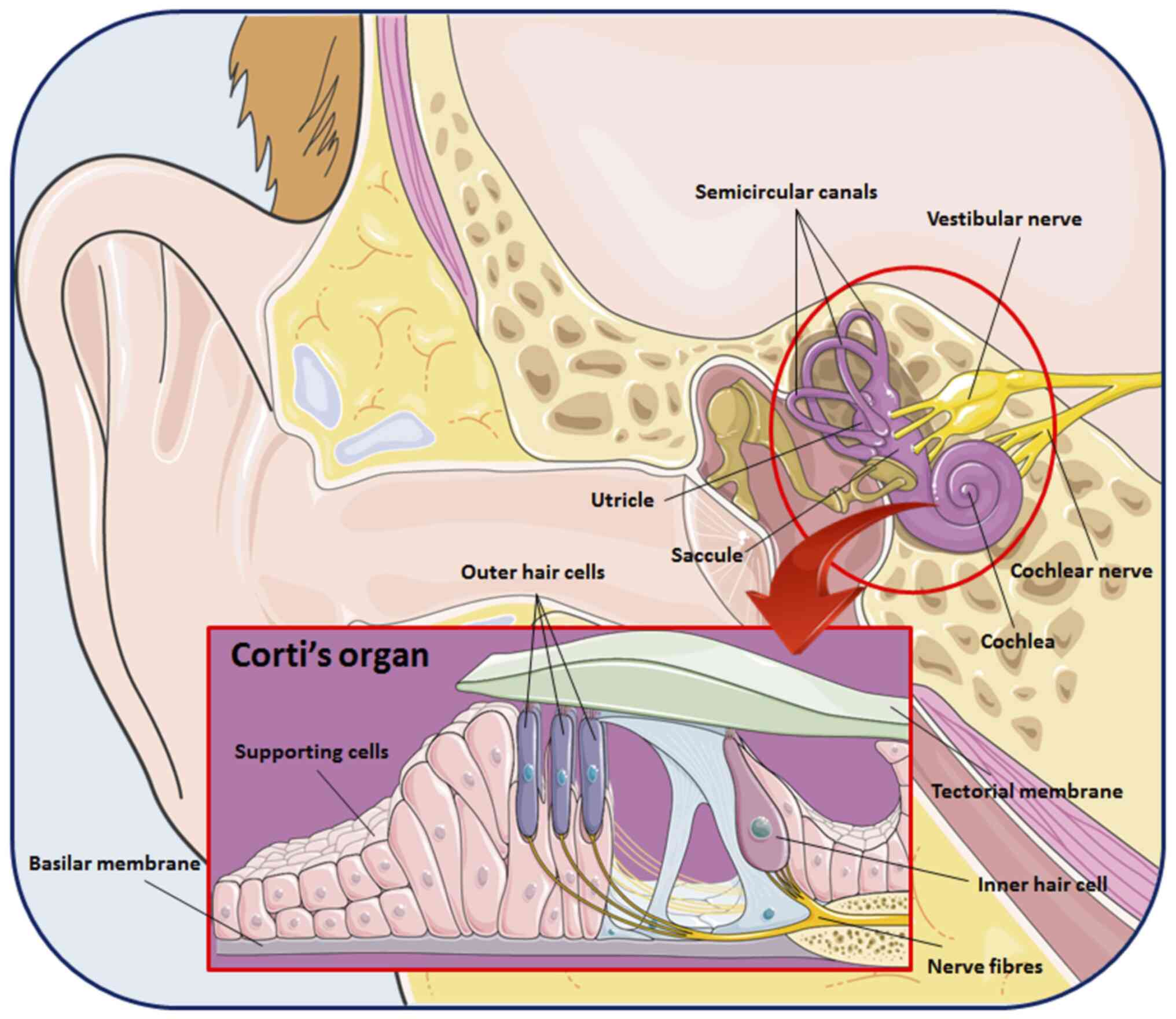
The ear is the organ of the human body responsible
for auditory function and maintaining balance. Anatomically, it
consists of external and internal parts (outer, middle and inner
ear), forming a complex structure. The auricle represents the
visible part of the outer ear, whose function is to collect
airborne sound waves and funnel them into the ear canal towards the
eardrum (1). From the tympanic
membrane, sound waves are transmitted to the oval window via an
ossicular chain (malleus, incus and stapes) suspended within the
air-filled middle ear cavity. The vibration of the three middle ear
bones plays a key role in amplifying the auditory stimuli, which
are conducted to a fluid-filled spiral structure, the cochlea
(2,3). Within the cochlear structure of the
inner ear, sound waves are transduced into electrochemical impulses
by the auditory hair cells of the organ of Corti that in turn send
these signals to the brain via the acoustic nerves (4,5). For a
proper function of the auditory system, the development of all the
parts must be parallel and in strict coordination from the outer to
the inner ear. Any alterations in this composite structure can lead
to ear defects and hearing loss (6).
Apart from hearing loss, other pathological
conditions can affect the ear, including dizziness, tinnitus, loss
of balance and inflammatory processes. These disorders occur in a
large percentage of the population worldwide and may have a
multi-factorial etiology (7,8). As
reported in the literature, aging and environmental factors (noise
exposure, chemical and physical factors) appear to be involved in
the onset of hearing-related disorders (9-11).
Moreover, even epigenetic alterations due to DNA methylation and
histone modifications may play critical roles in hearing
impairments and ear cancer (12).
In this context, microRNAs (miRNAs/miRs) have
recently attracted the attention of the scientific community for
their key role in gene regulation and expression, as well as for
the interaction with various environmental factors able to modulate
their expression levels (13-15).
miRNAs are a wide class of small non-coding single-stranded RNAs,
containing ~18-24 nucleotides, identified both in animals and
plants (16,17). These highly conserved RNAs present a
complex biogenesis that occurs not only in the nucleus, but also in
the cytoplasm (Fig. 1). At the
nuclear level, RNA polymerase II (RNA Pol II) transcribes the
majority of the miRNAs from the non-coding region of DNA to obtain
primary miRNAs (pri-miRNAs), while some miRNAs are transcribed by
another polymerase (RNA Pol III) (18). Specifically, pri-miRNAs have a
stem-loop structure and are characterized by a 5'-terminal
7-methylguanosine cap (m7G) and a 3'-terminal poly(A)
tail (19). Within the nucleus,
ribonuclease III Drosha modifies pri-miRNAs through a cleavage in
association with DiGeorge syndrome critical region gene 8 (DGcR8)
protein, to generate precursor-miRNAs (pre-miRNAs) (20). Subsequently, pre-miRNAs are
transported to the cytoplasm via exportin-5, another ribonuclease
III that acts synergistically with ras-related nuclear
protein-guanosine triphosphate (Ran-GTP), recognizing the
3'-terminal of pre-miRNAs (21,22).
In the cytoplasm, pre-miRNAs are cleaved by ribonuclease III Dicer
with the aid of transactivation response rna binding protein and
interferon-inducible double-stranded RNA-dependent activator
proteins to form a mature double-stranded miRNA, indicated as
miRNA-miRNA* duplex (23). Finally,
following the division of the miRNA-miRNA* duplex, the miRNA strand
is incorporated into the RNA-induced silencing complex (RISC),
while the miRNA* strand is degraded. Of note, the RISC is
characterized by several proteins involved in the mRNA silencing
process, such as Argonaute (Ago) proteins (24,25).
In particular, the miRNA strand associated with the RISC binds to
its complementary sequence in the 3'-untranslated region of the
target mRNA, either promoting the degradation of mRNA or inhibiting
protein translation (26,27).
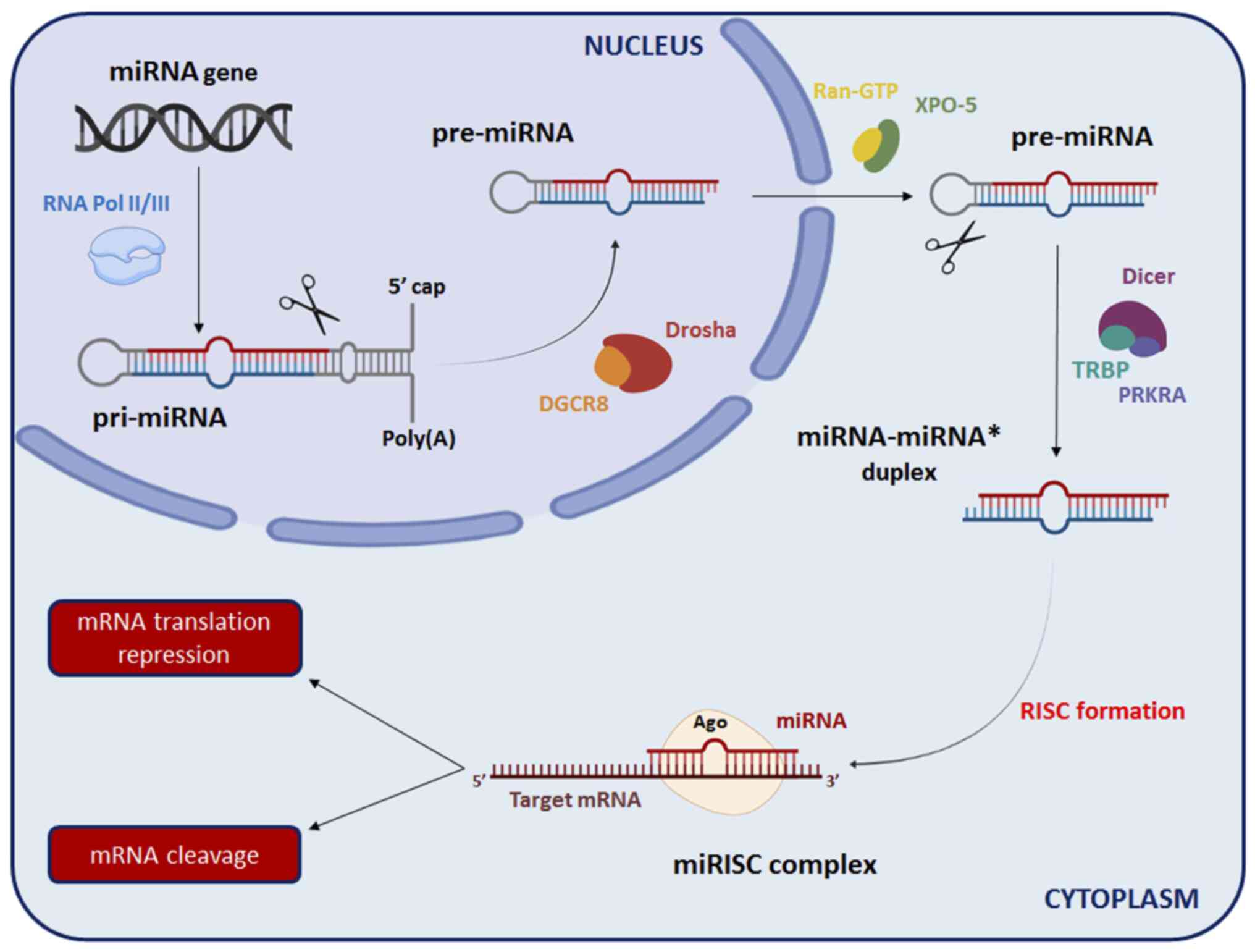 | Figure 1Schematic representation of miRNA
biosynthesis. Ago, Argonaute proteins; DGCR8, DiGeorge syndrome
critical region gene 8; miRNA, microRNA; pre-miRNA, precursor
miRNA; PRKRA, Interferon-Inducible double-stranded RNA-dependent
activator; pri-miRNA, primary miRNA; Ran-GTP, ras-related nuclear
protein-guanosine triphosphate; RISC, RNA-induced silencing
complex; RNA Pol II/III, RNA polymerase II/III; TRBP,
transactivation response RNA binding protein; XPO-5,
exportin-5. |
Various studies have demonstrated that miRNA
dysregulation is associated with a variety of pathological
conditions, including tumors, neurological disorders and other
acute and chronic diseases (28-31).
Of note, some studies have also demonstrated that a number of
miRNAs are involved in inner ear development, differentiation and
the survival of inner ear hair cells, suggesting that changes in
their expression levels may play a critical role in the onset of
hearing disorders (32-35).
On these bases, the aim of the present review article was to report
a summary of the studies that have been conducted over the past
years in order to provide a better understanding of the role of
miRNAs in inner ear development. In addition, the present review
article focused on the potential association between altered miRNA
expression and hearing loss, ear inflammatory processes,
drug-induced ototoxicity and cancers affecting the auditory system,
such as vestibular schwannoma.
2. Regulatory role of miRNAs in inner ear
development
The inner ear is a labyrinthine structure composed
of three semicircular canals, the utricle that connects these three
canals and the saccule. In addition, it is characterized by the
cochlea, a fluid-filled spiral structure able to convert sounds
into hearing through the organ of Corti (Fig. 2) (36,37).
The development of the inner ear is a complex process that requires
several steps, beginning from the transformation of the embryonic
ectoderm to the formation of functionally mature structures. This
process also includes an accurate histological organization of
supporting cells and mechanosensory hair cells, which transduce
hearing impulses to the neurons (38,39).
As widely described in the literature, a multitude
of genes and transcription factors, including atonal BHLH
transcription factor 1, neurogenin 1, neuronal differentiation 1,
fibroblast growth factor, Hedgehog, SRY-box transcription factor 2
(Sox2) and eyes absent 1, as well as the Notch and Wnt signaling
pathways, are responsible for this coordinated transition from
precursor cells to high differentiated cell types (40-43).
However, several studies have highlighted that inner ear
development is not exclusively regulated by proteins, indicating
that miRNAs may also play a key role in the development and
function of the auditory system.
For example, Jiang et al (44) conducted an in vitro study on
mouse neural stem cells to evaluate the expression levels of miRNAs
during neuronal differentiation. Notably, since miR-124 exhibited
the highest increase in expression among the considered miRNAs
(six-fold compared to baseline conditions), the research group
transfected the inner ear neural stem cells with RNA
oligoribonucleotides (non-specific miRNA, miR-124 mimics and
miR-124 inhibitor). Notably, the differentiation of neural cells
was reduced by the knockdown of miR-124, whereas the overexpression
of miR-124 resulted in a significant increase. In addition, the
expression levels of receptor kinase B (TrkB), a receptor involved
in the regulation of neurogenesis and survival of neurons, and cell
division control protein 42 homolog (Cdc42), a GTPase, which
controls the neurite extension of spiral ganglion neurons, were
also investigated (44). Notably,
Jiang et al (44) found that
the overexpression of miR-124 enhanced the levels of both these
factors, suggesting that this miRNA may regulate neural
differentiation by regulating TrkB and Cdc42.
Similarly, Du et al (45) assessed the spatial expression of
miRNAs during inner ear development using mouse embryos. In brief,
miR-183 family members (miR-183/96/182) and miR-15a were more
expressed in the sensory epithelium, while miR-194 was
predominantly detected in the spiral ganglion. Since miR-194
exhibited a dynamic expression during inner ear development, they
focused on this miRNA to investigate its function. Specifically,
the transfection of suspended cells from the spiral ganglion
highlighted that the overexpression of miR-194 affected neuron
morphology, including a disorganized somatodendritic cytoskeleton
(45). In addition, to further
elucidate the mechanisms of action, the research group evaluated
the expression of Ras homolog B (RhoB), a member of the Rho GTPase
family functionally connected to microtubule associated protein 1A
and actin regulatory proteins. Of note, miR-194 overexpression
induced a significant reduction in RhoB expression, while the
protein expression levels were enhanced following miRNA knockdown.
Overall, these results suggest that miR-194 may be crucial to the
morphogenesis of spiral ganglion neurons by targeting RhoB
(45).
Subsequently, the physiological functions of the
miR-183/96/182 cluster have also been investigated. For this
purpose, Geng et al (46)
used knockout mice in which the clustered gene had been
inactivated. As demonstrated by the auditory brain stem response
test, the loss of function of the miR-183/96/182 cluster was
strictly related to hearing loss in the knockout group compared to
the wild-type group. At the same time, the cochlear hair cells of
knockout mice exhibited severe morphological alterations, including
abnormal apices, lack of kinocilia and immature stereocilia of
equal length. Of note, due to severe defects that characterized the
hair cells, the organ of Corti of mutant mice exhibited no
functional mechanoelectrical transduction. Moreover, the research
group evaluated the expression of several predicted target genes
(46). Specifically, chloride
intracellular channel 5 protein, Radixin, ezrin, Rac family small
GTPase 1, myosin 1C and Sox2 levels were upregulated in the cochlea
of the knockout group compared to the controls. In summary, that
study highlighted that the inhibition of the miR-183/96/182 cluster
led to severe alterations in the morphology of hair cells and the
function of the organ of Corti in mice, confirming the importance
of miRNAs in the development of hearing functions (46).
3. Role of miRNAs in hearing loss
Hearing loss is a public health concern affecting a
large percentage of the worldwide population and is more frequent
in low- and middle-income countries than in industrialized ones
(47). According to the World
Health Organization (WHO), 466 million individuals (34 million
children) are currently affected by hearing loss, with a 50%
increase in hearing disorders expected for the year 2050 compared
to 2021 (~700 million individuals) (48).
It is possible to classify hearing loss in three
clinical forms: Conductive, sensorineural and mixed. The conductive
type is related to a disturbance in the transmission of sound waves
from the outer to the inner ear, which leads to difficulties in
speech comprehension and sound localization, as well as a negative
effect on balance (49,50). Sensorineural hearing loss represents
the most common clinical form among children and adults. It is due
to the functional damage of the inner ear hair cells or the
dysfunction of cochlear neural structures (51). As regards the mixed form, it occurs
when both conductive and sensorineural features are present
simultaneously (52).
Over the years, several factors have been implicated
in the development of hearing loss, whose co-occurrence can result
in severe impairments of the auditory system. Among the exogenous
factors, noise exposure, smoking and chemotherapeutic drugs are the
most well-known, while genetic mutations and aging represent the
main involved endogenous factors (53-57).
Of note, a number of recent studies have suggested that an altered
regulation of genes mediated by miRNA expression may also
contribute to hearing loss (58-61).
Xue et al (58) examined the potential association
between miR-29b and the apoptosis of cochlear hair cells, one of
the principal causes of age-related hearing loss. Through an in
vivo study on a mouse model (C57BL/6), they observed that aged
mice were characterized by a marked reduction in cochlear hair
cells and higher miR-29b expression levels compared to the controls
(young mice). At the same time, aged mice exhibited a significant
decrease in Sirtuin 1 (SIRT1) and proliferator-activated
receptor-gamma coactivator 1α (PGC-1α) expression levels, a
deacetylase that regulates the intracellular oxidative stress and a
coregulator involved in oxidative metabolism, respectively
(58). The hypothesis that miR-29b
may induce cell apoptosis and target SIRT1 and PGC-1α was confirmed
by the transfection of HEI-OC1 cells. Although further studies are
required to better elucidate the mechanisms of action, the
miR-29b/SIRT1/PGC-1α signaling pathway may represent a novel
potential target for the treatment of age-related hearing loss
(58).
Similarly, the role of miR-34a in the pathogenesis
of age-related hearing loss was evaluated in another study. Using
C57BL/6 mice, Pang et al (59) noted that aging was strictly related
not only to cochlear hair cell loss, but also to the upregulation
of miR-34a expression. Moreover, an altered autophagic flux was
observed in the cochlea of aged mice compared to young mice, as
demonstrated by the variation in the levels of autophagy markers
LC3-II and p62. To further investigate the mechanisms of action,
the authors transfected the HEI-OC1 mouse auditory cell line. Of
note, the overexpression of miR-34a reduced autophagy-related
protein 9A (ATG9A) expression, while the knockout of miR-34a
exerted the opposite effect. The obtained data highlight the
involvement of miRNAs in the progression of age-related hearing
loss, indicating that the downregulation of ATG9A by miR-34a may
play a crucial role in the impairment of the autophagic flux
(59).
Of note, Li et al (60) focused on miRNA serum levels in
subjects with occupational noise-induced hearing loss and healthy
controls. Among the considered miRNAs, hsa-miR-4652-3p expression
was downregulated in subjects with hearing isues compared to the
controls, while the levels of hsa-miR-3162-5p, hsa-miR-4484 and
hsa-miR-1229-5p were upregulated. However, only the hsa-miR-1229-5p
serum levels were significantly increased. These researchers then
conducted an in vitro study (293T cells) to validate the
predicted targets of miR-1229-5p. Notably, the overexpression of
miR-1229-5p markedly reduced mitogen-activated protein kinase 1
(MAPK1) levels (60). Taken
together, these results indicate that miR-1229-5p may represent a
novel diagnostic biomarker for noise exposure-related hearing loss.
In addition, since MAPK1 has been described as a key molecule in
regulating human genetic deafness, miR-1229-5p may be actively
involved in the progression of hearing disorders by inhibiting
MAPK1 expression (60).
Recently, the potential association between miRNAs
and sudden hearing loss was also investigated. In this regard,
Nunez et al (61) conducted
a prospective cohort study on a group of sudden sensorineural
hearing loss patients and a control group consisting of normal
hearing subjects. Briefly, the research group analyzed the
circulating miRNA expression profiles observing that eight miRNAs
(miR-375-3p, miR-195-5p, miR-128-3p, miR-30a-3p, miR-140-3p,
miR-590-5p, miR-132-3p and miR-186-5p) were differentially
expressed between the two groups examined. In addition, they noted
that the genes involved in phosphoinositide 3-kinase (PI3K)/protein
kinase B (AKT) and MAPK signaling pathways represented the putative
targets of the identified miRNAs. These findings confirm the
involvement of miRNAs in hearing disorders, supporting their
application as novel circulating biomarkers. However, further
studies are warranted in order to provide a better understanding of
the role of miRNAs in hearing loss (61).
A summary of miRNAs that have been investigated in
hearing loss is presented in Table
I.
 | Table ImiRNA expression in hearing loss. |
Table I
miRNA expression in hearing loss.
| Disease model | Study design/sample
type | Up/downregulated
miRNAs | Targets | (Refs.) |
|---|
| Age-related hearing
loss | In vivo
(mouse cochlea) In vitro (HEI-OC1 cells) | miR-29b (↑) | SIRT1, PGC-1α | (58) |
| Age-related hearing
loss | In vivo
(mouse cochlea) In vitro (HEI-OC1 cells) | miR-34a (↑) | ATG9A | (59) |
| Noise-induced
hearing loss | Cohort (blood
serum) In vitro (HEK293T cells) | miR-128-5p (↑) | MAPK1 | (60) |
| Sudden
sensorineural hearing loss | Cohort (blood
serum) | miR-375-3p,
miR-195-5p, miR-128-3p, miR-30a-3p, miR-140-3p, miR-590-5p,
miR-132-3p, miR-186-5p (differentially expressed) | PI3K/AKT,
MAPKs | (61) |
4. Involvement of miRNAs in ear infection
and inflammatory processes
Ear infection represents one of the most common
disorders affecting individuals of all ages. If left untreated,
these infections can cause hearing loss and other auditory system
complications, such as vestibular dysfunction. Depending on the
triggering pathogen, ear infections can be classified into viral,
bacterial and fungal (62).
Viruses frequently affect inner ear structures,
inducing the development of inflammatory processes and enhancing
the susceptibility to infection caused by other pathogens (63). Among viral infections, congenital
cytomegalovirus infection is the leading cause of hearing loss in
children (64). Other viruses, such
as Rubella, Herpes simplex virus and Zika virus, have been
described as pathogens strictly related to hearing loss in newborns
and adults (65-67).
As regards ear bacterial infections, Pseudomonas
aeruginosa, Streptococcus pneumonia, Haemophilus
influenzae, Staphylococcus aureus and Moraxella
catarrhalis are the most frequently involved pathogens
(68-72).
Of note, bacteria-induced meningitis has a cytotoxic effect on the
cochlea and may lead to neural damage (73). In addition, even the middle ear can
be affected by bacterial-induced inflammatory processes, of which
otitis media with effusion and chronic suppurative otitis media are
the most severe forms (74,75).
Compared to other ear infectious processes, fungi
are usually involved in otitis externa, a cutis/subcutis
inflammation of the external auditory canal. As reported in the
literature, Aspergillus and Candida are the main
causative genera of otomycosis (76,77).
Of note, several factors can promote the progression of otitis
externa, such as humidity, swimming and diving (78).
Over the past few years, the involvement of miRNAs
and epigenetic modifications in the pro-inflammatory and
anti-inflammatory processes have been investigated (79,80);
however, the role of miRNAs in the pathogenesis of ear inflammation
has not yet been clarified.
For example, Rudnicki et al (81) demonstrated that miR-224 plays a
crucial role in inner ear inflammation, downregulating pentraxin 3
(Ptx3), a protein involved in the immune response, whose release is
regulated by nuclear factor κB (NF-κB). Briefly, the transfection
of 293T and NIH3T3 cells revealed that the overexpression of
miR-224 was strictly related to the decrease in Ptx3 levels
(81). This inverse association was
also evaluated through an in vivo experiment on a mouse
model of inner ear inflammation (otitis interna). Notably, the
injection of lipopolysaccharide into the scala tympani of mice
induced an increase in miR-224 expression and Ptx3 mRNA levels,
while Ptx3 protein levels exhibited no significant variations
compared to the controls (81). As
was reported in that study, since even the miR-224 promoter has a
binding site for NF-κB, both miR-224 and Ptx3 may be recruited
during inflammation. Taken together, these findings suggest that
miR-224 and Ptx3 form a feedback loop, where miR-224 downregulates
Ptx3 reducing the inflammatory process (81).
Even miR-146 appears to be involved in ear
inflammatory processes. In this regard, Samuels et al
(82) conducted an in vitro
study on human middle ear epithelial cells (HMEECs) to evaluate the
expression of miR-146 following treatment with IL-1β or tumor
necrosis factor α (TNFα). Notably, miR-146a and miR-146b expression
levels were upregulated by treatment with these pro-inflammatory
cytokines (82). Moreover, they
evaluated miR-146 expression in middle ear biopsies of patients
with otitis media. Specifically, miR-146a was upregulated in
patients with otitis media with effusion and recurrent otitis
media, while miR-146b expression was significantly higher only in
the recurrent group compared to the controls (82). At the same time, that research group
noted that miR-146a and miR-146b expression was inversely related
to TNF receptor-associated factor (TRAF6) levels, a member of the
Toll-like receptor signaling pathway. In summary, that study
highlighted that miR-146a and miR-146b may play a key role in the
pathogenesis of otitis media by targeting TRAF6(82).
Recently, the role of miR-210 in otitis media was
also investigated. Specifically, Zhang et al (83) observed that miR-210 levels were
significantly downregulated in middle ear effusion in the serum of
patients affected by otitis media. On the contrary, the expression
levels of pro-inflammatory cytokines, as well as nitric oxide and
vascular endothelial growth factor (VEGF) were increased in the
otitis group compared to the controls (83). These authors then conducted a
functional assay to evaluate the potential regulatory role of the
aforementioned miRNA. Of note, the transfection of HMEECs with
miR-210 mimics led to a significant reduction in the expression
levels of pro-inflammatory cytokines and cell apoptosis, whereas
cell viability was increased. In addition, miR-210 overexpression
was also related to the downregulation of hypoxia-inducible
factor-1α, a transcriptional factor involved in pro-inflammatory
gene expression (83). Overall, the
obtained results suggested that miR-210 overexpression may
represent the starting point for a novel therapeutic approach with
which to reduce ear inflammatory processes. However, further
studies are required to better understand the mechanisms of action
of this miRNA (83). Of note,
inflammatory processes are not only sustained by microbial
infections. Indeed, it has been demonstrated that aging,
occupational or environmental noise and drugs can induce ototoxic
damage sustained by inflammatory processes (84-86).
As regards drug-induced ototoxicity, an in-depth description is
provided in the following chapter.
Aging is recognized as a main risk factor for
various pathologies. As already mentioned in the previous chapter,
miRNAs and inflammation play a pivotal role in age-related hearing
loss. In this context, the mechanisms through which aging is able
to alter the expression levels of miR-34a, which is involved in the
regulation of various processes, including inflammation and
autophagy, have been previously demonstrated in mice (59).
Similarly, it has also been demonstrated that
noise-related inflammation is responsible for miRNA dysregulation
and, in turn, hearing loss. As widely described in the previous
chapter, the dysregulation of different miRNAs has been associated
with noise-induced hearing loss (Table
I) (60). Among the miRNAs
dysregulated due to environments with high levels of noise,
miR-4652-3p, miR-3162-5p, miR-4484 and miR-1229-5p were recently
identified and associated with the regulation of pathways
notoriously involved in inflammatory processes, such as the MAPK
pathway (60).
Table II summarizes
the expression of miRNAs during ear inflammatory processes
(Table II).
 | Table IIAltered miRNA expression during ear
inflammation. |
Table II
Altered miRNA expression during ear
inflammation.
| Disease model | Study design/sample
type | Up/downregulated
miRNAs | Targets | (Refs.) |
|---|
| Otitis interna | In vivo
(mouse inner ear) In vitro (293T, NIH3T3 cells) | miR-224 (↑) | Ptx3 | (81) |
| Otitis media with
effusion and recurrent | Case-control
(middle ear biopsies) In vitro (HMEECs) | miR-146a, miR-146b
(↑) | TRAF6 | (82) |
| Otitis media with
effusion | Case-control
(middle ear effusion and blood serum) In vitro (HMEECs) | miR-210 (↓) | HIF-1α | (83) |
5. miRNAs and drug-induced ototoxicity
Ototoxicity is a pharmacological temporary or
permanent adverse event that affects the auditory system, causing a
dysfunction of the inner ear structures, particularly cochlear and
vestibular tissues (87,88). Over the years, several studies have
reported a wide spectrum of chemicals associated with ototoxicity
through both direct and indirect mechanisms mediated by miRNAs. In
this context, it has been demonstrated that chronic pesticide
exposure is associated with the dysregulation of various miRNAs
responsible for the regulation of key genes involved in
inflammatory processes or cellular homeostasis, and in turn, with
potential ototoxic effects (13,89-91).
Apart from pesticides, drugs used for the treatment of various
diseases may also exert ototoxic effects. Among platinum-based
anticancer drugs, cisplatin, carboplatin and oxaliplatin are the
most ototoxic (92,93). Moreover, aminoglycoside antibiotics
(neomycin, gentamicin, kanamycin, streptomycin and amikacin), loop
diuretics and macrolide antibiotics have been also described for
their ototoxic potential (94-96).
Typical symptoms of drug-induced ototoxicity are
represented by hearing loss, tinnitus, dizziness and loss of
balance. These hearing disorders can occur during or at the end of
the treatment period with a gradual or sudden appearance (97). Although ototoxic drugs can
compromise auditory function in all age groups, some studies have
reported that children are exposed to a higher risk of impaired
cognitive performance and language than adults (98-100).
Of note, the incidence of ototoxicity and severity is dependent on
several factors, including the selection of the therapeutic agent
and its pharmacokinetic, the route of administration and dose,
concomitant medications and genetic factors (101,102).
In this context, an increasing number of studies
have focused on miRNAs and their potential involvement in
drug-induced ototoxicity to discover novel early diagnostic
biomarkers and otoprotective therapeutic approaches.
For example, Kim et al (103) investigated the role of miRNAs in
neomycin-induced ototoxicity using zebrafish embryos. Specifically,
miRNA expression levels were evaluated during inner ear hair cell
regeneration following treatment with neomycin. Microarray analysis
revealed that the miR-183 cluster (miR-183/-96/-182) was
significantly upregulated at 12-24 h in the treatment group, while
the levels returned to basal levels after 48 h. The reported data
suggest that the overexpression of these miRNAs may be crucial in
repairing cochlear hair cell damage (103). Moreover, the same research group
noted that the knockdown of miR-183 partially affected the
regeneration process of hair cells. Taken together, these results
demonstrated that the miR-183 cluster exerts an otoprotective
effect able to contrast aminoglycoside antibiotics-induced hair
cell damage in zebrafish. This miRNA cluster may represent the
starting point for the development of novel therapeutic approaches
(103).
Circulating miRNA levels as novel diagnostic
biomarkers of drug-induced ototoxicity have also been investigated.
By using a mouse model of ototoxicity induced by the administration
of kanamycin and furosemide, Lee et al (104) detected the serum levels of
circulating miRNAs. Notably, they found that miR-205 expression was
higher in the ototoxic group compared to the controls (four-fold
increase until day 14). They then focused on the inner ear,
observing a significant difference in miR-205 levels among the
cochlear components (104).
Notably, the organ of Corti exhibited a slight increase only in the
first phase (day 5), while stria vascularis was characterized by a
gradual increase in miR-205 expression until day 14. Therefore, it
can be hypothesized that the increase in serum miR-205 levels may
be due to extravasate via stria vascularis (104). Overall, the obtained results
highlight that miR-205 expression is strictly related to
antibiotics-induced ototoxicity in mice, suggesting its potential
use as a novel diagnostic biomarker of drug-related hearing
impairments (104).
Subsequently, Li et al (105) conducted an in vivo study on
zebrafish embryos to evaluate the association between β-diketone
antibiotic-induced ototoxicity and the regulatory role of miRNAs in
hearing. Firstly, they noted a decreased response to acoustic
stimuli in the treatment group compared to the controls. At the
same time, treatment with β-diketone antibiotics also induced the
downregulation of miR-96 and miR-184 expression in zebrafish
otoliths (105). Subsequently, to
confirm the regulatory role of these miRNAs in hearing development,
the research group evaluated the effects of their inhibition and
overexpression. Of note, only the overexpression of miR-96 restored
hair cells following exposure to β-diketone antibiotics, indicating
that this miRNA plays a critical role in hearing development. On
the other hand, miR-184 overexpression exerted no effect on hair
cells, demonstrating that miR-184 was only involved in the
development of otic vesicles. These findings on miR-96 and miR-184
may provide the theoretical bases for the development of novel
intervention strategies against drug-induced ototoxicity (105).
Recently, the role of miR-182 on the hearing loss
induced by ototoxic drugs was also investigated. In this regard,
Chen et al (106)
demonstrated that miR-182 was able to reduce harmful effects on the
auditory system induced by kanamycin and furosemide. In brief,
pre-treatment with miR-182 attenuated the permanent threshold shift
caused by the concomitant administration of kanamycin and
furosemide in rats. At the same time, miR-182 overexpression
significantly reduced hair cell death, protecting stereocilia and
enhancing PI3K regulatory subunit p85α levels (106). In summary, that study suggested
that miR-182 played a key role in protecting the auditory function
against drug-induced deafness in rats. However, further in
vivo and in vitro studies are warranted in order to
better clarify the mechanisms of action and the potential targets
of this miRNA (106).
6. Altered miRNA expression in vestibular
schwannoma
Vestibular schwannoma, also known as acoustic
neuroma, is a slow-growing non-malignant tumor (average growth rate
of 1-2 mm/year) originating from the Schwann cells of the
vestibular nerve (107,108). This benign tumor, characterized by
a mortality rate of <1%, has been classified into two
typologies, sporadic and associated with neurofibromatosis type 2
(NF2) syndrome. Although vestibular schwannoma generally occurs
unilaterally, it may also appear bilaterally when related to NF2
(5%) (109-111).
The overall incidence of vestibular schwannoma is
1.4 per 100,000 individuals per year and the majority of cases
occur among middle-aged individuals of both sexes (112). Clinically, hearing loss and
tinnitus are the typical symptoms, observed in 94% and 83% of
patients, respectively. Other less frequent symptoms are
represented by vertigo (20%), facial numbness (12%) and facial
palsy (6%) (113,114).
Currently, the gold standard for the diagnosis of
vestibular schwannoma is represented by magnetic resonance imaging,
while surgical resection, fractionated radiotherapy and
radiosurgery represent the available treatment options (115,116). However, it is necessary to
identify novel therapeutic targets in order to develop less
invasive intervention strategies able to improve the management of
patient and hearing outcomes. Over the past decade, a growing body
of evidence has demonstrated that several miRNAs may be involved in
the progression of vestibular schwannoma, suggesting their
potential value as novel drug targets.
For example, miR-21 has been found to be
overexpressed in several tumor types, including vestibular
schwannoma. In this context, Cioffi et al (117) observed that the miR-21 expression
levels were higher in vestibular schwannoma samples than in the
controls. Moreover, the aberrant expression of miR-21 was strictly
related to low levels of phosphatase and tensin homolog (PTEN), a
protein that acts on the PI3K/AKT pathway, inhibiting cell
proliferation. The inverse association between miR-21 and PTEN
levels was confirmed through a functional analysis. Specifically,
the transfection of anti-miR-21 led to a significant reduction in
cell proliferation due to miR-21 knockdown (117). In summary, these data underline
that miR-21 overexpression is also involved in vestibular
schwannoma formation and growth by targeting PTEN, which results in
the hyperactivation of the PI3K/AKT pathway (117).
As previously reported by Saydam et al
(118), another miRNA that could
be involved in vestibular schwannoma is miR-7. In brief, they noted
that several miRNAs were deregulated in tumor samples compared to
normal peripheral nerve tissues derived from fresh autopsies. They
then investigated the functional significance of miR-7, the most
notably downregulated miRNA in vestibular schwannoma samples.
Specifically, the miR-7 levels were inversely related to epidermal
growth factor receptor (EGFR) and p21-activated kinase 1 (Pak1)
levels, both involved in cell division, survival and migration. In
addition, Saydam et al (118) found that miR-7 overexpression also
targeted activated Cdc42-associated kinase 1 (Ack1), a non-receptor
tyrosine kinase whose activation has been found in several tumor
types (prostate cancer, lung cancer, breast cancer, etc.) (119). These findings suggest that miR-7
could potentially act as a tumor suppressor in vestibular
schwannoma formation and growth by inhibiting the EGFR, Pak1 and
Ack1 signaling pathways (118).
Of note, Li et al (120) focused on miR-1, a small non-coding
RNA downregulated in various types of cancer (oral cancer,
colorectal cancer, lung cancer, etc.) (121-123).
As was expected, the miR-1 expression levels were significantly
decreased in vestibular schwannoma specimens compared to the
controls (normal vestibular nerve). The hypothesis that miR-1 may
play a suppressive role in this tumor was further confirmed by the
transfection of the HEI-193 cell line. Specifically, the
overexpression of miR-1 led to a significant reduction in cell
proliferation, enhancing the apoptotic process, while the knockout
of miR-1 exerted the opposite effect (120). At the same time, in vitro
experiments revealed that transfection with miR-1 mimic was
negatively related to VEGFA levels. Overall, the described results
highlight that miR-1 may play a crucial role in the progression of
vestibular schwannoma, providing a theoretical basis for the
development of novel effective treatments against this pathological
condition (120).
Recently, the potential inhibitory role of miR-205
in sporadic vestibular schwannoma was also investigated. Notably,
Yin et al (124) found that
miR-205 expression levels were lower in tumor tissues than in
normal great auricular nerves (controls). Secondly, the authors
transfected primary human vestibular schwannoma cell cultures and
the 293T cell line with miR-205 mimic to evaluate the effects of
miRNA overexpression on cell proliferation in vestibular
schwannoma. Of note, the functional assay revealed that miR-205
overexpression resulted in a significant decrease in cell growth.
Finally, Yin et al (124)
concentrated on the mechanisms of action. Specifically, they
observed that the in vitro overexpression of miR-205
significantly downregulated cyclin-dependent kinase 14 (CDK14)
levels, whose high expression was positively related to cell
growth. The obtained results demonstrate that miR-205 may play an
inhibitory role against vestibular schwannoma progression by
targeting CDK14. However, further studies are required to better
clarify the effects of miR-205 expression in this tumor and its
mechanisms of action (124).
The alterations of miRNAs involved in vestibular
Sschwannoma are summarized in Table
III.
 | Table IIIUpregulated and downregulated miRNAs
in vestibular schwannoma. |
Table III
Upregulated and downregulated miRNAs
in vestibular schwannoma.
| Study design/sample
type | Up/downregulated
miRNAs | Targets | (Refs.) |
|---|
| Case-control (tumor
tissue vs. normal vestibular nerve tissue) In vitro (primary
human VS cell cultures) | miR-21 (↑) | PTEN | (117) |
| Case-control (tumor
tissue vs. normal peripheral nerve tissue) In vitro (HEI-193
cells) | miR-7 (↓) | EGFR, Pak1,
Ack1 | (118) |
| Case-control (tumor
tissue vs. normal vestibular nerve tissue) In vitro (HEI-193
cells) | miR-1 (↓) | VEGFA | (120) |
| Case-control (tumor
tissue vs. normal great auricular nerve tissue) In vitro
(primary human VS cell cultures, 293T cells) | miR-205 (↓) | CDK14 | (124) |
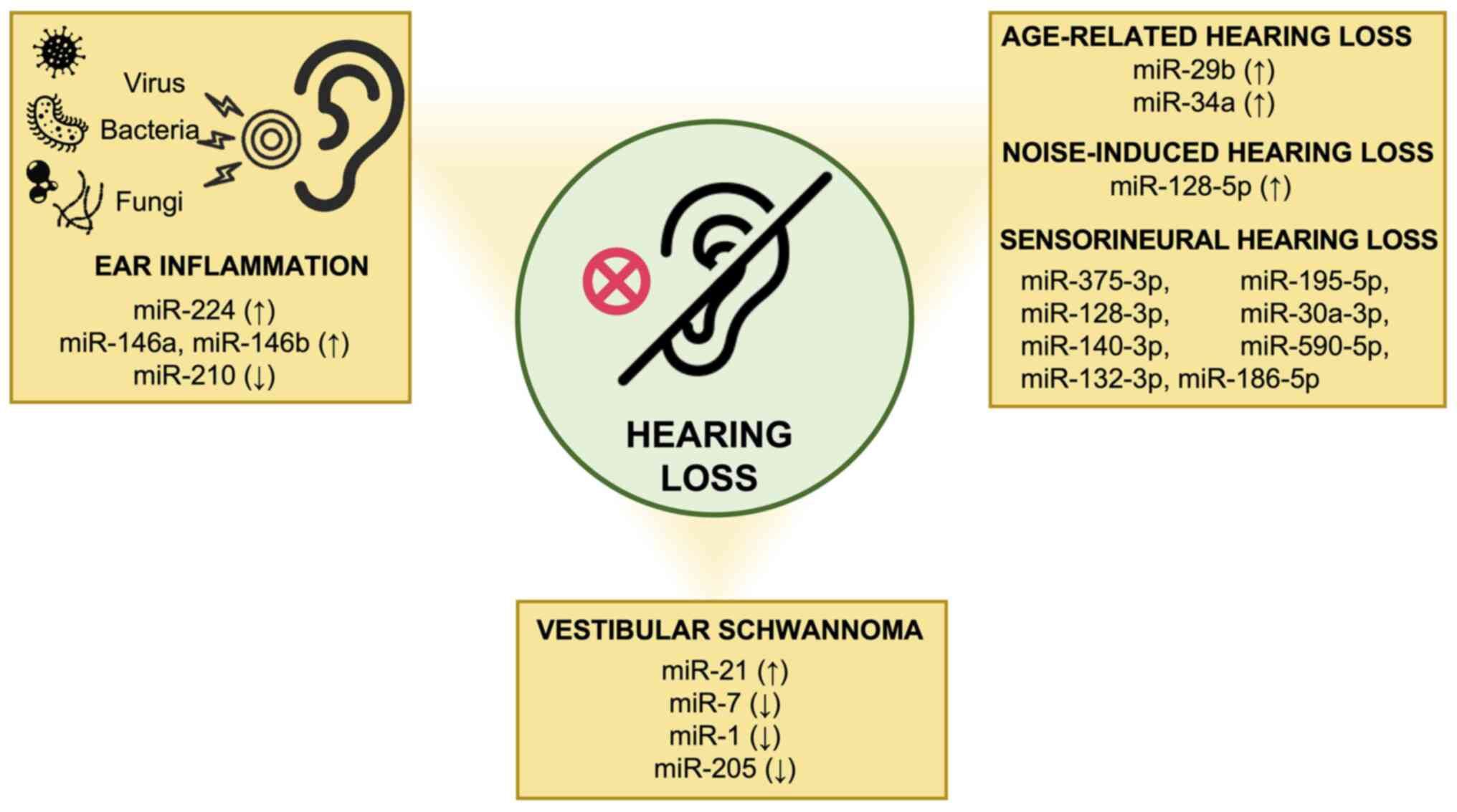
7. Conclusions and future perspectives
Hearing disorders affect an increasing number of
individuals worldwide, particularly during childhood, leading to a
significant reduction in the quality of life. Over the years, it
has been widely demonstrated that miRNAs play a key role in the
development and function of the auditory system. However, altered
miRNA expression levels appear to be involved in the progression of
hearing loss along with other factors, including aging, noise
exposure, ototoxic drugs, environmental and genetic factors
(Fig. 3). According to the studies
described in the present review article, miRNAs may be used as
novel diagnostic biomarkers for the early detection of hearing
loss. At the same time, miRNAs and related signaling pathways may
be considered the starting point for the development of novel
therapeutic approaches against hearing loss, ear inflammation and
vestibular schwannoma. Overall, although the current findings
represent a promising avenue, further in vitro and in
vivo studies are warranted in order to provide a better
knowledge of the mechanisms of action of miRNAs.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
Not applicable.
Authors' contributions
LF and AL conceptualized the manuscript. AL, CMG, GG
and SC wrote the original draft of the manuscript. LF, AL, DAS and
SC provided critical revisions. AL, MS and CL prepared the tables,
figures and critically analyzed the literature. All authors
contributed to manuscript revision and have read and approved the
final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
LF is an Editor of the journal, but had no personal
involvement in the reviewing process, or any influence in terms of
adjudicating on the final decision, for this article. The other
authors declare that they have no competing interests.
References
|
1
|
Anthwal N and Thompson H: The development
of the mammalian outer and middle ear. J Anat. 228:217–232.
2016.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Pfaff C, Schultz JA and Schellhorn R: The
vertebrate middle and inner ear: A short overview. J Morphol.
280:1098–1105. 2019.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Anthwal N, Joshi L and Tucker AS:
Evolution of the mammalian middle ear and jaw: Adaptations and
novel structures. J Anat. 222:147–160. 2013.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Goutman JD, Elgoyhen AB and Gómez-Casati
ME: Cochlear hair cells: The sound-sensing machines. FEBS Lett.
589:3354–3361. 2015.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Driver EC and Kelley MW: Development of
the cochlea. Development. 147(dev162263)2020.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Fuchs JC and Tucker AS: Development and
integration of the ear. Curr Top Dev Biol. 115:213–232.
2015.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Szmuilowicz J and Young R: Infections of
the Ear. Emerg Med Clin North Am. 37:1–9. 2019.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Sone M: Inner ear disturbances related to
middle ear inflammation. Nagoya J Med Sci. 79:1–7. 2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Uchida Y, Sugiura S, Nishita Y, Saji N,
Sone M and Ueda H: Age-related hearing loss and cognitive
decline-The potential mechanisms linking the two. Auris Nasus
Larynx. 46:1–9. 2019.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Themann CL and Masterson EA: Occupational
noise exposure: A review of its effects, epidemiology, and impact
with recommendations for reducing its burden. J Acoust Soc Am.
146(3879)2019.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Ohgami N, Iida M, Yajima I, Tamura H,
Ohgami K and Kato M: Hearing impairments caused by genetic and
environmental factors. Environ Health Prev Med. 18:10–15.
2013.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Provenzano MJ and Domann FE: A role for
epigenetics in hearing: Establishment and maintenance of auditory
specific gene expression patterns. Hear Res. 233:1–13.
2007.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Giambò F, Leone GM, Gattuso G, Rizzo R,
Cosentino A, Cinà D, Teodoro M, Costa C, Tsatsakis A, Fenga C and
Falzone L: Genetic and epigenetic alterations induced by pesticide
exposure: Integrated analysis of gene expression, microRNA
expression, and DNA methylation datasets. Int J Environ Res Public
Health. 18(8697)2021.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Filetti V, Loreto C, Falzone L, Lombardo
C, Cannizzaro E, Castorina S, Ledda C and Rapisarda V: Diagnostic
and prognostic value of three microRNAs in environmental
asbestiform fibers-associated malignant mesothelioma. J Pers Med.
11(1205)2021.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Filetti V, Falzone L, Rapisarda V,
Caltabiano R, Eleonora Graziano AC, Ledda C and Loreto C:
Modulation of microRNA expression levels after naturally occurring
asbestiform fibers exposure as a diagnostic biomarker of
mesothelial neoplastic transformation. Ecotoxicol Environ Saf.
198(110640)2020.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Macfarlane LA and Murphy PR: MicroRNA:
Biogenesis, function and role in cancer. Curr Genomics. 11:537–561.
2010.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Hammond SM: An overview of microRNAs. Adv
Drug Deliv Rev. 87:3–14. 2015.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Faller M and Guo F: MicroRNA biogenesis:
There's more than one way to skin a cat. Biochim Biophys Acta.
1779:663–667. 2008.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek
SH and Kim VN: MicroRNA genes are transcribed by RNA polymerase II.
EMBO J. 23:4051–4060. 2004.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Kwon SC, Nguyen TA, Choi YG, Jo MH, Hohng
S, Kim VN and Woo JS: Structure of Human DROSHA. Cell. 164:81–90.
2016.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Yi R, Qin Y, Macara IG and Cullen BR:
Exportin-5 mediates the nuclear export of pre-microRNAs and short
hairpin RNAs. Genes Dev. 17:3011–3016. 2003.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Wu K, He J, Pu W and Peng Y: The role of
exportin-5 in MicroRNA biogenesis and cancer. Genomics Proteomics
Bioinformatics. 16:120–126. 2018.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Koscianska E, Starega-Roslan J and
Krzyzosiak WJ: The role of Dicer protein partners in the processing
of microRNA precursors. PLoS One. 6(e28548)2011.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Kobayashi H and Tomari Y: RISC assembly:
Coordination between small RNAs and Argonaute proteins. Biochim
Biophys Acta. 1859:71–81. 2016.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Sarshad AA, Juan AH, Muler AIC,
Anastasakis DG, Wang X, Genzor P, Feng X, Tsai PF, Sun HW, Haase
AD, et al: Argonaute-miRNA complexes silence target mRNAs in the
nucleus of mammalian stem cells. Mol Cell. 71:1040–1050.e8.
2018.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Behm-Ansmant I, Rehwinkel J and Izaurralde
E: MicroRNAs silence gene expression by repressing protein
expression and/or by promoting mRNA decay. Cold Spring Harb Symp
Quant Biol. 71:523–530. 2006.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Fabian MR and Sonenberg N: The mechanics
of miRNA-mediated gene silencing: A look under the hood of miRISC.
Nat Struct Mol Biol. 19:586–593. 2012.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Crimi S, Falzone L, Gattuso G, Grillo CM,
Candido S, Bianchi A and Libra M: Droplet Digital PCR analysis of
liquid biopsy samples unveils the diagnostic role of
hsa-miR-133a-3p and hsa-miR-375-3p in oral cancer. Biology (Basel).
9(379)2020.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Falzone L, Grimaldi M, Celentano E,
Augustin LSA and Libra M: Identification of modulated MicroRNAs
associated with breast cancer, diet, and physical activity. Cancers
(Basel). 12(2555)2020.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Candido S, Lupo G, Pennisi M, Basile MS,
Anfuso CD, Petralia MC, Gattuso G, Vivarelli S, Spandidos DA, Libra
M and Falzone L: The analysis of miRNA expression profiling
datasets reveals inverse microRNA patterns in glioblastoma and
Alzheimer's disease. Oncol Rep. 42:911–922. 2019.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Yeh CH, Moles R and Nicot C: Clinical
significance of microRNAs in chronic and acute human leukemia. Mol
Cancer. 15(37)2016.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Mahmoodian Sani MR,
Hashemzadeh-Chaleshtori M, Saidijam M, Jami MS and Ghasemi-Dehkordi
P: MicroRNA-183 family in inner ear: Hair cell development and
deafness. J Audiol Otol. 20:131–138. 2016.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Sekine K, Matsumura T, Takizawa T, Kimura
Y, Saito S, Shiiba K, Shindo S, Okubo K and Ikezono T: Expression
profiling of MicroRNAs in the inner ear of elderly people by
real-time PCR quantification. Audiol Neurootol. 22:135–145.
2017.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Van den Ackerveken P, Mounier A, Huyghe A,
Sacheli R, Vanlerberghe PB, Volvert ML, Delacroix L, Nguyen L and
Malgrange B: The miR-183/ItgA3 axis is a key regulator of
prosensory area during early inner ear development. Cell Death
Differ. 24:2054–2065. 2017.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Cao H, Shi J, Du J, Chen K, Dong C, Jiang
D and Jiang H: MicroRNA-194 regulates the development and
differentiation of sensory patches and statoacoustic ganglion of
inner ear by Fgf4. Med Sci Monit. 24:1712–1723. 2018.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Khan S and Chang R: Anatomy of the
vestibular system: A review. NeuroRehabilitation. 32:437–443.
2013.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Ekdale EG: Form and function of the
mammalian inner ear. J Anat. 228:324–337. 2016.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Hudspeth AJ: Integrating the active
process of hair cells with cochlear function. Nat Rev Neurosci.
15:600–614. 2014.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Kopecky BJ, Jahan I and Fritzsch B:
Correct timing of proliferation and differentiation is necessary
for normal inner ear development and auditory hair cell viability.
Dev Dyn. 242:132–147. 2013.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Zhong C, Fu Y, Pan W, Yu J and Wang J:
Atoh1 and other related key regulators in the development of
auditory sensory epithelium in the mammalian inner ear: Function
and interplay. Dev Biol. 446:133–141. 2019.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Elliott KL, Pavlínková G, Chizhikov VV,
Yamoah EN and Fritzsch B: Development in the mammalian auditory
system depends on transcription factors. Int J Mol Sci.
22(4189)2021.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Shin JO, Ankamreddy H, Jakka NM, Lee S,
Kim UK and Bok J: Temporal and spatial expression patterns of
Hedgehog receptors in the developing inner and middle ear. Int J
Dev Biol. 61:557–563. 2017.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Żak M, Klis SF and Grolman W: The Wnt and
Notch signalling pathways in the developing cochlea: Formation of
hair cells and induction of regenerative potential. Int J Dev
Neurosci. 47:247–258. 2015.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Jiang D, Du J, Zhang X, Zhou W, Zong L,
Dong C, Chen K, Chen Y, Chen X and Jiang H: miR-124 promotes the
neuronal differentiation of mouse inner ear neural stem cells. Int
J Mol Med. 38:1367–1376. 2016.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Du J, Zhang X, Cao H, Jiang D, Wang X,
Zhou W, Chen K, Zhou J, Jiang H and Ba L: MiR-194 is involved in
morphogenesis of spiral ganglion neurons in inner ear by
rearranging actin cytoskeleton via targeting RhoB. Int J Dev
Neurosci. 63:16–26. 2017.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Geng R, Furness DN, Muraleedharan CK,
Zhang J, Dabdoub A, Lin V and Xu S: The microRNA-183/96/182 cluster
is essential for stereociliary bundle formation and function of
cochlear sensory hair cells. Sci Rep. 8(18022)2018.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Brown CS, Emmett SD, Robler SK and Tucci
DL: Global hearing loss prevention. Otolaryngol Clin North Am.
51:575–592. 2018.PubMed/NCBI View Article : Google Scholar
|
|
48
|
World Health Organization (WHO): Deafness
and hearing loss. WHO, Geneva, 2022. https://www.who.int/news-room/fact-sheets/detail/deafness-and-hearing-loss.
Accessed April 1, 2022.
|
|
49
|
Edmiston R and Mitchell C: Hearing loss in
adults. BMJ. 346(f2496)2013.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Horowitz G, Ungar OJ, Levit Y, Himmelfarb
M and Handzel O: The impact of conductive hearing loss on balance.
Clin Otolaryngol. 45:106–110. 2020.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Michels TC, Duffy MT and Rogers DJ:
Hearing loss in adults: Differential diagnosis and treatment. Am
Fam Physician. 100:98–108. 2019.PubMed/NCBI
|
|
52
|
Cunningham LL and Tucci DL: Hearing loss
in adults. N Engl J Med. 377:2465–2473. 2017.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Amanipour RM, Zhu X, Duvey G, Celanire S,
Walton JP and Frisina RD: Noise-Induced hearing loss in mice:
Effects of high and low levels of noise trauma in CBA mice. Annu
Int Conf IEEE Eng Med Biol Soc. 2018:1210–1213. 2018.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Lin BM, Wang M, Stankovic KM, Eavey R,
McKenna MJ, Curhan GC and Curhan SG: Cigarette smoking, smoking
cessation, and risk of hearing loss in women. Am J Med.
133:1180–1186. 2020.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Haugnes HS, Stenklev NC, Brydøy M, Dahl O,
Wilsgaard T, Laukli E and Fosså SD: Hearing loss before and after
cisplatin-based chemotherapy in testicular cancer survivors: A
longitudinal study. Acta Oncol. 57:1075–1083. 2018.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Yang T, Guo L, Wang L and Yu X: Diagnosis,
intervention, and prevention of genetic hearing loss. Adv Exp Med
Biol. 1130:73–92. 2019.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Bowl MR and Dawson SJ: Age-Related hearing
loss. Cold Spring Harb Perspect Med. 9(a033217)2019.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Xue T, Wei L, Zha DJ, Qiu JH, Chen FQ,
Qiao L and Qiu Y: miR-29b overexpression induces cochlear hair cell
apoptosis through the regulation of SIRT1/PGC-1α signaling:
Implications for age-related hearing loss. Int J Mol Med.
38:1387–1394. 2016.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Pang J, Xiong H, Lin P, Lai L, Yang H, Liu
Y, Huang Q, Chen S, Ye Y, Sun Y and Zheng Y: Activation of miR-34a
impairs autophagic flux and promotes cochlear cell death via
repressing ATG9A: Implications for age-related hearing loss. Cell
Death Dis. 8(e3079)2017.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Li YH, Yang Y, Yan YT, Xu LW, Ma HY, Shao
YX, Cao CJ, Wu X, Qi MJ, Wu YY, et al: Analysis of serum microRNA
expression in male workers with occupational noise-induced hearing
loss. Braz J Med Biol Res. 51(e6426)2018.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Nunez DA, Wijesinghe P, Nabi S, Yeh D and
Garnis C: microRNAs in sudden hearing loss. Laryngoscope.
130:E416–E422. 2020.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Gheorghe DC, Niculescu AG, Bîrcă AC and
Grumezescu AM: Nanoparticles for the treatment of inner ear
infections. Nanomaterials (Basel). 11(1311)2021.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Cohen BE, Durstenfeld A and Roehm PC:
Viral causes of hearing loss: A review for hearing health
professionals. Trends Hear. 18(2331216514541361)2014.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Palma S, Roversi MF, Bettini M, Mazzoni S,
Pietrosemoli P, Lucaccioni L, Berardi A and Genovese E: Hearing
loss in children with congenital cytomegalovirus infection: An
11-year retrospective study based on laboratory database of a
tertiary paediatric hospital. Acta Otorhinolaryngol Ital. 39:40–45.
2019.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Caroça C, Vicente V, Campelo P, Chasqueira
M, Caria H, Silva S, Paixão P and Paço J: Rubella in Sub-Saharan
Africa and sensorineural hearing loss: A case control study. BMC
Public Health. 17(146)2017.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Himmelein S, Lindemann A, Sinicina I, Horn
AKE, Brandt T, Strupp M and Hüfner K: differential involvement
during latent herpes simplex virus 1 infection of the superior and
inferior divisions of the vestibular Ganglia: Implications for
vestibular neuritis. J Virol. 91:e00331–17. 2017.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Yee KT, Neupane B, Bai F and Vetter DE:
Zika virus infection causes widespread damage to the inner ear.
Hear Res. 395(108000)2020.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Van Hoecke H, De Paepe AS, Lambert E, Van
Belleghem JD, Cools P, Van Simaey L, Deschaght P, Vaneechoutte M
and Dhooge I: Haemophilus influenzae biofilm formation in chronic
otitis media with effusion. Eur Arch Otorhinolaryngol.
273:3553–3560. 2016.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Rosenblut A, Napolitano C, Pereira A,
Moreno C, Kolhe D, Lepetic A and Ortega-Barria E: Etiology of acute
otitis media and serotype distribution of Streptococcus pneumoniae
and Haemophilus influenzae in Chilean children <5 years of age.
Medicine (Baltimore). 96(e5974)2017.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Emami A, Pirbonyeh N, Moattari A,
Bazargani A and Motamedifar M: Risk of otitis media with effusion
(OME) in children by Pseudomonas aeruginosa. Int J Pediatr
Otorhinolaryngol. 125:6–10. 2019.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Ozturk A, Cetintas İ, Bayraktar M and
İynen İ: Evaluation of microbial agents and their antibiotic
susceptibility profiles in patients with chronic suppurative otitis
media. Int J Clin Pract. 75(e14382)2021.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Sillanpää S, Oikarinen S, Sipilä M, Kramna
L, Rautiainen M, Huhtala H, Aittoniemi J, Laranne J, Hyöty H and
Cinek O: Moraxella catarrhalis might be more common than expected
in acute otitis media in young finnish children. J Clin Microbiol.
54:2373–2379. 2016.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Møller MN, Brandt C, Østergaard C and
Caye-Thomasen P: Bacterial invasion of the inner ear in association
with pneumococcal meningitis. Otol Neurotol. 35:e178–e186.
2014.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Niedzielski A, Chmielik LP and Stankiewicz
T: The formation of biofilm and bacteriology in otitis media with
effusion in children: A prospective cross-sectional study. Int J
Environ Res Public Health. 18(3555)2021.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Mofatteh MR, Shahabian Moghaddam F,
Yousefi M and Namaei MH: A study of bacterial pathogens and
antibiotic susceptibility patterns in chronic suppurative otitis
media. J Laryngol Otol. 132:41–45. 2018.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Ali K, Hamed MA, Hassan H, Esmail A and
Sheneef A: Identification of fungal pathogens in otomycosis and
their drug sensitivity: Our experience. Int Arch Otorhinolaryngol.
22:400–403. 2018.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Kiakojuri K, Mahdavi Omran S, Roodgari S,
Taghizadeh Armaki M, Hedayati MT, Shokohi T, Haghani I, Javidnia J,
Kermani F, Badali H and Abastabar M: Molecular identification and
antifungal susceptibility of yeasts and molds isolated from
patients with otomycosis. Mycopathologia. 186:245–257.
2021.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Hajioff D and MacKeith S: Otitis externa.
BMJ Clin Evid. 2015(0510)2015.PubMed/NCBI
|
|
79
|
Candido S, Tomasello BMR, Lavoro A,
Falzone L, Gattuso G and Libra M: Novel insights into epigenetic
regulation of IL6 pathway: In silico perspective on inflammation
and cancer relationship. Int J Mol Sci. 22(10172)2021.PubMed/NCBI View Article : Google Scholar
|
|
80
|
Sonkoly E and Pivarcsi A: microRNAs in
inflammation. Int Rev Immunol. 28:535–561. 2009.PubMed/NCBI View Article : Google Scholar
|
|
81
|
Rudnicki A, Shivatzki S, Beyer LA, Takada
Y, Raphael Y and Avraham KB: microRNA-224 regulates Pentraxin 3, a
component of the humoral arm of innate immunity, in inner ear
inflammation. Hum Mol Genet. 23:3138–3146. 2014.PubMed/NCBI View Article : Google Scholar
|
|
82
|
Samuels TL, Yan J, Khampang P, MacKinnon
A, Hong W, Johnston N and Kerschner JE: Association of microRNA 146
with middle ear hyperplasia in pediatric otitis media. Int J
Pediatr Otorhinolaryngol. 88:104–108. 2016.PubMed/NCBI View Article : Google Scholar
|
|
83
|
Zhang J, He J, Luo Y, Liu Y and Fan X:
miR-210 regulates the inflammation of otitis media with effusion by
inhibiting the expression of hypoxia-inducible factor (HIF)-1a.
Biochem Biophys Res Commun. 534:401–407. 2021.PubMed/NCBI View Article : Google Scholar
|
|
84
|
Frye MD, Ryan AF and Kurabi A:
Inflammation associated with noise-induced hearing loss. J Acoust
Soc Am. 146(4020)2019.PubMed/NCBI View Article : Google Scholar
|
|
85
|
Kociszewska D and Vlajkovic S: Age-Related
hearing loss: The link between inflammaging, immunosenescence, and
gut dysbiosis. Int J Mol Sci. 23(7348)2022.PubMed/NCBI View Article : Google Scholar
|
|
86
|
Lassale C, Vullo P, Cadar D, Batty GD,
Steptoe A and Zaninotto P: Association of inflammatory markers with
hearing impairment: The English Longitudinal study of ageing. Brain
Behav Immun. 83:112–119. 2020.PubMed/NCBI View Article : Google Scholar
|
|
87
|
Lanvers-Kaminsky C, Zehnhoff-Dinnesen AA,
Parfitt R and Ciarimboli G: Drug-induced ototoxicity: Mechanisms,
pharmacogenetics, and protective strategies. Clin Pharmacol Ther.
101:491–500. 2017.PubMed/NCBI View Article : Google Scholar
|
|
88
|
Kros CJ and Steyger PS: Aminoglycoside-
and cisplatin-induced ototoxicity: Mechanisms and otoprotective
strategies. Cold Spring Harb Perspect Med.
9(a033548)2019.PubMed/NCBI View Article : Google Scholar
|
|
89
|
Gattuso G, Falzone L, Costa C, Giambò F,
Teodoro M, Vivarelli S, Libra M and Fenga C: Chronic pesticide
exposure in farm workers is associated with the epigenetic
modulation of hsa-miR-199a-5p. Int J Environ Res Public Health.
19(7018)2022.PubMed/NCBI View Article : Google Scholar
|
|
90
|
Gatto MP, Fioretti M, Fabrizi G, Gherardi
M, Strafella E and Santarelli L: Effects of potential neurotoxic
pesticides on hearing loss: A review. Neurotoxicology. 42:24–32.
2014.PubMed/NCBI View Article : Google Scholar
|
|
91
|
Hoshino ACH, Pacheco-Ferreira H, Taguchi
CK, Tomita S and de Fátima Miranda M: Ototoxicity study in workers
exposed to organophosphate. Braz J Otorhinolaryngol. 74:912–918.
2008.PubMed/NCBI View Article : Google Scholar
|
|
92
|
DiSogra RM: Common aminoglycosides and
platinum-based ototoxic drugs: Cochlear/vestibular side effects and
incidence. Semin Hear. 40:104–107. 2019.PubMed/NCBI View Article : Google Scholar
|
|
93
|
Gersten BK, Fitzgerald TS, Fernandez KA
and Cunningham LL: Ototoxicity and platinum uptake following cyclic
administration of platinum-based chemotherapeutic agents. J Assoc
Res Otolaryngol. 21:303–321. 2020.PubMed/NCBI View Article : Google Scholar
|
|
94
|
Xie J, Talaska AE and Schacht J: New
developments in aminoglycoside therapy and ototoxicity. Hear Res.
281:28–37. 2011.PubMed/NCBI View Article : Google Scholar
|
|
95
|
Ding D, Liu H, Qi W, Jiang H, Li Y, Wu X,
Sun H, Gross K and Salvi R: Ototoxic effects and mechanisms of loop
diuretics. J Otol. 11:145–156. 2016.PubMed/NCBI View Article : Google Scholar
|
|
96
|
Ikeda AK, Prince AA, Chen JX, Lieu JEC and
Shin JJ: Macrolide-associated sensorineural hearing loss: A
systematic review. Laryngoscope. 128:228–236. 2018.PubMed/NCBI View Article : Google Scholar
|
|
97
|
Altissimi G, Colizza A, Cianfrone G, de
Vincentiis M, Greco A, Taurone S, Musacchio A, Ciofalo A, Turchetta
R, Angeletti D and Ralli M: Drugs inducing hearing loss, tinnitus,
dizziness and vertigo: An updated guide. Eur Rev Med Pharmacol Sci.
24:7946–7952. 2020.PubMed/NCBI View Article : Google Scholar
|
|
98
|
Landier W, Knight K, Wong FL, Lee J,
Thomas O, Kim H, Kreissman SG, Schmidt ML, Chen L, London WB, et
al: Ototoxicity in children with high-risk neuroblastoma:
Prevalence, risk factors, and concordance of grading scales-a
report from the Children's oncology group. J Clin Oncol.
32:527–534. 2014.PubMed/NCBI View Article : Google Scholar
|
|
99
|
Waissbluth S, Del Valle Á, Chuang A and
Becker A: Incidence and associated risk factors for
platinum-induced ototoxicity in pediatric patients. Int J Pediatr
Otorhinolaryngol. 111:174–179. 2018.PubMed/NCBI View Article : Google Scholar
|
|
100
|
Wei M and Yuan X: Cisplatin-induced
ototoxicity in children with solid tumor. J Pediatr Hematol Oncol.
41:e97–e100. 2019.PubMed/NCBI View Article : Google Scholar
|
|
101
|
Landier W: Ototoxicity and cancer therapy.
Cancer. 122:1647–1658. 2016.PubMed/NCBI View Article : Google Scholar
|
|
102
|
Ganesan P, Schmiedge J, Manchaiah V,
Swapna S, Dhandayutham S and Kothandaraman PP: Ototoxicity: A
challenge in diagnosis and treatment. J Audiol Otol. 22:59–68.
2018.PubMed/NCBI View Article : Google Scholar
|
|
103
|
Kim CW, Han JH, Wu L and Choi JY:
microRNA-183 is essential for hair cell regeneration after neomycin
injury in zebrafish. Yonsei Med J. 59:141–147. 2018.PubMed/NCBI View Article : Google Scholar
|
|
104
|
Lee SH, Ju HM, Choi JS, Ahn Y, Lee S and
Seo YJ: Circulating Serum miRNA-205 as a diagnostic biomarker for
ototoxicity in mice treated with aminoglycoside antibiotics. Int J
Mol Sci. 19(2836)2018.PubMed/NCBI View Article : Google Scholar
|
|
105
|
Li J, Ling Y, Huang W, Sun L, Li Y, Wang
C, Zhang Y, Wang X, Dahlgren RA and Wang H: Regulatory mechanisms
of miR-96 and miR-184 abnormal expressions on otic vesicle
development of zebrafish following exposure to β-diketone
antibiotics. Chemosphere. 214:228–238. 2019.PubMed/NCBI View Article : Google Scholar
|
|
106
|
Chen J, Liu Z, Yan H, Xing W, Mi W, Wang
R, Li W, Chen F, Qiu J and Zha D: miR-182 prevented ototoxic
deafness induced by co-administration of kanamycin and furosemide
in rats. Neurosci Lett. 723(134861)2020.PubMed/NCBI View Article : Google Scholar
|
|
107
|
Stangerup SE, Caye-Thomasen P, Tos M and
Thomsen J: The natural history of vestibular schwannoma. Otol
Neurotol. 27:547–552. 2006.PubMed/NCBI View Article : Google Scholar
|
|
108
|
Paldor I, Chen AS and Kaye AH: Growth rate
of vestibular schwannoma. J Clin Neurosci. 32:1–8. 2016.PubMed/NCBI View Article : Google Scholar
|
|
109
|
Sughrue ME, Yang I, Aranda D, Rutkowski
MJ, Fang S, Cheung SW and Parsa AT: Beyond audiofacial morbidity
after vestibular schwannoma surgery. J Neurosurg. 114:367–374.
2011.PubMed/NCBI View Article : Google Scholar
|
|
110
|
Halliday J, Rutherford SA, McCabe MG and
Evans DG: An update on the diagnosis and treatment of vestibular
schwannoma. Expert Rev Neurother. 18:29–39. 2018.PubMed/NCBI View Article : Google Scholar
|
|
111
|
Yao L, Alahmari M, Temel Y and Hovinga K:
Therapy of sporadic and NF2-Related vestibular schwannoma. Cancers
(Basel). 12(835)2020.PubMed/NCBI View Article : Google Scholar
|
|
112
|
Pandrangi VC, Han AY, Alonso JE, Peng KA
and St John MA: An update on epidemiology and management trends of
vestibular schwannomas. Otol Neurotol. 41:411–417. 2020.PubMed/NCBI View Article : Google Scholar
|
|
113
|
Andersen JF, Nilsen KS, Vassbotn FS,
Møller P, Myrseth E, Lund-Johansen M and Goplen FK: Predictors of
vertigo in patients with untreated vestibular schwannoma. Otol
Neurotol. 36:647–652. 2015.PubMed/NCBI View Article : Google Scholar
|
|
114
|
Kaul V and Cosetti MK: Management of
vestibular schwannoma (Including NF2): Facial nerve considerations.
Otolaryngol Clin North Am. 51:1193–1212. 2018.PubMed/NCBI View Article : Google Scholar
|
|
115
|
Dunn IF, Bi WL, Mukundan S, Delman BN,
Parish J, Atkins T, Asher AL and Olson JJ: Congress of neurological
surgeons systematic review and evidence-based guidelines on the
role of imaging in the diagnosis and management of patients with
vestibular schwannomas. Neurosurgery. 82:E32–E34. 2018.PubMed/NCBI View Article : Google Scholar
|
|
116
|
Goldbrunner R, Weller M, Regis J,
Lund-Johansen M, Stavrinou P, Reuss D, Evans DG, Lefranc F,
Sallabanda K, Falini A, et al: EANO guideline on the diagnosis and
treatment of vestibular schwannoma. Neuro Oncol. 22:31–45.
2020.PubMed/NCBI View Article : Google Scholar
|
|
117
|
Cioffi JA, Yue WY, Mendolia-Loffredo S,
Hansen KR, Wackym PA and Hansen MR: MicroRNA-21 overexpression
contributes to vestibular schwannoma cell proliferation and
survival. Otol Neurotol. 31:1455–1462. 2010.PubMed/NCBI View Article : Google Scholar
|
|
118
|
Saydam O, Senol O, Würdinger T, Mizrak A,
Ozdener GB, Stemmer-Rachamimov AO, Yi M, Stephens RM, Krichevsky
AM, Saydam N, et al: miRNA-7 attenuation in Schwannoma tumors
stimulates growth by upregulating three oncogenic signaling
pathways. Cancer Res. 71:852–861. 2011.PubMed/NCBI View Article : Google Scholar
|
|
119
|
Mahajan K and Mahajan NP: ACK1/TNK2
tyrosine kinase: Molecular signaling and evolving role in cancers.
Oncogene. 34:4162–4167. 2015.PubMed/NCBI View Article : Google Scholar
|
|
120
|
Li SL, Ma XH, Ji JF, Li H, Liu W, Lu FZ,
Wu ST and Zhang Y: miR-1 association with cell proliferation
inhibition and apoptosis in vestibular schwannoma by targeting
VEGFA. Genet Mol Res. 15(gmr15048923)2016.PubMed/NCBI View Article : Google Scholar
|
|
121
|
Peng CY, Liao YW, Lu MY, Yu CH, Yu CC and
Chou MY: Downregulation of miR-1 enhances tumorigenicity and
invasiveness in oral squamous cell carcinomas. J Formos Med Assoc.
116:782–789. 2017.PubMed/NCBI View Article : Google Scholar
|
|
122
|
Xu W, Zhang Z, Zou K, Cheng Y, Yang M,
Chen H, Wang H, Zhao J, Chen P, He L, et al: MiR-1 suppresses tumor
cell proliferation in colorectal cancer by inhibition of
Smad3-mediated tumor glycolysis. Cell Death Dis.
8(e2761)2017.PubMed/NCBI View Article : Google Scholar
|
|
123
|
Chen C, Zhou Y, Ding P and He L: miR-1
targeted downregulation of Bcl-2 increases chemosensitivity of lung
cancer cells. Genet Test Mol Biomarkers. 25:540–545.
2021.PubMed/NCBI View Article : Google Scholar
|
|
124
|
Yin X, Huo Z, Yan S, Wang Z, Yang T, Wu H
and Zhang Z: MiR-205 inhibits sporadic vestibular schwannoma cell
proliferation by targeting cyclin-dependent kinase 14. World
Neurosurg. 147:e25–e31. 2021.PubMed/NCBI View Article : Google Scholar
|