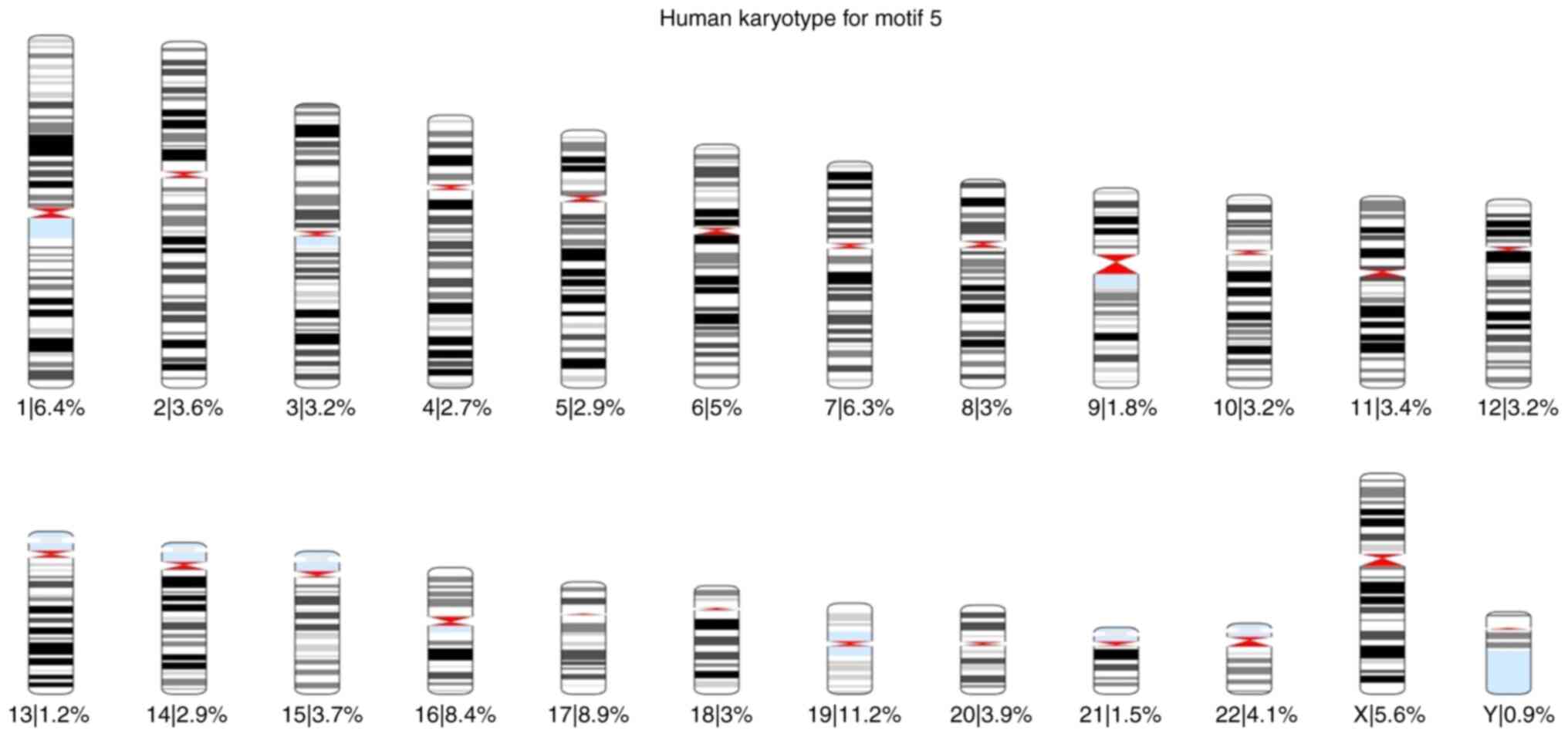
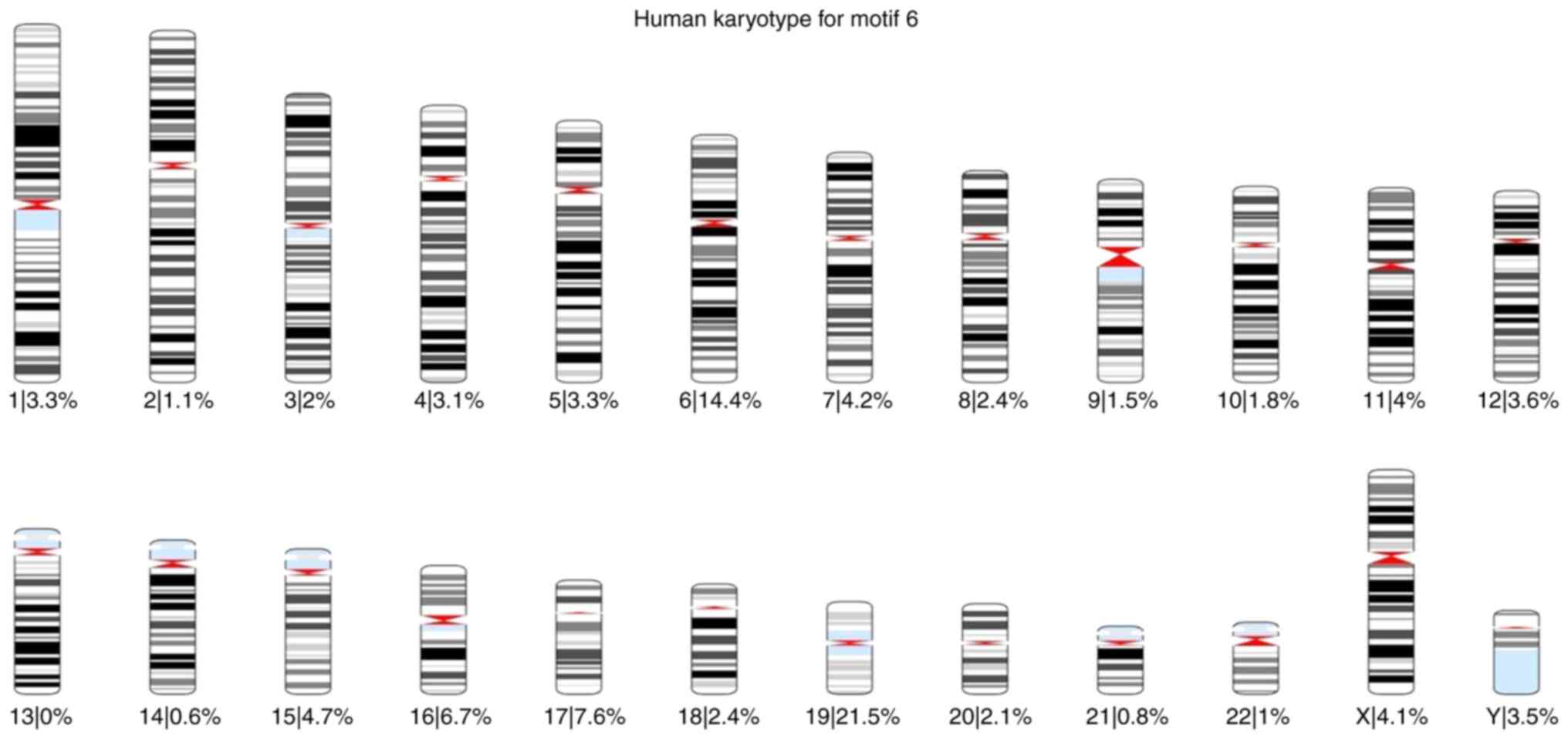
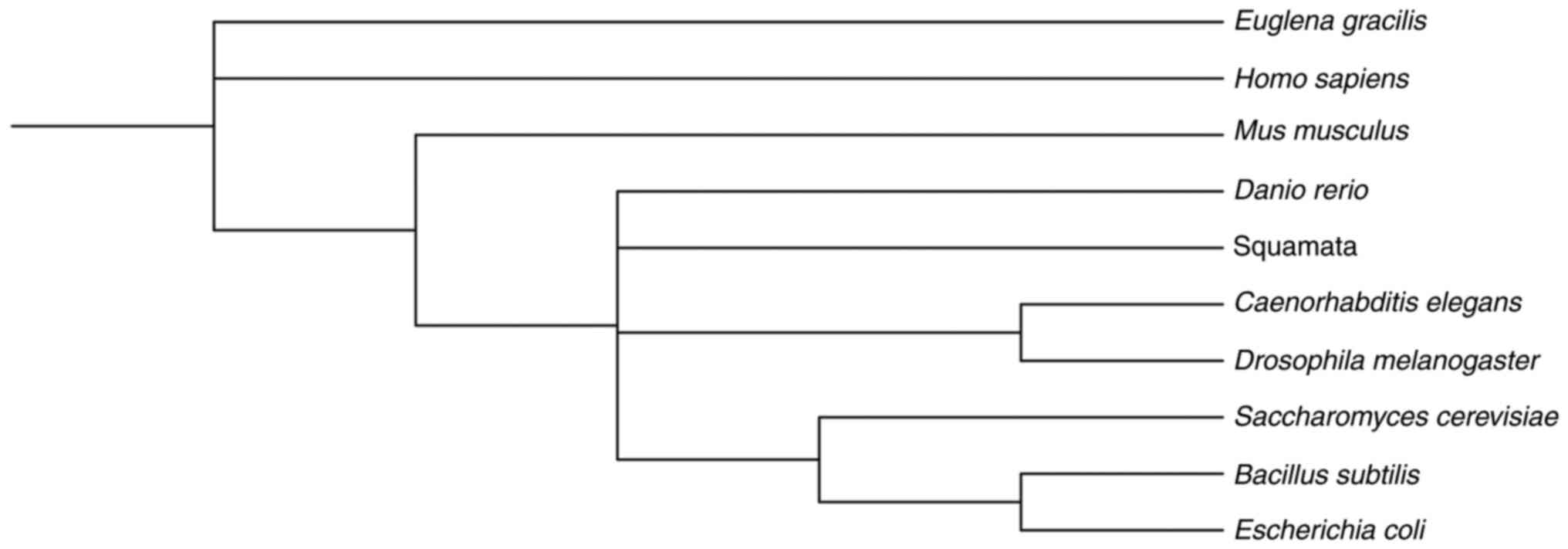
|
1
|
Carninci P, Kasukawa T, Katayama S, Gough
J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al:
The transcriptional landscape of the mammalian genome. Science.
309:1559–1563. 2005.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Mattick JS: RNA regulation: A new
genetics? Nat Rev Genet. 5:316–323. 2004.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Frith MC, Pheasant M and Mattick JS: The
amazing complexity of the human transcriptome. Eur J Hum Genet.
13:894–897. 2005.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Kaikkonen MU, Lam MTY and Glass CK:
Non-coding RNAs as regulators of gene expression and epigenetics.
Cardiovasc Res. 90:430–440. 2011.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Harrow J, Frankish A, Gonzalez JM,
Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa
A, Searle S, et al: GENCODE: The reference human genome annotation
for The ENCODE project. Genome Res. 22:1760–1774. 2012.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Salviano-Silva A, Lobo-Alves SC, Almeida
RC, Malheiros D and Petzl-Erler ML: Besides pathology: Long
non-coding RNA in cell and tissue homeostasis. Noncoding RNA.
4(3)2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Ma L, Bajic VB and Zhang Z: On the
classification of long non-coding RNAs. RNA Biol. 10:925–933.
2013.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Wang KC and Chang HY: Molecular mechanisms
of long noncoding RNAs. Mol Cell. 43:904–914. 2011.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Hombach S and Kretz M: Non-coding RNAs:
Classification, biology and functioning. Adv Exp Med Biol.
937:3–17. 2016.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Rinn JL and Chang HY: Genome regulation by
long noncoding RNAs. Annu Rev Biochem. 81:145–166. 2012.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Francis GA, Fayard E, Picard F and Auwerx
J: Nuclear receptors and the control of metabolism. Annu Rev
Physiol. 65:261–311. 2003.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Olivares AM, Moreno-Ramos OA and Haider
NB: Role of nuclear receptors in central nervous system development
and associated diseases. J Exp Neurosci. 9 (Suppl 2):S93–S121.
2016.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Foulds CE, Panigrahi AK, Coarfa C, Lanz RB
and O'Malley BW: Long noncoding RNAs as targets and regulators of
nuclear receptors. Curr Top Microbiol Immunol. 394:143–176.
2016.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Volders PJ, Helsens K, Wang X, Menten B,
Martens L, Gevaert K, Vandesompele J and Mestdagh P: LNCipedia: A
database for annotated human lncRNA transcript sequences and
structures. Nucleic Acids Res. 41 (Database Issue):D246–D251.
2013.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Bailey TL, Williams N, Misleh C and Li WW:
MEME: Discovering and analyzing DNA and protein sequence motifs.
Nucleic Acids Res. 34 (Web Server Issue):W369–W373. 2006.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Liu W and Wang X: Prediction of functional
microRNA targets by integrative modeling of microRNA binding and
target expression data. Genome Biol. 20(18)2019.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4(e05005)2015.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Garcia DM, Baek D, Shin C, Bell GW,
Grimson A and Bartel DP: Weak seed-pairing stability and high
target-site abundance decrease the proficiency of lsy-6 and other
microRNAs. Nat Struct Mol Biol. 18:1139–1146. 2011.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Grimson A, Farh KK, Johnston WK,
Garrett-Engele P, Lim LP and Bartel DP: MicroRNA targeting
specificity in mammals: Determinants beyond seed pairing. Mol Cell.
27:91–105. 2007.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Goecks J, Nekrutenko A and Taylor J:
Galaxy Team. Galaxy: A comprehensive approach for supporting
accessible, reproducible, and transparent computational research in
the life sciences. Genome Biol. 11(R86)2010.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Afgan E, Baker D, van den Beek M,
Blankenberg D, Bouvier D, Čech M, Chilton J, Clements D, Coraor N,
Eberhard C, et al: The Galaxy platform for accessible, reproducible
and collaborative biomedical analyses: 2016 update. Nucleic Acids
Res. 44 (W1):W3–W10. 2016.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Durbin R, Eddy SR, Krogh A and Mitchison
G: Biological sequence analysis: Probabilistic Models of Proteins
and Nucleic Acids. Cambridge University Press, 1998.
|
|
23
|
Ha M and Kim VN: Regulation of microRNA
biogenesis. Nat Rev Mol Cell Biol. 15:509–524. 2014.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Broughton JP, Lovci MT, Huang JL, Yeo GW
and Pasquinelli AE: Pairing beyond the seed supports microRNA
targeting specificity. Mol Cell. 64:320–333. 2016.PubMed/NCBI View Article : Google Scholar
|
|
25
|
John B, Enright AJ, Aravin A, Tuschl T,
Sander C and Marks DS: Human MicroRNA targets. PLoS Biol.
2(e363)2004.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Betel D, Koppal A, Agius P, Sander C and
Leslie C: Comprehensive modeling of microRNA targets predicts
functional non-conserved and non-canonical sites. Genome Biol.
11(R90)2010.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Krek A, Grün D, Poy MN, Wolf R, Rosenberg
L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M
and Rajewsky N: Combinatorial microRNA target predictions. Nat
Genet. 37:495–500. 2005.PubMed/NCBI View
Article : Google Scholar
|
|
28
|
Lewis BP, Shih IH, Jones-Rhoades MW,
Bartel DP and Burge CB: Prediction of mammalian microRNA targets.
Cell. 115:787–798. 2003.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Miranda KC, Huynh T, Tay Y, Ang YS, Tam
WL, Thomson AM, Lim B and Rigoutsos I: A pattern-based method for
the identification of MicroRNA binding sites and their
corresponding heteroduplexes. Cell. 126:1203–1217. 2006.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Kertesz M, Iovino N, Unnerstall U, Gaul U
and Segal E: The role of site accessibility in microRNA target
recognition. Nat Genet. 39:1278–1284. 2007.PubMed/NCBI View
Article : Google Scholar
|
|
32
|
Rehmsmeier M, Steffen P, Hochsmann M and
Giegerich R: Fast and effective prediction of microRNA/target
duplexes. RNA. 10:1507–1517. 2004.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Maragkakis M, Alexiou P, Papadopoulos GL,
Reczko M, Dalamagas T, Giannopoulos G, Goumas G, Koukis E, Kourtis
K, Simossis VA, et al: Accurate microRNA target prediction
correlates with protein repression levels. BMC Bioinformatics.
10(295)2009.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Karagkouni D, Paraskevopoulou MD,
Chatzopoulos S, Vlachos IS, Tastsoglou S, Kanellos I, Papadimitriou
D, Kavakiotis I, Maniou S, Skoufos G, et al: DIANA-TarBase v8: A
decade-long collection of experimentally supported miRNA-gene
interactions. Nucleic Acids Res. 46 (D1):D239–D245. 2018.PubMed/NCBI View Article : Google Scholar
|
|
35
|
McGeary SE, Lin KS, Shi CY, Pham TM,
Bisaria N, Kelley GM and Bartel DP: The biochemical basis of
microRNA targeting efficacy. Science. 366(eaav1741)2019.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Yang L, Shi CM, Chen L, Pang LX, Xu GF, Gu
N, Zhu LJ, Guo XR, Ni YH and Ji CB: The biological effects of
hsa-miR-1908 in human adipocytes. Mol Biol Rep. 42:927–935.
2015.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Kuang Q, Li J, You L, Shi C, Ji C, Guo X,
Xu M and Ni Y: Identification and characterization of NF-kappaB
binding sites in human miR-1908 promoter. Biomed Pharmacother.
74:158–163. 2015.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Bar M, Wyman SK, Fritz BR, Qi J, Garg KS,
Parkin RK, Kroh EM, Bendoraite A, Mitchell PS, Nelson AM, et al:
MicroRNA discovery and profiling in human embryonic stem cells by
deep sequencing of small RNA libraries. Stem Cells. 26:2496–2505.
2008.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Pencheva N, Tran H, Buss C, Huh D,
Drobnjak M, Busam K and Tavazoie SF: Convergent multi-miRNA
targeting of ApoE drives LRP1/LRP8-dependent melanoma metastasis
and angiogenesis. Cell. 151:1068–1082. 2012.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Xia X, Li Y, Wang W, Tang F, Tan J, Sun L,
Li Q, Sun L, Tang B and He S: MicroRNA-1908 functions as a
glioblastoma oncogene by suppressing PTEN tumor suppressor pathway.
Mol Cancer. 14(154)2015.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Chai Z, Fan H, Li Y, Song L, Jin X, Yu J,
Li Y, Ma C and Zhou R: miR-1908 as a novel prognosis marker of
glioma via promoting malignant phenotype and modulating SPRY4/RAF1
axis. Oncol Rep. 38:2717–2726. 2017.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Jiang X, Yang L, Pang L, Chen L, Guo X, Ji
C, Shi C and Ni Y: Expression of obesity-related miR-1908 in human
adipocytes is regulated by adipokines, free fatty acids and
hormones. Mol Med Rep. 10:1164–1169. 2014.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Grimwood J, Gordon LA, Olsen A, Terry A,
Schmutz J, Lamerdin J, Hellsten U, Goodstein D, Couronne O,
Tran-Gyamfi M, et al: The DNA sequence and biology of human
chromosome 19. Nature. 428:529–535. 2004.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Kumar S, Stecher G, Suleski M and Hedges
SB: TimeTree: A resource for timelines, timetrees, and divergence
times. Mol Biol Evol. 34:1812–1819. 2017.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Kim SH, Elango N, Warden C, Vigoda E and
Yi SV: Heterogeneous genomic molecular clocks in primates. PLoS
Genet. 2(e163)2006.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Kotin RM, Menninger JC, Ward DC and Berns
KI: Mapping and direct visualization of a region-specific viral DNA
integration site on chromosome 19q13-qter. Genomics. 10:831–834.
1991.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Samulski RJ, Zhu X, Xiao X, Brook JD,
Housman DE, Epstein N and Hunter LA: Targeted integration of
adeno-associated virus (AAV) into human chromosome 19. EMBO J.
10:3941–3950. 1991.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Kotin RM, Linden RM and Berns KI:
Characterization of a preferred site on human chromosome 19q for
integration of adeno-associated virus DNA by non-homologous
recombination. EMBO J. 11:5071–5078. 1992.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Aslanidis C, Jansen G, Amemiya C, Shutler
G, Mahadevan M, Tsilfidis C, Chen C, Alleman J, Wormskamp NG,
Vooijs M, et al: Cloning of the essential myotonic dystrophy region
and mapping of the putative defect. Nature. 355:548–551.
1992.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Feichtinger W and Schmid M: Increased
frequencies of sister chromatid exchanges at common fragile sites
(1)(q42) and (19)(q13). Hum Genet. 83:145–147. 1989.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Sievers F and Higgins DG: Clustal Omega
for making accurate alignments of many protein sequences. Protein
Sci. 27:135–145. 2018.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Waterhouse AM, Procter JB, Martin DMA,
Clamp M and Barton GJ: Jalview Version 2-a multiple sequence
alignment editor and analysis workbench. Bioinformatics.
25:1189–1191. 2009.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Deaton AM and Bird A: CpG islands and the
regulation of transcription. Genes Dev. 25:1010–1022.
2011.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Larsen F, Gundersen G, Lopez R and Prydz
H: CpG islands as gene markers in the human genome. Genomics.
13:1095–1107. 1992.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Yamashita R, Suzuki Y, Sugano S and Nakai
K: Genome-wide analysis reveals strong correlation between CpG
islands with nearby transcription start sites of genes and their
tissue specificity. Gene. 350:129–136. 2005.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Lamartina S, Sporeno E, Fattori E and
Toniatti C: Characteristics of the adeno-associated virus
preintegration site in human chromosome 19: Open chromatin
conformation and transcription-competent environment. J Virol.
74:7671–7677. 2000.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Bhagwan JR, Collins E, Mosqueira D, Bakar
M, Johnson BB, Thompson A, Smith JGW and Denning C: Variable
expression and silencing of CRISPR-Cas9 targeted transgenes
identifies the AAVS1 locus as not an entirely safe harbour.
F1000Res. 8(1911)2019.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Ogata T, Kozuka T and Kanda T:
Identification of an insulator in AAVS1, a preferred region for
integration of adeno-associated virus DNA. J Virol. 77:9000–9007.
2003.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Gaspar HB, Cooray S, Gilmour KC, Parsley
KL, Zhang F, Adams S, Bjorkegren E, Bayford J, Brown L, Davies EG,
et al: Hematopoietic stem cell gene therapy for adenosine
deaminase-deficient severe combined immunodeficiency leads to
long-term immunological recovery and metabolic correction. Sci
Transl Med. 3(97ra80)2011.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Cartier N, Hacein-Bey-Abina S, Bartholomae
CC, Veres G, Schmidt M, Kutschera I, Vidaud M, Abel U, Dal-Cortivo
L, Caccavelli L, et al: Hematopoietic stem cell gene therapy with a
lentiviral vector in X-linked adrenoleukodystrophy. Science.
326:818–823. 2009.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Hacein-Bey-Abina S, Pai SY, Gaspar HB,
Armant M, Berry CC, Blanche S, Bleesing J, Blondeau J, de Boer H,
Buckland KF, et al: A modified γ-retrovirus vector for X-linked
severe combined immunodeficiency. N Engl J Med. 371:1407–1417.
2014.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Martin DIK and Whitelaw E: The vagaries of
variegating transgenes. Bioessays. 18:919–923. 1996.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Bestor TH: Gene silencing as a threat to
the success of gene therapy. J Clin Invest. 105:409–411.
2000.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Papapetrou EP and Schambach A: Gene
insertion into genomic safe harbors for human gene therapy. Mol
Ther. 24:678–684. 2016.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Wu C and Dunbar CE: Stem cell gene
therapy: The risks of insertional mutagenesis and approaches to
minimize genotoxicity. Front Med. 5:356–371. 2011.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Lombardo A, Cesana D, Genovese P, Di
Stefano B, Provasi E, Colombo DF, Neri M, Magnani Z, Cantore A, Lo
Riso P, et al: Site-specific integration and tailoring of cassette
design for sustainable gene transfer. Nat Methods. 8:861–869.
2011.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Klatt D, Cheng E, Hoffmann D, Santilli G,
Thrasher AJ, Brendel C and Schambach A: Differential transgene
silencing of myeloid-specific promoters in the AAVS1 safe harbor
locus of induced pluripotent stem cell-derived myeloid cells. Hum
Gene Ther. 31:199–210. 2019.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Papapetrou EP, Lee G, Malani N, Setty M,
Riviere I, Tirunagari LM, Kadota K, Roth SL, Giardina P, Viale A,
et al: Genomic safe harbors permit high β-globin transgene
expression in thalassemia induced pluripotent stem cells. Nat
Biotechnol. 29:73–78. 2011.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Sadelain M, Papapetrou EP and Bushman FD:
Safe harbours for the integration of new DNA in the human genome.
Nat Rev Cancer. 12:51–58. 2011.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Smith SD, Morgan R, Gemmell R, Amylon MD,
Link MP, Linker C, Hecht BK, Warnke R, Glader BE and Hecht F:
Clinical and biologic characterization of T-cell neoplasias with
rearrangements of chromosome 7 band q34. Blood. 71:395–402.
1988.PubMed/NCBI
|
|
71
|
Maes OC, Chertkow HM, Wang E and Schipper
HM: MicroRNA: Implications for alzheimer disease and other human
CNS disorders. Curr Genomics. 10:154–168. 2009.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Garofalo M, Condorelli G and Croce CM:
MicroRNAs in diseases and drug response. Curr Opin Pharmacol.
8:661–667. 2008.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Juźwik CA, S Drake S, Zhang Y,
Paradis-Isler N, Sylvester A, Amar-Zifkin A, Douglas C, Morquette
B, Moore CS and Fournier AE: microRNA dysregulation in
neurodegenerative diseases: A systematic review. Prog Neurobiol.
182(101664)2019.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Venneri M and Passantino A: MiRNA: What
clinicians need to know. Eur J Intern Med. 113:6–9. 2023.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Zhang B, Pan X, Cobb GP and Anderson TA:
microRNAs as oncogenes and tumor suppressors. Dev Biol. 302:1–12.
2007.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Small EM and Olson EN: Pervasive roles of
microRNAs in cardiovascular biology. Nature. 469:336–342.
2011.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Romaine SPR, Tomaszewski M, Condorelli G
and Samani NJ: MicroRNAs in cardiovascular disease: An introduction
for clinicians. Heart. 101:921–928. 2015.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Bailey TL and Elkan C: Fitting a mixture
model by expectation maximization to discover motifs in
biopolymers. Proc Int Conf Intell Syst Mol Biol. 2:28–36.
1994.PubMed/NCBI
|
|
79
|
Tirosh I and Barkai N: Two strategies for
gene regulation by promoter nucleosomes. Genome Res. 18:1084–1091.
2008.PubMed/NCBI View Article : Google Scholar
|
|
80
|
Bird A: DNA methylation patterns and
epigenetic memory. Genes Dev. 16:6–21. 2002.PubMed/NCBI View Article : Google Scholar
|
|
81
|
Li E, Bestor TH and Jaenisch R: Targeted
mutation of the DNA methyltransferase gene results in embryonic
lethality. Cell. 69:915–926. 1992.PubMed/NCBI View Article : Google Scholar
|
|
82
|
Ulitsky I, Shkumatava A, Jan CH, Sive H
and Bartel DP: Conserved function of lincRNAs in vertebrate
embryonic development despite rapid sequence evolution. Cell.
147:1537–1550. 2011.PubMed/NCBI View Article : Google Scholar
|
|
83
|
Liu L, Wang Z, Jia J, Shi Y, Lian T and
Han X: Linc01230, transcriptionally regulated by PPARγ, is
identified as a novel modifier in endothelial function. Biochem
Biophys Res Commun. 507:369–376. 2018.PubMed/NCBI View Article : Google Scholar
|
|
84
|
Cunningham TJ, Kumar S, Yamaguchi TP and
Duester G: Wnt8a and Wnt3a cooperate in the axial stem cell niche
to promote mammalian body axis extension. Dev Dyn. 244:797–807.
2015.PubMed/NCBI View Article : Google Scholar
|
|
85
|
Okubo Y, Sugawara T, Abe-Koduka N, Kanno
J, Kimura A and Saga Y: Lfng regulates the synchronized oscillation
of the mouse segmentation clock via trans-repression of Notch
signalling. Nat Commun. 3(1141)2012.PubMed/NCBI View Article : Google Scholar
|
|
86
|
Pai SG, Carneiro BA, Mota JM, Costa R,
Leite CA, Barroso-Sousa R, Kaplan JB, Chae YK and Giles FJ:
Wnt/beta-catenin pathway: Modulating anticancer immune response. J
Hematol Oncol. 10(101)2017.PubMed/NCBI View Article : Google Scholar
|
|
87
|
Amsellem V, Dryden NH, Martinelli R,
Gavins F, Almagro LO, Birdsey GM, Haskard DO, Mason JC, Turowski P
and Randi AM: ICAM-2 regulates vascular permeability and N-cadherin
localization through ezrin-radixin-moesin (ERM) proteins and Rac-1
signalling. Cell Commun Signal. 12(12)2014.PubMed/NCBI View Article : Google Scholar
|
|
88
|
Overton HA, Fyfe MCT and Reynet C: GPR119,
a novel G protein-coupled receptor target for the treatment of type
2 diabetes and obesity. Br J Pharmacol. 153 (Suppl 1):S76–S81.
2008.PubMed/NCBI View Article : Google Scholar
|
|
89
|
Shang R, Wang M, Dai B, Du J, Wang J, Liu
Z, Qu S, Yang X, Liu J, Xia C, et al: Long noncoding RNA SLC2A1-AS1
regulates aerobic glycolysis and progression in hepatocellular
carcinoma via inhibiting the STAT3/FOXM1/GLUT1 pathway. Mol Oncol.
14:1381–1396. 2020.PubMed/NCBI View Article : Google Scholar
|
|
90
|
Kim JJ, Lee SB, Jang J, Yi SY, Kim SH, Han
SA, Lee JM, Tong SY, Vincelette ND, Gao B, et al: WSB1 promotes
tumor metastasis by inducing pVHL degradation. Genes Dev.
29:2244–2257. 2015.PubMed/NCBI View Article : Google Scholar
|
|
91
|
Shao X, Zhao T, Xi L, Zhang Y, He J, Zeng
J and Deng L: LINC00565 promotes the progression of colorectal
cancer by upregulating EZH2. Oncol Lett. 21(53)2021.PubMed/NCBI View Article : Google Scholar
|
|
92
|
Chen C, Feng Y, Wang J, Liang Y and Zou W:
Long non-coding RNA SNHG15 in various cancers: A meta and
bioinformatic analysis. BMC Cancer. 20(1156)2020.PubMed/NCBI View Article : Google Scholar
|
|
93
|
Qian C, Li H, Chang D, Wei B and Wang Y:
Identification of functional lncRNAs in atrial fibrillation by
integrative analysis of the lncRNA-mRNA network based on competing
endogenous RNAs hypothesis. J Cell Physiol. 234:11620–11630.
2019.PubMed/NCBI View Article : Google Scholar
|
|
94
|
Liu J, Xu R, Mai SJ, Ma YS, Zhang MY, Cao
PS, Weng NQ, Wang RQ, Cao D, Wei W, et al: LncRNA CSMD1-1 promotes
the progression of hepatocellular carcinoma by activating MYC
signaling. Theranostics. 10:7527–7544. 2020.PubMed/NCBI View Article : Google Scholar
|
|
95
|
Talwar D and Hammer MF: SCN8A epilepsy,
developmental encephalopathy, and related disorders. Pediatr
Neurol. 122:76–83. 2021.PubMed/NCBI View Article : Google Scholar
|
|
96
|
Chen XW, Feng YQ, Hao CJ, Guo XL, He X,
Zhou ZY, Guo N, Huang HP, Xiong W, Zheng H, et al: DTNBP1, a
schizophrenia susceptibility gene, affects kinetics of transmitter
release. J Cell Biol. 181:791–801. 2008.PubMed/NCBI View Article : Google Scholar
|
|
97
|
Chen CL, Ke Q, Luo M, Gao ZY, Li ZJ, Luo
ZG and Liu DB: Loss of LINC01939 expression predicts progression
and poor survival in gastric cancer. Pathol Res Pract.
214:1539–1543. 2018.PubMed/NCBI View Article : Google Scholar
|
|
98
|
Dosil M and Bustelo XR: Functional
characterization of Pwp2, a WD family protein essential for the
assembly of the 90 S pre-ribosomal particle. J Biol Chem.
279:37385–37397. 2004.PubMed/NCBI View Article : Google Scholar
|
|
99
|
Li G, Ruan X, Auerbach RK, Sandhu KS,
Zheng M, Wang P, Poh HM, Goh Y, Lim J, Zhang J, et al: Extensive
promoter-centered chromatin interactions provide a topological
basis for transcription regulation. Cell. 148:84–98.
2012.PubMed/NCBI View Article : Google Scholar
|
|
100
|
Li Z, Li X, Jian W, Xue Q and Liu Z: Roles
of long non-coding RNAs in the development of chronic pain. Front
Mol Neurosci. 14(760964)2021.PubMed/NCBI View Article : Google Scholar
|
|
101
|
Luo L, Martin SC, Parkington J, Cadena SM,
Zhu J, Ibebunjo C, Summermatter S, Londraville N, Patora-Komisarska
K, Widler L, et al: HDAC4 controls muscle homeostasis through
deacetylation of myosin heavy chain, PGC-1α, and Hsc70. Cell Rep.
29:749–763.e12. 2019.PubMed/NCBI View Article : Google Scholar
|
|
102
|
Mortison JD, Schenone M, Myers JA, Zhang
Z, Chen L, Ciarlo C, Comer E, Natchiar SK, Carr SA, Klaholz BP and
Myers AG: Tetracyclines modify translation by targeting key human
rRNA substructures. Cell Chem Biol. 25:1506–1518.e13.
2018.PubMed/NCBI View Article : Google Scholar
|
|
103
|
Nelson JK, Koenis DS, Scheij S, Cook EC,
Moeton M, Santos A, Lobaccaro JA, Baron S and Zelcer N: EEPD1 is a
novel LXR target gene in macrophages which regulates ABCA1
abundance and cholesterol efflux. Arterioscler Thromb Vasc Biol.
37:423–432. 2017.PubMed/NCBI View Article : Google Scholar
|
|
104
|
Zhang T, Xia W, Song X, Mao Q, Huang X,
Chen B, Liang Y, Wang H, Chen Y, Yu X, et al: Super-enhancer
hijacking LINC01977 promotes malignancy of early-stage lung
adenocarcinoma addicted to the canonical TGF-β/SMAD3 pathway. J
Hematol Oncol. 15(114)2022.PubMed/NCBI View Article : Google Scholar
|
|
105
|
Zeng J, Sun W, Chang J, Yi D, Zhu L, Zhang
Y, Pan X, Zhou Y, Lai M, Bian G, et al: HOXC4 up-regulates NF-κB
signaling and promotes the cell proliferation to drive development
of human hematopoiesis, especially CD43+ cells. Blood Sci.
2:117–128. 2020.PubMed/NCBI View Article : Google Scholar
|
|
106
|
Gao Y, Wang F, Zhang L, Kang M, Zhu L, Xu
L, Liang W and Zhang W: LINC00311 promotes cancer stem-like
properties by targeting miR-330-5p/TLR4 pathway in human papillary
thyroid cancer. Cancer Med. 9:1515–1528. 2020.PubMed/NCBI View Article : Google Scholar
|
|
107
|
Flosbach M, Oberle SG, Scherer S, Zecha J,
von Hoesslin M, Wiede F, Chennupati V, Cullen JG, List M, Pauling
JK, et al: PTPN2 deficiency enhances programmed T cell expansion
and survival capacity of activated T cells. Cell Rep.
32(107957)2020.PubMed/NCBI View Article : Google Scholar
|
|
108
|
Li Z, Chao TC, Chang KY, Lin N, Patil VS,
Shimizu C, Head SR, Burns JC and Rana TM: The long noncoding RNA
THRIL regulates TNFα expression through its interaction with
hnRNPL. Proc Natl Acad Sci USA. 111:1002–1007. 2014.PubMed/NCBI View Article : Google Scholar
|
|
109
|
Xu LB, Bo BX, Xiong J, Ren YJ, Han D, Wei
SH and Ren XP: Long non-coding RNA LINC00887 promotes progression
of lung carcinoma by targeting the microRNA-206/NRP1 axis. Oncol
Lett. 21(87)2021.PubMed/NCBI View Article : Google Scholar
|
|
110
|
Byun S, Affolter KE, Snow AK, Curtin K,
Cannon AR, Cannon-Albright LA, Thota R and Neklason DW:
Differential methylation of G-protein coupled receptor signaling
genes in gastrointestinal neuroendocrine tumors. Sci Rep.
11(12303)2021.PubMed/NCBI View Article : Google Scholar
|
|
111
|
Fang Z, Zhong M, Zhou L, Le Y, Wang H and
Fang Z: Low-density lipoprotein receptor-related protein 8
facilitates the proliferation and invasion of non-small cell lung
cancer cells by regulating the Wnt/β-catenin signaling pathway.
Bioengineered. 13:6807–6818. 2022.PubMed/NCBI View Article : Google Scholar
|
|
112
|
Huang R, Liu J, Chen X, Zhi Y, Ding S,
Ming J, Li Y, Wang Y and Na J: A long non-coding RNA LncSync
regulates mouse cardiomyocyte homeostasis and cardiac hypertrophy
through coordination of miRNA actions. Protein Cell. 14:153–157.
2023.PubMed/NCBI View Article : Google Scholar
|
|
113
|
Li N and Zhan X and Zhan X: The lncRNA
SNHG3 regulates energy metabolism of ovarian cancer by an analysis
of mitochondrial proteomes. Gynecol Oncol. 150:343–354.
2018.PubMed/NCBI View Article : Google Scholar
|
|
114
|
Zhen H, Du P, Yi Q, Tang X and Wang T:
LINC00958 promotes bladder cancer carcinogenesis by targeting
miR-490-3p and AURKA. BMC Cancer. 21(1145)2021.PubMed/NCBI View Article : Google Scholar
|
|
115
|
Majumdar R, Bandyopadhyay A, Deng H and
Maitra U: Phosphorylation of mammalian translation initiation
factor 5 (eIF5) in vitro and in vivo. Nucleic Acids Res.
30:1154–1162. 2002.PubMed/NCBI View Article : Google Scholar
|
|
116
|
Xu L, Wu Q, Yan H, Shu C, Fan W, Tong X
and Li Q: Long noncoding RNA KB-1460A1.5 inhibits glioma
tumorigenesis via miR-130a-3p/TSC1/mTOR/YY1 feedback loop. Cancer
Lett. 525:33–45. 2022.PubMed/NCBI View Article : Google Scholar
|
|
117
|
Wang F, Peters R, Jia J, Mudd M, Salemi M,
Allers L, Javed R, Duque TLA, Paddar MA, Trosdal ES, et al: ATG5
provides host protection acting as a switch in the atg8ylation
cascade between autophagy and secretion. Dev Cell. 58:866–884.e8.
2023.PubMed/NCBI View Article : Google Scholar
|
|
118
|
Cruickshank BM, Wasson MD, Brown JM,
Fernando W, Venkatesh J, Walker OL, Morales-Quintanilla F, Dahn ML,
Vidovic D, Dean CA, et al: LncRNA PART1 promotes proliferation and
migration, is associated with cancer stem cells, and alters the
miRNA landscape in triple-negative breast cancer. Cancers (Basel).
13(2644)2021.PubMed/NCBI View Article : Google Scholar
|
|
119
|
Vogler M: BCL2A1: The underdog in the BCL2
family. Cell Death Differ. 19:67–74. 2012.PubMed/NCBI View Article : Google Scholar
|
|
120
|
Barriocanal M, Carnero E, Segura V and
Fortes P: Long non-coding RNA BST2/BISPR is induced by IFN and
regulates the expression of the antiviral factor tetherin. Front
Immunol. 5(655)2015.PubMed/NCBI View Article : Google Scholar
|
|
121
|
Mohamed Haroon M, Lakshmanan V, Sarkar SR,
Lei K, Vemula PK and Palakodeti D: Mitochondrial state determines
functionally divergent stem cell population in planaria. Stem Cell
Reports. 16:1302–1316. 2021.PubMed/NCBI View Article : Google Scholar
|
|
122
|
Zhu W, Zhou BL, Rong LJ, Ye L, Xu HJ, Zhou
Y, Yan XJ, Liu WD, Zhu B, Wang L, et al: Roles of PTBP1 in
alternative splicing, glycolysis, and oncogensis. J Zhejiang Univ
Sci B. 21:122–136. 2020.PubMed/NCBI View Article : Google Scholar
|
|
123
|
Lee MY, Sumpter R Jr, Zou Z,
Sirasanagandla S, Wei Y, Mishra P, Rosewich H, Crane DI and Levine
B: Peroxisomal protein PEX13 functions in selective autophagy. EMBO
Rep. 18:48–60. 2017.PubMed/NCBI View Article : Google Scholar
|
|
124
|
L'Abbate A, Tolomeo D, De Astis F, Lonoce
A, Lo Cunsolo C, Mühlematter D, Schoumans J, Vandenberghe P, Van
Hoof A, Palumbo O, et al: t(15;21) translocations leading to the
concurrent downregulation of RUNX1 and its transcription factor
partner genes SIN3A and TCF12 in myeloid disorders. Mol Cancer.
14(211)2015.PubMed/NCBI View Article : Google Scholar
|
|
125
|
Stelzer G, Rosen N, Plaschkes I, Zimmerman
S, Twik M, Fishilevich S, Stein TI, Nudel R, Lieder I, Mazor Y, et
al: The GeneCards suite: From gene data mining to disease genome
sequence analyses. Curr Protoc Bioinformatics. 54:1.30.1–1.30.33.
2016.PubMed/NCBI View
Article : Google Scholar
|
|
126
|
Laudet V: Evolution of the nuclear
receptor superfamily: Early diversification from an ancestral
orphan receptor. J Mol Endocrinol. 19:207–226. 1997.PubMed/NCBI View Article : Google Scholar
|
|
127
|
Letunic I and Bork P: Interactive tree of
life (iTOL) v4: Recent updates and new developments. Nucleic Acids
Res. 47 (W1):W256–W259. 2019.PubMed/NCBI View Article : Google Scholar
|
|
128
|
Mouse Genome Sequencing Consortium.
Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal
P, Agarwala R, Ainscough R, Alexandersson M, et al: Initial
sequencing and comparative analysis of the mouse genome. Nature.
420:520–562. 2002.PubMed/NCBI View Article : Google Scholar
|
|
129
|
Castoe TA, de Koning APJ, Hall KT, Card
DC, Schield DR, Fujita MK, Ruggiero RP, Degner JF, Daza JM, Gu W,
et al: The Burmese python genome reveals the molecular basis for
extreme adaptation in snakes. Proc Natl Acad Sci USA.
110:20645–20650. 2013.PubMed/NCBI View Article : Google Scholar
|
|
130
|
Howe K, Clark MD, Torroja CF, Torrance J,
Berthelot C, Muffato M, Collins JE, Humphray S, McLaren K, Matthews
L, et al: The zebrafish reference genome sequence and its
relationship to the human genome. Nature. 496:498–503.
2013.PubMed/NCBI View Article : Google Scholar
|
|
131
|
Adams MD, Celniker SE, Holt RA, Evans CA,
Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF,
et al: The genome sequence of Drosophila melanogaster.
Science. 287:2185–2195. 2000.PubMed/NCBI View Article : Google Scholar
|
|
132
|
Hodgkin J, Plasterk RH and Waterston RH:
The nematode Caenorhabditis elegans and its genome. Science.
270:410–414. 1995.PubMed/NCBI View Article : Google Scholar
|
|
133
|
Blattner FR, Plunkett G III, Bloch CA,
Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK,
Mayhew GF, et al: The complete genome sequence of Escherichia
coli K-12. Science. 277:1453–1462. 1997.PubMed/NCBI View Article : Google Scholar
|
|
134
|
Weng S, Dong Q, Balakrishnan R, Christie
K, Costanzo M, Dolinski K, Dwight SS, Engel S, Fisk DG, Hong E, et
al: Saccharomyces genome database (SGD) provides biochemical and
structural information for budding yeast proteins. Nucleic Acids
Res. 31:216–218. 2003.PubMed/NCBI View Article : Google Scholar
|
|
135
|
Petersohn A, Brigulla M, Haas S, Hoheisel
JD, Völker U and Hecker M: Global analysis of the general stress
response of Bacillus subtilis. J Bacteriol. 183:5617–5631.
2001.PubMed/NCBI View Article : Google Scholar
|
|
136
|
Ebenezer TE, Carrington M, Lebert M, Kelly
S and Field MC: Euglena gracilis genome and transcriptome:
Organelles, nuclear genome assembly strategies and initial
features. Adv Exp Med Biol. 979:125–140. 2017.PubMed/NCBI View Article : Google Scholar
|
|
137
|
Owen GI and Zelent A: Origins and
evolutionary diversification of the nuclear receptor superfamily.
Cell Mol Life Sci. 57:809–827. 2000.PubMed/NCBI View Article : Google Scholar
|
|
138
|
Holzer G, Markov GV and Laudet V:
Evolution of nuclear receptors and ligand signaling: Toward a soft
key-lock model? Curr Top Dev Biol. 125:1–38. 2017.PubMed/NCBI View Article : Google Scholar
|
|
139
|
Penvose A, Keenan JL, Bray D, Ramlall V
and Siggers T: Comprehensive study of nuclear receptor DNA binding
provides a revised framework for understanding receptor
specificity. Nat Commun. 10(2514)2019.PubMed/NCBI View Article : Google Scholar
|
|
140
|
Cotnoir-White D, Laperrière D and Mader S:
Evolution of the repertoire of nuclear receptor binding sites in
genomes. Mol Cell Endocrinol. 334:76–82. 2011.PubMed/NCBI View Article : Google Scholar
|
|
141
|
Hanly D, Esteller M and Berdasco M:
Altered long non-coding RNA expression in cancer: Potential
biomarkers and therapeutic targets? In: Chemical Epigenetics. Mai A
(ed). Topics in Medicinal Chemistry. Vol. 33. Springer, Cham,
pp401-428, 2019.
|
|
142
|
Fu D, Shi Y, Liu JB, Wu TM, Jia CY, Yang
HQ, Zhang DD, Yang XL, Wang HM and Ma YS: Targeting long non-coding
RNA to therapeutically regulate gene expression in cancer. Mol Ther
Nucleic Acids. 21:712–724. 2020.PubMed/NCBI View Article : Google Scholar
|
|
143
|
Zhang L, Peng D, Sood AK, Dang CV and
Zhong X: Shedding light on the dark cancer genomes: Long noncoding
RNAs as novel biomarkers and potential therapeutic targets for
cancer. Mol Cancer Ther. 17:1816–1823. 2018.PubMed/NCBI View Article : Google Scholar
|
|
144
|
Fatima R, Akhade VS, Pal D and Rao SM:
Long noncoding RNAs in development and cancer: Potential biomarkers
and therapeutic targets. Mol Cell Ther. 3(5)2015.PubMed/NCBI View Article : Google Scholar
|