Introduction
As per the central dogma of biology, RNA functions
as an intermediate molecule, assisting transfer genetic information
from the DNA level to the protein synthesis level. For a number of
years, this process was considered the primary pathway for gene
expression. However, over time, the functional importance of RNA
transcripts and other non-coding regions, that are not directly
encoded to proteins, has been recognized. Various of these
non-coding RNAs (ncRNAs) are involved in several cellular
functions, including the regulation of gene expression, alternative
splicing, RNA processing, the inhibition of translation and mRNA
degradation. In fact, RNA plays a crucial role in almost every
level of genome regulation (1-3).
ncRNAs are typically divided into two main classes:
Structural and regulatory. Structural ncRNAs include ribosomal
RNAs, transfer RNAs and small nuclear RNAs. On the other hand,
regulatory ncRNAs are further classified into several
subcategories, such as microRNAs (miRNAs/miRs), Piwi-interacting
RNAs, small interfering RNAs and long ncRNAs (lncRNAs). In
addition, a subclass of promoter-associated RNAs that interacts
with RNA promoters and is produced by genomic enhancer regions
(enhancer RNAs) has previously been described (4).
lncRNAs are a highly diverse and heterogeneous class
of ncRNAs, varying in their characteristics, locations in the
genome and functional roles. They are the most abundant class of
non-coding transcripts, and they typically have low expression
levels and are >200 nucleotides in length (5). These RNAs can be categorized based on
their location in the genome, into intronic, intergenic (lincRNAs),
divergent, sense and antisense lncRNAs (6,7).
Unlike small ncRNAs, lncRNAs are less conserved across species and
their roles in the regulation of gene expression are not yet fully
understood (8).
Even though the limited number of functional lncRNAs
have been well characterized, they play essential roles in the
regulation of gene expression at various stages. lncRNAs are
involved in post-transcriptional regulation by controlling protein
synthesis, RNA maturation and RNA transport, as well as in gene
silencing through modulation of chromatin structure. Due to the
vast variety of lncRNAs, categorizing them is challenging. However,
several studies have classified them based on their molecular
functions, as signaling molecules that relay temporal, spatial and
developmental information; as decoys that isolate various RNA
molecules and proteins to inhibit their activity; as guides that
direct epigenetic regulators and transcriptional factors to
specific genomic loci; and as scaffolds that facilitate the
formation of macromolecular complexes with a various functions
(8). Notably, individual lncRNAs
may function through more than one mechanism, potentially leading
to the combination of functions that create more complex functions
in cellular procedures. Thus, understanding the shared features of
these mechanisms is crucial for elucidating the functions of
lncRNAs and their crucial roles in cellular regulation (8).
In addition to the aforementioned functions, several
lncRNAs regulate gene expression by binding to miRNAs, functioning
as ‘miRNA sponges’. These lncRNAs contain multiple binding sites
for one or more miRNAs and prevent their interaction with target
mRNAs. As a result, the target mRNAs of the miRNAs are upregulated,
increasing their expression (8,9).
The direct and indirect interactions between lncRNAs
and transcription factors are well-documented. For instance, lncRNA
Gas5, which is activated in the absence of growth factors, contains
a hairpin structure sequence pattern that resembles the DNA-binding
site of the glucocorticoid receptor (GR), a member of the family of
nuclear receptors (NRs). By acting as a decoy, Gas5 blocks the
access of GR to DNA, thus inhibiting the transcription of GR
metabolic target genes (10). NRs
are transcription factors that bind to DNA and regulate the
expression of genes involved in processes, such as homeostasis,
stress response and metabolism (11). The dysregulation of NRs has been
reported in various diseases, including neurodegenerative diseases,
cancer, metabolic syndromes, diabetes, obesity and autism (12). Thus, understanding the mechanisms of
interaction between lncRNAs and NRs is critical for determining the
complex molecular pathways involved and could lead to the
development of more effective and specialized therapies for plenty
of diseases.
To further explore the regulatory roles of lncRNAs
in gene expression, the present study examined lncRNAs whose
expression levels were altered following the activation of an NR.
These lncRNAs were classified into two groups as follows: Those
that are upregulated and those that are downregulated. The aim was
to explore whether any lncRNAs fail to exhibit a regulatory
function. Considering the evolutionary conservation of lncRNAs, it
was hypothesized that the presence of sequence motifs within lncRNA
sequences could provide insight into their regulatory roles. These
motifs, if present with non-random frequency, may serve as
signatures that contribute to the regulatory functions of
lncRNAs.
Using the MEME algorithm, the lncRNA sequences from
the aforementioned groups were analyzed and six specific sequence
motifs (three in each group) were identified. The potential roles
of these motifs were investigated, providing insight into how
lncRNAs containing these motifs may regulate a wide range of
biological processes. According to the results obtained, one of the
discovered motifs was found to be complementary to the miRNA
hsa-miR-1908-3p, suggesting that lncRNAs containing this motif may
act as sponges for this miRNA, regulating the expression of its
target mRNAs. Additionally, all six motifs were detected in
specific genomic regions of human genome, particularly on
chromosome 19, with a high percentage of occurrence. Among these
regions on chromosome 19, the motifs were aligned with the
adeno-associated virus (AAV) integration site 1 (AAVS1), a known
hotspot for the AAV integration. Notably, a number of lncRNAs
containing at least one of these motifs, participate in various
biological pathways such as development, immune response,
inflammation, cell cycle, regulation of gene expression and
homeostasis, highlighting the diverse roles of lncRNAs in cellular
pathways. To explore the evolutionary significance of these motifs,
the present study investigated their presence across various
species, including Mus musculus, Squamata (lizard), Danio
rerio (zebrafish), Drosophila melanogaster, Caenorhabditis elegans,
Escherichia coli (K12 MG1655), Saccharomyces cerevisiae,
Bacillus subtilis (PY79) and Euglena gracilis, which
differ both evolutionarily and genetically. This cross-species
analysis revealed that these motifs are evolutionary conserved,
suggesting their potential importance in fundamental biological
processes. The conservation of these motifs across a wide range of
species highlights their potential functional relevance, warranting
further investigation into their roles in gene regulation and
interaction with NRs.
Data and methods
The present study developed an in-house pipeline for
the analysis of the collected dataset of lncRNAs with known
expression levels, as documented in the literature. These lncRNAs
are related to the activation and expression levels of NRs. This
pipeline includes the following key steps: i) Literature review and
data retrieval: A comprehensive search in current literature was
conducted to identify lncRNAs whose expression levels are known to
be associated with the activation of NRs. In this stage, the
corresponding RNA sequences were also retrieved for further
analysis. ii) Classification of lncRNAs: The identified lncRNAs
were classified into two groups. The first group included lncRNAs
whose expression was upregulated following the activation of an NR,
while the second group included lncRNAs whose expression was
downregulated in response to the activation of an NR. Subsequently,
conserved sequence motifs were identified within these two lncRNA
sub-groups. iii) Statistical evaluation of motif significance: The
identified motifs were statistically evaluated to determine their
significance. This involved estimating the probability of these
motifs occurring randomly within the entire human DNA. iv)
Exploration of motif functionality: In this step, we explored the
potential functions of the identified motifs, focusing on their
role in gene regulation. This included investigating their
potential to function as binding sites for miRNAs, examining their
presence within genome regulatory regions and exploring their
conservation through evolution. These analyses helped broaden the
understanding of the roles of these lncRNAs in cellular processes.
A detailed description of the in-house pipeline is provided
below.
Data collection and filtering and annotation.
The IDs of lncRNAs related to the NRs were collected from the
literature according to the study by Foulds et al (13). The IDs of lncRNAs had the following
formats: i) Name of lncRNA (e.g., MALAT-1); ii) ID such as
AC145110.1; iii) ID such as NR_026765; iv) ID such as
RP1-148H17.1.
The first three ID formats correspond to different
types of IDs detected at GenBank, a comprehensive genetic sequence
database available at NCBI (ncbi.nlm.nih.gov). The fourth ID format derived from
LNCipedia database (14), a novel
database for human lncRNA transcripts and genes.
Based on these ID formats, the dataset of lncRNAs
was classified into four subcategories. To retrieve the
corresponding sequences for each subcategory, an in-house algorithm
was developed using the MATLAB bioinformatics toolbox. The details
of the four algorithms used for sequence retrieval are presented in
Data S1.
Following the creation of the four individual
algorithms for each type of lncRNA ID, we developed a unified
algorithm designed to retrieve the sequences of the entire dataset,
containing all ID formats simultaneously. The aggregate algorithm
used for retrieving all lncRNA sequences is presented in Data S2.
Dataset classification and motif exploration.
In order to further classify the lncRNAs within the two
sub-datasets, the present study investigated the presence of joint
motifs across the lncRNA sequences in each subgroup. Following the
collection, filtering and annotation of the data, the Multiple EM
for Motif Elicitation (MEME) software, version 4.10.0(15), was employed to identify joint motifs
within the lncRNA sequences of each subgroup. MEME is a part of the
ΜEME Suite (https://meme-suite.org/meme/), which is a
comprehensive set of tools for motif-based sequence analysis. This
tool is designed to detect motifs or sequence patterns that occur
repeatedly in a group of related nucleotide or protein sequences,
finding common biological functions and structural features across
different sequences. The term ‘pattern’ or ‘motif’ refers to a
short, sequence that repeats within a group of similar sequences.
The motifs have no gaps, while the MEME algorithm calculates how
often each motif appears across sequences, allowing the detection
of multiple motifs (15). For the
purposes of the present study, the MEME algorithm was run on a
local server using the Linux operating system.
Statistical evaluation of the randomness of the
occurrence of the discovered motifs. To calculate the
probability of finding specific nucleotide bases of each motif at a
random position within the 3.2 billion bases of the human DNA, it
was assumed that the occurrence of a nucleotide at one position in
the motif's sequence is independent of the occurrence of
nucleotides at other positions. This assumption represents the
simplest hypothesis, involving the fewest parameters. Thus, the
rule of probabilities for independent events was applied, which
defines the probability of multiple events occurring independently.
Specifically, two events are considered independent if the
occurrence or non-occurrence of one does not influence the
probability of the other event's occurrence or non-occurrence
(16,17). To calculate the probability of both
events A and B occurring together, the formula for independent
events was used: P (A and B)=P (A) x P (B). This approach was
applied to assess the randomness of each motif existence within
human DNA.
Exploration of the potential role of the
identified motifs. A total of six motifs were discovered, three
in each subgroup of lncRNAs. To explore the potential role of the
collected lncRNAs in the regulation of gene expression, the present
study focused on the lncRNAs that contained at least one of the
discovered motifs. Since motifs are usually conserved through
evolution and contribute crucial functions to the regions they
occupy, the authors proceeded to explore the potential roles of
these motifs in genome regulation, which could reflect the
functions of the lncRNAs that contain them. For this purpose, two
different approaches were pursued. The first included querying the
miRBase database (https://www.mirbase.org/) to identify mature miRNAs
with complementary sequences to these motifs, suggesting the
possibility of miRNA-lncRNA interaction. The second approach
included the exploration of the presence of these motif sequences
across whole human DNA in order to determine their existence in
other genomic regions, assess whether they were associated with
regulatory regions, and explore potential roles in genomic
regulation. In addition, the evolutionary conservation of the
motifs was explored to determine the maintenance of their functions
through evolution.
Exploration of the interactions of miRNA target
genes. The main purpose of this approach was to identify the
potential role of lncRNAs containing the identified motifs in
functioning as ‘sponges’ for miRNAs, where the motifs serve as
interaction/binding regions between lncRNAs and miRNAs. For this
purpose, the blastn algorithm from miRbase was used, with the
complement sequence of each motif as input. This allowed this
algorithm to search for similarities between the motif sequences
and known miRNA sequences in the miRBase. Subsequently, the authors
further investigated the predicted target genes of the identified
miRNA using the bioinformatic tool miRTarget from the miRDB
database (16), and the TargetScanS
database (Release 8.0) (17-19).
Finally, the location of the binding sites in the
target mRNAs was examined by using the multiple alignment algorithm
ClustalW, using as input the mRNA sequence of the targets and the
complementary sequence of miR-1908-3p.
Exploration of motif sequences in whole human
DNA. To investigate whether these motif sequences also appear
in other regions of the genome, the online tool Find Individual
Motif Occurrences (FIMO) from the MEME Suite was used. This tool is
an algorithm designed to identify specific matches of a given motif
within a set of sequences, specialized for analyzing shorter
sequences such as motifs. Similar to a BLAST algorithm, FIMO
searches for matches to each motif. When employed locally, motifs
must be in MEME Motif Format; however, the online version of FIMO
supports additional motif formats.
Subsequently, an algorithm was developed using the
MATLAB Bioinformatics Toolbox to create a chromosome plot for each
identified motif. This plot displayed the locations of the motif
sequences by marking them with a red dot next to the corresponding
regions on the chromosome where they were detected. The input for
this algorithm was the output from the FIMO tool in tsv format. The
specific algorithm we developed is presented in Data S3.
The FIMO tool was also used to examine the presence
of the identified sequence motifs in the genomes of various
organisms, including Mus musculus, Squamata (lizard),
Danio rerio (zebrafish), Drosophila melanogaster, Caenorhabditis
elegans, Escherichia coli (K12 MG1655), Saccharomyces
cerevisiae and Bacillus subtilis (PY79), to assess their
evolutionary conservation. Additionally, the present study
investigated the presence of these motifs in the genome of
Euglena gracilis, a protist believed to possess an ancient
form of NRs. Due to the unavailability of Euglena's genome
in the database of the FIMO tool, the ‘Matcher’ tool available on
the Galaxy platform (usegalaxy.org)
was employed to check for potential sequence matches (20).
Galaxy is an open-source, web-based platform that
aids computational biology research by providing a comprehensive
suite of tools for data analysis. The ‘Matcher’ tool within this
platform compares two different sequences to identify regions of
similarity, supporting both local and global alignments. This tool
enables the identification of conserved genomic regions across
species and helps in detecting functionally important motifs that
have been preserved through evolution. This tool is widely used in
genomics and bioinformatics for tasks such as gene annotation,
mutation analysis, and the discovery of conserved regulatory
regions (21).
Results and Discussion
Data collection, filtering and
annotation
The initial dataset, collected from the study by
Foulds et al (13),
contained 1,823 lncRNA IDs, with sequences for 1,788 lncRNAs
successfully identified and retrieved. Following the removal of the
duplicate sequences, the final dataset consisted of 1,753 lncRNAs.
These were then classified into two subgroups as follows: The first
subgroup contained 778 upregulated lncRNAs and the second subgroup
contained 975 downregulated lncRNAs (Table SI).
Dataset classification and motif
identification
The MEME algorithm was employed to identify
potential motifs within the lncRNA sequences of each subgroup. The
results of this analysis led to the identification of six motifs:
Three motifs in the upregulated lncRNA group and three in the
downregulated lncRNA group.
In the upregulated lncRNAs group following NR
activation, the first identified motif was
GATCCGCCCGCCTCGGCCTCCCAAAGTGCTGGGATTACAGG, found in 140 lncRNAs
from the total 778 (Fig. S1). The
second motif discovered in this group was
TGGCCAGGCTGGTCTCGAACTCCTGACCT, present in 127 lncRNAs (Fig. S2), while the third motif was
TAATCCCAGCACTTTGGGAGGCCGAGG, which occurs in 115 lncRNAs (Fig. S3).
For the downregulated lncRNA group following NR
activation, the first motif discovered was
GTGGCTCACGCCTGTAATCCCAGCACTTTGGGAGGCCGAGG, found in 145 lncRNA out
of the total of 975 (Fig. S4). The
second motif discovered was
AGGTCAGGAGTTCGAGACCAGCCTGGCCAACATGGTGAAAC, which is present in 128
lncRNAs (Fig. S5), and the third
motif was CCTCAGCCTCCCGAGTAGCTGGGACTACAGGCGC, found in 129 lncRNAs
from this group (Fig. S6).
Statistical evaluation
To calculate the probability of each motif appearing
randomly at a position within the 3.2 billion base pairs of human
DNA, the following steps were taken:
The first motif from the upregulated lncRNAs
subgroup consists of 41 nucleotide bases. Since each base (A, C, G,
or T) has a ¼ probability of being in a specific position in the
sequence, the probability of finding the full sequence of these 41
nucleotide bases is:
(¼)41=2.23517418x10-25.
Taking into consideration the total number of human
DNA base pairs (3.2 billion or 3.2x109), the probability
of this sequence occurring randomly at any position in the genome
is:
3.2x109x2.23517418x10-25=7.15255974x10-16.
Accordingly, for the second motif in the upregulated
lncRNA subgroup, consisting of 29 nucleotides, the probability
calculation is as follows:
(¼)29=1.8626451x10-18. The probability of
finding this specific sequence randomly in human genome is:
3.2x109x1.8626451x10-18=5.96174432x10-10.
For the third motif in the upregulated lncRNAs subgroup, which
consists of 27 nucleotides, the results are as follows:
(¼)27=7.8886x10-17. Thus, the probability of
its random occurrence is:
3.2x109x7.8886x10-17=2.52435x10-7.
In the downregulated lncRNA subgroup, both first and
second motifs consist of 41 nucleotides; thus, the same probability
calculations apply: (¼)41=2.23517418x10-25.
Thus, the probability of their random occurrence is:
3.2x109x2.23517418x10-25=7.15255974x10-16.
The third motif in the downregulated lncRNA subgroup, consists of
34 nucleotides: (¼)34=1.421085x10-21. Thus,
the probability of its random occurrence is:
3.2x109x1.421085x10-21=4.5474755x10-13.
The calculated probabilities are extraordinarily low
compared to common biological standards and are consistent with the
hypothesis that these motifs are not randomly occurring in the
genome (22).
The above-calculated probabilities suggest that the
motifs discovered are non-random occurrences in the human genome,
even when assuming independent events. However, the nucleotide
sequence of DNA is linked to biological functions. The order of the
bases is strictly determined by the way nucleotides bind each other
in the double helix structure. The complementary pairing of
nucleotides restricts the possible sequences that DNA can form,
meaning that the probability of these motifs occurring randomly in
the genome would likely be even lower than the values calculated
above.
Exploration of the potential role of
the identified motifs. Exploration of the miRNA-target gene
interactions
As previously mentioned, lncRNAs can function as
miRNA sponges, regulating the expression of mRNA targets (8,9).
Considering this regulatory role, we searched for potential miRNAs
complementary to the six identified motifs in miRBase. In the
present study, the search revealed that the motif
GATCCGCCCGCCTCGGCCTCCCAAAGTGCTGGGATTACAGG was complementary to
hsa-miR-1908-3p.
miRNAs typically bind to complementary sequences in
mRNAs, mainly targeting the 3' untranslated regions (UTRs)
(23), while there is evidence to
indicate that they can also target the 5' UTRs and coding regions,
thus inhibiting mRNA expression (24). To predict the potential targets of
miRNAs, several computational approaches have been developed, such
as miRanda (25), mirSVR (26), PicTar (27), TargetScan (28), TargetScanS (29), RNA22(30), PITA (31), RNAhybird (32) and DIANA-microT (33). In addition, there are miRNA target
prediction databases such as TarBase (34), PicTar (27) and TargetScan (28).
Thus, to identify mRNAs targeted by hsa-miR-1908-3p,
whose expression may be affected by the interaction between lncRNAs
and this miRNA, the miRDB database was used. This database, through
its miRTarget bioinformatics tool, predicts the mRNA based on
high-throughput sequencing experiments. By this analysis, 11
targets were identified, including METTL21A, CHFR, MTA1, CAVIN4,
IGFBP3, GRB2, DYNLL2, H2AFX, TNRC6C, C1orf229 and ELOVL2
(Table I). In addition, the
TargetScanS (Release 8.0) database (17-19,35)
was also searched, and 487 predicted targets were found (Table SII), including the
aforementioned 11 targets.
 | Table IThe target genes of mir-1908-3p were
predicted using the miRTarget tool of miRDB. |
Table I
The target genes of mir-1908-3p were
predicted using the miRTarget tool of miRDB.
| Gene symbol | Gene
description | miRNA name | Target score |
|---|
| METTL21A | Methyltransferase
like 21A |
hsa-miR-1908-3p | 80 |
| CHFR | Checkpoint with
forkhead and ring finger domains |
hsa-miR-1908-3p | 62 |
| MTA1 | Metastasis
associated 1 |
hsa-miR-1908-3p | 62 |
| CAVIN4 | Caveolae associated
protein 4 |
hsa-miR-1908-3p | 61 |
| IGFBP3 | Insulin like growth
factor binding protein 3 |
hsa-miR-1908-3p | 61 |
| GRB2 | Growth factor
receptor bound protein 2 |
hsa-miR-1908-3p | 56 |
| DYNLL2 | Dynein light chain
LC8-type 2 |
hsa-miR-1908-3p | 55 |
| H2AFX | H2A histone family
member X |
hsa-miR-1908-3p | 53 |
| TNRC6C | Trinucleotide
repeat containing 6C |
hsa-miR-1908-3p | 53 |
| C1orf229 | Chromosome 1 open
reading frame 229 |
hsa-miR-1908-3p | 51 |
| ELOVL2 | ELOVL fatty acid
elongase 2 |
hsa-miR-1908-3p | 50 |
miR-1908 is encoded by the first intron of the
FADS1 gene and is highly expressed in mature human
adipocytes, where it is likely involved in regulating the
preadipocyte differentiation (36).
The promoter region of miR-1908 contains two binding sites of
NF-κB, suggesting its transcription is regulated by this
transcription factor (37).
miR-1908 was first discovered in human embryonic stem cells in
2008(38), and its functional role
in melanoma metastasis and angiogenesis was explored in
2012(39). More specifically,
miR-1908, in conjunction with miR-199a-3p and miR-199a-5p, inhibits
apolipoprotein E and DNAJ heat shock protein family 23 member A4,
which reduces the interaction of Apo-E with low-density lipoprotein
receptor-related protein (LRP)-1 and LRP-8. This interaction
promotes melanoma cell invasion and endothelial cell recruitment,
resulting in poor prognosis (39).
Moreover, miR-1908 dysregulation has been shown to be associated
with various types of cancer, where its dysregulation tends to
enhance cell proliferation, invasion, migration and angiogenesis,
thus reducing the survival rate of affected patients (40,41).
Finally, free fatty acids and adipokines, such as resistin and
leptin, are considered to downregulate the expression of miR-1908,
suggesting a role of this miRNA in regulating obesity-related
insulin resistance (42).
Exploration of motif sequences in
whole human DNA
For the analysis of motif sequences across the whole
human genome using the FIMO tool, the following results were
obtained for each of the six motifs. In the upregulated lncRNA
subgroup, the first motif appeared 53,524 times, the second motif
was detected 51,673 times and the third motif was found 53,936
times. In the downregulated lncRNA subgroup, the first motif
(fourth in total) was found 63,126 times, the second motif (fifth
in total) was detected 58,442 times and the third motif (sixth in
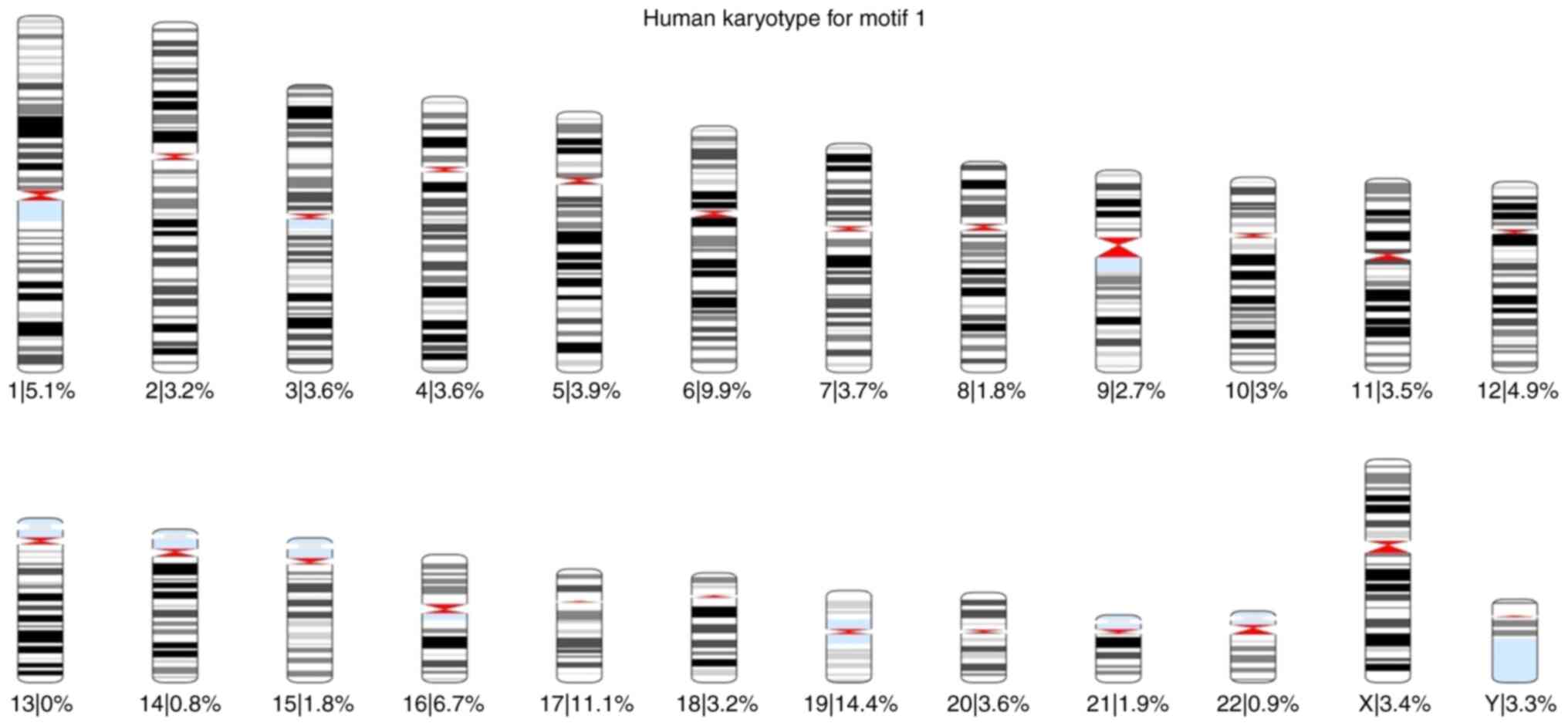
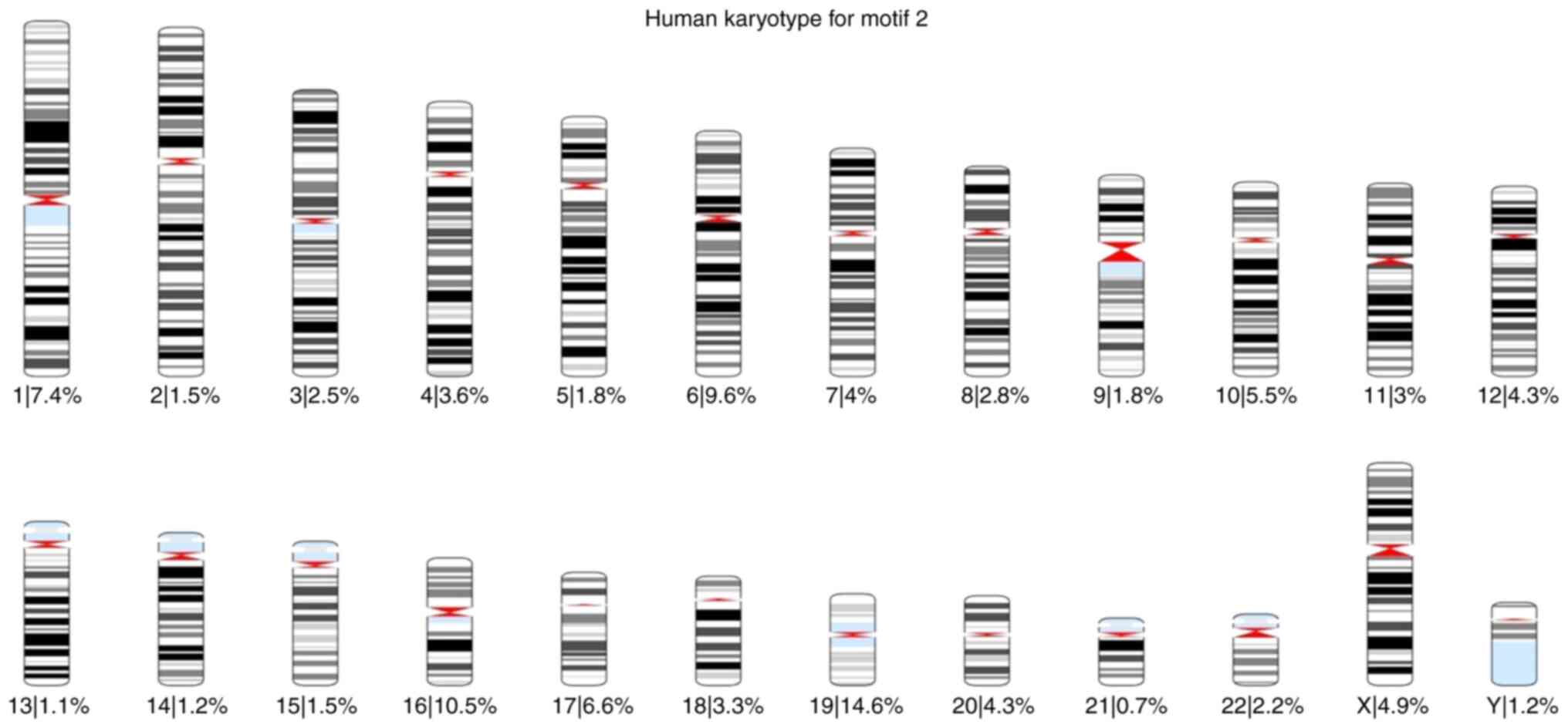
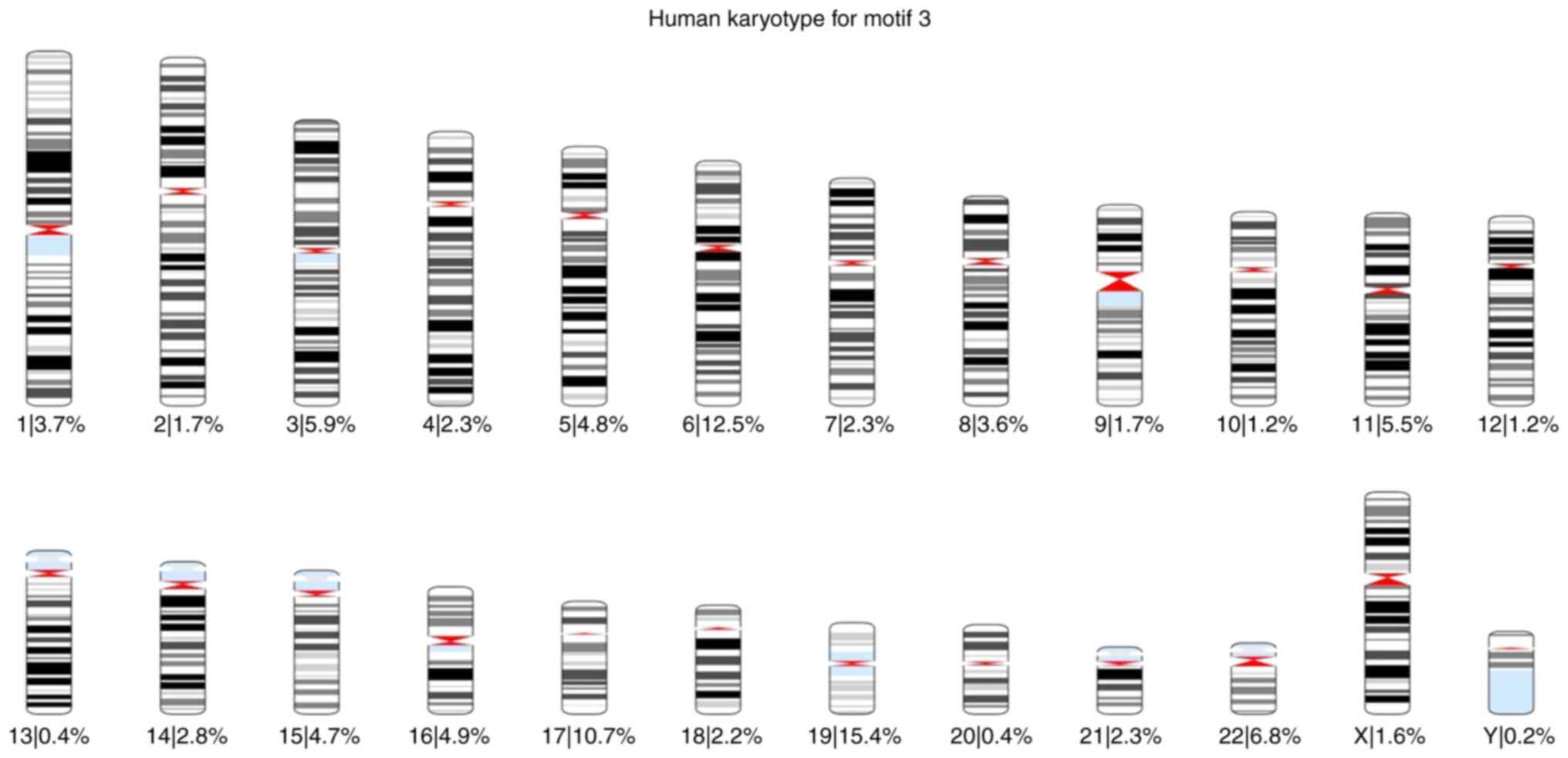
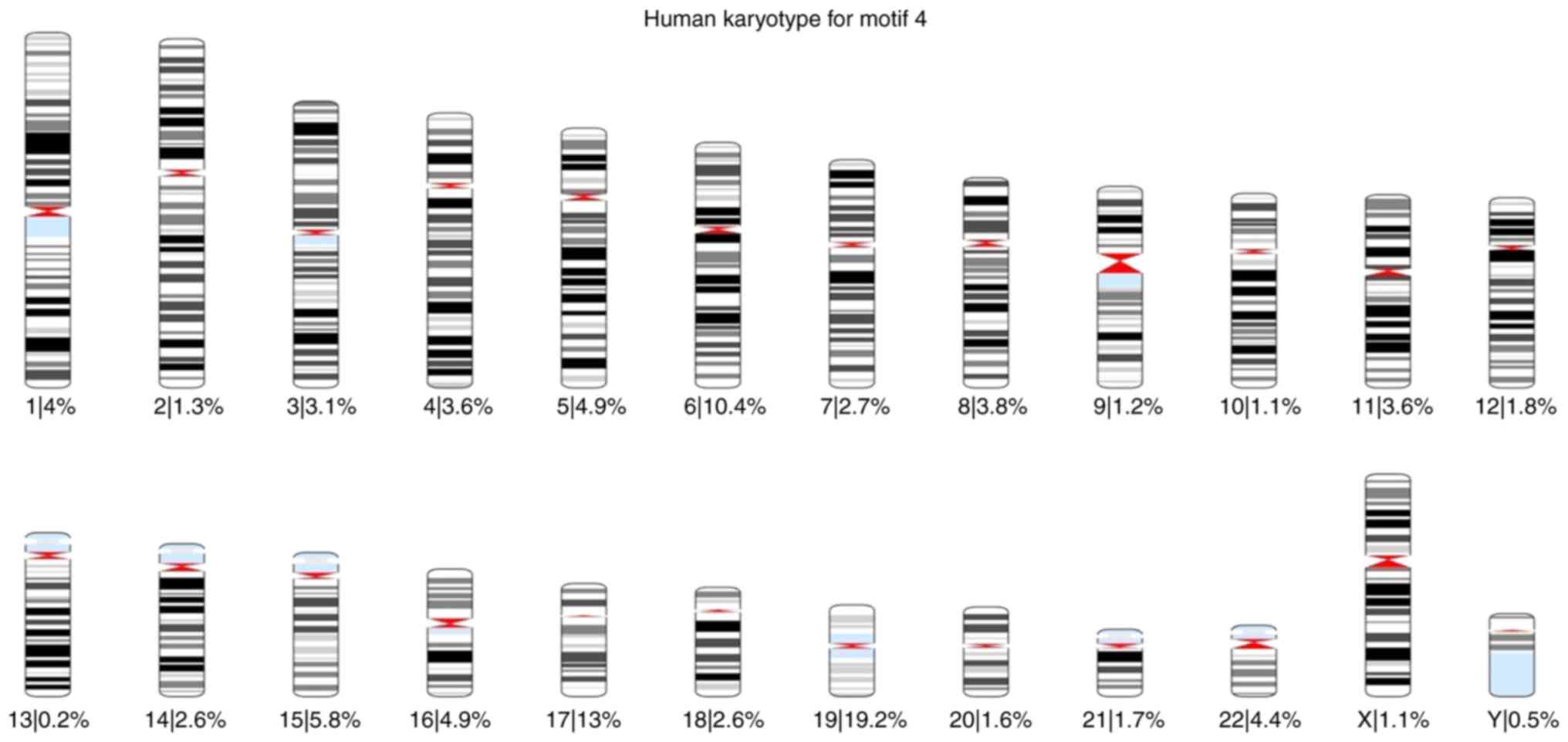
total) was found 57,943 times. Using these results, karyotypes were
created for each motif, and the percentage of the occurrences of
each motif across all chromosomes was calculated. The karyotypes
are presented in Fig. S7, Fig. S8, Fig.
S9, Fig. S10, Fig. S11 and Fig. S12.
Subsequently, the present study focused on the 1,000
highest-scoring loci which occurred using the FIMO tool for each
motif across the human genome. Using these 1,000 loci, karyotypes
for each motif were also created, and the percentage of occurrences
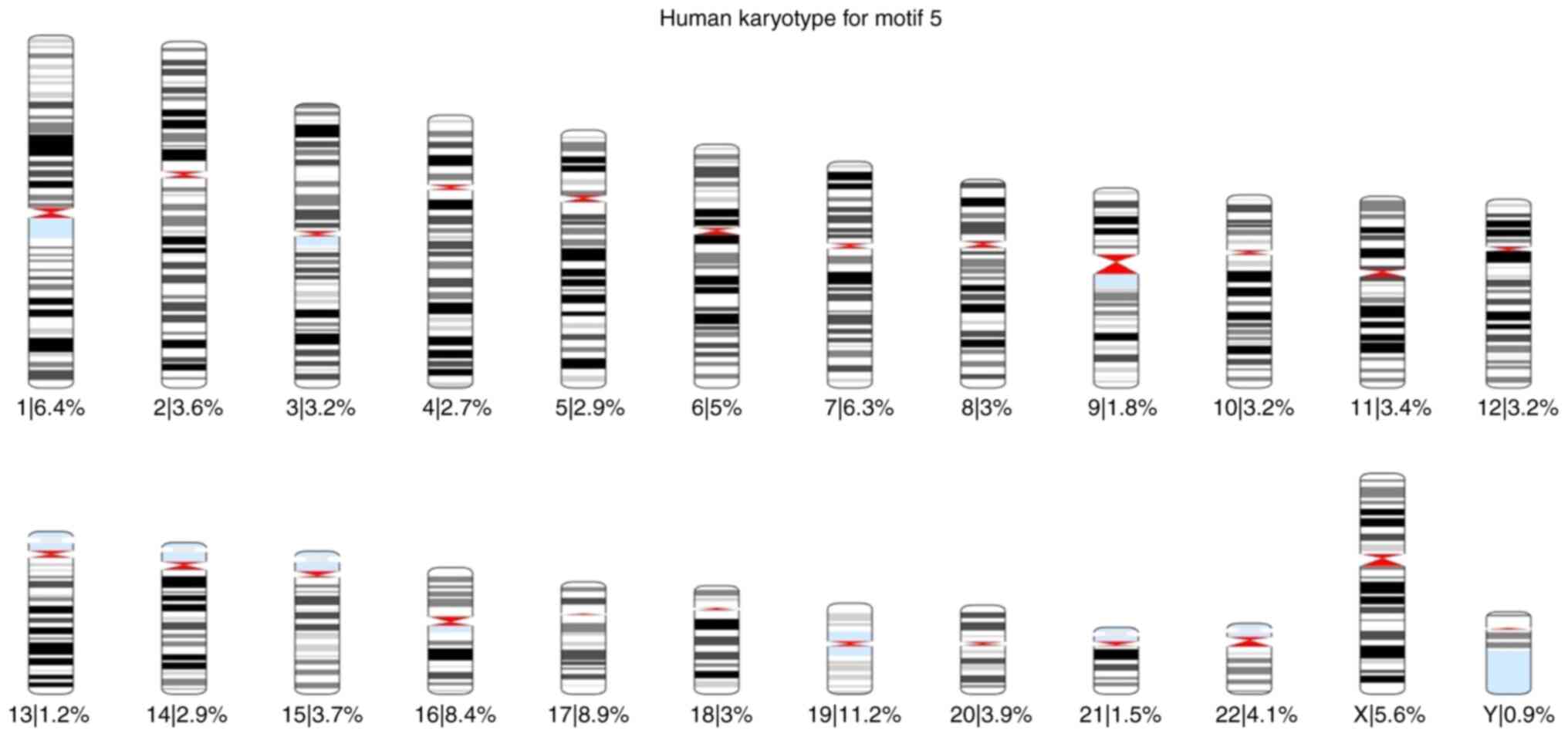
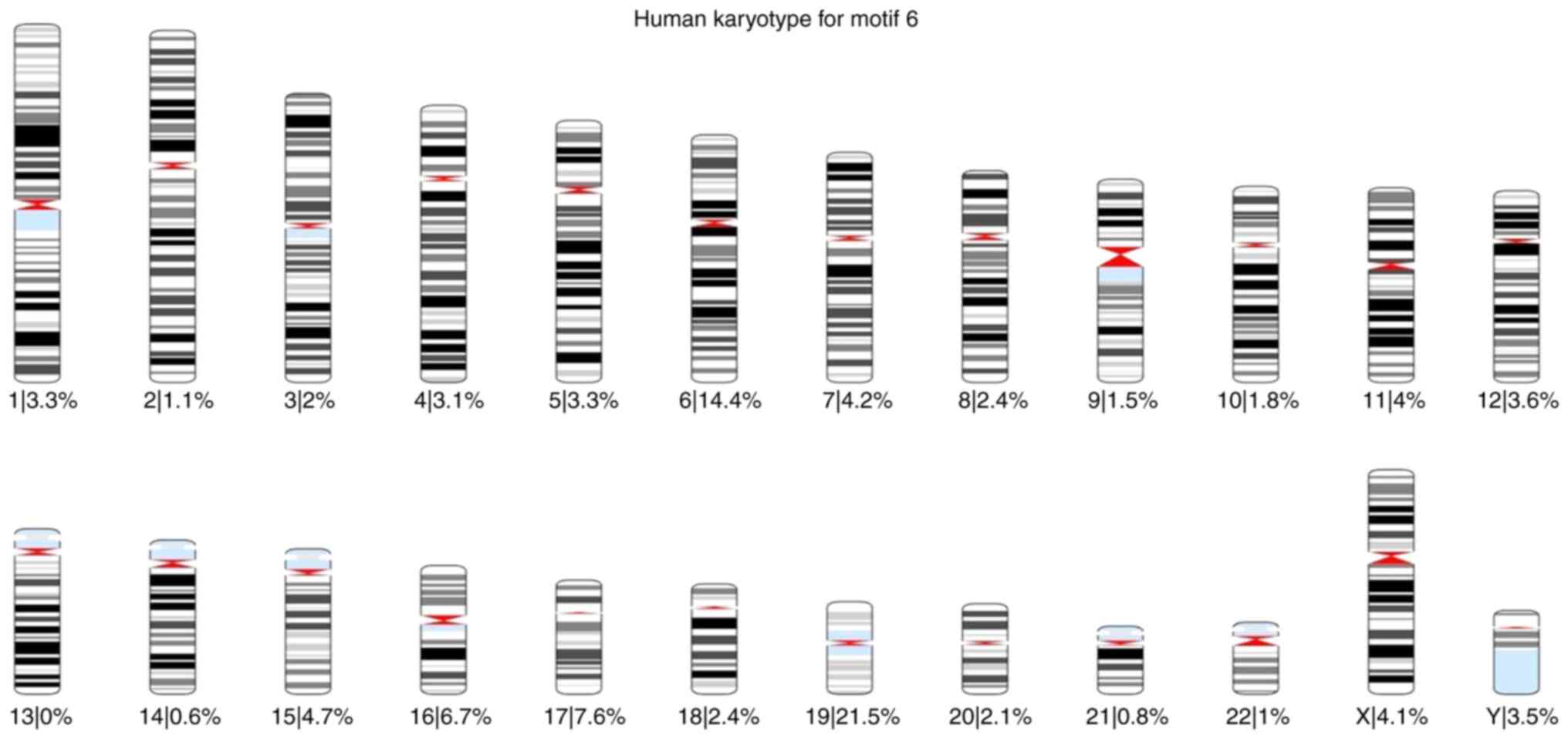
of each motif was also calculated for each chromosome (Fig. 1, Fig.
2, Fig. 3, Fig. 4, Fig.
5 and Fig. 6).
In addition, the chromosome plots for each motif
were created, where the 1,000 loci are represented as red dots
beside the corresponding region. These chromosome plots are
presented in Fig. S13, Fig. S14, Fig. S15, Fig. S16, Fig. S17 and Fig. S18.
Based on these karyotypes and chromosome plots, it
was observed that chromosome 19 exhibited a fairly high percentage
of motif occurrence relative to its size. In fact, for all six
motifs, the percentage of occurrence in chromosome 19 was higher
than that of the longer chromosomes.
According to the literature, human chromosome 19 is
recognized for its unusual nature, which was noted even before the
initial publication of its DNA sequence (43). One of its unusual features is the
considerably high gene density, which exceeds the genome-wide
average by >2-fold and includes 20 large, tandemly clustered
gene families (43). In addition,
chromosome 19 contains a considerable number of segmental
duplications, with 6.2% of its sequence lying within
intrachromosomal segmental duplications (43). The sequence divergence within these
duplications implies that these events occurred between 30 and 40
million years ago (44). These
duplications likely played a pivotal role in the evolution of
phenotypic characteristics that occurred by genes on chromosome 19
across primates, including humans. Moreover, chromosome 19 presents
an unusually high repeat content, comprising 55%, with Alu repeats
comprising 26% of the sequence of the chromosome (44). Another notable feature is the GC
content (48%) of chromosome 19, the highest of any human
chromosome, compared to the GC content of the whole human genome of
41%. This elevated GC content enhances the potential for wide gene
regulation via DNA methylation, particularly at CpG islands, as
well as in CpG regions located within promoters and enhancers
(45).
According to the chromosome plots, the six
identified motifs were detected in various regions of chromosome
19. Between those regions, all six motifs aligned with a specific
locus, AAVS1, located on chromosome 19q13.42. This site was first
mapped in 1991 Kotin et al (46) and Samulski et al (47), who were investigating the
integration mechanism of the AAV into the host genome. According to
these studies, the AAV preferentially intergrades into the AAVS1
region of human chromosome 19q13.42, a unique phenomenon among
eukaryotic DNA viruses (48).
Several features make the AAVS1 region particularly
noteworthy. Notably, the locus for myotonic dystrophy (49), as well as a common breakpoint
related to chronic B-cell leukemia (BCL-3) (47,48),
are located at this region. In addition, AAVS1 undergoes frequent
sister chromatid exchanges (50).
The alignments of motifs with the AAVS1 region were developed using
the Clustal Omega program (51) and
visualized with Jalview version 2.11.2.6 program (52) (Fig.
S19, Fig. S20, Fig. S21, Fig. S22, Fig. S23 and Fig. S24).
The AAVS1 region is located within the first intron
of the phosphatase 1 regulatory subunit 12C (PPP1R12C) gene,
which encodes a protein with a poorly defined function. This region
exhibits several unique features that contribute to its unique
nature. First, it includes a tetrad GACT repeat, which functions as
an AAV Rep-binding element, also detected in the AAV2 inverted
terminal repeats, facilitating the exclusive integration of the AAV
genome in the presence of AAV replication protein. Second,
chromosome 19 is GC-rich, with a higher number of CpG islands
relative to GpC islands, a characteristic linked to regions of
transcriptional activity in vertebrates, which may be relevant to
gene expression (53-55).
Third, several putative transcription factor-binding sites are
present in AAVS1, including CREB, AP-1, AP-2 and Sp1 (48,56).
Fourth, topoisomerase I is predicted to cleave at least two sites
within AAVS1(48). Fifth, Lamartina
et al (56) demonstrated an
open chromatin conformation of AAVS1, accompanied by promoter
activity on chromosome 19 in cultured cells. All these features
position AAVS1 as an approachable region, subject to regulation by
both host and viral regulatory elements.
The potential of the AAVS1 site as a target for gene
therapy applications has been investigated and well-characterized.
This site is known for its ability to integrate exogenous DNA with
limited disruption to endogenous genes, providing a preferred site
for stable transgene expression (57). A key feature that facilitates the
usage of the AAVS1 site in gene therapy is its open chromatin
structure, which is accompanied by endogenous insulator elements
that protect the integrated DNA sequences from trans-activation or
repression (58).
The insertion of exogenous genes into the human
genome is conducted mainly using lentiviral and gamma-retroviral
vectors. However, these vectors tend to integrate randomly within
the genome (59-61),
which can lead to unpredictable interactions between the transgene
and the host genome. Such random integration may result in the
attenuation or complete silencing of the transgene (62-64)
or, more critically, in the dysregulation of host gene expression
(64,65), which poses significant risks in gene
therapy application. One promising strategy to mitigate these risks
involves the use of gene-editing tools that enable site-specific
insertion of transgenes into genomic safe harbors (GSHs) (64,66,67).
GSHs are genomic locus that support a stable environment for
transgene expression without interfering the integrity of
endogenous genes (64,68,69).
The AAVS1 site is considered a GSH that provides a stable transgene
expression (70) due to the
presence of flanking insulator regions (58). According to the study by Lombardo
et al (66), the GSH AAVS1
can support transgene expression across different human cell types,
facilitated by the active and open chromatin structure of the
PPP1R12C gene located in this region (66).
The regulatory role of lncRNAs has been
well-characterized. Numerous studies have detected that lncRNAs
function as crucial regulators of gene expression, participating in
various pathways by acting as decoys, guides and scaffolds for
molecular interactions. In the present study, it was hypothesized
that the regulatory roles of lncRNAs are indisputable, and to
investigate this hypothesis, an in-house pipeline was developed to
analyze a collection of lncRNAs. This pipeline enabled the
identification of conserved motifs within their sequences that may
contribute to their regulatory functions. The analysis revealed
that one of the motifs may function as a sponge for a specific
miRNA, thereby suppressing the function of the miRNA and leading to
the upregulation of its mRNA targets. The function of miRNAs in the
suppression of the translation of mRNA targets is well known, with
implications for numerous pathological conditions, such as
neurodegeneration (71-73),
cancer (72,74,75)
and cardiovascular diseases (74,76,77).
Consequently, the interaction between lncRNAs and miRNAs could have
significant consequences, either promoting disease prevention and
treatment or exacerbating disease progression and severity.
Furthermore, all six motifs identified in the
present study were detected across numerous regions of the human
genome, with chromosome 19 presenting a markedly high frequency of
motif occurrence relative to other chromosomes. Focusing on
chromosome 19, it was found that these motifs were aligned with the
AAVS1 regulatory region, a genomic locus known for its unique
features, including the specific integration of the AAV, the
presence of transcription factor binding sites, an AAV Rep-binding
element, high GC-content, open chromatin, and two cleavage sites
for topoisomerase I. It is noteworthy that the identified motifs
demonstrate a pronounced GC content, a feature commonly associated
with key regulatory regions, including transcription factor binding
sites, polymerase binding sites, and loci involved in chromatin
modification and DNA methylation. GC-rich sequences are often
enriched at genomic sites that regulate transcription and chromatin
remodeling due to their structural properties, such as enhanced
stability and the presence of CpG sites that are key for epigenetic
regulation (78-81).
The existence of these motifs in regions integral to the regulation
of the human genome, combined with their GC-rich nature, suggests
their role in regulating gene expression through DNA methylation
processes. Consequently, these motifs and the lncRNAs containing
them, likely function as pivotal regulatory elements in gene
expression.
In addition to the potential functions of lncRNAs
that contain the identified motifs-based on the presence of these
motif sequences in well-characterized regulatory regions of DNA and
their interactions with other molecules, including miRNAs, the
present study further investigated the documented roles of the
lncRNAs containing at least one of these motifs in the literature
(82-124)
and relevant databases such as NCBI (ncbi.nlm.nih.gov) and GeneCards (125). Despite the incomplete annotation
and characterization of these lncRNAs, the majority of the examined
lncRNAs are located either within intergenic space, introns of
coding genes, or are antisense to coding genes. These lncRNAs are
believed to influence the functions of their associated genes,
mainly by suppressing gene functions. The lncRNAs examined are
involved in various biological processes, such as development,
cellular proliferation, immune responses, inflammation,
transmembrane transport, gene expression regulation, chromatin and
chromosome structure modulation, differentiation, cell cycle
control, spermatogenesis, metabolic regulation, and several
functions related to the nervous and cardiovascular systems. The
results from the present study, as depicted in the diagrams shown
in Fig. S25, Fig. S26, Fig. S27, Fig. S28, Fig. S29 and Fig. S30, illustrate the involvement of
lncRNAs with each specific motif in these diverse biological
pathways.
The emergence of NRs dates back >1.5 billion
years, originating in early eukaryotic organisms as
ligand-dependent transcription factors. These receptors were
responsive to environmental cues such as hormones, light, and
metabolic shifts. The earliest NRs likely played a critical role in
regulating gene expression, helping primitive eukaryotes adapt to
dynamic external conditions, thus aiding their survival and
proliferation. Of note, unicellular organisms such as Euglena
gracilis, a photosynthetic protist, contain an ancestral
version of the NR, providing evidence that these mechanisms were
present well before the evolution of multicellular organisms. This
discovery enhances the understanding of the origins of NRs and
their evolutionary significance (126).
As evolution progressed, NRs diversified into a
complex protein family with distinct functions. Different
subfamilies evolved to regulate a wide variety of biological
processes across animals, plants, fungi and even bacteria. In
modern organisms, NRs govern critical processes such as metabolism,
immune response, reproduction, and development. The diversity of
these receptors is reflected in the large number of subfamilies,
including steroid hormone receptors, thyroid hormone receptors,
peroxisome proliferator-activated receptors and orphan NRs. Despite
their varied roles, these receptors retain a conserved
ligand-binding domain that allows them to interact with specific
signaling molecules to regulate gene expression (126).
The present study discovered six novel sequence
motifs within the lncRNA sequences under investigation. These
lncRNAs exhibited altered expression levels upon the activation of
NRs, either increasing or decreasing in abundance. Notably, several
of these lncRNAs, which contain at least one of the identified
motifs, are regulated by multiple NRs. This suggests that these
motifs may be associated with NR function but do not show
specificity to any single receptor, underscoring the complexity of
the regulatory network between NRs and ncRNAs. This finding
highlights the importance of further research to explore their
broader biological implications.
To investigate the evolutionary conservation of
these motifs, the FIMO tool was applied to examine their presence
in various organisms. The present study analyzed species such as
Mus musculus, Squamata (lizard), Danio rerio
(zebrafish), Drosophila melanogaster, Caenorhabditis elegans,
Escherichia coli (K12 MG1655), Saccharomyces cerevisiae
and Bacillus subtilis (PY79). Additionally, the present
study explored the presence of these motifs in the genome of
Euglena gracilis using the ‘Matcher’ tool on the Galaxy
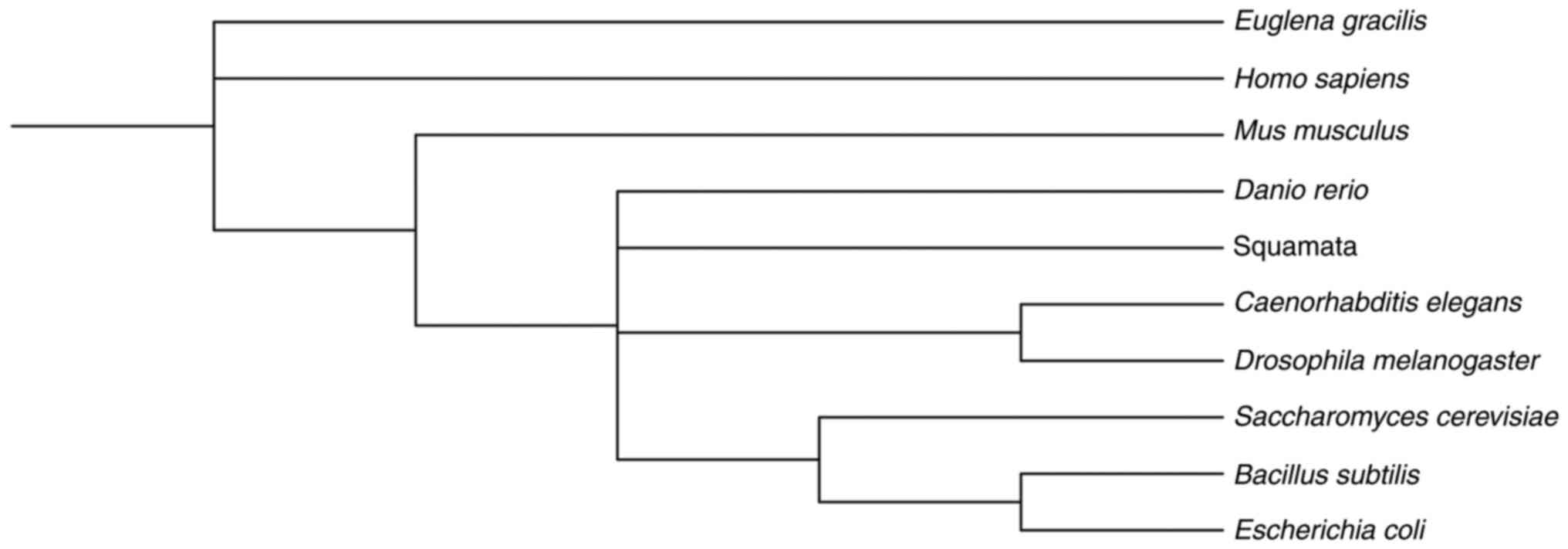
platform. A phylogenetic tree was subsequently constructed using
the iTOL tool (Interactive Tree Of Life), which enabled the
visualization and annotation of the evolutionary associations among
the organisms and the conservation of the motifs across species
(127). The resulting phylogenetic
tree (Fig. 7) clearly illustrates
the evolutionary conservation of these motifs, which were
identified repeatedly in the studied organisms (Table II).
 | Table IIOccurrence frequency of the motifs in
the genome sequences of the species Mus musculus, Squamata
(lizard), Danio rerio (zebrafish), Drosophila
melanogaster, Caenorhabditis elegans, Escherichia coli,
Saccharomyces cerevisiae, Bacillus subtilis (PY79) and
Euglena gracilis. |
Table II
Occurrence frequency of the motifs in
the genome sequences of the species Mus musculus, Squamata
(lizard), Danio rerio (zebrafish), Drosophila
melanogaster, Caenorhabditis elegans, Escherichia coli,
Saccharomyces cerevisiae, Bacillus subtilis (PY79) and
Euglena gracilis.
| Species | Motif 1 | Motif 2 | Motif 3 | Motif 4 | Motif 5 | Motif 6 |
|---|
| Mus
musculus | 61,755 | 83,484 | 56,870 | 64,325 | 81,207 | 81,748 |
| Squamata
(lizard) | 50,267 | 53,382 | 98,626 | 52,822 | 51,308 | 59,874 |
| Danio
rerio | 66,238 | 91,870 | 54,896 | 91,910 | 97,171 | 42,731 |
| Drosophila
melanogaster | 41,658 | 35,397 | 29,320 | 33,960 | 44,610 | 60,662 |
| Caenorhabditis
elegans | 42,982 | 20,446 | 21,768 | 29,076 | 27,321 | 40,169 |
| Escherichia
coli | 903 | 682 | 644 | 679 | 1269 | 760 |
| Saccharomyces
cerevisiae | 3,060 | 2,386 | 3,416 | 3,110 | 3,770 | 5,483 |
| Bacillus
subtilis | 912 | 663 | 662 | 638 | 865 | 830 |
| Euglena
gracilis | 364 | 1,657 | 1,278 | 541 | 522 | 1,711 |
The genetic similarities between humans and the
other organisms examined in the present study exhibit significant
variation. For instance, humans and Mus musculus (mouse)
share ~85% genetic similarity, reflecting a divergence of ~70
million years ago, making the mouse an essential model for human
genetic research, particularly in disease mechanisms and
therapeutic development (128).
Squamata, a diverse group of reptiles, includes species that serve
as critical models for studying thermoregulation, behavior and
reproductive strategies. Research on lizards and snakes has
provided important insights into immunity, skin regeneration, and
other physiological processes crucial for survival in varied
environments (129). The genetic
similarity between humans and different Squamata species varies
considerably, with lizards sharing ~70% genetic similarity with
humans, diverging ~250 million years ago. The six motifs identified
in the present study were also present in Squamata, suggesting
their ancient evolutionary origins. Danio rerio (zebrafish),
with a genetic similarity to humans of ~70%, diverged ~400 million
years ago and is an critical model organism for developmental
biology and gene function studies (130). Drosophila melanogaster
(fruit fly), sharing ~60% of its genes with humans and diverging
~600 million years ago, is extensively used in genetic regulation,
neurobiology, and cellular research (131). Caenorhabditis elegans
(nematode), with ~40% genetic similarity to humans, has been
instrumental in research on aging, developmental biology and
cellular processes (132).
Escherichia coli K12 MG1655, although lacking direct gene
homologs in humans, plays a crucial role in understanding
fundamental gene regulation and bacterial genetics (133). Saccharomyces cerevisiae
(yeast) shares ~23% of its genes with humans and diverged from
humans >1 billion years ago, rendering it a key organism for
cell biology, genomics and metabolism research (134). Bacillus subtilis PY79, a
soil bacterium, has markedly contributed to the understanding of
bacterial gene regulation and stress responses (135). Finally, Euglena gracilis, a
single-celled eukaryote, is considered to share ancient NR
mechanisms with humans, offering insights into the early evolution
of eukaryotic gene regulation (136).
The presence of the discovered motifs in species
that diverged long before humans, including organisms with distinct
genomes, such as Euglena gracilis and Saccharomyces
cerevisiae, combined with the lack of specificity in these
motifs, suggests they may represent ancestral binding sites for the
precursors of NRs. The conservation of these motifs across a broad
evolutionary spectrum emphasizes their potential role in regulating
gene expression via NR binding. Given that these motifs do not
exhibit specificity for any particular NR, it was hypothesized that
they may have functioned as general binding sites for ancestral NR
precursors (137). Moreover, these
motifs play a crucial role in the evolution of NR function by
enabling NRs to interact with a broader range of binding sites,
facilitating gene regulation in response to environmental and
internal cues (126,138).
The widespread presence of these motifs across
different evolutionary lineages points to their ancient origins,
likely predating the highly specific NRs found in modern organisms.
In early eukaryotes, these motifs could have functioned as genomic
binding sites for precursor forms of NRs, which were less specific
in their binding preferences compared to the more specialized NRs
currently observed (139). This
generalization would have been essential in the early stages of NR
evolution, allowing for flexible and robust gene regulation. These
early, versatile genomic binding sites could have allowed
primordial NR-like proteins to modulate gene expression
effectively, even in the absence of the highly refined
receptor-genome interactions seen in contemporary systems.
The conservation of these motifs across such
diverse species also suggests that they may have functioned as
backup mechanisms, ensuring the continued regulation of essential
genes when canonical NR binding sites, such as the glucocorticoid
response element for GR, were unavailable or dysfunctional
(139). In this sense, these
motifs may have served as a safety net, allowing the NR system to
maintain gene expression and cellular functions even in the absence
of optimal binding site availability. This backup role would have
been particularly important in the early evolutionary stages, when
the molecular machinery for precise gene regulation was still
evolving (140).
As NRs became more specialized through evolution,
the ancestral motifs likely persisted due to their role in
maintaining regulatory flexibility. In modern organisms, these
motifs may still contribute to the robustness of gene regulation,
particularly in cases where the canonical NR-binding sites are
compromised or unable to function. Their ability to interact with
multiple NRs enhances the adaptability of the organism to different
signaling environments, providing a more versatile and resilient
regulatory system (140).
The preservation of these motifs across such a
broad evolutionary spectrum underscores their importance in the
evolutionary trajectory of gene regulation. These motifs may
represent a foundational aspect of NR functionality, offering a
mechanism that allowed early organisms to adapt to changing
environments by ensuring continued gene regulation, even when
specific receptor-binding sites were not fully evolved. The fact
that these motifs can still be found in modern organisms suggests
that they continue to play a role in gene regulation, particularly
in situations where canonical NR interactions are suboptimal or
unavailable (126).
This broader, more flexible role in gene regulation
may have allowed organisms to thrive in variable environments and
adapt to new challenges. The conservation of motifs across both
ancient and modern species highlights their functional importance
and their potential as key elements in maintaining the adaptability
of the NR signaling system throughout evolutionary history.
To date, numerous studies have examined and
described the potential function of lncRNAs in the regulation of
gene expression and their abilities to act as biomarkers or as
pharmaceutical targets, or even therapeutic molecules (141-144).
In the present study, six conserved motifs were detected for the
first time in the sequences of the lncRNAs studied which are
suggested to participate in the regulatory function of lncRNAs that
contain them. However, to fully understand the role of these
motifs, and the lncRNAs containing them, in gene regulation, future
research is required to explore their functional significance in
greater detail. Experimental studies could confirm whether these
motifs act as alternative binding sites for NRs or other
transcription factors. Additionally, further investigations into
the mechanisms through which these motifs contribute to the
regulation of gene expression in both ancient and modern species
are warranted to provide valuable insight into the evolution of
NR-mediated gene regulation and its ongoing role in cellular
adaptability.
Supplementary Material
The first discovered motif in the
upregulated lncRNA subgroup.
The second discovered motif in the
upregulated lncRNA subgroup.
The third discovered motif in the
upregulated lncRNA subgroup.
The first discovered motif in the
downregulated lncRNA subgroup.
The second discovered motif in the
downregulated lncRNA subgroup.
The third discovered motif in the
downregulated lncRNA subgroup.
Human karyotype in which the
percentage of the occurrence of motif 1 in each chromosome is
presented.
Human karyotype in which the
percentage of the occurrence of motif 2 in each chromosome is
presented.
Human karyotype in which the
percentage of the occurrence of motif 3 in each chromosome is
presented.
Human karyotype in which the
percentage of the occurrence of motif 4 in each chromosome is
presented.
Human karyotype in which the
percentage of the occurrence of motif 5 in each chromosome is
presented.
Human karyotype in which the
percentage of the occurrence of motif 6 in each chromosome is
presented.
Chromosome plot for the 1,000
higher-scored loci of motif 1 in the human genome which are
presented as red dots next to the corresponding chromosomal region
where they are located.
Chromosome plot for the 1,000
higher-scored loci of motif 2 in the human genome which are
presented as red dots next to the corresponding chromosomal region
where they are located.
Chromosome plot for the 1,000
higher-scored loci of motif 3 in the human genome which are
presented as red dots next to the corresponding chromosomal region
where they are located.
Chromosome plot for the 1,000
higher-scored loci of motif 4 in the human genome which are
presented as red dots next to the corresponding chromosomal region
where they are located.
Chromosome plot for the 1,000
higher-scored loci of motif 5 in the human genome which are
presented as red dots next to the corresponding chromosomal region
where they are located.
Chromosome plot for the 1,000
higher-scored loci of motif 6 in the human genome which are
presented as red dots next to the corresponding chromosomal region
where they are located.
Multiple alignment of the sequences of
motif 1 with AAVS1 site using Clustal Omega (51) visualized with JalView (52).
Multiple alignment of the sequences of
motif 2 with AAVS1 site using Clustal Omega (51) visualized with JalView (52).
Multiple alignment of the sequences of
motif 3 with AAVS1 site using Clustal Omega (51) visualized with JalView (52).
Multiple alignment of the sequences of
motif 4 with AAVS1 site using Clustal Omega (51) visualized with JalView (52).
Multiple alignment of the sequences of
motif 5 with AAVS1 site using Clustal Omega (51) visualized with JalView (52).
Multiple alignment of the sequences of
motif 6 with AAVS1 site using Clustal Omega (51) visualized with JalView (52).
The biological pathways in which the
lncRNAs that contain the motif 1 participate and have a regulatory
function, according to GeneCards (https://www.genecards.org/) and NCBI Gene (https://www.ncbi.nlm.nih.gov/gene/)
databases.
The biological pathways in which the
lncRNAs that contain the motif 2 participate and have a regulatory
function, according to GeneCards (https://www.genecards.org/) and NCBI Gene (https://www.ncbi.nlm.nih.gov/gene/)
databases.
The biological pathways in which the
lncRNAs that contain the motif 3 participate and have a regulatory
function, according to GeneCards (https://www.genecards.org/) and NCBI Gene (https://www.ncbi.nlm.nih.gov/gene/)
databases.
The biological pathways in which the
lncRNAs that contain the motif 4 participate and have a regulatory
function, according to GeneCards (https://www.genecards.org/) and NCBI Gene (https://www.ncbi.nlm.nih.gov/gene/)
databases.
The biological pathways in which the
lncRNAs that contain the motif 5 participate and have a regulatory
function, according to GeneCards (https://www.genecards.org/) and NCBI Gene (https://www.ncbi.nlm.nih.gov/gene/)
databases.
The biological pathways in which the
lncRNAs that contain the motif 6 participate and have a regulatory
function, according to GeneCards (https://www.genecards.org/) and NCBI Gene (https://www.ncbi.nlm.nih.gov/gene/)
databases.
Data S1
Data S2
Data S1
The dataset of the lncRNAs that are
either upregulated or downregulated following the activation of an
NR.
List of 487 transcripts which are
targets of hsa-miR-1908-3p according to the TargetScanS (Release
8.0) database.
Acknowledgements
Not applicable.
Funding
Funding: The authors would like to acknowledge funding from the
following organizations: i) AdjustEBOVGP-Dx (RIA2018EF-2081):
Biochemical Adjustments of native EBOV Glycoprotein in Patient
Sample to Unmask target Epitopes for Rapid Diagnostic Testing. A
European and Developing Countries Clinical Trials Partnership
(EDCTP2) under the Horizon 2020 ‘Research and Innovation Actions’
DESCA; and ii) ‘MilkSafe: A novel pipeline to enrich formula milk
using omics technologies’, a research co-financed by the European
Regional Development Fund of the European Union and Greek national
funds through the Operational Program Competitiveness,
Entrepreneurship and Innovation, under the call
RESEARCH-CREATE-INNOVATE (project code: T2EDK-02222).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
DV conceived the study. DV, GPC, EE and KP
contributed to the overall study design. KP and LP developed the
in-house algorithms in MATLAB. KP, EE and DV performed and
evaluated the analysis using bioinformatics tools, as well as the
statistical analysis. KP, LP, GPC, EE and DV wrote, drafted and
critically revised the manuscript. DV, LP and KP confirm the
authenticity of all the raw data. KP, LP, GPC, EE and DV wrote,
drafted, revised, edited and reviewed the manuscript. All authors
have read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
GPC is the Editor in Chief of the journal, and DV
and EE are Editors of the journal. However, they had no personal
involvement in the reviewing process, or any influence in terms of
adjudicating on the final decision, for this article. The other
authors declare that they have no competing interests.
References
|
1
|
Carninci P, Kasukawa T, Katayama S, Gough
J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al:
The transcriptional landscape of the mammalian genome. Science.
309:1559–1563. 2005.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Mattick JS: RNA regulation: A new
genetics? Nat Rev Genet. 5:316–323. 2004.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Frith MC, Pheasant M and Mattick JS: The
amazing complexity of the human transcriptome. Eur J Hum Genet.
13:894–897. 2005.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Kaikkonen MU, Lam MTY and Glass CK:
Non-coding RNAs as regulators of gene expression and epigenetics.
Cardiovasc Res. 90:430–440. 2011.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Harrow J, Frankish A, Gonzalez JM,
Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa
A, Searle S, et al: GENCODE: The reference human genome annotation
for The ENCODE project. Genome Res. 22:1760–1774. 2012.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Salviano-Silva A, Lobo-Alves SC, Almeida
RC, Malheiros D and Petzl-Erler ML: Besides pathology: Long
non-coding RNA in cell and tissue homeostasis. Noncoding RNA.
4(3)2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Ma L, Bajic VB and Zhang Z: On the
classification of long non-coding RNAs. RNA Biol. 10:925–933.
2013.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Wang KC and Chang HY: Molecular mechanisms
of long noncoding RNAs. Mol Cell. 43:904–914. 2011.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Hombach S and Kretz M: Non-coding RNAs:
Classification, biology and functioning. Adv Exp Med Biol.
937:3–17. 2016.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Rinn JL and Chang HY: Genome regulation by
long noncoding RNAs. Annu Rev Biochem. 81:145–166. 2012.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Francis GA, Fayard E, Picard F and Auwerx
J: Nuclear receptors and the control of metabolism. Annu Rev
Physiol. 65:261–311. 2003.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Olivares AM, Moreno-Ramos OA and Haider
NB: Role of nuclear receptors in central nervous system development
and associated diseases. J Exp Neurosci. 9 (Suppl 2):S93–S121.
2016.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Foulds CE, Panigrahi AK, Coarfa C, Lanz RB
and O'Malley BW: Long noncoding RNAs as targets and regulators of
nuclear receptors. Curr Top Microbiol Immunol. 394:143–176.
2016.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Volders PJ, Helsens K, Wang X, Menten B,
Martens L, Gevaert K, Vandesompele J and Mestdagh P: LNCipedia: A
database for annotated human lncRNA transcript sequences and
structures. Nucleic Acids Res. 41 (Database Issue):D246–D251.
2013.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Bailey TL, Williams N, Misleh C and Li WW:
MEME: Discovering and analyzing DNA and protein sequence motifs.
Nucleic Acids Res. 34 (Web Server Issue):W369–W373. 2006.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Liu W and Wang X: Prediction of functional
microRNA targets by integrative modeling of microRNA binding and
target expression data. Genome Biol. 20(18)2019.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4(e05005)2015.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Garcia DM, Baek D, Shin C, Bell GW,
Grimson A and Bartel DP: Weak seed-pairing stability and high
target-site abundance decrease the proficiency of lsy-6 and other
microRNAs. Nat Struct Mol Biol. 18:1139–1146. 2011.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Grimson A, Farh KK, Johnston WK,
Garrett-Engele P, Lim LP and Bartel DP: MicroRNA targeting
specificity in mammals: Determinants beyond seed pairing. Mol Cell.
27:91–105. 2007.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Goecks J, Nekrutenko A and Taylor J:
Galaxy Team. Galaxy: A comprehensive approach for supporting
accessible, reproducible, and transparent computational research in
the life sciences. Genome Biol. 11(R86)2010.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Afgan E, Baker D, van den Beek M,
Blankenberg D, Bouvier D, Čech M, Chilton J, Clements D, Coraor N,
Eberhard C, et al: The Galaxy platform for accessible, reproducible
and collaborative biomedical analyses: 2016 update. Nucleic Acids
Res. 44 (W1):W3–W10. 2016.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Durbin R, Eddy SR, Krogh A and Mitchison
G: Biological sequence analysis: Probabilistic Models of Proteins
and Nucleic Acids. Cambridge University Press, 1998.
|
|
23
|
Ha M and Kim VN: Regulation of microRNA
biogenesis. Nat Rev Mol Cell Biol. 15:509–524. 2014.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Broughton JP, Lovci MT, Huang JL, Yeo GW
and Pasquinelli AE: Pairing beyond the seed supports microRNA
targeting specificity. Mol Cell. 64:320–333. 2016.PubMed/NCBI View Article : Google Scholar
|
|
25
|
John B, Enright AJ, Aravin A, Tuschl T,
Sander C and Marks DS: Human MicroRNA targets. PLoS Biol.
2(e363)2004.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Betel D, Koppal A, Agius P, Sander C and
Leslie C: Comprehensive modeling of microRNA targets predicts
functional non-conserved and non-canonical sites. Genome Biol.
11(R90)2010.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Krek A, Grün D, Poy MN, Wolf R, Rosenberg
L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M
and Rajewsky N: Combinatorial microRNA target predictions. Nat
Genet. 37:495–500. 2005.PubMed/NCBI View
Article : Google Scholar
|
|
28
|
Lewis BP, Shih IH, Jones-Rhoades MW,
Bartel DP and Burge CB: Prediction of mammalian microRNA targets.
Cell. 115:787–798. 2003.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Miranda KC, Huynh T, Tay Y, Ang YS, Tam
WL, Thomson AM, Lim B and Rigoutsos I: A pattern-based method for
the identification of MicroRNA binding sites and their
corresponding heteroduplexes. Cell. 126:1203–1217. 2006.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Kertesz M, Iovino N, Unnerstall U, Gaul U
and Segal E: The role of site accessibility in microRNA target
recognition. Nat Genet. 39:1278–1284. 2007.PubMed/NCBI View
Article : Google Scholar
|
|
32
|
Rehmsmeier M, Steffen P, Hochsmann M and
Giegerich R: Fast and effective prediction of microRNA/target
duplexes. RNA. 10:1507–1517. 2004.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Maragkakis M, Alexiou P, Papadopoulos GL,
Reczko M, Dalamagas T, Giannopoulos G, Goumas G, Koukis E, Kourtis
K, Simossis VA, et al: Accurate microRNA target prediction
correlates with protein repression levels. BMC Bioinformatics.
10(295)2009.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Karagkouni D, Paraskevopoulou MD,
Chatzopoulos S, Vlachos IS, Tastsoglou S, Kanellos I, Papadimitriou
D, Kavakiotis I, Maniou S, Skoufos G, et al: DIANA-TarBase v8: A
decade-long collection of experimentally supported miRNA-gene
interactions. Nucleic Acids Res. 46 (D1):D239–D245. 2018.PubMed/NCBI View Article : Google Scholar
|
|
35
|
McGeary SE, Lin KS, Shi CY, Pham TM,
Bisaria N, Kelley GM and Bartel DP: The biochemical basis of
microRNA targeting efficacy. Science. 366(eaav1741)2019.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Yang L, Shi CM, Chen L, Pang LX, Xu GF, Gu
N, Zhu LJ, Guo XR, Ni YH and Ji CB: The biological effects of
hsa-miR-1908 in human adipocytes. Mol Biol Rep. 42:927–935.
2015.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Kuang Q, Li J, You L, Shi C, Ji C, Guo X,
Xu M and Ni Y: Identification and characterization of NF-kappaB
binding sites in human miR-1908 promoter. Biomed Pharmacother.
74:158–163. 2015.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Bar M, Wyman SK, Fritz BR, Qi J, Garg KS,
Parkin RK, Kroh EM, Bendoraite A, Mitchell PS, Nelson AM, et al:
MicroRNA discovery and profiling in human embryonic stem cells by
deep sequencing of small RNA libraries. Stem Cells. 26:2496–2505.
2008.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Pencheva N, Tran H, Buss C, Huh D,
Drobnjak M, Busam K and Tavazoie SF: Convergent multi-miRNA
targeting of ApoE drives LRP1/LRP8-dependent melanoma metastasis
and angiogenesis. Cell. 151:1068–1082. 2012.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Xia X, Li Y, Wang W, Tang F, Tan J, Sun L,
Li Q, Sun L, Tang B and He S: MicroRNA-1908 functions as a
glioblastoma oncogene by suppressing PTEN tumor suppressor pathway.
Mol Cancer. 14(154)2015.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Chai Z, Fan H, Li Y, Song L, Jin X, Yu J,
Li Y, Ma C and Zhou R: miR-1908 as a novel prognosis marker of
glioma via promoting malignant phenotype and modulating SPRY4/RAF1
axis. Oncol Rep. 38:2717–2726. 2017.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Jiang X, Yang L, Pang L, Chen L, Guo X, Ji
C, Shi C and Ni Y: Expression of obesity-related miR-1908 in human
adipocytes is regulated by adipokines, free fatty acids and
hormones. Mol Med Rep. 10:1164–1169. 2014.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Grimwood J, Gordon LA, Olsen A, Terry A,
Schmutz J, Lamerdin J, Hellsten U, Goodstein D, Couronne O,
Tran-Gyamfi M, et al: The DNA sequence and biology of human
chromosome 19. Nature. 428:529–535. 2004.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Kumar S, Stecher G, Suleski M and Hedges
SB: TimeTree: A resource for timelines, timetrees, and divergence
times. Mol Biol Evol. 34:1812–1819. 2017.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Kim SH, Elango N, Warden C, Vigoda E and
Yi SV: Heterogeneous genomic molecular clocks in primates. PLoS
Genet. 2(e163)2006.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Kotin RM, Menninger JC, Ward DC and Berns
KI: Mapping and direct visualization of a region-specific viral DNA
integration site on chromosome 19q13-qter. Genomics. 10:831–834.
1991.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Samulski RJ, Zhu X, Xiao X, Brook JD,
Housman DE, Epstein N and Hunter LA: Targeted integration of
adeno-associated virus (AAV) into human chromosome 19. EMBO J.
10:3941–3950. 1991.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Kotin RM, Linden RM and Berns KI:
Characterization of a preferred site on human chromosome 19q for
integration of adeno-associated virus DNA by non-homologous
recombination. EMBO J. 11:5071–5078. 1992.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Aslanidis C, Jansen G, Amemiya C, Shutler
G, Mahadevan M, Tsilfidis C, Chen C, Alleman J, Wormskamp NG,
Vooijs M, et al: Cloning of the essential myotonic dystrophy region
and mapping of the putative defect. Nature. 355:548–551.
1992.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Feichtinger W and Schmid M: Increased
frequencies of sister chromatid exchanges at common fragile sites
(1)(q42) and (19)(q13). Hum Genet. 83:145–147. 1989.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Sievers F and Higgins DG: Clustal Omega
for making accurate alignments of many protein sequences. Protein
Sci. 27:135–145. 2018.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Waterhouse AM, Procter JB, Martin DMA,
Clamp M and Barton GJ: Jalview Version 2-a multiple sequence
alignment editor and analysis workbench. Bioinformatics.
25:1189–1191. 2009.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Deaton AM and Bird A: CpG islands and the
regulation of transcription. Genes Dev. 25:1010–1022.
2011.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Larsen F, Gundersen G, Lopez R and Prydz
H: CpG islands as gene markers in the human genome. Genomics.
13:1095–1107. 1992.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Yamashita R, Suzuki Y, Sugano S and Nakai
K: Genome-wide analysis reveals strong correlation between CpG
islands with nearby transcription start sites of genes and their
tissue specificity. Gene. 350:129–136. 2005.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Lamartina S, Sporeno E, Fattori E and
Toniatti C: Characteristics of the adeno-associated virus
preintegration site in human chromosome 19: Open chromatin
conformation and transcription-competent environment. J Virol.
74:7671–7677. 2000.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Bhagwan JR, Collins E, Mosqueira D, Bakar
M, Johnson BB, Thompson A, Smith JGW and Denning C: Variable
expression and silencing of CRISPR-Cas9 targeted transgenes
identifies the AAVS1 locus as not an entirely safe harbour.
F1000Res. 8(1911)2019.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Ogata T, Kozuka T and Kanda T:
Identification of an insulator in AAVS1, a preferred region for
integration of adeno-associated virus DNA. J Virol. 77:9000–9007.
2003.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Gaspar HB, Cooray S, Gilmour KC, Parsley
KL, Zhang F, Adams S, Bjorkegren E, Bayford J, Brown L, Davies EG,
et al: Hematopoietic stem cell gene therapy for adenosine
deaminase-deficient severe combined immunodeficiency leads to
long-term immunological recovery and metabolic correction. Sci
Transl Med. 3(97ra80)2011.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Cartier N, Hacein-Bey-Abina S, Bartholomae
CC, Veres G, Schmidt M, Kutschera I, Vidaud M, Abel U, Dal-Cortivo
L, Caccavelli L, et al: Hematopoietic stem cell gene therapy with a
lentiviral vector in X-linked adrenoleukodystrophy. Science.
326:818–823. 2009.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Hacein-Bey-Abina S, Pai SY, Gaspar HB,
Armant M, Berry CC, Blanche S, Bleesing J, Blondeau J, de Boer H,
Buckland KF, et al: A modified γ-retrovirus vector for X-linked
severe combined immunodeficiency. N Engl J Med. 371:1407–1417.
2014.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Martin DIK and Whitelaw E: The vagaries of
variegating transgenes. Bioessays. 18:919–923. 1996.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Bestor TH: Gene silencing as a threat to
the success of gene therapy. J Clin Invest. 105:409–411.
2000.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Papapetrou EP and Schambach A: Gene
insertion into genomic safe harbors for human gene therapy. Mol
Ther. 24:678–684. 2016.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Wu C and Dunbar CE: Stem cell gene
therapy: The risks of insertional mutagenesis and approaches to
minimize genotoxicity. Front Med. 5:356–371. 2011.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Lombardo A, Cesana D, Genovese P, Di
Stefano B, Provasi E, Colombo DF, Neri M, Magnani Z, Cantore A, Lo
Riso P, et al: Site-specific integration and tailoring of cassette
design for sustainable gene transfer. Nat Methods. 8:861–869.
2011.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Klatt D, Cheng E, Hoffmann D, Santilli G,
Thrasher AJ, Brendel C and Schambach A: Differential transgene
silencing of myeloid-specific promoters in the AAVS1 safe harbor
locus of induced pluripotent stem cell-derived myeloid cells. Hum
Gene Ther. 31:199–210. 2019.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Papapetrou EP, Lee G, Malani N, Setty M,
Riviere I, Tirunagari LM, Kadota K, Roth SL, Giardina P, Viale A,
et al: Genomic safe harbors permit high β-globin transgene
expression in thalassemia induced pluripotent stem cells. Nat
Biotechnol. 29:73–78. 2011.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Sadelain M, Papapetrou EP and Bushman FD:
Safe harbours for the integration of new DNA in the human genome.
Nat Rev Cancer. 12:51–58. 2011.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Smith SD, Morgan R, Gemmell R, Amylon MD,
Link MP, Linker C, Hecht BK, Warnke R, Glader BE and Hecht F:
Clinical and biologic characterization of T-cell neoplasias with
rearrangements of chromosome 7 band q34. Blood. 71:395–402.
1988.PubMed/NCBI
|
|
71
|
Maes OC, Chertkow HM, Wang E and Schipper
HM: MicroRNA: Implications for alzheimer disease and other human
CNS disorders. Curr Genomics. 10:154–168. 2009.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Garofalo M, Condorelli G and Croce CM:
MicroRNAs in diseases and drug response. Curr Opin Pharmacol.
8:661–667. 2008.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Juźwik CA, S Drake S, Zhang Y,
Paradis-Isler N, Sylvester A, Amar-Zifkin A, Douglas C, Morquette
B, Moore CS and Fournier AE: microRNA dysregulation in
neurodegenerative diseases: A systematic review. Prog Neurobiol.
182(101664)2019.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Venneri M and Passantino A: MiRNA: What
clinicians need to know. Eur J Intern Med. 113:6–9. 2023.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Zhang B, Pan X, Cobb GP and Anderson TA:
microRNAs as oncogenes and tumor suppressors. Dev Biol. 302:1–12.
2007.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Small EM and Olson EN: Pervasive roles of
microRNAs in cardiovascular biology. Nature. 469:336–342.
2011.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Romaine SPR, Tomaszewski M, Condorelli G
and Samani NJ: MicroRNAs in cardiovascular disease: An introduction
for clinicians. Heart. 101:921–928. 2015.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Bailey TL and Elkan C: Fitting a mixture
model by expectation maximization to discover motifs in
biopolymers. Proc Int Conf Intell Syst Mol Biol. 2:28–36.
1994.PubMed/NCBI
|
|
79
|
Tirosh I and Barkai N: Two strategies for
gene regulation by promoter nucleosomes. Genome Res. 18:1084–1091.
2008.PubMed/NCBI View Article : Google Scholar
|
|
80
|
Bird A: DNA methylation patterns and
epigenetic memory. Genes Dev. 16:6–21. 2002.PubMed/NCBI View Article : Google Scholar
|
|
81
|
Li E, Bestor TH and Jaenisch R: Targeted
mutation of the DNA methyltransferase gene results in embryonic
lethality. Cell. 69:915–926. 1992.PubMed/NCBI View Article : Google Scholar
|
|
82
|
Ulitsky I, Shkumatava A, Jan CH, Sive H
and Bartel DP: Conserved function of lincRNAs in vertebrate
embryonic development despite rapid sequence evolution. Cell.
147:1537–1550. 2011.PubMed/NCBI View Article : Google Scholar
|
|
83
|
Liu L, Wang Z, Jia J, Shi Y, Lian T and
Han X: Linc01230, transcriptionally regulated by PPARγ, is
identified as a novel modifier in endothelial function. Biochem
Biophys Res Commun. 507:369–376. 2018.PubMed/NCBI View Article : Google Scholar
|
|
84
|
Cunningham TJ, Kumar S, Yamaguchi TP and
Duester G: Wnt8a and Wnt3a cooperate in the axial stem cell niche
to promote mammalian body axis extension. Dev Dyn. 244:797–807.
2015.PubMed/NCBI View Article : Google Scholar
|
|
85
|
Okubo Y, Sugawara T, Abe-Koduka N, Kanno
J, Kimura A and Saga Y: Lfng regulates the synchronized oscillation
of the mouse segmentation clock via trans-repression of Notch
signalling. Nat Commun. 3(1141)2012.PubMed/NCBI View Article : Google Scholar
|
|
86
|
Pai SG, Carneiro BA, Mota JM, Costa R,
Leite CA, Barroso-Sousa R, Kaplan JB, Chae YK and Giles FJ:
Wnt/beta-catenin pathway: Modulating anticancer immune response. J
Hematol Oncol. 10(101)2017.PubMed/NCBI View Article : Google Scholar
|
|
87
|
Amsellem V, Dryden NH, Martinelli R,
Gavins F, Almagro LO, Birdsey GM, Haskard DO, Mason JC, Turowski P
and Randi AM: ICAM-2 regulates vascular permeability and N-cadherin
localization through ezrin-radixin-moesin (ERM) proteins and Rac-1
signalling. Cell Commun Signal. 12(12)2014.PubMed/NCBI View Article : Google Scholar
|
|
88
|
Overton HA, Fyfe MCT and Reynet C: GPR119,
a novel G protein-coupled receptor target for the treatment of type
2 diabetes and obesity. Br J Pharmacol. 153 (Suppl 1):S76–S81.
2008.PubMed/NCBI View Article : Google Scholar
|
|
89
|
Shang R, Wang M, Dai B, Du J, Wang J, Liu
Z, Qu S, Yang X, Liu J, Xia C, et al: Long noncoding RNA SLC2A1-AS1
regulates aerobic glycolysis and progression in hepatocellular
carcinoma via inhibiting the STAT3/FOXM1/GLUT1 pathway. Mol Oncol.
14:1381–1396. 2020.PubMed/NCBI View Article : Google Scholar
|
|
90
|
Kim JJ, Lee SB, Jang J, Yi SY, Kim SH, Han
SA, Lee JM, Tong SY, Vincelette ND, Gao B, et al: WSB1 promotes
tumor metastasis by inducing pVHL degradation. Genes Dev.
29:2244–2257. 2015.PubMed/NCBI View Article : Google Scholar
|
|
91
|
Shao X, Zhao T, Xi L, Zhang Y, He J, Zeng
J and Deng L: LINC00565 promotes the progression of colorectal
cancer by upregulating EZH2. Oncol Lett. 21(53)2021.PubMed/NCBI View Article : Google Scholar
|
|
92
|
Chen C, Feng Y, Wang J, Liang Y and Zou W:
Long non-coding RNA SNHG15 in various cancers: A meta and
bioinformatic analysis. BMC Cancer. 20(1156)2020.PubMed/NCBI View Article : Google Scholar
|
|
93
|
Qian C, Li H, Chang D, Wei B and Wang Y:
Identification of functional lncRNAs in atrial fibrillation by
integrative analysis of the lncRNA-mRNA network based on competing
endogenous RNAs hypothesis. J Cell Physiol. 234:11620–11630.
2019.PubMed/NCBI View Article : Google Scholar
|
|
94
|
Liu J, Xu R, Mai SJ, Ma YS, Zhang MY, Cao
PS, Weng NQ, Wang RQ, Cao D, Wei W, et al: LncRNA CSMD1-1 promotes
the progression of hepatocellular carcinoma by activating MYC
signaling. Theranostics. 10:7527–7544. 2020.PubMed/NCBI View Article : Google Scholar
|
|
95
|
Talwar D and Hammer MF: SCN8A epilepsy,
developmental encephalopathy, and related disorders. Pediatr
Neurol. 122:76–83. 2021.PubMed/NCBI View Article : Google Scholar
|
|
96
|
Chen XW, Feng YQ, Hao CJ, Guo XL, He X,
Zhou ZY, Guo N, Huang HP, Xiong W, Zheng H, et al: DTNBP1, a
schizophrenia susceptibility gene, affects kinetics of transmitter
release. J Cell Biol. 181:791–801. 2008.PubMed/NCBI View Article : Google Scholar
|
|
97
|
Chen CL, Ke Q, Luo M, Gao ZY, Li ZJ, Luo
ZG and Liu DB: Loss of LINC01939 expression predicts progression
and poor survival in gastric cancer. Pathol Res Pract.
214:1539–1543. 2018.PubMed/NCBI View Article : Google Scholar
|
|
98
|
Dosil M and Bustelo XR: Functional
characterization of Pwp2, a WD family protein essential for the
assembly of the 90 S pre-ribosomal particle. J Biol Chem.
279:37385–37397. 2004.PubMed/NCBI View Article : Google Scholar
|
|
99
|
Li G, Ruan X, Auerbach RK, Sandhu KS,
Zheng M, Wang P, Poh HM, Goh Y, Lim J, Zhang J, et al: Extensive
promoter-centered chromatin interactions provide a topological
basis for transcription regulation. Cell. 148:84–98.
2012.PubMed/NCBI View Article : Google Scholar
|
|
100
|
Li Z, Li X, Jian W, Xue Q and Liu Z: Roles
of long non-coding RNAs in the development of chronic pain. Front
Mol Neurosci. 14(760964)2021.PubMed/NCBI View Article : Google Scholar
|
|
101
|
Luo L, Martin SC, Parkington J, Cadena SM,
Zhu J, Ibebunjo C, Summermatter S, Londraville N, Patora-Komisarska
K, Widler L, et al: HDAC4 controls muscle homeostasis through
deacetylation of myosin heavy chain, PGC-1α, and Hsc70. Cell Rep.
29:749–763.e12. 2019.PubMed/NCBI View Article : Google Scholar
|
|
102
|
Mortison JD, Schenone M, Myers JA, Zhang
Z, Chen L, Ciarlo C, Comer E, Natchiar SK, Carr SA, Klaholz BP and
Myers AG: Tetracyclines modify translation by targeting key human
rRNA substructures. Cell Chem Biol. 25:1506–1518.e13.
2018.PubMed/NCBI View Article : Google Scholar
|
|
103
|
Nelson JK, Koenis DS, Scheij S, Cook EC,
Moeton M, Santos A, Lobaccaro JA, Baron S and Zelcer N: EEPD1 is a
novel LXR target gene in macrophages which regulates ABCA1
abundance and cholesterol efflux. Arterioscler Thromb Vasc Biol.
37:423–432. 2017.PubMed/NCBI View Article : Google Scholar
|
|
104
|
Zhang T, Xia W, Song X, Mao Q, Huang X,
Chen B, Liang Y, Wang H, Chen Y, Yu X, et al: Super-enhancer
hijacking LINC01977 promotes malignancy of early-stage lung
adenocarcinoma addicted to the canonical TGF-β/SMAD3 pathway. J
Hematol Oncol. 15(114)2022.PubMed/NCBI View Article : Google Scholar
|
|
105
|
Zeng J, Sun W, Chang J, Yi D, Zhu L, Zhang
Y, Pan X, Zhou Y, Lai M, Bian G, et al: HOXC4 up-regulates NF-κB
signaling and promotes the cell proliferation to drive development
of human hematopoiesis, especially CD43+ cells. Blood Sci.
2:117–128. 2020.PubMed/NCBI View Article : Google Scholar
|
|
106
|
Gao Y, Wang F, Zhang L, Kang M, Zhu L, Xu
L, Liang W and Zhang W: LINC00311 promotes cancer stem-like
properties by targeting miR-330-5p/TLR4 pathway in human papillary
thyroid cancer. Cancer Med. 9:1515–1528. 2020.PubMed/NCBI View Article : Google Scholar
|
|
107
|
Flosbach M, Oberle SG, Scherer S, Zecha J,
von Hoesslin M, Wiede F, Chennupati V, Cullen JG, List M, Pauling
JK, et al: PTPN2 deficiency enhances programmed T cell expansion
and survival capacity of activated T cells. Cell Rep.
32(107957)2020.PubMed/NCBI View Article : Google Scholar
|
|
108
|
Li Z, Chao TC, Chang KY, Lin N, Patil VS,
Shimizu C, Head SR, Burns JC and Rana TM: The long noncoding RNA
THRIL regulates TNFα expression through its interaction with
hnRNPL. Proc Natl Acad Sci USA. 111:1002–1007. 2014.PubMed/NCBI View Article : Google Scholar
|
|
109
|
Xu LB, Bo BX, Xiong J, Ren YJ, Han D, Wei
SH and Ren XP: Long non-coding RNA LINC00887 promotes progression
of lung carcinoma by targeting the microRNA-206/NRP1 axis. Oncol
Lett. 21(87)2021.PubMed/NCBI View Article : Google Scholar
|
|
110
|
Byun S, Affolter KE, Snow AK, Curtin K,
Cannon AR, Cannon-Albright LA, Thota R and Neklason DW:
Differential methylation of G-protein coupled receptor signaling
genes in gastrointestinal neuroendocrine tumors. Sci Rep.
11(12303)2021.PubMed/NCBI View Article : Google Scholar
|
|
111
|
Fang Z, Zhong M, Zhou L, Le Y, Wang H and
Fang Z: Low-density lipoprotein receptor-related protein 8
facilitates the proliferation and invasion of non-small cell lung
cancer cells by regulating the Wnt/β-catenin signaling pathway.
Bioengineered. 13:6807–6818. 2022.PubMed/NCBI View Article : Google Scholar
|
|
112
|
Huang R, Liu J, Chen X, Zhi Y, Ding S,
Ming J, Li Y, Wang Y and Na J: A long non-coding RNA LncSync
regulates mouse cardiomyocyte homeostasis and cardiac hypertrophy
through coordination of miRNA actions. Protein Cell. 14:153–157.
2023.PubMed/NCBI View Article : Google Scholar
|
|
113
|
Li N and Zhan X and Zhan X: The lncRNA
SNHG3 regulates energy metabolism of ovarian cancer by an analysis
of mitochondrial proteomes. Gynecol Oncol. 150:343–354.
2018.PubMed/NCBI View Article : Google Scholar
|
|
114
|
Zhen H, Du P, Yi Q, Tang X and Wang T:
LINC00958 promotes bladder cancer carcinogenesis by targeting
miR-490-3p and AURKA. BMC Cancer. 21(1145)2021.PubMed/NCBI View Article : Google Scholar
|
|
115
|
Majumdar R, Bandyopadhyay A, Deng H and
Maitra U: Phosphorylation of mammalian translation initiation
factor 5 (eIF5) in vitro and in vivo. Nucleic Acids Res.
30:1154–1162. 2002.PubMed/NCBI View Article : Google Scholar
|
|
116
|
Xu L, Wu Q, Yan H, Shu C, Fan W, Tong X
and Li Q: Long noncoding RNA KB-1460A1.5 inhibits glioma
tumorigenesis via miR-130a-3p/TSC1/mTOR/YY1 feedback loop. Cancer
Lett. 525:33–45. 2022.PubMed/NCBI View Article : Google Scholar
|
|
117
|
Wang F, Peters R, Jia J, Mudd M, Salemi M,
Allers L, Javed R, Duque TLA, Paddar MA, Trosdal ES, et al: ATG5
provides host protection acting as a switch in the atg8ylation
cascade between autophagy and secretion. Dev Cell. 58:866–884.e8.
2023.PubMed/NCBI View Article : Google Scholar
|
|
118
|
Cruickshank BM, Wasson MD, Brown JM,
Fernando W, Venkatesh J, Walker OL, Morales-Quintanilla F, Dahn ML,
Vidovic D, Dean CA, et al: LncRNA PART1 promotes proliferation and
migration, is associated with cancer stem cells, and alters the
miRNA landscape in triple-negative breast cancer. Cancers (Basel).
13(2644)2021.PubMed/NCBI View Article : Google Scholar
|
|
119
|
Vogler M: BCL2A1: The underdog in the BCL2
family. Cell Death Differ. 19:67–74. 2012.PubMed/NCBI View Article : Google Scholar
|
|
120
|
Barriocanal M, Carnero E, Segura V and
Fortes P: Long non-coding RNA BST2/BISPR is induced by IFN and
regulates the expression of the antiviral factor tetherin. Front
Immunol. 5(655)2015.PubMed/NCBI View Article : Google Scholar
|
|
121
|
Mohamed Haroon M, Lakshmanan V, Sarkar SR,
Lei K, Vemula PK and Palakodeti D: Mitochondrial state determines
functionally divergent stem cell population in planaria. Stem Cell
Reports. 16:1302–1316. 2021.PubMed/NCBI View Article : Google Scholar
|
|
122
|
Zhu W, Zhou BL, Rong LJ, Ye L, Xu HJ, Zhou
Y, Yan XJ, Liu WD, Zhu B, Wang L, et al: Roles of PTBP1 in
alternative splicing, glycolysis, and oncogensis. J Zhejiang Univ
Sci B. 21:122–136. 2020.PubMed/NCBI View Article : Google Scholar
|
|
123
|
Lee MY, Sumpter R Jr, Zou Z,
Sirasanagandla S, Wei Y, Mishra P, Rosewich H, Crane DI and Levine
B: Peroxisomal protein PEX13 functions in selective autophagy. EMBO
Rep. 18:48–60. 2017.PubMed/NCBI View Article : Google Scholar
|
|
124
|
L'Abbate A, Tolomeo D, De Astis F, Lonoce
A, Lo Cunsolo C, Mühlematter D, Schoumans J, Vandenberghe P, Van
Hoof A, Palumbo O, et al: t(15;21) translocations leading to the
concurrent downregulation of RUNX1 and its transcription factor
partner genes SIN3A and TCF12 in myeloid disorders. Mol Cancer.
14(211)2015.PubMed/NCBI View Article : Google Scholar
|
|
125
|
Stelzer G, Rosen N, Plaschkes I, Zimmerman
S, Twik M, Fishilevich S, Stein TI, Nudel R, Lieder I, Mazor Y, et
al: The GeneCards suite: From gene data mining to disease genome
sequence analyses. Curr Protoc Bioinformatics. 54:1.30.1–1.30.33.
2016.PubMed/NCBI View
Article : Google Scholar
|
|
126
|
Laudet V: Evolution of the nuclear
receptor superfamily: Early diversification from an ancestral
orphan receptor. J Mol Endocrinol. 19:207–226. 1997.PubMed/NCBI View Article : Google Scholar
|
|
127
|
Letunic I and Bork P: Interactive tree of
life (iTOL) v4: Recent updates and new developments. Nucleic Acids
Res. 47 (W1):W256–W259. 2019.PubMed/NCBI View Article : Google Scholar
|
|
128
|
Mouse Genome Sequencing Consortium.
Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal
P, Agarwala R, Ainscough R, Alexandersson M, et al: Initial
sequencing and comparative analysis of the mouse genome. Nature.
420:520–562. 2002.PubMed/NCBI View Article : Google Scholar
|
|
129
|
Castoe TA, de Koning APJ, Hall KT, Card
DC, Schield DR, Fujita MK, Ruggiero RP, Degner JF, Daza JM, Gu W,
et al: The Burmese python genome reveals the molecular basis for
extreme adaptation in snakes. Proc Natl Acad Sci USA.
110:20645–20650. 2013.PubMed/NCBI View Article : Google Scholar
|
|
130
|
Howe K, Clark MD, Torroja CF, Torrance J,
Berthelot C, Muffato M, Collins JE, Humphray S, McLaren K, Matthews
L, et al: The zebrafish reference genome sequence and its
relationship to the human genome. Nature. 496:498–503.
2013.PubMed/NCBI View Article : Google Scholar
|
|
131
|
Adams MD, Celniker SE, Holt RA, Evans CA,
Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF,
et al: The genome sequence of Drosophila melanogaster.
Science. 287:2185–2195. 2000.PubMed/NCBI View Article : Google Scholar
|
|
132
|
Hodgkin J, Plasterk RH and Waterston RH:
The nematode Caenorhabditis elegans and its genome. Science.
270:410–414. 1995.PubMed/NCBI View Article : Google Scholar
|
|
133
|
Blattner FR, Plunkett G III, Bloch CA,
Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK,
Mayhew GF, et al: The complete genome sequence of Escherichia
coli K-12. Science. 277:1453–1462. 1997.PubMed/NCBI View Article : Google Scholar
|
|
134
|
Weng S, Dong Q, Balakrishnan R, Christie
K, Costanzo M, Dolinski K, Dwight SS, Engel S, Fisk DG, Hong E, et
al: Saccharomyces genome database (SGD) provides biochemical and
structural information for budding yeast proteins. Nucleic Acids
Res. 31:216–218. 2003.PubMed/NCBI View Article : Google Scholar
|
|
135
|
Petersohn A, Brigulla M, Haas S, Hoheisel
JD, Völker U and Hecker M: Global analysis of the general stress
response of Bacillus subtilis. J Bacteriol. 183:5617–5631.
2001.PubMed/NCBI View Article : Google Scholar
|
|
136
|
Ebenezer TE, Carrington M, Lebert M, Kelly
S and Field MC: Euglena gracilis genome and transcriptome:
Organelles, nuclear genome assembly strategies and initial
features. Adv Exp Med Biol. 979:125–140. 2017.PubMed/NCBI View Article : Google Scholar
|
|
137
|
Owen GI and Zelent A: Origins and
evolutionary diversification of the nuclear receptor superfamily.
Cell Mol Life Sci. 57:809–827. 2000.PubMed/NCBI View Article : Google Scholar
|
|
138
|
Holzer G, Markov GV and Laudet V:
Evolution of nuclear receptors and ligand signaling: Toward a soft
key-lock model? Curr Top Dev Biol. 125:1–38. 2017.PubMed/NCBI View Article : Google Scholar
|
|
139
|
Penvose A, Keenan JL, Bray D, Ramlall V
and Siggers T: Comprehensive study of nuclear receptor DNA binding
provides a revised framework for understanding receptor
specificity. Nat Commun. 10(2514)2019.PubMed/NCBI View Article : Google Scholar
|
|
140
|
Cotnoir-White D, Laperrière D and Mader S:
Evolution of the repertoire of nuclear receptor binding sites in
genomes. Mol Cell Endocrinol. 334:76–82. 2011.PubMed/NCBI View Article : Google Scholar
|
|
141
|
Hanly D, Esteller M and Berdasco M:
Altered long non-coding RNA expression in cancer: Potential
biomarkers and therapeutic targets? In: Chemical Epigenetics. Mai A
(ed). Topics in Medicinal Chemistry. Vol. 33. Springer, Cham,
pp401-428, 2019.
|
|
142
|
Fu D, Shi Y, Liu JB, Wu TM, Jia CY, Yang
HQ, Zhang DD, Yang XL, Wang HM and Ma YS: Targeting long non-coding
RNA to therapeutically regulate gene expression in cancer. Mol Ther
Nucleic Acids. 21:712–724. 2020.PubMed/NCBI View Article : Google Scholar
|
|
143
|
Zhang L, Peng D, Sood AK, Dang CV and
Zhong X: Shedding light on the dark cancer genomes: Long noncoding
RNAs as novel biomarkers and potential therapeutic targets for
cancer. Mol Cancer Ther. 17:1816–1823. 2018.PubMed/NCBI View Article : Google Scholar
|
|
144
|
Fatima R, Akhade VS, Pal D and Rao SM:
Long noncoding RNAs in development and cancer: Potential biomarkers
and therapeutic targets. Mol Cell Ther. 3(5)2015.PubMed/NCBI View Article : Google Scholar
|