Introduction
Breast cancer is classified into 5 distinct
molecular subgroups: Normal-like, basal-like, luminal A, luminal B
and human epidermal growth factor receptor (HER) 2-enriched
(1-3).
The majority of basal-like breast cancer cells fail to express
estrogen receptor, progesterone receptor and HER2, and are
therefore termed triple-negative breast cancer (TNBC) (3,4).
Patients with TNBC do not benefit from hormonal (tamoxifen) and
anti-HER2 antibody (trastuzumab) therapies (1,5). The
lack of targeted therapies together with the high chance of lymph
node involvement, high rates of metastasis, high tumor grade/size
at the time of diagnosis, and high rates of relapse leads to a poor
prognosis for TNBC (5).
Despite advances in treatment strategies and early
detection, metastases remain as the primary cause of cancer-related
mortality (5). Metastasis is a
complex multi-step process involving genetic and/or epigenetic
alterations that allow individual cancer cells to leave the primary
tumor site, degrade and migrate through the extracellular matrix
(ECM), intravasate into nearby blood and lymphatic vessels, then
exit from the circulation into the parenchyma of distant tissue to
form a metastatic lesion (6).
Breast cancer primarily metastasizes to bone, brain, lung and liver
(7). Metastasis is promoted by
matrix metalloproteinases (MMPs) that degrade the ECM (8). Epithelial-to mesenchymal transition
(EMT) also plays a central role in metastasis by allowing cancer
cells to acquire a motile mesenchymal phenotype (9,10).
The Wnt/β-catenin and transforming growth factor (TGF) β/Smad
signaling pathways initiate the expression of a cascade of
EMT-promoting transcription factors that include Snail, Slug,
Twist1 and 2, and ZEB1 and 2 (9,10).
Although E-cadherin was originally described as a tumor suppressor
and the decreased expression of E-cadherin is a hallmark of EMT,
recent findings indicate that E-cadherin both suppresses and
promotes cancer (11). The
hypermethylation of DNA by DNA methyltransferase 1 (DNMT1) results
in the dysregulation of genes involved in TNBC tumorigenesis and
progression, including EMT-promoting and metastasis-associated
genes (12). Metastasis
suppressors, such as n-Myc downstream regulated gene 1 (NDRG1), are
involved in the inhibition of TGF-β/Smad and Wnt/β-catenin
pathways, thereby suppressing EMT (13,14).
Novel therapeutics that target EMT and the metastatic process are
required to reduce TNBC-associated mortality.
Certain bioactive dietary phytochemicals have
attracted interest over the past few decades due to their potential
for use in cancer prevention and treatment (15). Piperlongumine (PL), also known as
piplartine, is an amide alkaloid found in long pepper plant
(Piper longum) fruits that has a long history of use as a
culinary spice, as well as in Ayurvedic medicine for the treatment
of a variety of illnesses, including cancer (16,17).
Laboratory studies have demonstrated that micromolar concentrations
of PL are cytotoxic for multiple cancer cell types, including
breast cancer cells (18-23).
In TNBC cells, PL has been reported to induce apoptosis via the
suppression of signal transducer and activator of transcription
(STAT) 3 activation and the inhibition of phosphatidylinositol
3-kinase/Akt/mammalian target of rapamycin signaling (22,23).
Biocompatible nanoparticle (NP)-based drug delivery systems
increase the effectiveness of anticancer drugs (24). NP delivery has the potential to
increase the aqueous solubility and bioavailability of PL, as well
as to further decrease its already low toxicity in preclinical
models and clinical application (25,26).
In this regard, the poly(ethylene glycol)-poly(lactide-co-glycolic)
acid (PEG-PLGA) polymer-based NP delivery of luteolin, a flavonoid
with potent anticancer activity but poor pharmacokinetics, has been
shown to result in more potent tumor growth inhibitory effects than
the administration of free luteolin in a preclinical model of head
and neck cancer (27). In
addition, the authors have recently demonstrated that piperine, a
major alkaloid of black pepper that is structurally similar to PL,
retains its cytotoxic effect on TNBC cells when encapsulated in
biocompatible methoxy poly(ethylene
glycol)-poly(lactide-co-glycolic) acid (mPEG-PLGA) NPs (28).
The present study compared the effects of free PL
and PL-loaded mPEG-PLGA NPs (PL-NPs) on the in vitro growth
of 3 different TNBC cell lines, as well as the metastasis-promoting
activities of MDA-MB-231 TNBC cells. Cell growth/viability,
migration/invasion, and the expression levels of MMP2, NDRG1,
EMT-associated molecules, and the epigenetic regulator, DNMT1,
associated with cancer progression were determined by western blot
analysis and/or reverse transcription-quantitative polymerase chain
reaction (RT-qPCR). Free PL and PL-NPs exerted a similar inhibitory
effect on the growth and metastatic activities of TNBC cells,
supporting the feasibility of the use of NPs to deliver PL for
prevention or reduction of TNBC metastasis.
Materials and methods
Chemicals and reagents
Dimethyl sulfoxide (DMSO),
3-(4,5-demethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT),
phenylmethylsulfonyl fluoride, Triton X-100, sodium deoxycholate,
aprotinin, leupeptin, sodium fluoride, pepstatin A, dithiothreitol,
polyamine oxidase, dichloromethane and gelatin were purchased from
Sigma-Aldrich; Merck KGaA. L-glutamine, 10,000 units/ml
penicillin/10,000 µg/ml streptomycin solution, 1M N-2
hydroxyethylpiperazine-N-2-ethane sulfonic acid (HEPES) buffer
solution, fetal bovine serum (FBS), and Dulbecco's modified Eagle
medium (DMEM) were from Invitrogen; Thermo Fisher Scientific, Inc.
mPEG-PLGA (5-10 kDa) was from Akina Inc. Acrylamide/bis-acrylamide
(29: 1, 30% solution), ammonium persulfate, sodium dodecyl sulfate,
Tris-base, Tween-20, tetramethylethelyenediamide,
ethylenediaminetetraacetic acid (EDTA) and ethylene
glycol-bis(β-aminoethyl ether)-N,N,N',N'-tetraacetic acid (EGTA)
were purchased from BioShop Canada Inc. Bio-Rad protein assay dye
reagent concentrate and SsoFast EvaGreen™ Supermix® were
from Bio-Rad Laboratories, Inc. PL, fibronectin and sodium
orthovanadate were from EMD Millipore (Etobicoke, ON). ARP 100 was
from Santa Cruz Biotechnology Inc..
Antibodies
Horseradish peroxidase (HRP)-conjugated rabbit
anti-human β-actin monoclonal antibodies (Abs; cat. no. 12620), and
rabbit monoclonal Abs against human β-catenin (cat. no. 8480), Slug
(cat. no. 9585), ZEB1 (cat. no. 3396), pan-cadherin (cat. no.
4073), Smad3 (cat. no. 9520), NDRG1 (cat. no. 9485), and DNMT-1
(cat. no. 5032), were purchased from Cell Signaling Technology,
Inc. Donkey anti-rabbit HRP-conjugated Abs (cat. no. sc-2313) were
from Santa Cruz Biotechnology Inc. All Abs were diluted in 5% w/v
fat-free milk or 5% w/v BSA, in Tween-TBS [20 mM Tris-HCl (pH 7.6),
200 mM NaCl, 0.05% Tween-20].
Cell lines and culture conditions
MDA-MB-231 human breast adenocarcinoma cells were
kindly provided by Dr S. Drover (Memorial University of
Newfoundland, St. John's, NL, Canada). MDA-MB-468 human breast
adenocarcinoma cells were a generous gift from Dr P. Lee (Dalhousie
University, Halifax, NS, Canada). BT-549 human breast ductal
carcinoma cells were kindly provided by Dr P. Marcato (Dalhousie
University, Halifax, NS, Canada). All breast cancer cell lines were
free of mycoplasma contamination and were authenticated by the
American Type Culture Collection (ATCC) using the short tandem
repeat method. Breast cancer cells were cultured in DMEM that was
supplemented with 10% heat-inactivated FBS, 100 U/ml penicillin,
100 µg/ml streptomycin, 2 mM of L-glutamine, and 5 mM of HEPES;
henceforth, known as complete DMEM. Cells were maintained at 37˚C
in a humidified 10% CO2 incubator.
NP formulation
NPs were prepared from mPEG-PLGA polymers using the
thin-film hydration method, as previously described (28). In brief, 5 mg PL and 45 mg
mPEG-PLGA were co-dissolved in 5 ml dichloromethane and transferred
to a 250 ml round-bottom flask. The mixture was evaporated under
vacuum using a rotary evaporator (Büchi Labortechnik) at 60˚C. The
co-evaporation of PL and mPEG-PLGA resulted in a homogenous mixture
in a form of a thin film coating the inner surface of the flask.
The thin-film material was re-dissolved in 5 ml 0.9% w/v NaCl
solution and stirred at 60˚C to allow the self-assembly of polymers
into PL-containing micelles. The mixture was placed in a 14 kDa
cut-off dialysis membrane and dialyzed against 0.9% NaCl solution
at room temperature to remove any encapsulated PL. The 0.9% NaCl
solution was replaced after 30 min, 2 and 4 h. The PL-NP
preparation was then flash-frozen in liquid nitrogen and
lyophilized. PL-NPs were reconstituted in sterile water and
sonicated for 5 min using a Q125 ultrasonic probe at 50W output
(QSonica L.L.C.) to obtain NPs of the desired size and further
improve PL entrapment.
A JEM 1230 transmission electron microscope (JEOL
Ltd.) and AMT Image Capture Engine (version 7.0; AMT Imaging) was
used to image and measure 90 random particles, yielding an average
NP size of 52.8±1.2 nm. Prior to use, the PL-NPs were
filter-sterilized using a 0.20 µm syringe filter, which also
removed polymer aggregates and any remaining PL crystals. The
encapsulation efficiency was 20%, as determined by
spectrophotometric analysis and a standard curve based on the
absorbance of PL at 346 nm.
MTT assay
MDA-MB-231, MDA-MB-468 and BT549 cells were seeded
into quadruplicate wells of a 96-well flat bottom cell culture
plate at a concentration of 5x103 cells/well and
incubated overnight to allow cell attachment. Cells were then
cultured for 48 h in the presence of medium alone, vehicle (DMSO)
alone, 2.5-10 µM free PL (dissolved in DMSO) or PL-NPs, or empty
NPs. MTT solution was added to each well to a final concentration
of 0.5 µg/ml. Cell-free supernatant was removed and formazan
crystals were solubilized in DMSO. The absorbance was measured at
570 nm using an Expert 96 microplate reader (Biochrom ASYS) and the
percentage metabolic activity was determined.
Transwell migration and invasion
assays
MDA-MB-231 cell monolayers were cultured for 36 h in
the presence of the vehicle (DMSO) alone, 2.5 µM of free PL or
PL-NPs, or empty NPs. The cells were then serum-starved for 12 h,
harvested and resuspended at 1x106 cells/ml in 1 ml of
appropriate treatment made in serum-free DMEM. A 50 µl aliquot of
the cell suspension was loaded into the upper chamber of a
Transwell migration apparatus. The cells migrated through an 8 µm
porous membrane that was uncoated for migration assays or coated
with fibronectin (0.05% w/v) or gelatin (0.01% w/v) for invasion
assays. Growth medium containing 10% FBS was used as a
chemoattractant. Migrated cells were stained for 45 sec at room
temperature with a Diff-Quik™ staining kit (Siemens Inc.),
photographed using a Nikon Eclipse TS 100 phase contrast microscope
and Infinity 1 camera (Nikon Canada Inc.), and quantified using
ImageJ software (version 1.51; National Institutes of Health).
Western blot analysis
MDA-MB-231 cell monolayers were cultured for 48 h in
the presence of the vehicle (DMSO) alone, 2.5 or 5 µM of free PL or
PL-NPs, or empty NPs. Cells were collected and resuspended in 50 µl
of cold lysis buffer [0.1% v/v NP-40, 0.25% w/v sodium
deoxycholate, 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 5 mM EDTA, 5 mM
EGTA pH 7.5] with a mixture of 1 mM phenylmethylsulfonyl fluoride,
5 µg/ml leupeptin, 5 µg/ml pepstatin, 10 µg/ml apotinin, 100 µM
sodium orthovanadate, 1 mM dithiothreitol, 10 mM sodium fluoride
and 10 µM polyamine oxidase. Following 15 min of incubation at 4˚C,
debris was removed by centrifugation at 14,000 x g for 10 min at
4˚C. The supernatant containing total cellular proteins was
collected and quantified by Bradford protein assay. Protein samples
(30 µg) were loaded into wells of a 10% sodium dodecyl
sulfate-polyacrylamide gel and proteins were separated by
electrophoresis. Proteins were transferred to a nitrocellulose
membrane and blocked with 5% fat-free milk for 1 h at room
temperature to avoid non-specific binding. Blots were incubated
overnight at 4˚C with primary Abs (anti-Slug, anti-ZEB1, and
anti-pan-cadherin, 1:500; anti-β-catenin, anti-Smad3, anti-NDRG1,
anti-DMNT1, anti-β-actin, 1:1,000). The membranes were thoroughly
washed for 30 min with Tweet-TBS, and then incubated for 2 h at
room temperature with the HRP-conjugated secondary Ab (1:1,000),
followed by washing with Tween-TBS. Equal protein loading was
confirmed by probing for β-actin expression. Protein bands were
visualized with chemiluminescent HRP substrate Luminata™ (Merck
KGaA) and Amersham high performance chemiluminescence film (GE
Healthcare). Image Lab software (version 5.2; Bio-Rad) was used for
densitometric analysis.
RT-qPCR
MDA-MB-231 cell monolayers were cultured for 48 h in
the presence of the vehicle (DMSO) alone, the indicated
concentrations of free PL or PL-NPs, or empty NPs. Total RNA was
isolated from cells using the RNeasy mini kit (Qiagen Inc.)
according to the manufacturer's instructions. RNA quantity and
purity were determined by spectrophotometric analysis.
Approximately 500 ng of RNA were reverse transcribed to cDNA using
the iScript™ cDNA synthesis kit (Bio-Rad Laboratories, Inc.),
according to the manufacturer's instructions. Appropriately diluted
cDNA samples were combined with primer mix (10 µM forward and
reverse primers), nuclease-free water, and SsoFast EvaGreen™
Supermix® (Bio-Rad Laboratories, Inc.) or
SYBR®-Green PCR master mix (Qiagen Inc.) at a 1: 1: 3: 5
ratio, respectively. Samples were transferred to a Multiplate™
96-well unskirted polypropylene PCR plate in triplicates and placed
in the CFX Connect™ RT-PCR detection system (B Bio-Rad
Laboratories, Inc.). The reaction steps were as follows: 10 min of
activation at 95˚C, 40 cycles of 10 sec of denaturation at 95˚C,
and 20 sec at the primer-specific annealing temperature. β-actin
was also amplified at the same time and used as a reference gene.
Data obtained from the RT-qPCR reaction were analyzed using CFX
Manager software (version 3.1, Bio-Rad Laboratories, Inc.). The
sequences of the primers were as follows: MMP2 (63˚C) forward,
5'-TGGCAAGTACGGCTTCTGTC-3' and reverse, 5'-TTCTTGTCGCGGTCGTAGTC-3';
E-cadherin (64˚C) forward, 5'-CAGCCACAGACGCGGACGAT-3' and reverse,
5'-CTCTCGGTCCAGCCCAGTGGT-3'; and β-actin forward,
5'-AAGATCAAGATCATTGCTCCTC-3' and reverse,
5'-CAACTAAGTCATAGTCCGCC-3'.
Statistical analysis
Statistical analysis was performed using one-way
analysis of variance (ANOVA) with the Tukey-Kramer or Bonferroni
multiple comparisons post hoc test, where appropriate, using
GraphPad Prism analysis software (version 5.0, GraphPad Software
Inc.). Differences were considered statistically significant at
P<0.05.
Results
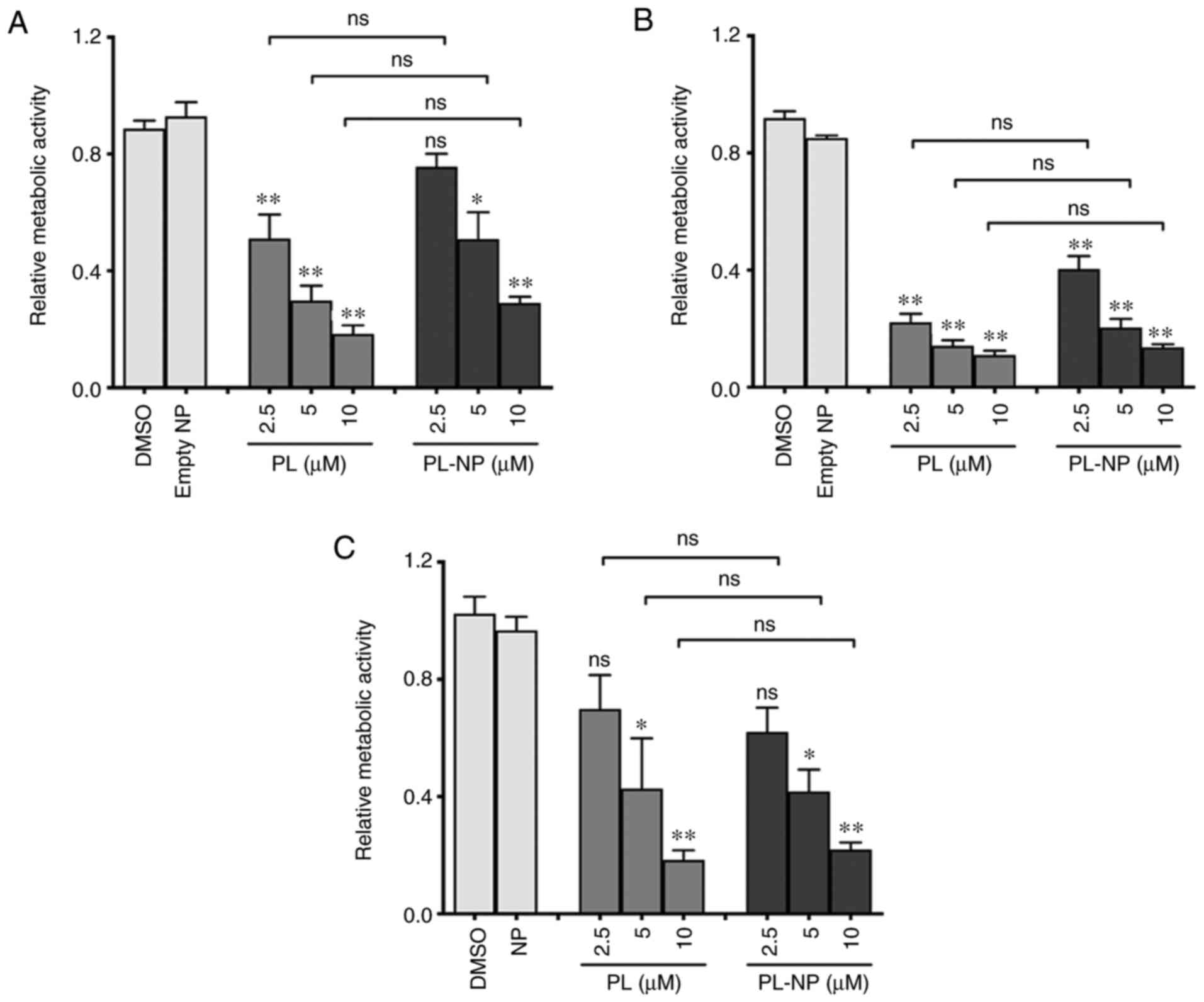
PL and PL-NPs are equally cytotoxic to
TNBC cells
The cytotoxicity of free PL and PL-NPs was compared
using 3 different TNBC cell lines (Fig. 1). Following culture for 48 h in the
presence of various concentrations of free PL (2.5-10 µM) or
equivalent concentrations of PL-NPs, MTT assays revealed a similar
concentration-dependent decrease in the number of TNBC cells
(MDA-MB-231, MDA-MB-468 and BT-549) following treatment with PL and
PL-NPs. Since the inhibitory effects of 2.5 and 5 µM of PL in
PL-NPs on the growth of MDA-MB-231 cells did not differ
significantly and approximated the IC50, these
concentrations of PL-NPs and free PL were used in all subsequent
experiments with MDA-MB-231 TNBC cells.
PL-NPs and free PL are effective
inhibitors of TNBC cell migration and invasion
Transwell assays revealed that 2.5 µM of free PL or
an equivalent concentration of PL-NPs significantly reduced the
chemoattractant-induced migration of MDA-MB-231 TNBC cells through
an uncoated membrane (Fig. 2A).
Free PL and PL-NPs exerted a similar inhibitory effect on the
ability of MDA-MB-231 to migrate through fibronectin- and
gelatin-coated membranes (Fig. 2B
and C, respectively) used to
assess tumor cell invasiveness, suggesting the capacity to suppress
the degradation of ECM components during metastasis. The results of
RT-qPCR analysis revealed that the mRNA expression of ECM-degrading
MMP2 in MDA-MB-231 cells was suppressed by approximately 70% in the
presence of free PL or PL-NPs (Fig.
2D). MMP2 expression was determined by RT-qPCR as we were not
able to identify a good anti-MMP2 Ab for western blot analysis. In
addition, MDA-MB-231 cell migration through a gelatin-coated
membrane was also markedly decreased in the presence of the
selective MMP2 inhibitor, ARP 100 (Fig. S1), suggesting that the inhibitory
effect of free PL and PL-NPs on MDA-MB-231 cell invasiveness was at
least in part due to reduced MMP-2 expression.
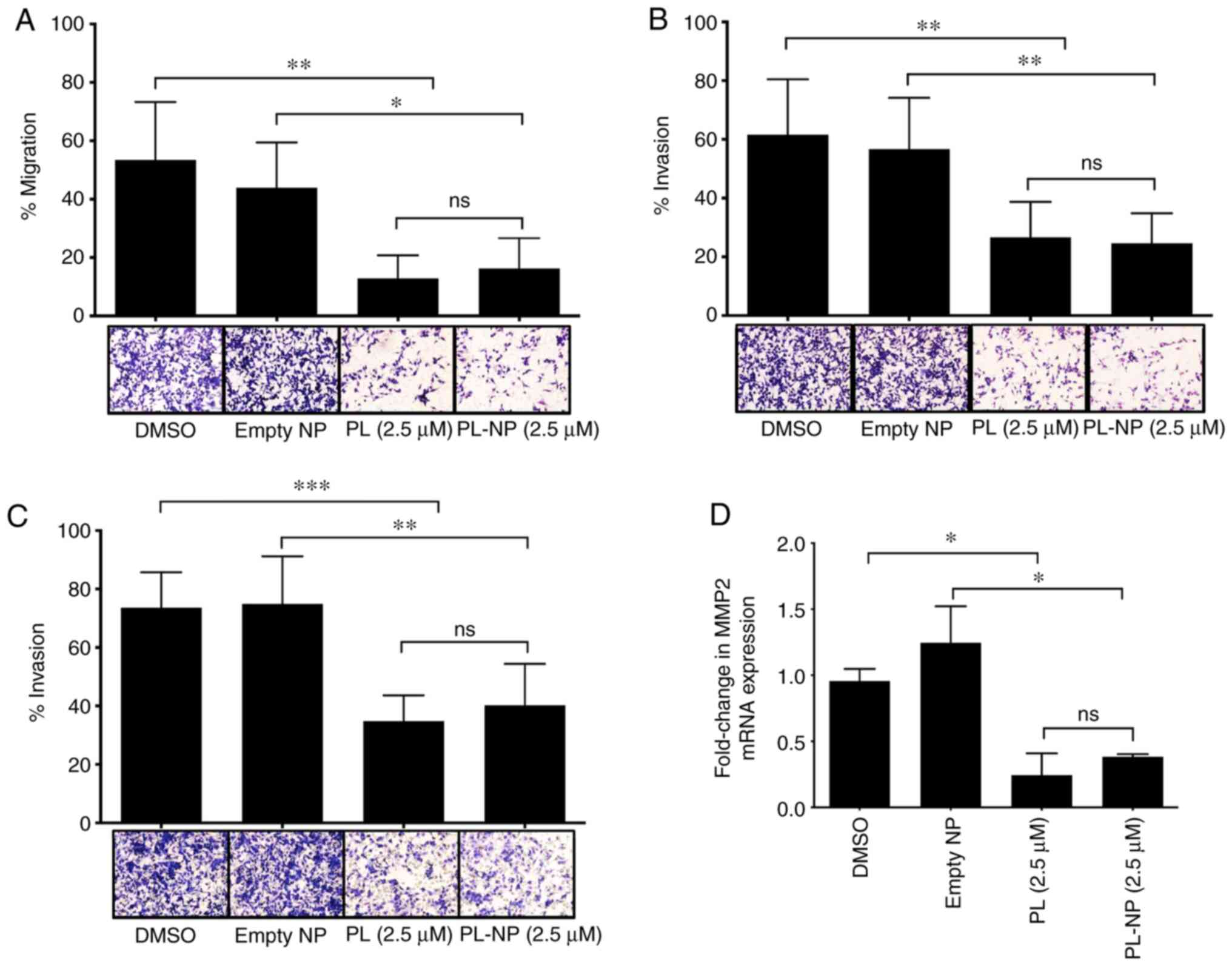 | Figure 2PL and PL-NPs inhibit the migration
and invasiveness of MDA-MB-231 TNBC cells and their expression of
MMP2. MDA-MB-231 cells were cultured in the presence of vehicle
(DMSO), empty NPs, or 2.5 µM free PL or PL-NPs in
serum-supplemented complete DMEM for 36 h followed by washing and
culture for 12 h in serum-free complete DMEM. Cells were loaded
into the upper chamber of a Transwell migration apparatus. Cells
that moved through (A) uncoated 8 µm porous membranes used to
assess migration, or (B) fibronectin-coated, and (C) gelatin-coated
8 µm porous membranes used to assess invasiveness, were stained and
membranes were photographed at x20 magnification. Data shown are
the mean number of migrating cells ± SEM of 3 (uncoated), 5
(fibronectin-coated) and 4 (gelatin-coated) independent
experiments. (D) MDA-MB-231 cells were cultured for 48 h in the
presence of vehicle (DMSO), empty NPs, or 2.5 µM free PL or PL-NPs.
Total RNA was isolated from cells and mRNA was converted to cDNA.
MMP2 mRNA expression was determined by RT-qPCR. β-actin was used as
the reference gene. Data shown are mean MMP2 mRNA expression ± SEM
of 3 independent experiments. (A-D) Statistical significance was
determined by one-way ANOVA with the Bonferroni multiple
comparisons post-test; *P<0.05,
**P<0.01, ***P<0.001; ns, not
significant. TNBC, triple-negative breast cancer; PL,
piperlongumine; NPs, nanoparticles. |
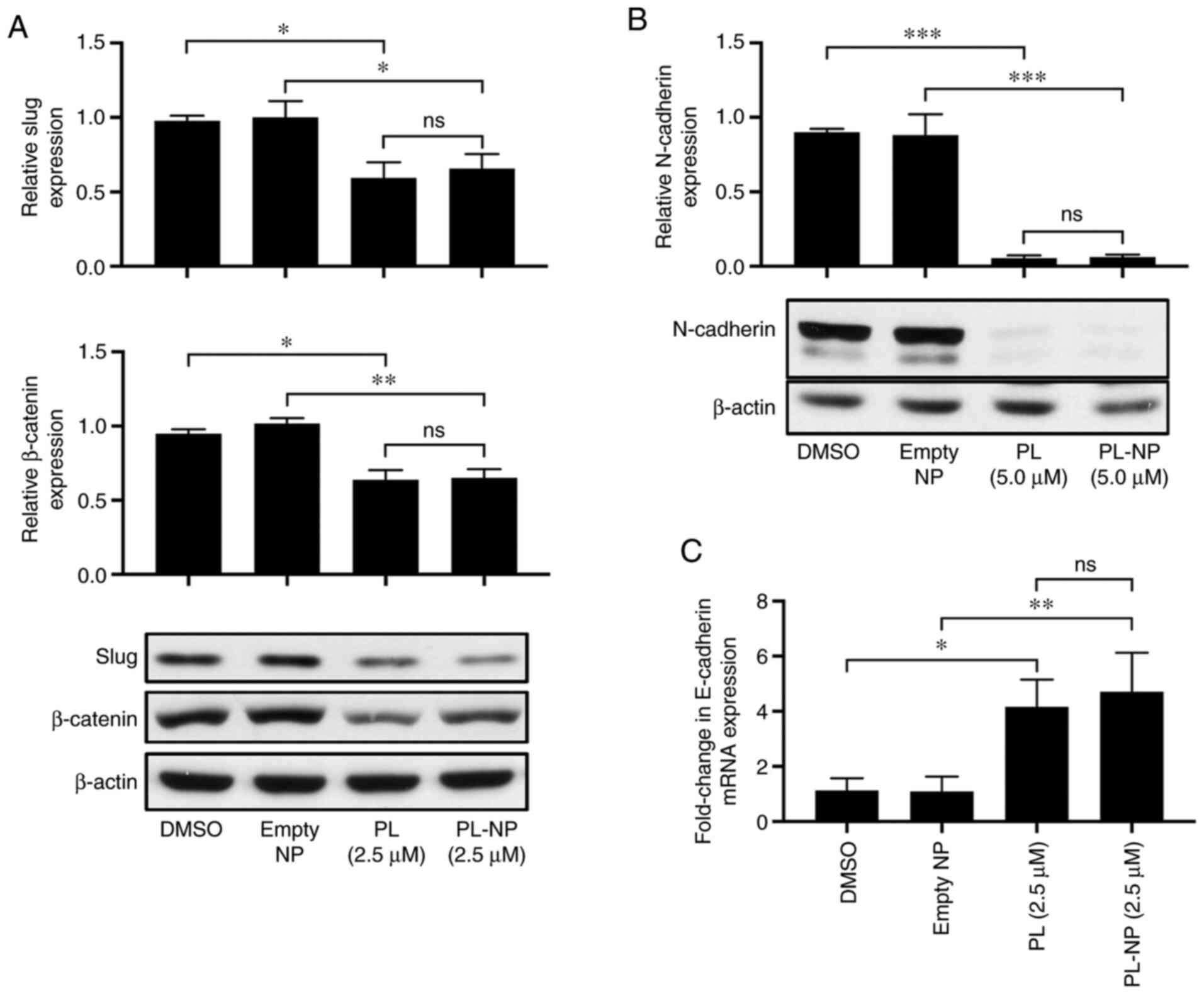
Free PL and PL-NPs downregulate the
expression of mesenchymal markers and upregulate E-cadherin
expression in TNBC cells
Western blot analysis revealed that culture in the
presence of 2.5 µM free PL or an equivalent concentration of PL-NPs
significantly reduced the MDA-MB-231 TNBC cell expression of the
EMT-promoting transcription factor, Slug (Fig. 3A). The expression of ZEB1, another
EMT-promoting transcription factor, was also reduced in the
MDA-MB-231 cells following treatment with 2.5 µM free PL or an
equivalent concentration of PL-NPs (Fig. S2). Consistent with the inhibition
of EMT, exposure to free PL and PL-NPs reduced the expression of
the mesenchymal markers, β-catenin (Fig. 3A) and N-cadherin (Fig. 3B). By contrast, RT-qPCR revealed
that expression of mRNA coding for the epithelial marker,
E-cadherin, was increased 4-fold in the presence of free PL or
PL-NPs (Fig. 3C). E-cadherin
expression was determined by RT-qPCR as we were not able to
identify a good anti-E-cadherin Ab for western blot analysis. Taken
together, these findings indicate an equivalent inhibitory effect
of free PL and PL-NPs on EMT of TNBC cells.
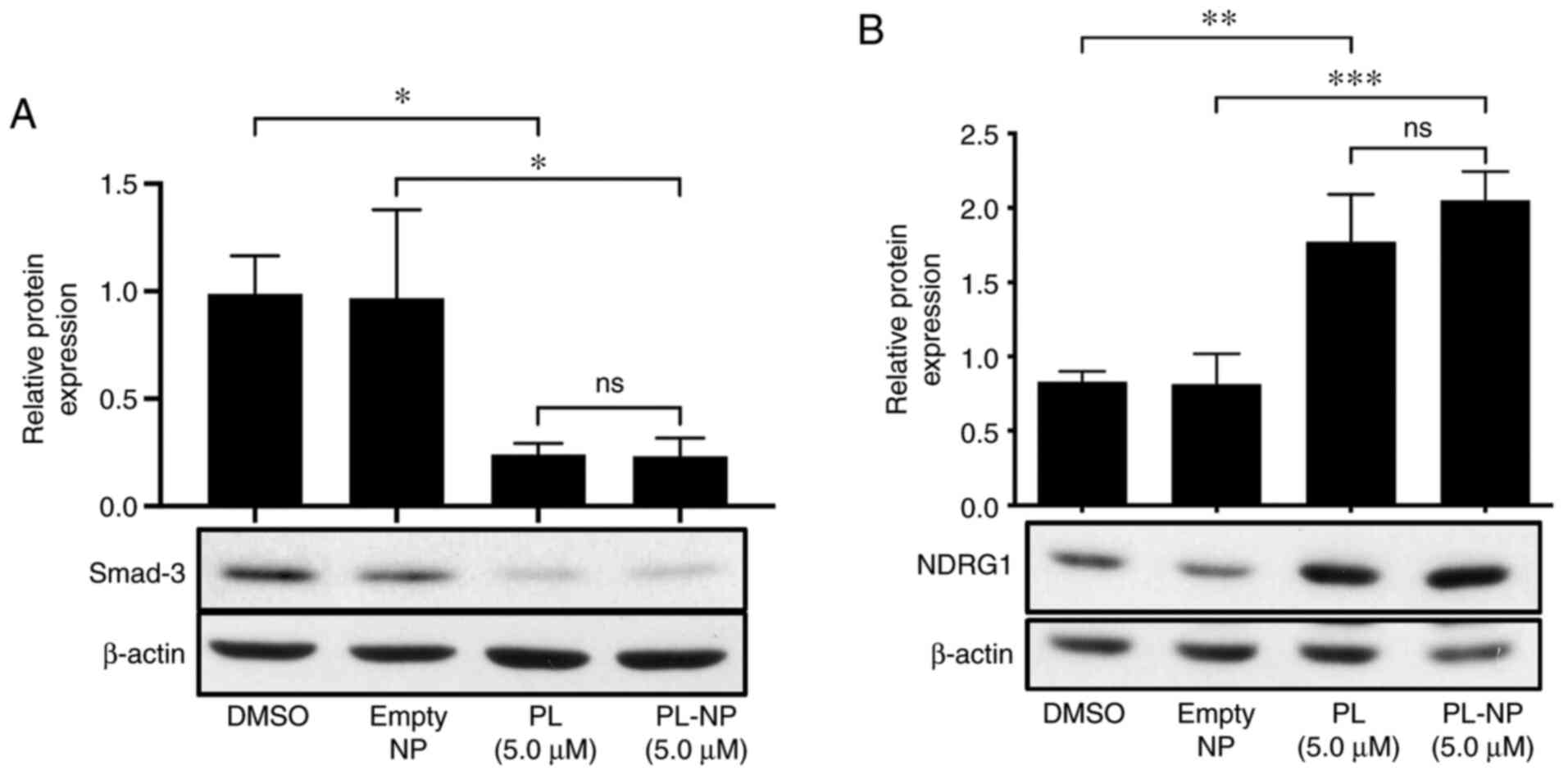
Free PL and PL-NPs inhibit Smad3
expression and increase NDRG1 expression in TNBC cells
The effects of free PL and PL-NPs on the expression
of TGFβ/Smad signaling pathway-associated Smad-3 and
anti-metastatic NDRG1 in MDA-MB-231 cells were then determined.
MDA-MB-231 cells that were cultured in the presence of 5 µM free PL
or an equivalent concentration of PL-NPs exhibited decreased Smad3
expression (Fig. 4A) and an
increased expression of NDRG1 (Fig.
4B).
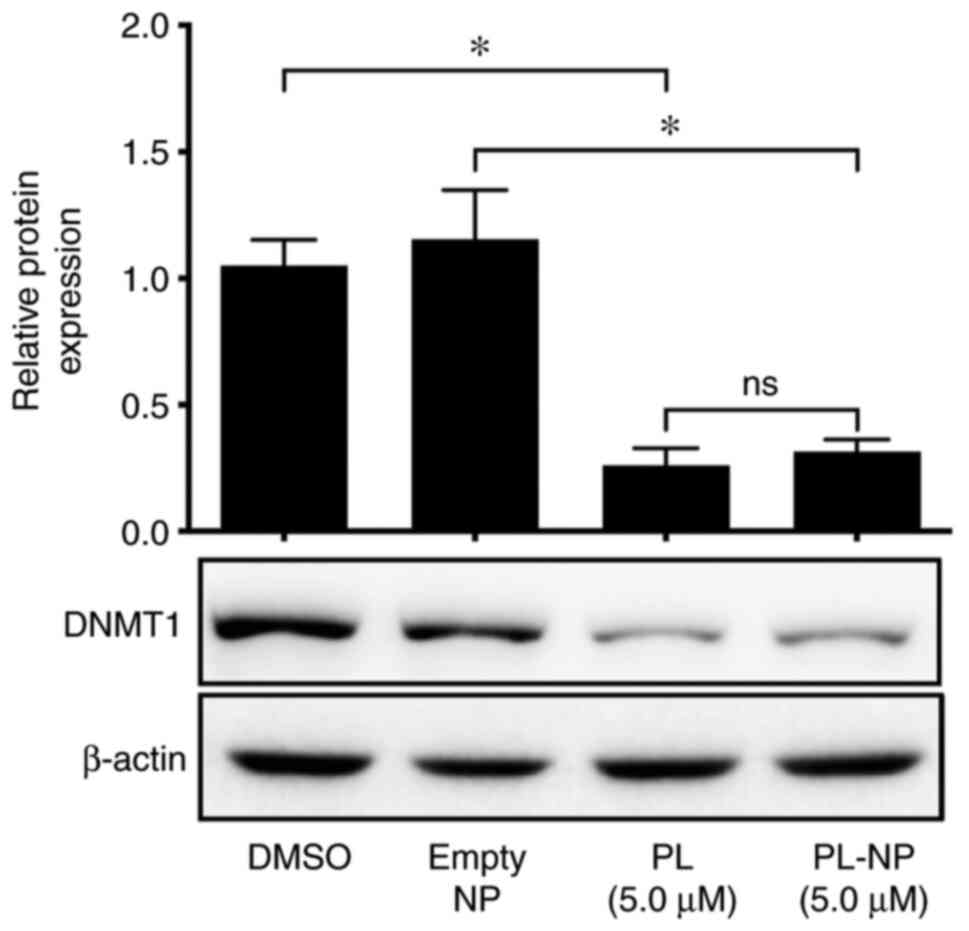
TNBC cell expression of DNMT1 is
inhibited by free PL and PL-NPs
Subsequently, the expression of DNMT1 was examined
following treatment of the MDA-MB-231 TNBC cells with free PL or
PL-NPs to determine whether there may be an effect on the
hypermethylation of TNBC cell DNA. As shown in Fig. 5, the DNMT1 levels were markedly
reduced in the presence of 5 µM free PL or an equivalent
concentration of PL-NPs, suggesting that PL has the potential to
affect the epigenetic regulation of EMT.
Discussion
PL has been shown to exert potent cytotoxic effects
on TNBC cells (22,23); however, the anti-metastatic effects
of PL on TNBC cells have not yet been fully elucidated. Moreover,
the feasibility of using biodegradable NPs to deliver PL to TNBCs
has not yet been demonstrated. NP delivery enhances the bioefficacy
of phytochemicals, such as PL, by overcoming the barriers posed by
low solubility and poor bioavailability, as well as reducing the
potential for undesirable toxicity to healthy tissues (25,26).
To the best of our knowledge, the present study demonstrates for
the first time that PL encapsulated in biocompatible NPs formed
from mPEG-PLGA was as effective as free PL for the inhibition of
TNBC (MDA-MB-231, MDA-MB-468 and BT-549) cell growth in monolayer
cultures. Neoplastic cells take up PEG-PLGA NPs by endocytosis,
after which the NPs release their cargo within the acidic
environment of lysosomes (29).
The present findings are in line with those of other studies
showing the reduced in vitro growth of other cancer cell
types following the NP-based delivery of other tumoricidal
phytochemicals (27,28,30).
Tumor cell migration and invasion through the
basement membrane and ECM are essential components of tumor
metastasis (31,32). Drugs that interfere with these
processes may therefore be successful in preventing or reducing
metastasis. In the present study, both free PL and PL-NP inhibited
the chemoattractant-directed migration of MDA-MB-231 TNBC cells
across cell-permeable membranes, including membranes that were
coated with the ECM components gelatin and fibronectin. The reduced
migration of PL-treated TNBC cells across membranes coated with ECM
components could, at least in part, result from the decreased
expression of ECM-degrading MMPs that are involved in metastasis
(8). Indeed, the present study
demonstrated that free PL and PL-NPs downregulated the MDA-MB-231
cell expression of MMP2, which degrades both fibronectin and
gelatin (33). MMP2 inhibition by
ARP 100, a selective inhibitor of MMP2, also decreased the
invasiveness of MDA-MB-231 cells. Taken together, these findings
suggest that the decreased expression of MMP2 in the presence of PL
contributed to the reduced invasiveness of PL-treated MDA-MB-231
cells. The reduced expression of MMP2 in the presence of PL was
most likely due to the PL-mediated inhibition of
phosphatidylinositol 3-kinase/Akt/mammalian target of rapamycin
signaling (23), as this signaling
pathway is known to regulate MMP2 expression in malignant gliomas
(34).
The present study also demonstrated that free PL and
PL-NPs interfered with MDA-MB-231 TNBC cell expression of Slug and
ZEB1, which are EMT-promoting transcription factors (9,10).
The expression of β-catenin by MDA-MB-231 cells was also suppressed
in the presence of PL, suggesting the inhibition of the
Wnt/β-catenin signaling pathway. This is consistent with the
PL-induced downregulation of Slug expression, since Wnt/β-catenin
signaling promotes Slug expression by breast cancer cells (35). The inhibition of the
phosphatidylinositol 3-kinase/Akt/mammalian target of rapamycin
signaling pathway by PL (23) may
also account for the observed decrease in β-catenin expression,
since Akt inhibition blocks the nuclear localization of β-catenin,
leading to its phosphorylation and proteosomal degradation
(36). In addition, epithelial and
mesenchymal marker expression was altered following the PL and
PL-NP treatment of MDA-MB-231 cells. PL upregulated the expression
of the epithelial marker, E-cadherin, and downregulated the
expression of the mesenchymal marker, N-cadherin, which is
associated with an invasive phenotype of breast cancer cells
(37). Interestingly, ectopic
expression of E-cadherin by MDA-MB-231 cells causes a shift from
mesenchymal-like to epithelial-like morphology (38). E-cadherin re-expression and
N-cadherin suppression following the PL and PL-NP treatment of
MDA-MB-231 cells may be the result of the decreased Slug
expression, since E-cadherin expression by MDA-MB-231 cells is
suppressed by Slug via the upregulation of miR-221(39), and there is an inverse association
between E-cadherin and N-cadherin expression (40).
In the present study, the MDA-MB-231 TNBC cells
treated with free PL or PL-NPs exhibited a reduced expression of
Smad3, which is a key component of the TGFβ/Smad signaling pathway
involved in the initiation of EMT (9,10).
Although the present study did not determine the effects of PL
treatment on Smad3 phosphorylation, it is likely that a decrease in
available Smad3 would have a negative effect on Smad3 signaling, as
it has been shown in colon cancer cells following Smad3 knockdown,
that also resulted in decreased overall levels of Smad3
phosphorylation (41). The
expression of NDRG1, which is a tumor metastasis suppressor that
abrogates the TGFβ/Smad-induced upregulation of Slug and other
EMT-promoting transcription factors (13), was upregulated in the present study
when MDA-MB-231 cells were treated with PL. NDRG1 also inhibits the
Wnt/β-catenin signaling pathway (14); thus, the increased NDRG1 expression
would be expected to suppress Wnt/β-catenin signaling. Taken
together, these findings suggest that PL may promote the
acquisition of an epithelial phenotype by mesenchymal-like
MDA-MB-231 cells at least in part by modulating TGFβ/Smad and
Wnt/β-catenin signaling pathways. At this time, the mechanism
through which exposure to PL downregulates Smad3 expression and
upregulates NDRG1 expression remains to be elucidated. However, PL
has been reported to inhibit extracellular signal-regulated kinase
1/2 activation in colorectal cancer cells (42). A similar effect in MDA-MB-231 cells
may account for the reduced Smad3 expression, since the inhibition
of extracellular signal-regulated kinase signaling suppresses Smad3
expression in epithelial cells (43). The PL-induced downregulation of
DNMT1 expression in MDA-MB-231 cells may account for the increased
NDRG1 expression, since the inhibition of DNA methylation
upregulates NDRG1 expression (44), and epigenetic silencing of NDRG1 in
breast cancer cells is the result of DNA hypermethylation (45).
EMT and the metastasis of TNBC cells is associated
with the DNMT1-mediated hypermethylation of DNA (12). For example, E-cadherin expression
by breast cancer cells is silenced by DNMT1-mediated DNA
hypermethylation (46,47). The present study demonstrates that
free PL and PL-NPs inhibits DNMT1 expression in MDA-MB-231 cells,
which may account for the re-expression of E-cadherin by PL-treated
MDA-MB-231 cells. Although, to the best of our knowledge, this is
the first study to investigate the effects of PL on the epigenetic
machinery of TNBC cells, previous studies have demonstrated the
effects of other dietary phytochemicals on the epigenome. For
example, epigallocatechin gallate has been shown to inhibit DNMT1
activity in human esophageal, colon and prostate cancer cells,
resulting in the re-expression of several tumor suppressor genes
(48).
In conclusion, the present study demonstrates, for
the first time, to the best of our knowledge, that NPs formed from
biocompatible mPEG-PGLA can deliver PL to cultures of TNBC cells
without any loss of efficacy in comparison to free PL. In this
regard, the growth of TNBC cells in monolayer cultures was
inhibited by PL-NPs to the same extent as free PL. In addition,
TNBC migration/invasion and the expression of EMT-promoting
proteins was markedly decreased in the presence of PL-NPs. By
contrast, TNBC cell expression of the tumor suppressor, NDRG1, and
E-cadherin, which is associated with a less invasive epithelial
phenotype, was upregulated by PL-NP treatment. Moreover, PL-NPs
have the potential to prevent the hypermethylation of DNA via the
PL-mediated inhibition of DNMT1 expression. Further analysis of the
effects of PL-NPs on the epigenome is important, considering the
interest in compounds that block the epigenetic modification of DNA
as chemotherapeutic agents (49).
Collectively, these findings reveal an inhibitory effect of PL-NPs
on the metastatic potential of TNBC cells that warrant further
investigation in preclinical models of TNBC.
Supplementary Material
Inhibition of MMP2 by ARP 100
interferes with MDA-MB-231 invasiveness. MDA-MB-231 cells were
cultured in the presence of the vehicle (DMSO) or the indi-cated
concentrations of the MMP2 inhibitor ARP 100 in serum-supplemented
complete DMEM for 36 h followed by washing and culture for 12 h in
serum-free complete DMEM. Cells were then loaded into the upper
chamber of a Transwell migration apparatus. Cells that migrated
through a 0.05% w/v gelatin-coated 8 μm porous membrane were
stained and membranes were photographed at x20 magnification.
Representative images are shown. MMP2, matrix metalloproteinase
2.
PL and PL-NPs suppress MDA-MB-231 TNBC
cell expression of ZEB1. MDA-MB-231 cells were cultured for 48 h in
the presence of medium alone, the vehicle (DMSO), empty NPs, or 2.5
μM free PL or PL-NPs. Total protein was isolated from lysed cells
and subjected to western blot analysis of ZEB1 expression. Equal
protein loading was confirmed by probing for β-actin. PL,
piperlongumine; NPs, nanoparticles; TNBC, triple-negative breast
cancer.
Acknowledgements
The authors would like to thank Ms. Mary Ann Trevors
(Faculty of Medicine Electron Microscopy Core Facility, Dalhousie
University) for providing assistance with NP characterization.
Funding
The present study was funded by a grant (no. 314347)
to DWH from the Canadian Cancer Society. JGR was funded by a Nova
Scotia Graduate Scholarship and a Cancer Research Training program
award.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article or are available from the
corresponding author on reasonable request.
Authors' contributions
JGR performed all the assays. JGR and WF performed
data analysis. JGR and WF drafted the manuscript. DWH designed the
research. DWH and WF revised the manuscript. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Perou CM, Sørlie T, Eisen MB, van de Rijn
M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA,
et al: Molecular portraits of human breast tumours. Nature.
406:747–752. 2000.PubMed/NCBI View
Article : Google Scholar
|
|
2
|
Sørlie T, Perou CM, Tibshirani R, Aas T,
Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey
SS, et al: Gene expression patterns of breast carcinomas
distinguish tumor subclasses with clinical implications. Proc Natl
Acad Sci USA. 98:10869–10874. 2001.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Eroles P, Bosch A, Pérez-Fidalgo JA and
Lluch A: Molecular biology in breast cancer: Intrinsic subtypes and
signaling pathways. Cancer Treat Rev. 38:698–707. 2012.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Schnitt SJ: Classification and prognosis
of invasive breast cancer: From morphology to molecular taxonomy.
Mod Pathol. 23 (Suppl 2):S60–S64. 2010.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Sørlie T: Molecular portraits of breast
cancer: Tumour subtypes as distinct disease entities. Eur J Cancer.
40:2667–2675. 2004.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Chambers AF, Groom AC and MacDonald IC:
Dissemination and growth of cancer cells in metastatic sites. Nat
Rev Cancer. 2:563–572. 2002.PubMed/NCBI View
Article : Google Scholar
|
|
7
|
Kennecke H, Yerushalmi R, Woods R, Cheang
MC, Voduc D, Speers CH, Nielsen TO and Gelmon K: Metastatic
behavior of breast cancer subtypes. J Clin Oncol. 28:3271–3277.
2010.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Radisky ES and Radisky DC: Matrix
metalloproteinase-induced epithelial-mesenchymal transition in
breast cancer. J Mammary Gland Biol Neoplasia. 15:201–212.
2010.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Garg M: Epithelial-mesenchymal
transition-activating transcription factors-multifunctional
regulators in cancer. World J Stem Cells. 5:188–195.
2013.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Kalluri R and Weinberg RA: The basics of
epithelial-mesenchymal transition. J Clin Invest. 119:1420–1428.
2009.PubMed/NCBI View
Article : Google Scholar
|
|
11
|
Hu QP, Kuang JY, Yang QK, Bian XW and Yu
SC: Beyond a tumor suppressor: Soluble E-cadherin promotes the
progression of cancer. Int J Cancer. 138:2804–2812. 2016.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Wong KK: DNMT1: A key drug target in
triple-negative breast cancer. Sem Cancer Biol, May 24, 2020
(Online ahead of print).
|
|
13
|
Chen Z, Zhang D, Yue F, Zheng M, Kovacevic
Z and Richardson DR: The iron chelators Dp44mT and DFO inhibit
TGF-β-induced epithelial-mesenchymal transition via up-regulation
of N-Myc downstream-regulated gene 1 (NDRG1). J Biol Chem.
287:17016–17028. 2012.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Liu W, Xing F, Iiizumi-Gairani M, Okuda H,
Watabe M, Pai SK, Pandey PR, Hirota S, Kobayashi A, Mo YY, et al:
N-myc downstream regulated gene 1 modulates Wnt-β-catenin
signalling and pleiotropically suppresses metastasis. EMBO Mol Med.
4:93–108. 2012.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Logan J and Bourasssa MW: The rationale
for a role for diet and nutrition in the prevention and treatment
of cancer. Eur J Cancer Prev. 27:406–410. 2018.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Bezerra DP, Pessoa C, de Moraes MO,
Saker-Neto N, Silveira ER and Costa-Lutufo LV: Overview of the
therapeutic potential of piplartine (piperlongumine). Eur J Pharm
Sci. 48:453–463. 2013.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Prasad S and Tyagi A: Historical spice as
a future drug: Therapeutic potential of piperlongumine. Curr Pharm
Des. 22:4151–4159. 2016.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Tripathi SK and Biswal BK: Piperlongumine,
a potent anticancer phytotherapeutic: Perspectives on contemporary
status and future possibilities as an anticancer agent. Pharmacol
Res. 156(104772)2020.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Liu JM, Pan F, Li L, Liu QR, Chen Y, Xiong
XX, Cheng K, Yu SB, Shi Z, Yu AC and Chen XQ: Piperlongumine
selectively kills glioblastoma multiforme cells via reactive oxygen
species accumulation dependent JNK and p38 activation. Biochem
Biophy Res Commun. 437:87–93. 2013.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Roh JL, Kim EH, Park JY, Kim JW, Kwon M
and Lee BH: Piperlongumine selectively kills cancer cells and
increases cisplatin antitumor activity in head and neck cancer.
Oncotarget. 5:9227–9238. 2014.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Park JA, Na HH, Jin HO and Kim KC:
Increased expression of FosB through reactive oxygen species
accumulation functions as pro-apoptotic protein in piperlongumine
treataed MCF7 breast cancer cells. Mol Cells. 42:884–892.
2019.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Chen D, Ma Y, Li P, Liu M, Fang Y, Zhang
J, Zhang B, Hui Y and Yin Y: Piperlongumine induces apoptosis and
synergizes with doxorubicin by inhibiting the JAK2-STAT3 pathway in
triple-negative breast cancer. Molecules. 24(2338)2019.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Shrivastava S, Kulkarni P, Thummuri D,
Jeengar MK, Naidu VG, Alvala M, Redddy GB and Ramakrishna S:
Piperlongumine, an alkaloid causes inhibition of PI3 K/Akt/mTOR
signaling axis to induce caspase-dependent apoptosis in human
triple-negative breast cancer cells. Apoptosis. 19:1148–1164.
2014.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Ashfaq UA, Riaz M, Yasmeen E and Yousaf
MZ: Recent advances in nanoparticle-based targeted drug-delivery
systems against cancer an role of tumor microenvironment. Crit Rev
Ther Drug Carrier Syst. 34:317–353. 2017.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Fofaria NM, Qhattal HS, Liu X and
Srivastava SK: Nanoemulsion formulations for anti-cancer agent
piplartine-characterization, toxicological, pharmacokinetics and
efficacy studies. Int J Pharm. 498:12–22. 2016.PubMed/NCBI View Article : Google Scholar
|
|
26
|
de Lima Moreira F, Habenschus MD, Barth T,
Marques LM, Pilon AC, da Silva Bolzani V, Vessecchi R, Lopes NP and
de Oliveira AR: Metabolic profile and safety of piperlongumine. Sci
Rep. 6(33646)2016.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Majumdar D, Jung KH, Zhang H, Nannapaneni
S, Wang X, Amin AR, Chen Z, Chen ZG and Shin DM: Luteolin
nanoparticle in chemoprevention: In vitro and in vivo anticancer
activity. Cancer Prev Res (Phila). 7:65–73. 2014.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Rad JG and Hoskin DW: Delivery of
apoptosis-inducing piperine to triple-negative breast cancer cells
via co-polymeric nanoparticles. Anticancer Res. 40:689–694.
2020.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Danhier F, Ansorena E, Silva JM, Coco R,
Le Breton A and Préat V: PLGA-based nanoparticles: An overview of
biomedical applications. J Control Release. 161:505–522.
2012.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Yang Q, Liao J, Deng X, Liang J, Long C,
Xie C, Chen X, Zhang L, Sun J, Peng J, et al: Anti-tumor activity
and safety evaluation of fisetin-loaded methoxy poly(ethylene
glycol)-poly(epsilon-caprolactone) nanoparticles. J Biomed
Nanotechnol. 10:580–591. 2014.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Valastyan S and Weinberg RA: Tumor
metastasis: Molecular insights and evolving paradigms. Cell.
147:275–292. 2011.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Tester AM, Ruangpanit N, Anderson RL and
Thompson EW: MMP-9 secretion and MMP-2 activation distinguish
invasive and metastatic sublines of a mouse mammary carcinoma
system showing epithelial-mesenchymal transition traits. Clin Exp
Metastasis. 18:553–560. 2000.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Nagase H, Visse R and Murphy G: Structure
and function of matrix metalloproteinases and TIMPs. Cardiovasc
Res. 69:562–573. 2006.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Kubiatowski T, Jang T, Lachyankar MB,
Salmonsen R, Nabi RR, Quesenberry PJ, Litofsky NS, Ross AH and
Recht LD: Association of increased phosphatidylinositol 3-kinase
signaling with increased invasiveness and gelatinase activity in
malignant gliomas. J Neurosurg. 95:480–488. 2001.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Wu ZQ, Li XY, Hu CY, Ford M, Kleer CG and
Weiss SJ: Canonical Wnt signaling regulates Slug activity and links
epithelial-mesenchymal transition with epigenetic breast cancer 1,
early onset (BRCA1) repression. Proc Natl Acad Sci USA.
109:16654–16659. 2012.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Zhou BP, Deng J, Xia W, Xu J, Li YM,
Gunduz M and Hung MC: Dual regulation of Snail by
GSK-3beta-mediated phosphorylation in control of
epithelial-mesenchymal transition. Nat Cell Biol. 6:931–940.
2004.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Nagi C, Guttman M, Jaffer S, Qiao R, Keren
R, Triana A, Li M, Godbold J, Bleiweiss IJ and Hazan RB: N-cadherin
expression in breast cancer: Correlation with an aggressive
histologic variant-invasive micropapillary carcinoma. Breast Cancer
Res Treat. 94:225–235. 2005.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Chao YL, Shepard CR and Wells A: Breast
carcinoma cells re-express E-cadherin during mesenchymal to
epithelial reverting transition. Mol Cancer. 9(179)2010.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Pan Y, Li J, Zhang Y, Wang N, Liang H, Liu
Y, Zhang CY, Zen K and Gu H: Slug-upregulated miR-221 promotes
breast cancer progression through suppressing E-cadherin
expression. Sci Rep. 6(25798)2016.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Loh CY, Chai JY, Tang TF, Wong WF, Sethi
G, Shanmugam MK, Chong PP and Looi CY: The E-cadherin and
N-cadherin switch in epithelial-to-mesenchymal transition:
Signaling, therapeutic implications, and challenges. Cells.
8(118)2019.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Bailey KL, Agarwal E, Chowdhury S, Luo J,
Brattain MG, Black JD and Wang J: TGFβ/Smad3 regulates
proliferation and apoptosis through IRS-1 inhibition in colon
cancer cells. PLoS One. 12(e0176096)2017.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Gao F, Zhou L, Li M, Liu W, Yang S and Li
W: Inhibition of ERKs/Akt-mediated c-Fos expression is required for
piperlongumine-induced cyclin D1 downregulation and tumor
suppression in colorectal cancer cells. Onco Targets Ther.
13:5591–5603. 2020.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Ross KR, Corey DA, Dunn JM and Kelley TJ:
SMAD3 expression is regulated by mitogen-activated protein kinase
kinase-1 in epithelial and smooth muscle cells. Cell Signal.
19:923–931. 2007.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Guan RJ, Ford HL, Fu Y, Li Y, Shaw LM and
Pardee AB: Drg-1 as a differentiation-related, putative metastatic
suppressor gene in human colon cancer. Cancer Res. 60:749–755.
2000.PubMed/NCBI
|
|
45
|
Han LL, Hou L, Zhou MJ, Ma ZL, Lin DL, Wu
L and Ge YL: Aberrant NDRG1 methylation associated with its
decreased expression and clinicopathological significance in breast
cancer. J Biomed Sci. 20(52)2013.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Graff JR, Gabrielson E, Fujii H, Baylin SB
and Herman JG: Methylation patterns of the E-cadherin 5' CpG island
are unstable and reflect the dynamic, heterogeneous loss of
E-cadherin expression during metastatic progression. J Biol Chem.
275:2727–2732. 2000.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Graff JR, Herman JG, Lapidus RG, Chopra H,
Xu R, Jarrard DF, Isaacs WB, Pitha PM, Davidson NE and Baylin SB:
E-cadherin expression is silenced by DNA hypermethylation in human
breast and prostate carcinomas. Cancer Res. 55:5195–5199.
1995.PubMed/NCBI
|
|
48
|
Fang MZ, Wang Y, Ai N, Hou Z, Sun Y, Lu H,
Welsh W and Yang CS: Tea polyphenol (-)-epigallocatechin-3-gallate
inhibits DNA methyltransferase and reactivates methylation-silenced
genes in cancer cell lines. Cancer Res. 63:7563–7570.
2003.PubMed/NCBI
|
|
49
|
Jones PA, Issa JP and Baylin S: Targeting
the cancer epigenome for therapy. Nat Rev Genet. 17:630–641.
2016.PubMed/NCBI View Article : Google Scholar
|