Introduction
Despite advances in technology and medicine in terms
of diagnosis and therapy, cancer is now the leading cause of death
worldwide and thus remains a serious health issue and an economic
burden (1). With the exception of
non-melanoma skin cancer, prostate tumour is the most frequent
cancer in men, while breast tumour is the most frequent cancer in
women.
Tumour surgical excision, radiotherapy,
immunotherapy and chemotherapy are the gold standard approaches for
cancer treatment; however, current therapies still present critical
points, mainly in terms of safety, efficacy and efficiency. The
existing chemotherapy alternatives are often associated with
non-complete tumour eradication and/or high toxicity and
non-specificity. Additionally, several reports highlight a
remarkable and crescent induction of cellular multiresistance to
chemotherapeutic agents. Thus, there is a need to develop new
alternative anticancer drugs with fewer side effects. One important
strategy to develop effective anticancer agents relies on the study
of bioactive compounds derived from natural sources. Many of these,
as well their derivatives, have been extensively studied and are
proven to be effective for cancer prevention and therapy (2).
Alkaloids are nitrogenated pharmacologically active
compounds, which have been reported to be one of the most important
groups of phytocompounds obtained from natural sources (3). β-carbolines belongs to the group of
indolic alkaloids (4). These
alkaloids are mainly found in plants of the Zygophyllaceae,
Malpighiaceae, Fabaceae, Myristicaceae, Elaeagnaceae and
Passifloraceae families (5).
Harmine (7-methoxy-1methyl-9H-pyrido[3,4-b]indole),
a tricyclic β-carboline alkaloid, was originally isolated from
seeds of Peganum harmala L. (Zygophyllaceae) (6). However, there are also reports of
this alkaloid as an important metabolite present in Passiflora
incarnata L. and Passiflora edulis f. flavicarpa
Degener, fruits of these plants are highly consumed in tropical
countries and are popularly known as passion fruit or maracujá in
the Brazilian indigenous language (6,7). The
emphasis on the ‘maracujá’ fruit and seed is due to the extensive
consumption of passion fruit juice (fresh or processed) and on
ongoing investigations into its potential as a functional food.
Many substances present in the fruits, mainly in the pulp and seed,
can contribute to beneficial effects, such as antioxidant,
anti-hypertension, as well as decreasing blood glucose and
cholesterol. Commercial varieties of passion fruit are also rich in
alkaloids, flavonoids, carotenoids, minerals and vitamins A and C,
substances responsible for a variety of functional effects on other
foods (8,9).
Previous studies have also shown that harmine have
multiple pharmacological activities, including antiplasmodial,
antileishmanial, antiviral, antimicrobial, anti-inflammatory and
anticancer effects (6,10). In vitro studies have
demonstrated that the planar structure of harmine leads to
inhibition of topoisomerase activity (11). In addition, harmine has been shown
to exert an anti-proliferative effect, in a dose and time-dependent
manner, on several types of cancer cells, such as C33A, HL60,
MGC-803, SW620 and SW480 cell lines (10,12).
Although previous studies have reported that
β-carboline alkaloids can activate both the intrinsic and extrinsic
pathways of apoptosis in tumor cells (13), the mechanisms of action of harmine
are not fully elucidated. Therefore, the aim of this study was to
perform biological screening for the antitumour activity of
β-carboline alkaloid harmine and its interaction with DNA, using
in silico, in vitro and in vivo models.
Materials and methods
Chemicals and antibodies
Agarose, propidium iodide (PI), acridine orange,
calf thymus DNA (CT-DNA), tetrazolium salt (MTT), dimethylsulfoxide
(DMSO), bovine serum albumin (BSA), tetramethylrhodamine methyl
ester (TMRE), 5,5'-dithiobis(2-nitrobenzoic acid) (DTNB) and the
protease inhibitor cocktail were purchased from Sigma-Aldrich;
Merck KGaA. Antibiotics, Dulbecco's modified Eagle's medium (DMEM),
Dulbecco's phosphate-buffered saline (PBS) and fetal bovine serum
(FBS) were purchased from Gibco; Thermo Fisher Scientific, Inc. The
kit of PI/ribonuclease A (RNAse) were acquired from Immunostep.
Phosphatase inhibitor cocktail was purchased from Calbiochem (Merck
Biosciences). Rabbit polyclonal antibodies raised against CDK2
(cat. no. sc-163), cyclin A (cat. no. sc-596), cyclin B1 (cat. no.
sc-752), PARP1 (cat. no. sc-7150), actin (cat. no. sc-7210) and
mouse monoclonal antibodies raised against Bax (cat. no. sc-7480),
Bcl-xL (cat. no. sc-8392) and p53 (cat. no. sc-126) were from Santa
Cruz Biotechnology. Rabbit polyclonal antibody raised against
phospho-Rb (cat. no. 9308) was acquired from Cell Signalling
Technology. Polyclonal goat secondary antibodies for anti-rabbit
IgG (cat. no. AP132P) and for anti-mouse IgG (cat. no. AP181P), as
well as the chemiluminescence detection kit of HRP-coupled
antibodies were from Merck Millipore. All other chemicals used were
American Chemical Society (ACS) grade reagents.
Cell culture
Human breast carcinoma (MCF-7), cervix
adenocarcinoma (HeLa) and normal mouse fibroblast (McCoy) cells
were purchased from the Rio de Janeiro cell bank, Brazil. The cells
were maintained at constant temperature (37˚C) under a 5%
CO2 atmosphere with 95% air humidity. The culture medium
used was DMEM supplemented with 10% FBS, penicillin (100 U/ml) and
streptomycin (100 µg/ml).
Cytotoxicity assay
The cell viability was assessed using the MTT assay
(14). Cells were plated at a
density of 1x104 cells/well in 96-well plates. After
reaching 80% confluence, the cells were treated with different
concentrations of harmine (0.1 to 1,000 µM) for 48 and 72 h. The
negative control was treated only with standard culture medium
containing 0.1% DMSO. After the treatment, cells were washed with
PBS and incubated for 2 h with MTT (0.5 mg/ml). The formazan
crystals were solubilized by adding DMSO (100 µl/well), and the
colored solutions were read at 550 nm. Three independent
experiments were conducted and the results are presented as the
half maximal inhibition concentration (IC50), calculated
using Graph Pad Prism 6 (GraphPad Software Inc.). The selectivity
index (SI) of the compounds under study are expressed as previously
reported by Koch et al (15), with a minor modification: SI
(selectivity index) = IC50 of the compound in a normal
cell line/IC50 of the same compound in the cancer cell
line.
Assessment of cell death type
The type of cell death induced by harmine was
evaluated by a staining method, using PI and acridine orange
(16). This method allows the
differentiation of viable cells (green) from those that are dying
through apoptosis (orange) or necrosis (dark red). MCF-7 cells were
seeded (2x105/well) in 6-well plates. After confluence
was reached, the cells were treated with IC30 and
IC50 concentrations of harmine for 72 h. Fresh medium
was used as a negative control. After treatment, the cells were
stained with PI (100 mg/ml) and acridine orange (100 mg/ml) (5 µl;
1:1 v/v) and then visualized using an Olympus microscope, model
BX41 (Olympus Corporation). Results are expressed as the
percentages of cells that were viable, apoptotic or necrotic.
Mitochondrial membrane potential
(MMP)
Mitochondrial membrane depolarization was determined
using the fluorescent probe, TMRE, which accumulates in the
mitochondrial matrix in proportion to mitochondrial membrane
potential (ΔΨm) (17). MCF-7 cells
were seeded at a density of 1x106/well on 6-well plates.
After confluence the cells were treated with IC30 and
IC50 concentrations of harmine for 48 h. Thereafter, the
cells were incubated in the dark with TMRE (25 nM) for 20 min at
37˚C. Cell fluorescence was determined using a BD FACS Canto II
flow cytometer (BD Biosciences) and results are presented as the
percentages of cells with high TMRE fluorescence.
Cell cycle distribution
The DNA content of the cells was measured by flow
cytometry using a PI (50 µg/ml) and RNAse (0.2 mg/ml) solution kit.
MCF-7 cells were seeded (2x105/well) in 6-well plates.
After confluence was reached, the cells were synchronized using
nocodazole (30 ng/ml) for 14 h and then treated with harmine
(IC30) for 72 h. Subsequently, the cells were fixed
overnight with ethanol (70%) at -20˚C, washed with a PBS/albumin
(2%) solution, and later incubated with the PI/RNAse solution for
15 min at room temperature. The cells were evaluated using the BD
FACS Canto II flow cytometer (BD Biosciences) and categorized into
each phase of the cell cycle, according to the content of DNA.
Flowing Software 2.5.0 (Perttu Terho; Cell Imaging Core, Turku
Center for Biotechnology, University of Turku) was used to process
the data.
DNA interaction and intercalation
The potential interaction of harmine with
calf-thymus DNA (CT-DNA) was assessed through Spectrophotometric
UV-Visible Scanning Titration (18). Increasing concentrations of harmine
(0-250 µM) were used for an absorption titration assay with a
constant concentration of CT-DNA (150 µM). To obtain the spectra of
the samples, a Hitachi U-2910® spectrophotometer was
used; and UV-Visible scanning was performed from 200 to 800 nm. The
changes in CT-DNA absorbance, after incubation with harmine, as
well as the maximum absorption wavelength shift, were
determined.
Potential DNA intercalation was evaluated by
fluorescence titration measurements, using the DNA-intercalating
agent PI, according to Da Silveira et al (19). CT-DNA (150 µM) was saturated with
PI (300 µM) in a phosphate buffer (50 mM) containing 0.1 M NaCl (pH
7.4). Increasing concentrations of harmine (0-250 µM) were then
incubated with fixed concentrations of CT-DNA and PI for 10 min at
room temperature. Doxorubicin, a standard antitumor intercalating
drug, was used as a positive control. The SpectraMax
Paradigma® Multileader was used to measure the variation
of the sample fluorescence. Excitation and emission wavelengths of
492 and 620 nm were used, respectively.
The circular dichroism (CD) DNA interaction assay
was performed as described by Bertoldo et al (20). The CD spectra were measured in a
JASCO-810 spectropolarimeter (JASCO). A fixed concentration of
CT-DNA (150 µM), prepared in a phosphate buffer (50 mM) containing
0.1 M NaCl (pH 7.4), was titrated with increasing concentrations of
harmine to achieve a molar ratio of 0.04, 0.05 and 0.07
DNA/harmine. All CD spectra of DNA and DNA/harmine were recorded
over the range of 220-350 nm and the final data were expressed in
millidegrees (mdegs).
In silico study: DNA docking and
molecular dynamic
The B-DNA template structure sequence d
(CGTGAATTCACG) (PDB ID1G3X) was retrieved from Protein Data Bank
(21). The ligand INE.mol2 file
came from the ZINC database (http://zinc.docking.org/substance/18847046) and was
minimized after removal of original co-precipitated ligands and
water with UCSF Chimera 1.13(22)
using the AMBER99bsc1 force field (23). The ACPYPE tool, based on Python
(24), was used as an Antechamber
to generate INE topology for molecular simulations (25). Molecular docking simulations were
applied to AutoDock MGLTools 1.5.6rc3 to generate a DNA-INE complex
input file (.pdbqt) (26).
Geometries and binding affinity energies were then calculated
through AutoDock Vina 1.2.2 algorithm, using default parameters
(27). H-bonds and/or hydrophobic
interactions predicted between INE and DNA nucleotides were
visualized with PyMOL open source 1.8.7.0’ for Python 3.6(28) and with LigPlot+
2.1(29). However, subsequent
molecular dynamic (MD) simulations used only the ligand pose
(pdbqt) that had the lowest RMSD and binding affinity. Avogadro
(30) was used to convert this
file to .pdb format, before MD simulations with GROMACS version
2018.4 (31,32) using an AMBER99bsc1 force field
(23). The TIP3P water model was
applied in order to solvate and neutralize the system with two Na+
ions inside an octahedron box (33). The runs made for energy
minimization and solvent equilibration kept grid space under NVP
and NPT ensemble conditions (T=300 K, P=1 bar). All simulations
used the same time step (1 fs). Position restricted heavy atoms of
DNA accessible to the solvent, without disturbing the complex
structure.
Molecular Mechanics Poisson-Boltzmann
surface area (MM-PBSA) energies calculations
MM-PBSA calculations performed after MD simulations
showed some conformational fluctuations and residue energy
contributions for free binding energy (34). The MM-PBSA method calculated the
energies and trajectories related to DNA and the ligand INE
complex. This method, using explicit solvent, also enables the
calculation of non-bonded potentials (electrostatic and van der
Waals) under default parameters. Results were visualized with
Visual Molecular Dynamics (VMD) (35) for LINUXAMD64, version 1.9.4a12
(2017). Calculations of trajectories used Verlet through MD
neighbor searching and the vdW cutoff-scheme (36). Additionally, the ‘readHBmap.py’
(Python 2) program showed frames with H-bonds. Additionally, H bond
occupancy graphics showed frames (1 frame equal to 2 ps) where this
interaction contributed to the macromolecule - ligand complex
stability (37). Finally, frames
selected were visualized (Chimera and LigPlot+ 2.1) and
key intermolecular interactions between DNA-ligand were
evaluated.
DNA damage
The DNA fragmentation was evaluated by the comet
assay, according to Singh et al (38). MCF-7 cells were seeded
(2.5x104/well) in 24-well plates. After confluence, the
cells were treated with IC30 and IC50
concentrations of harmine for 72 h. After resuspending the cells in
low-melting point agarose (0.75%), they were placed on slides
pre-covered with agarose (1.5%) and incubated for 10 min at -8˚C.
The cells were then lysed for 7 days using the lysis solution (2.5
M NaCl; 10 mM Tris; 100 mM EDTA; 1% Triton X-100 and 10% DMSO; pH
10.0) and then submitted to horizontal electrophoresis (25 V and
300 mA) in alkaline buffer (300 mM NaOH; 1 mM EDTA; pH 13.0) for 20
min at 8˚C. Subsequently, the slides were neutralized and fixed
with the respective solutions; neutralizing solution (0.4 M Tris
HCl; pH 7.5) and fixing solution (15% TCA; 5% ZnSO4; 5%
glycerol). These procedures were intercalated with water washing.
After drying at room temperature, the slides were stained (0.1%
NH4NO3, 0.1% AgNO3, 0.25%
tungstosilicic acid, 0.15% formaldehyde, 5%
Na2CO3, v/v), washed with acetic acid (0.01%)
and visualized on an Olympus microscope, model BX41 (Japan). The
comets were classified according to Ross et al (39), in which the score 0 represented the
undamaged nuclei and a score of 4 represents maximally damaged
nuclei.
Western blotting
MCF-7 cells were treated with the IC30
concentration of harmine for 48 h and then washed with PBS and
lysed in RIPA buffer (50 mM Tris-HCl, pH 7.4; 150 mM NaCl; 1% NP40;
0.25% Na-deoxycholate and 1 mM phenylmethylsulfonyl fluoride),
supplemented with protease inhibitor (1%) and phosphatase inhibitor
(3%) cocktails. Laemmli buffer (60 mM Tris-HCl, 2% sodium dodecyl
sulphate (SDS), 10% glycerol, 5% β mercaptoethanol and 0.01%
bromophenol blue, pH 6.8) was used to denature the proteins.
Subsequently, 25 µg of denatured protein were submitted to sodium
dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE),
followed by electrotransfer to polyvinylidene fluoride (PVDF)
membranes (40). Following
blocking and washing, the membranes were incubated with the primary
antibodies overnight at - 8˚C (Rabbit polyclonal antibodies raised
against phospho-Rb (1:1,000, v/v), CDK2 (1:1,000, v/v), cyclin A
(1:1,000, v/v), cyclin B1 (1:1,000, v/v) and PARP1 (1:1,000, v/v);
Mouse monoclonal antibodies raised against Bax (1:200, v/v), Bcl-xL
(1:200, v/v) and p53 (1:200, v/v). Lastly, the membranes were
washed and incubated with secondary antibodies for at least 1 h.
The polyclonal goat anti-rabbit IgG antibody (1:5,000, v/v) and
polyclonal goat anti-mouse IgG antibody (1:3,000, v/v) are both
peroxidase conjugated. A chemiluminescence detection kit for
HRP-coupled antibodies was used for immunodetection. Actin
(1:1,000, v/v) was used as a loading control. Images acquired
(ChemiDoc MP System; Bio-Rad Laboratories, Inc.) were normalized
with actin.
In vivo antitumour activity
The antitumor activity of harmine was evaluated
using male Balb/c mice (20±2 g, n=12) kept under controlled
conditions (12 h light-dark cycle, 22±2˚C, 60% air humidity),
receiving water and food ad libitum. Previous tests were
done to select the maximal safe dose of harmine with the optimal
dilution and the doses of 10 mg/kg/day and 20 mg/kg/day were chosen
to continue the experiments. The induction of Ehrlich ascitic
carcinoma (EAC) was carried out according to a method previously
reported by Kviecinski et al (41). Before tumour induction, all mice
were weighed (g) and their abdominal circumferences (cm) were
measured. The mice were then inoculated intraperitoneally with EAC
cells (5x106 cells/200 µl). After 24 h, the animals were
divided into 4 groups (n=12). The negative control group was
treated with saline containing 1% DMSO. The positive control group
received doxorubicin (0.6 mg/kg/day) as previously reported
(42). Test groups received
harmine at 10 or 20 mg/kg/day, respectively. The treatments were
administered intraperitoneally for 9 consecutive days. Twenty-four
hours later, all mice were weighed and their abdominal
circumferences measured again. Using the formula reported by Felipe
et al (43), the inhibition
(%) of tumour growth was calculated as follow: [(variation in waist
circumference of the treated group x 100)/variation in waist
circumference of the control group] - 100. The variation in body
weight of the treated animals was calculated as reported by
Kviecinski et al (41):
Final weight (after 9 days of treatment) - Initial weight (day
zero, day of tumour inoculation). Then, the animals (n=12) were
kept alive and assisted on a daily basis to evaluate survival
(lifespan), according to Kaplan and Meier (44). Lastly, the treatment was repeated
and the animals were euthanised after 9 days of treatment. The
ascitic fluid was collected in graduated falcon conical tubes and
centrifuged at 5,000 x g for 5 min to measure packed tumour cells
volume (41). Viability of the
tumour cells was assessed through the trypan blue assay (45).
Statistical analyses
All in vitro assays were performed in
technical triplicates, whereas the in vivo experiments were
performed in biological replications (n=12; α=0.05; Power (1 -
β)=0.90; critical t=1.81; df (degree of freedom)=10; Effect size
|ρ|=0.67). The results were presented as means ± standard
deviations or as percentages. Data were analysed using analysis of
variance one-way ANOVA followed by the Bonferroni post-hoc test or
log-rank test, when necessary. Comparisons were performed using the
software GraphPad Prism 6 (GraphPad Software Inc.). P<0.05 was
considered to indicate a statistically significant difference.
Results
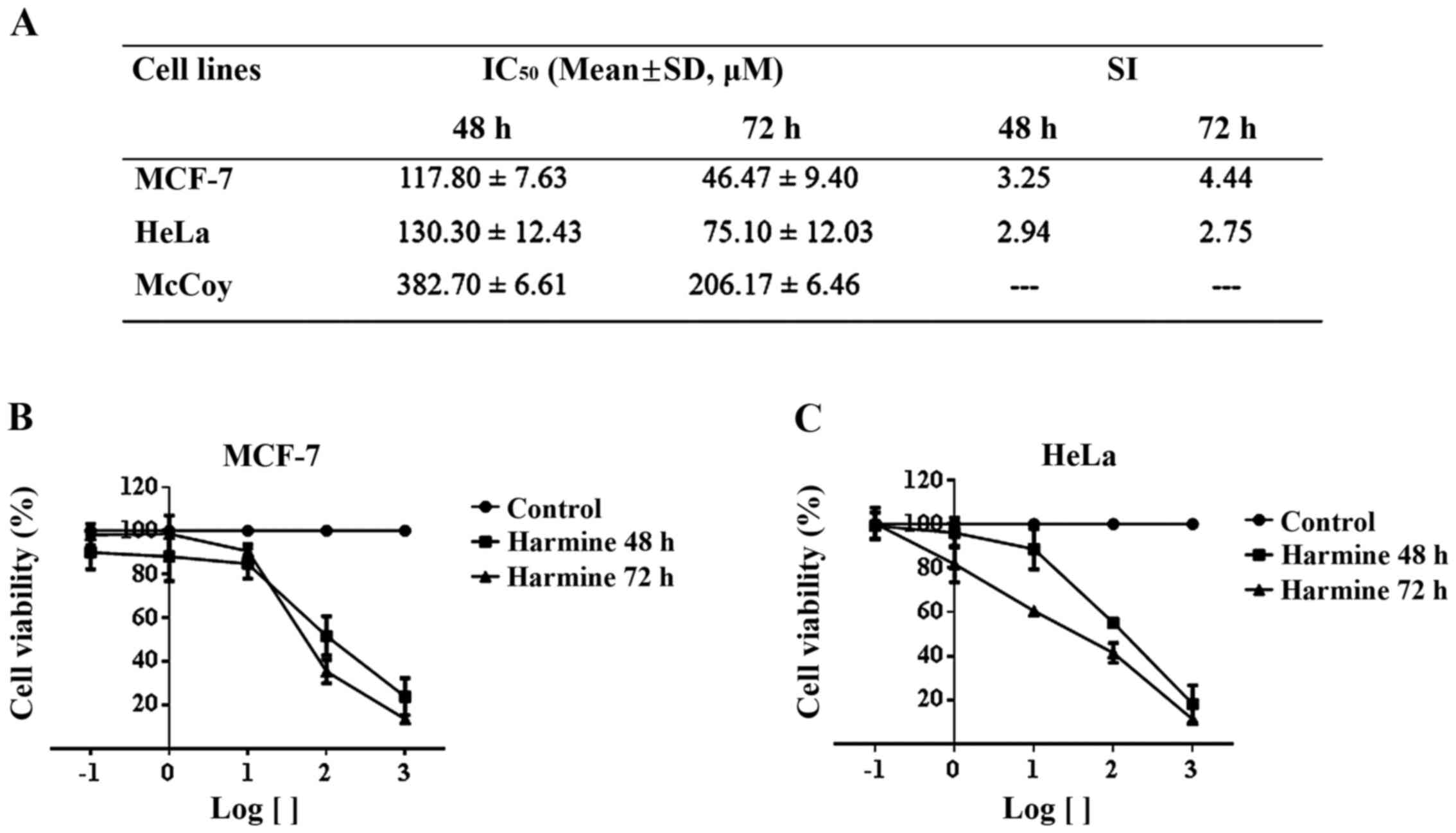
Harmine is cytotoxic with selectivity
to tumour cells
The cytotoxic effect of β-carboline alkaloid harmine
was evaluated against two tumour cell lines (MCF-7 and HeLa) and a
normal cell line (McCoy). The values obtained for the
IC50 and SI for harmine in 48 and 72 h are shown in
Fig. 1A. Harmine significantly
decreased tumour cell viability in a time and dose-dependent manner
(Fig. 1B and C). Additionally, harmine showed
selectivity for all tumour cell lines tested (Fig. 1A) and it is worth noting that this
alkaloid presented the highest selectivity for MCF-7 cells
(SI=4.44).
Harmine induces predominantly
apoptosis and modulates the expression of Bcl-2 and p53
proteins
Results indicated that harmine was able to induce
significant cell death (Fig. 2A)
and the numbers of apoptotic cells in the presence of this alkaloid
(27.88 and 46.47 µM) were significant. However, only the
concentration of 46.47 µM harmine caused an increase in necrotic
cells (8.67±3.47%), when compared to the negative control
(1.81±0.70%). Also, harmine (46.47 µM) decreased the number of
viable cells by approximately 32.9% while increasing the number of
apoptotic (82.3%) and necrotic (79.1%) cells, compared to the
negative control.
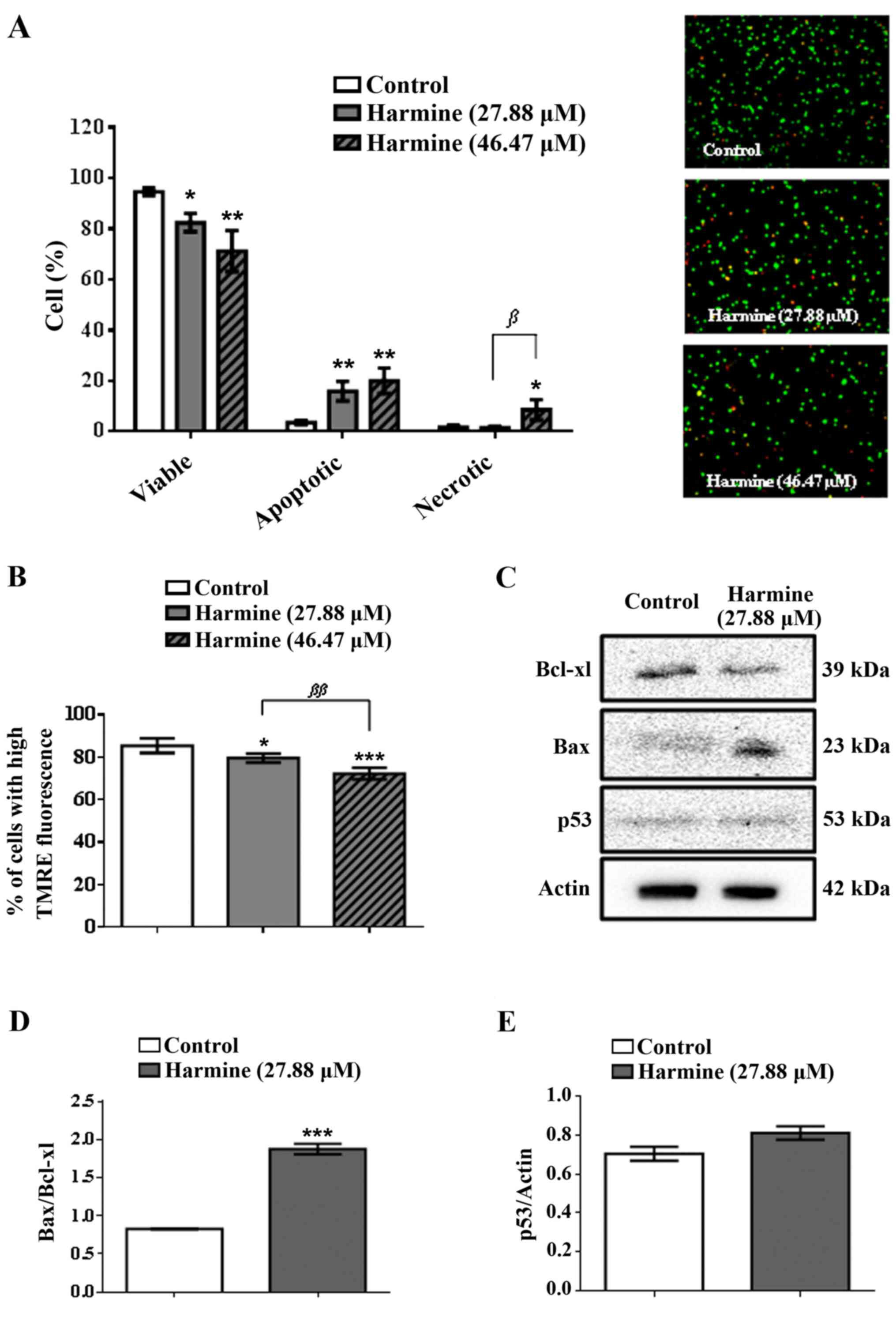 | Figure 2(A) Morphology of MCF-7 cells and the
type of cell death induced by harmine (27.88 and 46.47 µM, 72 h),
evaluated by a staining method with propidium iodide and acridine
orange and visualized by fluorescence microscopy (x400); red,
orange and green stains denote necrotic, apoptotic and viable
cells, respectively. (B) Effects of harmine (27.88 and 46.47 µM, 48
h) on the mitochondrial membrane potential (ΔΨm) of MCF-7 cells.
(C-E) Imunoblotting and quantitative data of proteins that modulate
apoptosis (Bcl-xL, Bax and p53) of MCF-7 cells treated with harmine
(27.88 µM, 48 h). Results are expressed as the means ± standard
deviation of three independent experiments. Data were analysed by
analysis of variance one-way ANOVA and Bonferroni test.
*, ** and *** denote statistical
differences compared to negative control data, when P<0.05,
P<0.01 and P<0.001, respectively. β and
ββ denote statistical differences comparing the
concentration of 27.88 µM to 46.47 µM harmine, with P<0.05 and
P<0.01. |
The modulators effects of harmine on pro- and
anti-apoptotic proteins were investigated by exposing MCF-7 cells
to harmine (27.88 µM) for 48 h. Fig.
2C and D show that harmine
increased the expression of Bax protein, while reducing the
expression of Bcl-xL protein. Accordingly, the Bax/Bcl-xL ratio was
significantly increased when MCF-7 cells were treated with harmine,
suggesting cell death through apoptosis via the mitochondrial
pathway. However, harmine induced apoptotic cell death
independently of p53, since MCF-7 cells show underexpression of
this pro-apoptotic protein (Fig.
2C and E).
Harmine induces mitochondrial membrane
depolarization
Our results showed that harmine induced a
dose-dependent depletion of ΔΨm in the MCF-7 cells (Fig. 2B). At concentrations of 27.88 and
46.47 µM harmine showed a ΔΨm of 79.65±1.85 and 72.26±2.41%,
respectively, whereas the negative control was 85.45±3.08%. Thus,
harmine at the concentration of 46.47 µM was capable of decreasing
the ΔΨm of MCF-7 cells by 15.4%, when compared to the negative
control.
Harmine induces cell cycle arrest
through inhibition of phosphorylation of pRb and decreases
expression of CDK2, cyclin A and B1
Fig. 3A and
B show the changes in the MCF-7
cell cycle after treatment with harmine. Cells from the negative
control had 65.50±0.96, 13.41±1.03 and 21.01±1.59% of cells in
G1, S and G2/M phase, respectively. The
treated cells presented 67.70±0.85, 3.53±0.58 and 26.12±0.38% of
cells in G1, S and G2/M phase, respectively.
Harmine (27.88 µM) promoted a significant decrease (73.7%) in the
number of cells in the S phase and a significant increase
(approximately 19.6%) in the number of cells in the G2/M
phase, when compared with the negative control. However, MCF-7
cells treated with harmine was not statistically different in
relation to the number of cells in G1 phase, when
compared to untreated cells, suggesting a G2/M arrest of
cells. In addition, after the treatment there was an increase of
cells in the sub-G1 area (2.67±0.11%), compared to the
negative control (0.08±0.08%), which is indicative of cell death
through apoptosis, confirming the previous result (Fig. 2A).
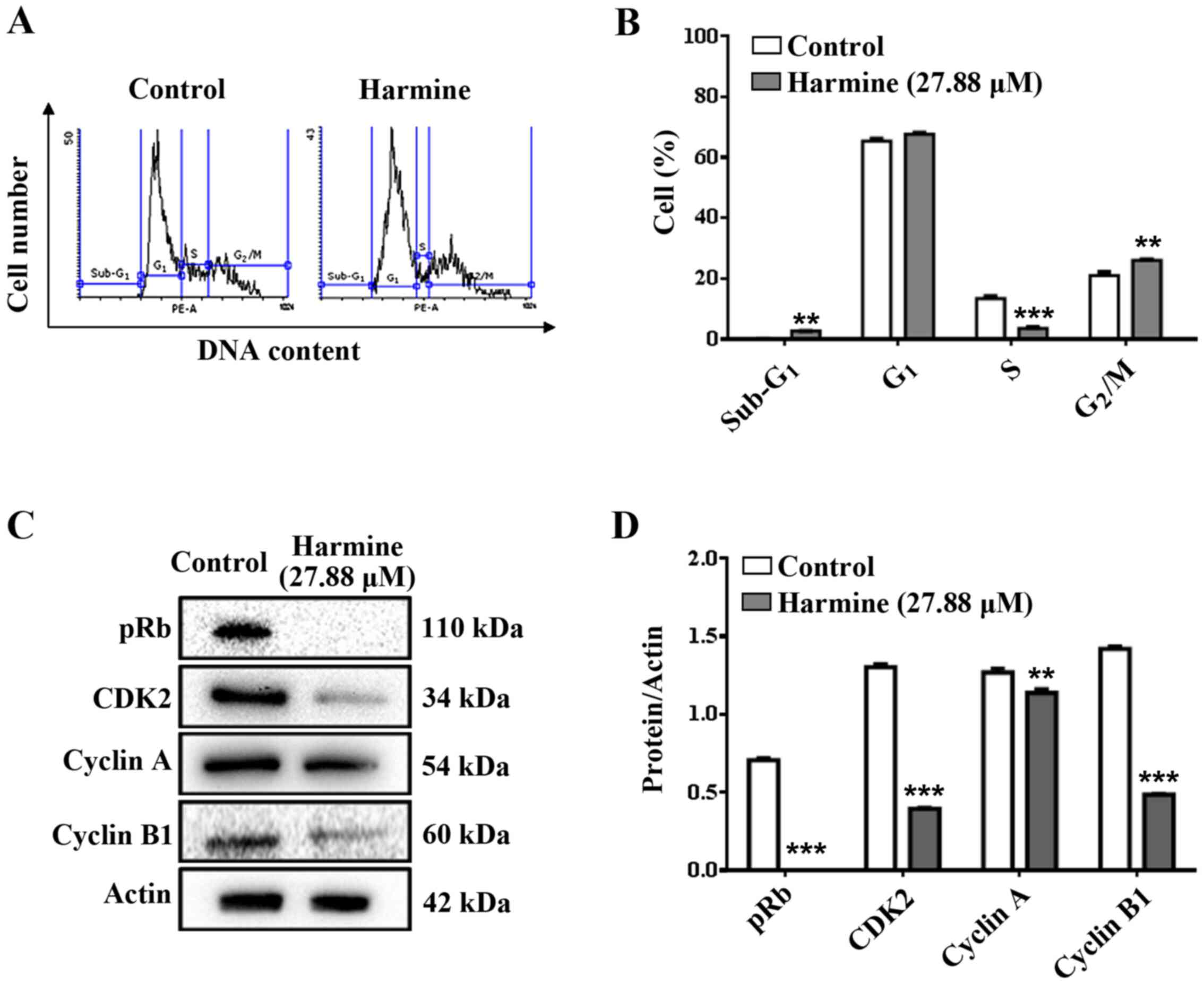 | Figure 3(A) Histogram and (B) percentages of
MCF-7 cells in each phase of the cell cycle (Sub-G1,
G1, S and G2/M). Cells were treated with
harmine (27.88 µM, 72 h) and analysed by flow cytometry. (C)
Imunoblotting and (D) Quantitative data of regulatory proteins of
the cell cycle; pRb, CDK2, cyclin A and cyclin B1, respectively, in
MCF-7 cells treated with harmine (27.88 µM, 48 h).
Sub-G1: Debris and dead cells; G1: cells not
presenting DNA duplication; S: cells presenting intermediate DNA
content; G2/M: cells presenting duplicated DNA. Results
are expressed as the means ± standard deviation of three
independent experiments. Data were analysed by analysis of variance
one-way ANOVA and Bonferroni test. ** and ***
denote statistical differences compared to the negative control,
when P<0.01 and P<0.001, respectively. |
Treatment of MCF-7 cells with harmine (27.88 µM) for
48 h totally inhibited the phosphorylation of pRb (Fig. 3C and D). Additionally, harmine caused a
significant decrease in the expression of cell cycle proteins,
CDK2, cyclin A and cyclin B1 (Fig.
3C and D), which may explain
the significant reduction of cells present in the S phase and cell
cycle arrest at the G2/M phase (Fig. 3A and B).
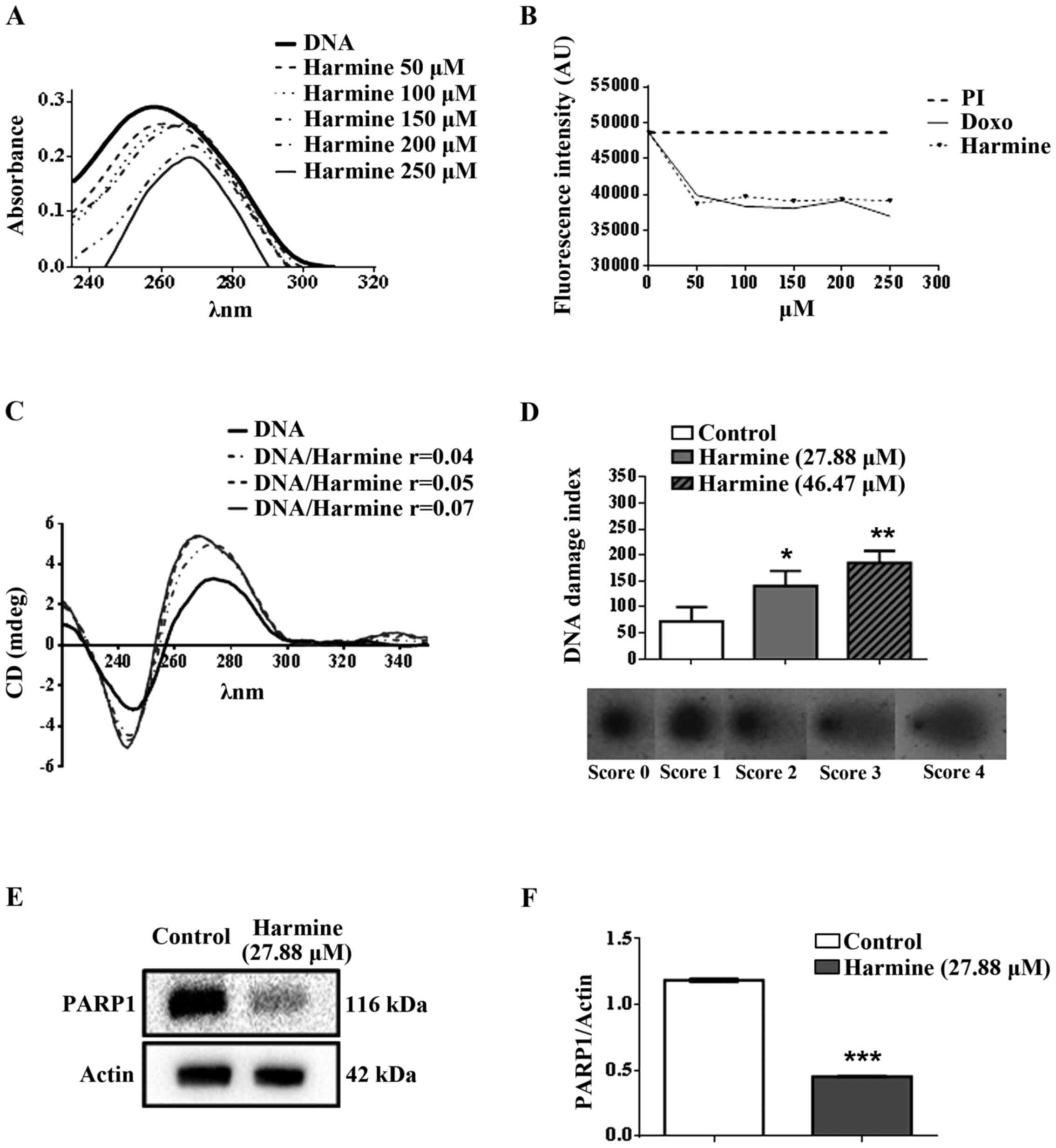
Harmine interacts with CT-DNA
Firstly, the interaction between harmine and CT-DNA
was analysed by spectrophotometric UV-visible scanning titration,
causing both hypochromic and bathochromic effects (Fig. 4A); thereby suggesting that
β-carboline alkaloid binds to DNA by intercalation.
In order to find further evidence of the
intercalative interaction of harmine with CT-DNA,
spectrophotometric titrations were carried out using the
DNA-intercalating agent, PI. When the concentration of harmine was
increased (Fig. 4B), PI
fluorescence decreased, suggesting that harmine succeed in
intercalating between the nucleobase pairs of DNA and to displace
PI bound to DNA, consequently causing a reduction in the
fluorescence intensity of the sample.
CD was also employed to gain insight into the DNA
conformational alterations that occur after harmine was bound to
DNA. A typical CD spectrum of CT-DNA in its B form shows a positive
band with a maximum at 275 nm, due to base stacking, and a negative
band with a minimum at 248 nm, due to right-handed helicity
(46). Therefore, alterations in
the B-DNA secondary structure lead to a change in the CD spectrum
(47). Our results showed that
harmine was able to induce changes in the CD spectrum by increasing
the intensity of the negative and the positive bands of CT-DNA
(Fig. 4C). Accordingly, the CD
spectrum results from the DNA-harmine complex were consistent with
those of the intercalation model.
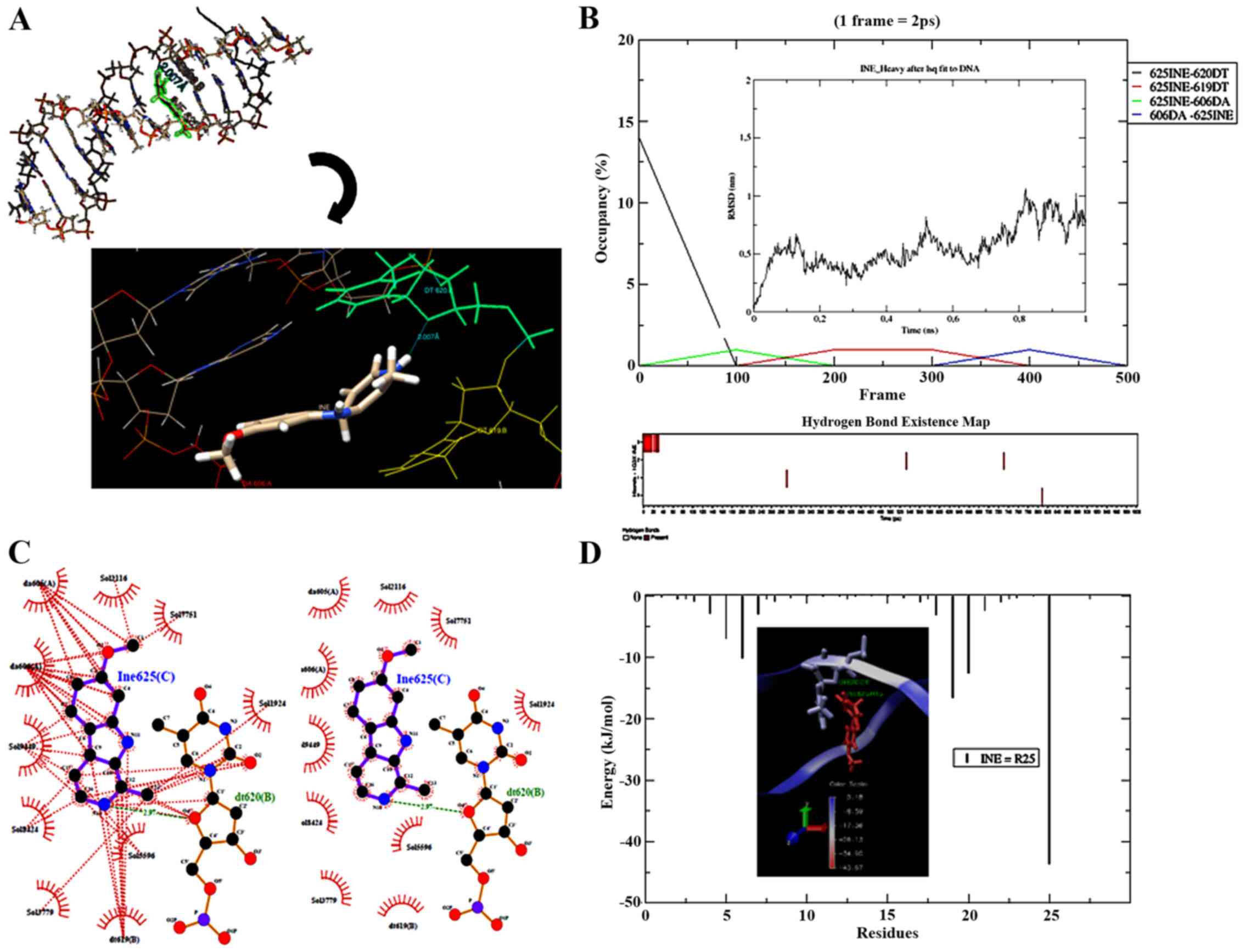
In silico prediction
Molecular docking and dynamic studies of the binding
model of harmine with DNA were used to gather further structural
details of the DNA-harmine complex. The pose that had the lowest
binding energy, predicted by AutoDock Vina (-7.6 kcal/mol; RMSD
0.00000) after submission to MD simulations (1 ns), showed harmine
to be intercalated between the hydrophobic portions of nucleotide
residues dt620 and dt619 in the strand B of DNA (Fig. 5A). The frame correspondent to 10 ps
showed one H-bond between the harmine donor atom N14, which shares
the H atom (H15) with the acceptor atom (O4') of residue dt620
(2.007 Å) (Fig. 5A), although this
H-bond presents only 2.8% occupancy. Fig. 5B (bottom) also has other H-bonds,
which act as acceptors to nucleotides dt619 (0.4%) and da606
(0.2%). The nucleotide da606, however, formed one H-bond with
harmine (0.2%) and, for this reason, acted as an H-bond donor.
Additionally, variations in the RMSD of harmine (Fig. 5B, insert) indicated an initial
stability of the complex that was briefly maintained by H-bonding.
Probably fluctuations of RMSD (~5Å) come from the DNA-harmine
hydrophobic interactions (residues da605, da606, dt619 and dt620)
initially predicted by AutoDock Vina (Fig. S1A), which had the highest
hydrophobic contribution value (11.78219) among other predicted
poses of harmine. The same hydrophobic interactions remain after MD
simulations (Fig. 5C).
Additionally, the H-bonds predicted by GROMACS showed a
predominance of pairs with distances of up to 3.5Å (Fig. S2A), confirming the H-bond distance
progression variations of close to 3.5Å (Fig. S2B). Indeed, the H-bond angle
distribution from the ideal of 150˚ to a less favourable angle
(50˚) (Fig. S2C) confirmed the
minor role of H-bonds in the maintenance of harmine inside the DNA
hydrophobic cavity.
The MM-PBSA method predicted some energies in the
full DNA-harmine complex. The non-ligated potential corresponding
to van der Waals energy was -121.93±0.81 kJ/mol, while the
electrostatic energy was 14.55±0.28 kJ/mol. The value of complex
binding energy (-106.42±0.82 kJ/mol) corresponded to the polar
solvation energy (14.54±0.28 kJ/mol) and non-polar solvation energy
related to the Solvent Accessible Surface Area (SASA) (-10.55±0.06
kJ/mol) plus molecular mechanic energy (Fig. S2D). Additionally, MM-PBSA
calculated the van der Waals energy that represented the main
contribution to the molecular energy of the complex at vacuum
(Fig. S2E). The interior of the
harmine complex had the lowest energy value, which contributed to
its total energy (Fig. 5D).
Harmine also interacted with the water molecules associated with
the solvation shell (Fig. 5C).
Finally, explicit solvent modelling (TIP3P) from GROMACS
simulations allowed MM-PBSA to calculate the DNA-harmine complex
SASA Free Energy of Solvation (ΔGsolv) (Fig. S2F). The ΔGsolv was maintained at
approximately -44 kJ/mol throughout the assay. This confirmed that
the energy required to move harmine from the aqueous solvent to the
non-polar environment (DNA) was very low, which favoured the
DNA-harmine interactions.
Harmine induces DNA damage and
decreases the expression of PARP1
After treating MCF-7 cells with 27.88 or 46.47 µM,
harmine caused DNA damage indexes of 140.61±28.66 and
184.35±23.66%, respectively, while the negative control index was
72.26±27.72% (Fig. 4D). In
addition, 27.88 and 46.47 µM harmine increased DNA damage in MCF-7
cells by 48.6 and 60.8% respectively, when compared to the
untreated cells.
Furthermore, harmine (27.88 µM, 48 h) significantly
decreased the expression of PARP1 (Fig. 4E and F), suggesting a possible
reduction of the PARP1-dependent DNA repair mechanism.
Harmine inhibits tumour growth and
increases animal lifespan
The treatment of Balb/c mice with harmine was able
to significantly inhibit tumour growth, in a dose-dependent manner,
compared to the negative control (Fig.
6A). At the concentration of 10 mg/kg/day, harmine inhibited
tumour growth by 16.77±3.88%, whereas the concentration of 20
mg/kg/day had a significantly greater effect, inhibiting tumour
growth by 31.10±4.19%. Doxorubicin (0.6 mg/kg/day) used as a
positive control was able to inhibit tumour growth by
53.20±5.02%.
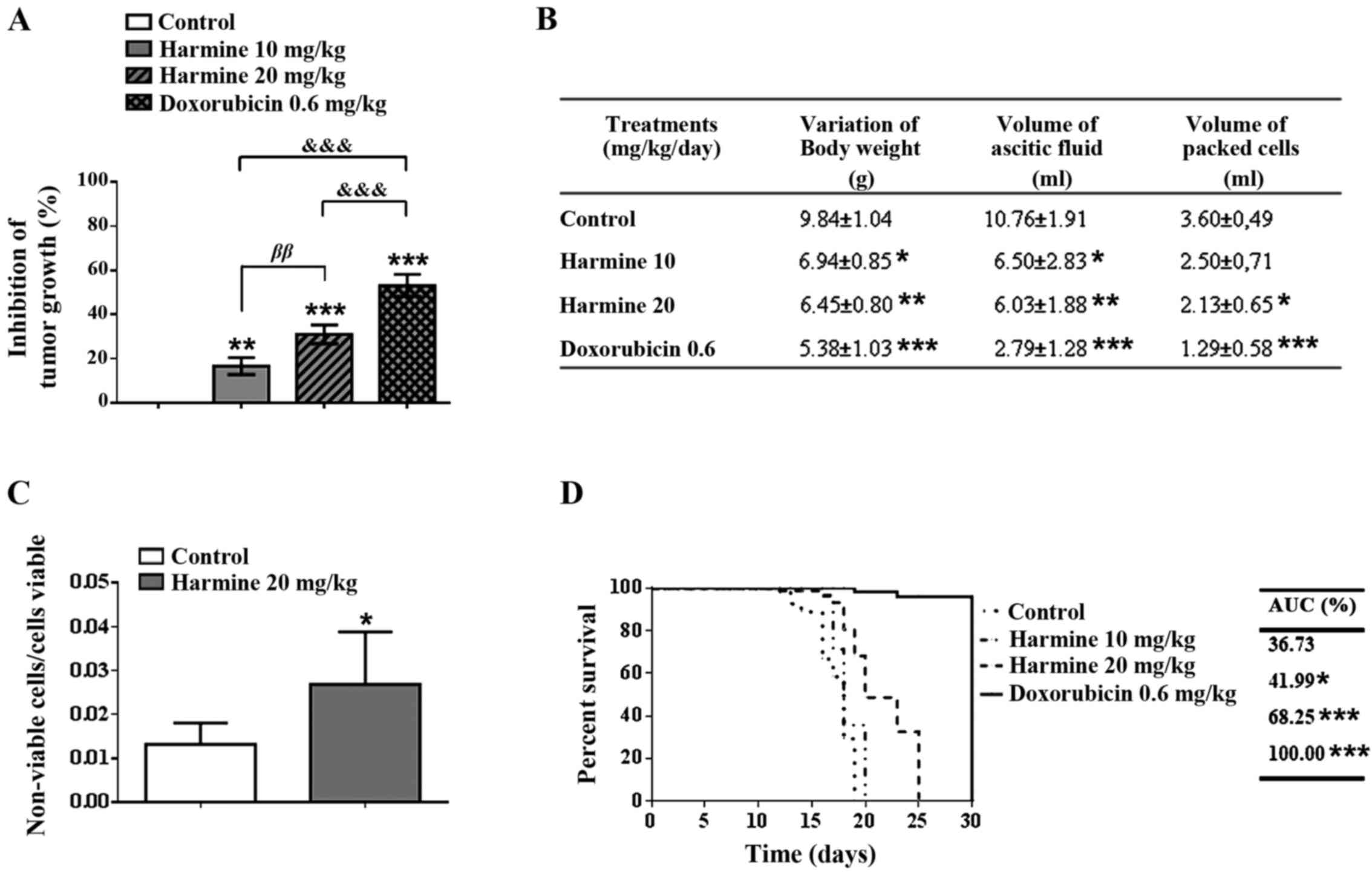 | Figure 6Antitumor effect of harmine (10 and
20 mg/kg/day, 9 days) in male Balb/c mice bearing Ehrlich ascitic
carcinoma (EAC) cells. (A) Inhibition of tumour growth, based on
the variation in abdominal circumference. (B) Body weight, volume
of ascitic fluid and volume of packed tumour cells in mice treated
with harmine. (C) Viability of EAC cells assessed by trypan blue
assay. (D) Survival rate of mice treated with harmine, according to
the Kaplan-Meier method. The results are expressed as the means ±
standard deviation of biological replication, n=12. Data were
analysed by analysis of variance one-way ANOVA followed by
Bonferroni test or log-rank test, when necessary. *,
** and *** denote statistical differences
compared to data of the negative control, when P<0.05, P<0.01
and P<0.001, respectively. ββ denote statistical
differences comparing mice that received 10 mg/kg/day to those that
received 20 mg/kg/day (P<0.01). &&&
denote statistical differences compared to the positive control
(P<0.001). |
Harmine also reduced significantly the variation of
body weight and the volume of ascitic fluid, in a dose-dependent
manner, when compared to the negative control. However, it is noted
that only harmine at concentration of 20 mg/kg/day was able to
decrease significantly the volume of packed tumour cells
(2.13±0.65), compared to the negative control (3.60±0.49) (Fig. 6B). Furthermore, the proportion of
non-viable/viable tumour cells increased when the treatment was
done with 20 mg/kg/day of harmine (Fig. 6C). Doxorubicin (0.6 mg/kg/day) also
reduced significantly the evaluated parameters.
The effect of harmine on mouse lifespan was
evaluated using the method of Kaplan and Meier (1958) (Fig. 6D). Animals receiving 10 mg/kg/day
of harmine demonstrated a small prolongation of lifespan (42.0%),
when compared to the negative control group (36.7%). In contrast,
animals receiving 20 mg/kg/day of harmine presented a significantly
longer lifespan (68.3%), when compared with the negative control.
No mortality occurred in animals treated with doxorubicin (0.6
mg/kg/day).
Discussion
The selective cytotoxic effect to MCF-7 and HeLa
cells was clearly observed in the treatment with harmine at
different treatment times. The SI demonstrates the potential of a
compound to target tumour cell lines preferentially, instead of
normal cell lines (48).
Therefore, a SI value of greater than two indicates low toxicity
for normal cells and high toxicity against tumour cell lines.
Observing the results harmine showed a higher selectivity to the
MCF-7 cells, where SI values of 3.25 and 4.44 were obtained in 48
and 72 h of treatment, respectively.
Harmine was able to induce significant cell death in
MCF-7 cells, which could be associated with an increase in the
numbers of apoptotic and necrotic cells, while decreasing the
numbers of viable cells. Furthermore, it was capable of decreasing
the mitochondrial membrane potential (ΔΨm) in a dose-dependent
manner. In this regard, it is already known that ΔΨm, which is
established by the proton pumps of the electron transport chain, is
crucial for mitochondrial function. Loss of ΔΨm can decrease ATP
generation and Ca2+ uptake, in addition to triggering
the apoptotic process for being related to the transient activation
of the mitochondrial permeability transition pore, in which diverse
pro-apoptotic proteins are released from the mitochondria into the
cytoplasm, e.g., cytochrome c and Bax (49-51).
The results obtained by the assays of the type of cell death and
mitochondrial depolarization corroborate each other. Therefore, our
findings suggest that harmine caused mitochondrial dysfunction,
accompanied by a loss of mitochondrial membrane potential and
induced cell death via apoptosis.
Bcl-2 family proteins are key regulators of
apoptosis via the mitochondrial pathway. Proteins such as Bcl-2 and
Bcl-xL prevent apoptosis, whereas proteins associated to Bcl-2,
such as Bax and Bak, promote apoptosis (52). The results showed that harmine
significantly increased the Bax/Bcl-xL ratio, suggesting cell death
through apoptosis via the mitochondrial pathway. Such findings
confirmed previous results regarding the type of cell death and
mitochondrial depolarization. However, it should be noted that
harmine induced apoptotic cell death independently of p53. Luo and
collaborators (53) observed
similar results in a human colon cancer cell line treated with the
anti-cancer compound
methoxy-1-styryl-9Hpyrido-[3,4-b]-indole (JKA97), a
synthetic harmine analogue. Their results demonstrated that
JKA97-inducing cell death occurred via the Bax-dependent and
p53-independent pathway.
By assessing the effect of harmine on each phase of
the cell cycle in MCF-7 cells, it was possible to verify that
harmine had the ability to promote a significant decrease in the
number of cells in the S phase, while significantly increasing the
number of cells in the G2/M phase, thereby suggesting a
G2/M arrest of cells. In fact, cancer cells exhibiting a
faulty G1 checkpoint due to loss of p53 or pRb may show
a greater sensitivity to G2/M checkpoint inhibitors.
This allows cells with damaged DNA to enter through aberrant
mitosis and undergo apoptosis. Additionally, the ability of harmine
to cause cell cycle arrest at G2/M could be indicative
of its inhibitory effect on proteins involved in cell cycle
regulation.
Each stage of the cell cycle is tightly regulated by
cyclin-dependent kinases (CDKs) and their regulatory partners
cyclins, which are expressed in a periodic manner. Activated CDKs
phosphorylate several key substrates that promote cell cycle
progression (54). Upon mitogenic
stimuli, the cells receive signals to enter the G1 phase and
initiate the cell cycle progression. Subsequently, CDK4/CDK6/cyclin
D and cyclin E/CDK2 complexes initiate a phosphorylation cascade of
retinoblastoma protein (RB1, also known as p105-RB) with consequent
dissociation of the entire repressive complex (RB1-E2F). This
inactivates the function of protein RB1 (pRb) as a transcriptional
repressor and enables E2F transcription factors (E2Fs) to increase
the transcription of genes involved in cell cycle progression,
notably CDK2, cyclin A and cyclin B proteins. Finally, cyclin
A/CDK2, cyclin A/CDK1 and cyclin B/CDK1 complexes become
responsible for keeping pRb in a hyperphosphorylated state until
the end of cell cycle (55).
Consequently, inactivation of pRb and permanent activation of E2F
can cause unrestrained proliferation. Harmine was capable of
totally inhibiting pRb phosphorylation, which suggests that pRb was
fully activated and repressing the E2F transcription factors,
thereby leading to decreased expression of various regulatory
proteins of the cell cycle, in such a way demonstrating the pRb
anti-proliferative function. Furthermore, harmine caused a
significant decrease in the cell cycle proteins, CDK2, cyclin A and
cyclin B1, confirming the results regarding the significant
reduction of cells present in the S phase and cell cycle arrest at
the G2/M phase. It is well known that the cyclin E/CDK2
complex regulates G1/S phase transition, whereas the
cyclin A/CDK2 complex is required during the S phase. The cyclin
A/CDK1 complex regulates G2/M phase transition, while
the cyclin B/CDK1 complex regulates the M phase progression
(54).
It is important to point out here that harmine
caused an intense inhibition of the S phase (73.7%) in the MCF-7
cells. This S phase inhibitory effect could arise from the
synergistic effect of pRb anti-proliferative function, which
decreased CDK2 expression and is also due to the fact that harmine
is an ATP-competitive CDK inhibitor (56).
Hilgendorf and collaborators (57) reported that pRb can promote DNA
damage-induced apoptosis in the highly proliferative tumour cells
by transcriptionally co-activating pro-apoptotic genes, such as
caspase 7 and p73. Moreover, pRb can also directly bind to and
activate Bax protein. These findings indicate that both the
transcriptional as well as the mitochondrial pro-apoptotic
functions of pRb occurred independently of p53, thereby
corroborating our results.
Depending on how severe the DNA damage is, the basic
molecular mechanisms of the response against DNA damage can switch
between cell-cycle arrest (giving the appropriate cell time to
attempt the repair of the DNA lesion) to induction of death
programs, such as apoptosis or necrosis (58). As DNA damage can induce cell-cycle
arrest and/or cell death, DNA is an important molecular target of
many antitumor agents (59).
To achieve a greater understanding regarding the
binding of harmine to DNA, we investigated the DNA-harmine
interaction. Firstly, the results of UV-Visible spectroscopy
suggested that harmine binds to CT-DNA by intercalation, since
harmine caused a hypochromic and bathochromic effect. According to
Villanueva and collaborators (47), hypochromic and bathochromic effects
are evidence of stacking interactions between conjugated aromatic
systems that intercalate within the DNA nucleobases. The chemical
structure of harmine is characterized by a pyridine ring fused to
an indolyl ring, besides a methyl group and a methoxy group at
positions 1 and 7, respectively (60). As such, the molecular geometry of
harmine (tricyclic planar) facilitates its binding to DNA by
intercalation.
The ability of harmine to intercalate between DNA
nucleobases was further confirmed by fluorescence spectroscopy and
CD spectroscopy. The results from molecular simulation indicated
that van der Waals and hydrophobic interactions, plus the water
solvation-harmine interactions, facilitated its binding into the
adenine-thymine specific DNA structure by intercalation. Molecular
docking confirmed the hypochromic and bathochromic effects of
harmine, as well as displacement of intercalated PI and CD spectrum
changes, which proved experimentally that harmine indeed
intercalated between subsequent nucleobase pairs of DNA. Finally,
harmine caused significant DNA damage in MCF-7 cells in a
dose-dependent manner, which confirmed the results obtained from
spectrometry and molecular simulation.
Structural DNA damage triggers a variety of DNA
repair mechanisms, collectively termed DNA damage response (DDR),
and one of the pillars of DDR is the activity of poly (adenosine
diphosphate ribose) polymerase (PARP) (61). PARP1, one of the best-studied
members of this family of enzymes, is overexpressed in a variety of
cancers. Its expression has been linked to the poor prognosis of
cancers, most notably breast cancer (62). Our results showed that harmine
significantly decreased the expression of PARP1, indicating that it
has the ability to decrease the PARP1-dependent DNA repair
mechanism, in addition to inducing DNA damage itself.
According to Cseh and collaborators (61), PARP1 has another function beyond
DNA repair; i.e., PARP-1 controls the integrity and function of
mitochondria, a critical source of PARP-1 co-enzyme
NAD+. Among other important nuclear-encoded
mitochondrial genes, PARP-1 contributes to the trans-activation of
genes that encode critical components of the mitochondrial electron
transport chain, such as cytochrome c and complex I subunit NADH
dehydrogenase 2 (ND2). Thus, it is possible that the loss of ΔΨm
observed in MCF-7 cells treated with harmine may occur, at least in
part, due to the decrease in PARP1 expression, also contributing to
the cell death by apoptosis. However, more studies are necessary to
further elucidate this hypothesis.
Finally, after the evaluation of the in
silico and in vitro cytotoxic effects of harmine, we
determined its antitumour activity in vivo, using Balb/c
mice bearing Ehrlich ascitic carcinoma (EAC). Accordingly, harmine
was able to significantly inhibit the tumour growth besides
decreasing the variation of body weight, volume of ascitic fluid
and volume of packed tumour cells. Moreover, harmine increased the
proportion of non-viable/viable tumour cells, which consequently
increased the lifespan of treated animals, thus demonstrating its
anti-tumour effects in vivo. It should be also noted that,
based on our search of the related literature, this is the first
report of the effects of the β-carboline alkaloid harmine on the
EAC model.
In conclusion, the current study identified that the
β-carboline alkaloid harmine has a cytotoxic selective effect to
tumour cell lines. In addition, our findings showed that harmine
intercalated between nucleobase pairs of DNA rich in
adenine-thymine, induced a dose-dependent DNA damage and decreasing
the PARP1-dependent DNA repair mechanism, which caused cell cycle
arrest at the G2/M phase via inhibition of
phosphorylation of pRb and reduction of the expression of CDK2,
cyclin A and cyclin B1. Harmine also induced mitochondrial-related
cellular apoptosis by modulating the expression of Bcl-2 family
proteins and decreasing the mitochondrial membrane potential.
Moreover, harmine presented in vivo antitumour effects on
Balb/c mice bearing Ehrlich ascitic carcinoma. Taken together, the
results suggest that harmine, present in the pulp and seed of the
passion fruit, could be used in the future as a promising
coadjutant anticancer agent. In addition, we suggest to address
this fruit as a functional food.
Supplementary Material
Figure S1. (A) AutoDock Vina and
GROMACS (10 ps) predicted the interactions between harmine and the
DNA molecule (PDB 1G3X). (B) AutoDock Vina predicted Pose 1 (pose
with the lowest binding energy) of harmine which was docked inside
of the DNA molecule. There are no H.bonds only hydrophobic
interactions between harmine and DNA nucleotides. (C) Hydrophobic
interactions and H.bond formed between the original ligand
9.acridine.peptide (acridine.tetra arginine; 9ac) and DNA
nucleotides. RMSD, root.mean.square distance between the docking
pose and the binding configuration in the crystallographic model;
Ine, harmine.
Figure S2. MD simulations with GROMACS
predicted the intercalation of harmine inside the hydrophobic
cavity of DNA: (A) Number of H‑bonds formed between ligand and
nucleotides. (B) Harmine and dt620 progression of H‑bond distances.
(C) Progression of H‑bond angles (average angle 49.4175˚). (D)
Energies of complex (ΔE) calculated using MM‑PBSA method. (E) van
der Waals Energy (blue line) represented the major contribution to
the total Molecular Mechanics Energy calculated in vacuum (red
line). (F) Complex Solvation Free Energy (ΔGsolv) related to SASA
calculated with MM‑PBSA. INE, harmine.
Acknowledgements
Not applicable.
Funding
The authors are grateful to CEBIME-UFSC, LAMEB-UFSC
and for the financial support provided by the Brazilian
governmental agencies Conselho Nacional de Pesquisa (CNPq) and
Coordenação de Aperfeiçoamento de Pessoal de Nível Superior
(CAPES). RC Pedrosa (Proc. 302404/2011-2) and MR Kviecinski (Proc.
420084/2018-5) are recipients of research grants from CNPq. Nádia
S.R.S. Mota, Valdelúcia M.A.S. Grinevicius, Tâmila Siminski,
Gabriela M. Almeida, Rodrigo C. Zeferino are fellows of
CAPES/CNPq-Brazil.
Availability of data and materials
Not applicable.
Authors' contributions
NSRSM, TS and GMA performed the in vitro
assays. CTP performed the circular dichroism DNA interaction assay.
The other DNA interaction assays were performed by NSRSM. VMASG
performed the in silico simulation (AutoDock Vina and
GROMACS). NSRSM and KBF performed the western blotting. NSRSM and
RCZ performed the in vivo assays. RCP designed the research
and revised the manuscript. MRK and DWF also revised the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
All animal procedures were carried out according to
internationally accepted principles for laboratory animal use and
care (NIH publication no. 85-23, revised in 1985). The experimental
protocol received approval from the ethics committee of the
Universidade Federal de Santa Catarina, Brazil (CEUA-PP00784).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
World Health Organization (WHO): Cancer.
WHO, Geneva, 2018. https://www.who.int/cancer/en/.
Accessed September 12, 2018.
|
|
2
|
Ali R, Mirza Z, Ashraf GMD, Kamal MA,
Ansari SA, Damanhouri GA, Abuzenadah AM, Chaudhary AG and Sheikh
IA: New anticancer agents: Recent developments in tumor therapy.
Anticancer Res. 32:2999–3005. 2012.PubMed/NCBI
|
|
3
|
Patel K, Gadewar M, Tripathi R, Prasad SK
and Patel DK: A review on medicinal importance, pharmacological
activity and bioanalytical aspects of beta-carboline alkaloid
‘Harmine’. Asian Pac J Trop Biomed. 2:660–664. 2012.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Piechowska P, Zawirska-Wojtasiak R and
Mildner-Szkudlarz S: Bioactive β-carbolines in food: a review.
Nutrients. 11:1–10. 2019.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Tsuchiya H, Shimizu H and Iinuma M:
Beta-carboline alkaloids in crude drugs. Chem Pharm Bull (Tokyo).
47:440–443. 1999.
|
|
6
|
Li S, Teng L, Liu W, Cheng X, Jianga B,
Wang Z and Wang CH: Pharmacokinetic study of harmane and its 10
metabolites in rat after intravenous and oral administration by
UPLC-ESI-MS/MS. Pharm Biol. 54:1768–1781. 2016.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Pierson JT, Dietzgen RG, Shaw PN,
Roberts-Thomson SJ, Monteith GR and Gidley MJ: Major Australian
tropical fruits biodiversity: Bioactive compounds and their
bioactivities. Mol Nutr Food Res. 56:357–387. 2012.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Lam SK and Ng TB: Passiflin, a novel
dimeric antifungal protein from seeds of the passion fruit.
Phytomedicine. 16:172–180. 2009.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Ingale AG and Hivrale AU: Pharmacological
studies of Passiflora sp. and their bioactive compounds. Afr
J Plant Sci. 4:417–426. 2010.
|
|
10
|
Zhang P, Huang CR, Wang W, Zhang XK, Chen
JJ, Wang JJ, Lin C and Jiang JW: Harmine hydrochloride triggers G2
phase arrest and apoptosis in MGC-803 cells and SMMC-7721 cells by
upregulating p21, activating caspase-8/Bid, and downregulating
ERK/Bad pathway. Phytother Res. 30:31–40. 2016.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Sobhani AM, Ebrahimi SA and Mahmoudian M:
An in vitro evaluation of human DNA topoisomerase I inhibition by
Peganum harmala L. seeds extract and its β-carboline
alkaloids. J Pharm Pharm Sci. 5:19–23. 2002.PubMed/NCBI
|
|
12
|
Liu J, Li Q, Liu Z, Lin L, Zhang X, Cao M
and Jiang J: Harmine induces cell cycle arrest and mitochondrial
pathway-mediated cellular apoptosis in SW620 cells via inhibition
of the Akt and ERK signaling pathways. Oncol Rep. 35:3363–3370.
2016.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Hamsa TP and Kuttan G: Harmine activates
intrinsic and extrinsic pathways of apoptosis in B16F-10 melanoma.
Chin Med. 6(11)2011.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Mosmann T: Rapid colorimetric assay for
cellular growth and survival: Application to proliferation and
cytotoxicity assays. J Immunol Methods. 65:55–63. 1983.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Koch A, Tamez P, Pezzuto J and Soejarto D:
Evaluation of plants used for antimalarial treatment by the Maasai
of Kenya. J Ethnopharmacol. 101:95–99. 2005.PubMed/NCBI View Article : Google Scholar
|
|
16
|
McGahon AJ, Martin SJ, Bissonnette RP,
Mahboubi A, Shi Y, Mogil RJ, Nishioka WK and Green DR: The end of
the (cell) line: Methods for the study of apoptosis in vitro.
Methods Cell Biol. 46:153–185. 1995.PubMed/NCBI View Article : Google Scholar
|
|
17
|
O'Reilly CM, Fogarty KE, Drummond RM, Tuft
RA and Walsh JV Jr: Quantitative analysis of spontaneous
mitochondrial depolarizations. Biophys J. 85:3350–3357.
2003.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Navarro M, Cisneros-Fajardo EJ,
Fernandez-Mestre M, Arrieche D and Marchan E: Synthesis,
characterization, DNA binding study and biological activity against
Leishmania mexicana of [Cu(dppz)2]BF4. J Inorg Biochem. 97:364–369.
2003.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Da Silveira VC, Benezra H, Luz JS, Georg
RC, Oliveira CC and Ferreira AMDC: Binding of oxindole-Schiff base
copper (II) complexes to DNA and its modulation by the ligand. J
Inorg Biochem. 105:1692–1703. 2011.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Bertoldo JB, Razzera G, Vernal J, Brod
FCA, Arisi ACM and Terenzi H: Structural stability of
Staphylococcus xylosus lipase is modulated by Zn(2+) ions. Biochim
Biophys Acta. 1814:1120–1126. 2011.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Malinina L, Soler-López M, Aymamí J and
Subirana JÁ: Intercalation of an acridine-peptide drug in an AA/TT
base step in the crystal structure of [d(CGCGAATTCGCG)](2) with six
duplexes and seven Mg(2+) ions in the asymmetric unit.
Biochemistry. 41:9341–9348. 2002.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Pettersen EF, Goddard TD, Huang CC, Couch
GS, Greenblatt DM, Meng EC and Ferrin TE and Ferrin TE: UCSF
Chimera - a visualization system for exploratory research and
analysis. J Comput Chem. 25:1605–1612. 2004.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Hornak V, Abel R, Okur A, Strockbine B,
Roitberg A and Simmerling C: Comparison of multiple AMBER force
fields and development of improved protein backbone parameters.
Proteins. 65:712–725. 2006.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Sousa da Silva AW and Vranken WF: ACPYPE -
AnteChamber PYthon Parser interfacE. BMC Res Notes.
5(367)2012.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Wang J, Wang W, Kollman PA and Case DA:
Automatic atom type and bond type perception in molecular
mechanical calculations. J Mol Graph Model. 25:247–260.
2006.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Forli S, Huey R, Pique ME, Sanner MF,
Goodsell DS and Olson AJ and Olson AJ: Computational protein-ligand
docking and virtual drug screening with the AutoDock suite. Nat
Protoc. 11:905–919. 2016.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Trott O and Olson AJ: AutoDock Vina:
Improving the speed and accuracy of docking with a new scoring
function, efficient optimization, and multithreading. J Comput
Chem. 31:455–461. 2010.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Schrödinger LCC: The PyMOL Molecular
Graphics System, Version 1.8. Schrödinger. LCC, New York, NY,
2015.
|
|
29
|
Laskowski RA and Swindells MB: LigPlot+:
Multiple ligand-protein interaction diagrams for drug discovery. J
Chem Inf Model. 51:2778–2786. 2011.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Hanwell MD, Curtis DE, Lonie DC,
Vandermeersch T, Zurek E and Hutchison GR: Avogadro: An advanced
semantic chemical editor, visualization, and analysis platform. J
Cheminform. 4(17)2012.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Van Der Spoel D, Lindahl E, Hess B,
Groenhof G, Mark AE and Berendsen HJC: GROMACS: Fast, Flexible, and
Free. J Comput Chem. 26:1701–1718. 2005.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Abraham M, Hess B, Van Der Spoel D,
Lindahl E and the GROMACS development team: GROMACS User Manual
version 4, 2018. uriwww.gromacs.orghttps://www.gromacs.org.
|
|
33
|
Jorgensen WL, Chandrasekhar J, Madura JD,
Impey RW and Klein ML: Comparison of simple potential functions for
simulating liquid water. J Chem Phys. 79:926–935. 1983.
|
|
34
|
Kumari R, Kumar R and Lynn A: Open Source
Drug Discovery Consortium: g_mmpbsa - a GROMACS tool for
high-throughput MM-PBSA calculations. J Chem Inf Model.
54:1951–1962. 2014.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Hsin J, Arkhipov A, Yin Y, Stone JE and
Schulten K: Using VMD: an introductory tutorial. Curr Protoc
Bioinformatics: CHAPTER: Unit–5.7, 2008.
|
|
36
|
Verlet L: Computer ‘Experiments’ on
Classical Fluids. I. Thermodynamical Properties of Lennard-Jones
Molecules. Phys Rev. 159:98–103. 1967.
|
|
37
|
Lemkul JA, Allen WJ and Bevan DR:
Practical considerations for building GROMOS-compatible
small-molecule topologies. J Chem Inf Model. 50:2221–2235.
2010.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Singh NP, McCoy MT, Tice RR and Schneider
EL: A simple technique for quantitation of low levels of DNA damage
in individual cells. Exp Cell Res. 175:184–191. 1988.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Ross GM, McMillan TJ, Wilcox P and Collins
AR: The single cell microgel electrophoresis assay (comet assay):
Technical aspects and applications. Report on the 5th LH Gray Trust
Workshop, Institute of Cancer Research, 1994. Mutat Res. 337:57–60.
1995.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Laemmli UK: Cleavage of structural
proteins during the assembly of the head of bacteriophage T4.
Nature. 227:680–685. 1970.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Kviecinski MR, Benelli P, Felipe KB,
Correia JFG, Pich CT, Ferreira SRS and Pedrosa RC: SFE from
Bidens pilosa Linné to obtain extracts rich in cytotoxic
polyacetilenes with antitumor activity. J Supercrit Fluids.
56:243–248. 2011.
|
|
42
|
Hossain MA, Kim D, Jang JY, Kang YJ, Yoon
JH, Moon JK, Chung HY, Kim GY, Choi YH, Copple BL and Kim ND:
Aspirin enhances doxorubicin-induced apoptosis and reduces tumor
growth in human hepatocellular carcinoma cells in vitro and
in vivo. Int J Oncol. 40:1636–1642. 2012.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Felipe KB, Kviecinski MR, Ourique F,
Bücker NF, Farias MS, Castro LSEPW, Grinevicius VMAS, Motta NS,
Correia JFG, Rossi MH and Pedrosa RC: Inhibition of tumor
proliferation associated with cell cycle arrest caused by extract
and fraction from Casearia sylvestris (Salicaceae). J
Ethnopharmacol. 155:1492–1499. 2014.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Kaplan EL and Meier P: Nonparametric
estimation from incomplete observations. J Am Stat Assoc.
53:457–481. 1958.
|
|
45
|
Strober W: Trypan blue exclusion test of
cell viability. Curr Protoc Immunol 3 (Appendix): 3B, 2001.
|
|
46
|
Fox K: Drug-DNA interaction protocols. 2nd
edition. Humana Press, Southampton, 1977.
|
|
47
|
Villanueva PJ, Martinez A, Baca ST,
DeJesus RE, Larragoity M, Contreras L, Gutierrez DA, Varela-Ramirez
A and Aguilera RJ: Pyronaridine exerts potent cytotoxicity on human
breast and hematological cancer cells through induction of
apoptosis. PLoS One. 13(e0206467)2018.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Badisa RB, Darling-Reed SF, Joseph P,
Cooperwood JS, Latinwo LM and Goodman CB: Selective cytotoxic
activities of two novel synthetic drugs on human breast carcinoma
MCF-7 cells. Anticancer Res. 29:2993–2996. 2009.PubMed/NCBI
|
|
49
|
Duchen MR: Contributions of mitochondria
to animal physiology: From homeostatic sensor to calcium signalling
and cell death. J Physiol. 516:1–17. 1999.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Tan ML, Ooi JP, Ismail N, Moad AI and
Muhammad TS: Programmed cell death pathways and current antitumor
targets. Pharm Res. 26:1547–1560. 2009.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Nakagawa Y, Suzuki T, Ishii H, Ogata A and
Nakae D: Mitochondrial dysfunction and biotransformation of
β-carboline alkaloids, harmine and harmaline, on isolated rat
hepatocytes. Chem Biol Interact. 188:393–403. 2010.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Brown R: The bcl-2 family of proteins. Br
Med Bull. 53:466–477. 1997.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Luo W, Liu J, Li J, Zhang D, Liu M, Addo
JK, Patil S, Zhang L, Yu J, Buolamwini JK, et al: Anti-cancer
effects of JKA97 are associated with its induction of cell
apoptosis via a Bax-dependent and p53-independent pathway. J Biol
Chem. 283:8624–8633. 2008.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Bai J, Li Y and Zhang G: Cell cycle
regulation and anticancer drug discovery. Cancer Biol Med.
14:348–362. 2017.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Johnson J, Thijssen B, McDermott U,
Garnett M, Wessels LFA and Bernards R: Targeting the RB-E2F pathway
in breast cancer. Oncogene. 35:4829–4835. 2016.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Song Y, Kesuma D, Wang J, Deng Y, Duan J,
Wang JH and Qi RZ: Specific inhibition of cyclin-dependent kinases
and cell proliferation by harmine. Biochem Biophys Res Commun.
317:128–132. 2004.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Hilgendorf KI, Leshchiner ES, Nedelcu S,
Maynard MA, Calo E, Ianari A, Walensky LD and Lees JA: The
retinoblastoma protein induces apoptosis directly at the
mitochondria. Genes Dev. 27:1003–1015. 2013.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Surova O and Zhivotovsky B: Various modes
of cell death induced by DNA damage. Oncogene. 32:3789–3797.
2013.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Hurley LH: DNA and its associated
processes as targets for cancer therapy. Nat Rev Cancer. 2:188–200.
2002.PubMed/NCBI View
Article : Google Scholar
|
|
60
|
Pagano B, Caterino M, Filosa R and
Giancola C: Binding of Harmine Derivatives to DNA: A Spectroscopic
Investigation. Molecules. 22(1831)2017.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Cseh AM, Fábián Z, Sümegi B and Scorrano
L: Poly(adenosine diphosphate-ribose) polymerase as therapeutic
target: Lessons learned from its inhibitors. Oncotarget.
8:50221–50239. 2017.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Rojo F, García-Parra J, Zazo S, Tusquets
I, Ferrer-Lozano J, Menendez S, Eroles P, Chamizo C, Servitja S,
Ramírez-Merino N, et al: Nuclear PARP-1 protein overexpression is
associated with poor overall survival in early breast cancer. Ann
Oncol. 23:1156–1164. 2012.PubMed/NCBI View Article : Google Scholar
|