Introduction
Lung adenocarcinoma accounts for about half of all
non-small cell lung cancer (NSCLC) cases and is one of the major
causes of death in developed countries (1). Epidermal growth factor receptor
(EGFR) tyrosine kinase inhibitors (TKIs) have been intensively
assessed over the past several years as targeted agents for
advanced NSCLC. Whereas EGFR-TKIs are highly effective in the
treatment of adenocarcinoma associated with specific EGFR mutations
that cause sustained receptor activity, drug effectiveness is
significantly lower in patients without the activating mutations,
and even patients with the mutations frequently develop resistance
to EGFR-TKI (2). Therefore, new
therapeutic targets that can overcome inherent or acquired
resistance to EGFR-TKIs are highly desirable. Recently, it has been
suggested that acquired resistance to EGFR-TKIs may be related to
amplification of a hepatocyte growth factor (HGF) receptor, termed
MET (3). HGF expression can
induce EGFR-TKI resistance to lung adenocarcinoma cells with
EGFR-activating mutations (4),
and MET inhibition can reduce proliferation of lung adenocarcinoma
cell lines that show resistance to EGFR-TKIs (3). MET amplification occurs in about 20%
of NSCLC patients and is associated with poor survival.
The lipid metabolism pathway may also modulate the
effectiveness of EGFR-TKIs in lung adenocarcinoma patients. It has
been suggested that lipid-lowering drug statins may reduce cancer
risk (5), and a large
case-control study of US veterans found that this may be true for
lung cancer (6), although some
reports claim otherwise (7,8).
In vitro studies have shown that inhibition of the
mevalonate pathway by statins reduces EGFR autophosphorylation
(9), downstream AKT signaling
(10), and EGF-induced RhoA
translocation to the plasma membrane (11). Enhancement of EGFR-TKI
effectiveness by statins seems to occur not only in cells with
EGFR-activating mutations but also in EGFR-TKI-resistant NSCLC cell
lines (12). The mechanism of
EGFR signaling inhibition is not fully characterized, but reduced
prenylation of small GTP-binding proteins may be of importance
(13). However, depletion of
cholesterol in the plasma membrane is known to increase EGFR
signaling activity, perhaps by releasing EGFR from lipid rafts and
inhibiting receptor internalization (14,15). This suggests that the lipid
metabolism pathway can influence EGFR signaling in both a positive
and negative manner.
This study sought to characterize the lipid
metabolism pathway in lung adenocarcinoma using gene expression
correlation analysis of microarray data. More specifically, pathway
genes that show associations with EGFR or MET were examined in
detail, because EGFR and MET are among the best-studied growth
signals in lung cancer patients. Gene expression profiles have been
used to classify lung cancer (16), to discover gene sets which are
predictive of disease prognosis (17), and to investigate molecular
mechanisms of disease progression (18). However, large-scale analysis of
the association between metabolic and growth factor signaling
pathways has not been conducted in lung cancer tissue. In the
present study, a set of lipid metabolism pathway genes, the
expression of which are highly correlated with EGFR or MET, were
first selected. Next, genes in the microarray dataset showing
significant correlation with selected genes were examined in terms
of functional properties. Finally, possible regulatory mechanisms
of correlated expression were inferred using known transcription
factor target sequences. This type of analysis predicts how the
lipid metabolic pathway may functionally interact with EGFR, MET,
and other biological processes in lung cancer cells, and offers an
insight into the roles of EGFR and MET inhibition in lung cancer
therapeutics.
Materials and methods
Microarray data
The microarray dataset GSE10072 (19) from the Gene Expression Omnibus
(20) was used for analysis. The
dataset contains expression profiles of 58 tumor and 49 non-tumor
tissues. The information was originally obtained using the
Affymetrix Human Genome U133A Array. The data from 22,215 probes in
the array were normalized using the quantile normalization function
(quantilenorm) of the Matlab Bioinformatics Toolbox (MathWorks,
Natick, MA).
Classification of genes by Gene
Ontology
The DAVID functional annotation tool [version 6.7b
(21,22)] was used to classify gene sets by
Gene Ontology identifiers or using UCSC transcription factor
binding sites (23). Functional
categories with a Benjamini-Hochberg statistic (24) of <0.025 were considered
statistically significant.
Statistical analysis
Pearson correlation coefficients were calculated
using the ‘corr’ function from Matlab. The 2.5th and 97.5th
percentiles of coefficients for 100,000 pairwise combinations
between randomly selected genes in the dataset were −0.379 and
0.428, respectively, and these were used as threshold values for
significantly negative and positive correlations. Two-sample
t-testing was achieved using the ‘ttest2’ function from Matlab.
Results
Correlation of lipid metabolism genes
with EGFR expression
A total of 301 genes classified as ‘lipid metabolic
process’ (GO:0006629) by gene ontology were selected and Pearson
correlation coefficients were calculated between the expression of
such genes and EGFR and MET. Although no gene showed a positive
correlation with EGFR or MET expression, eight and nine such genes
displayed a negative correlation with EGFR and MET expression,
respectively, in cancer samples (Table I). The negative correlations were
not evident in normal lung samples, except for MVK, which showed a
significant negative correlation with MET in both cancerous and
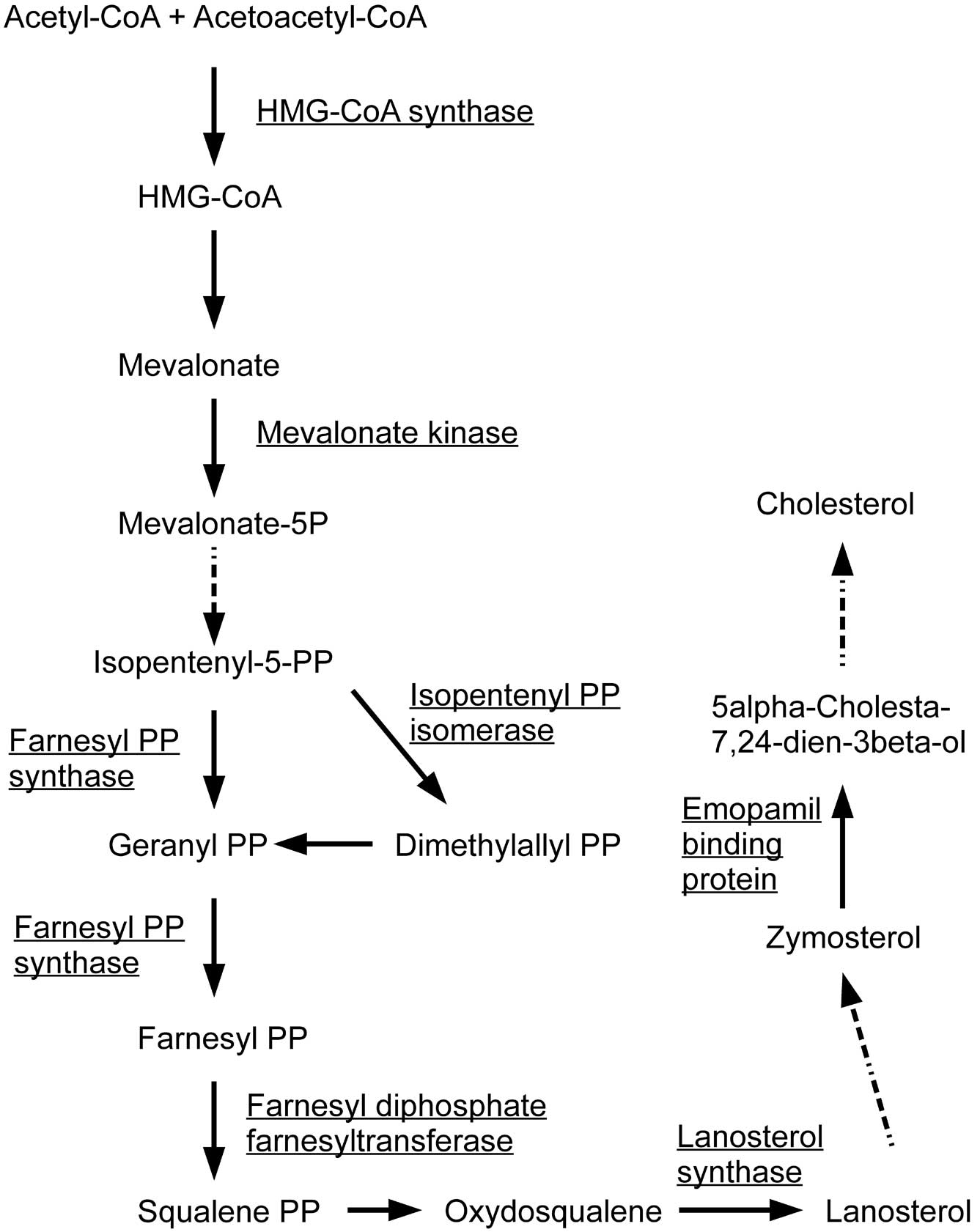
normal cells. Among the negatively correlated genes, HMG-coenzyme A
synthase 1 (HMGCS1), farnesyl-diphosphate farnesyltransferase 1
(FDFT1), farnesyl diphosphate synthase (FDPS),
isopentenyl-diphosphate δ isomerase 1 (IDI1), lanosterol synthase
(LSS), emopamil binding protein (EBP) and mevalonate kinase (MVK)
are known to be involved in the first steps of steroid biosynthesis
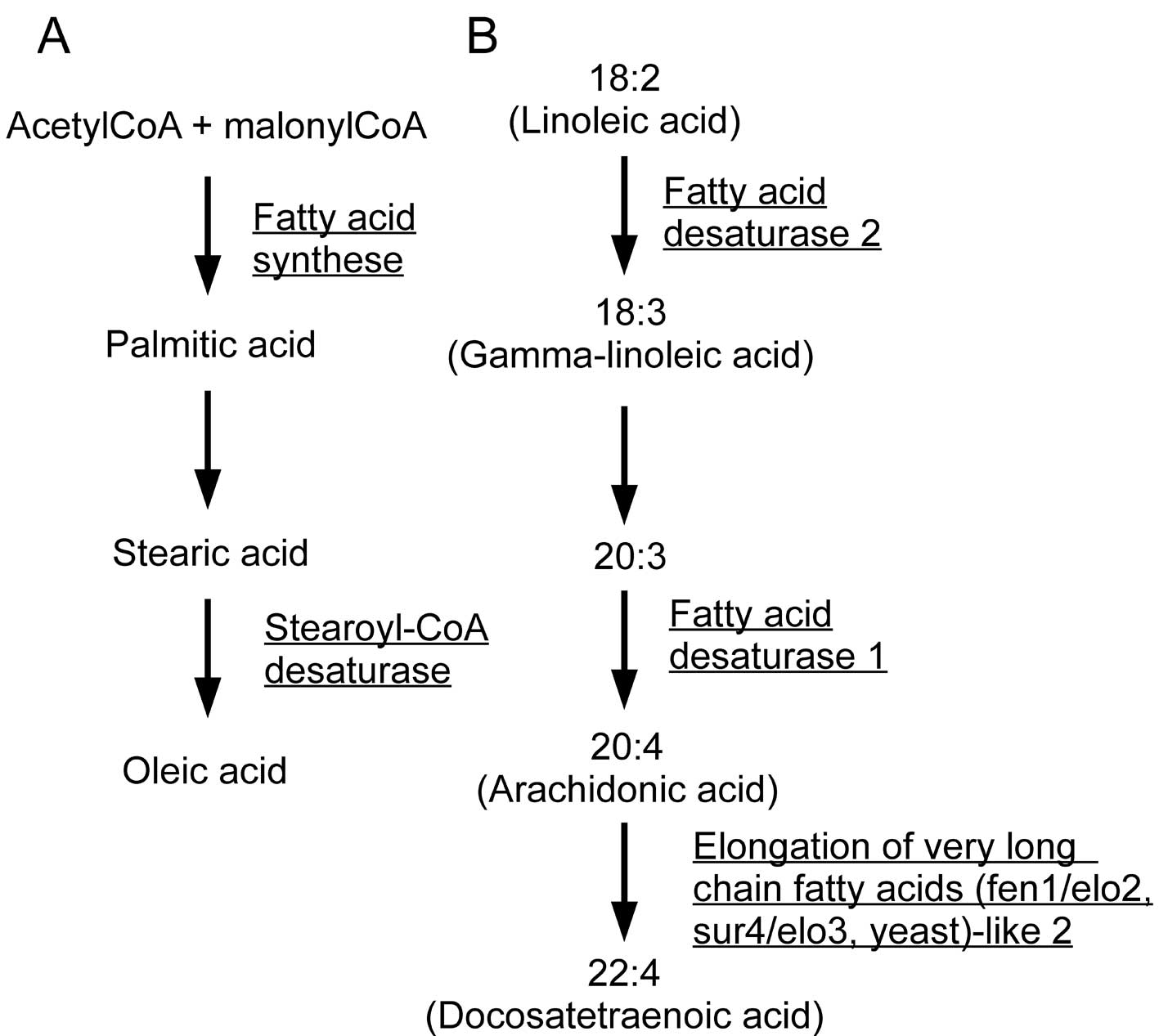
(Fig. 1). FAS and stearoyl-CoA
desaturase (SCD) mediate the synthesis of monounsaturated fatty
acids from acetyl-CoA, and fatty acid desaturase 1 (FADS1), fatty
acid desaturase 2 (FADS2), and elongation of very long chain fatty
acids (fen1/elo2, sur4/elo3, yeast)-like 2 (ELOVL2) catalyze the
production of polyunsaturated fatty acids, including arachidonic
acid (Fig. 2). Fatty acid
2-hydroxylase (FA2H) is involved in sphingolipid metabolism and
mutations in this gene are known to cause leukodystrophy, whereas
phosphatidylglycerophosphate synthase 1 (PGS1) is involved in
glycerophospholipid metabolism, synthesizing
phosphatidyl-glycerophosphate from CDP-diacylglycerol. These
results suggest that EGFR and MET are closely, but negatively,
associated with the expression of a variety of fatty acid
biosynthesis genes in lung adenocarcinoma tissue.
 | Table ILipid metabolic process genes
negatively correlated with EGFR or MET. |
Table I
Lipid metabolic process genes
negatively correlated with EGFR or MET.
| Symbol | Name | Cancera | Normala |
|---|
| EGFR |
| FA2H | Fatty acid
2-hydroxylase | −0.442 | −0.073 |
| FADS2 | Fatty acid
desaturase 2 | −0.419 | 0.181 |
| FDFT1 |
Farnesyl-diphosphate farnesyltransferase
1 | −0.414 | −0.093 |
| FDPS | Farnesyl
diphosphate synthase | −0.402 | −0.321 |
| HMGCS1 | HMG-coenzyme A
synthase 1 | −0.559 | −0.224 |
| IDI1 |
Isopentenyl-diphosphate δ isomerase 1 | −0.438 | −0.084 |
| LSS | Lanosterol
synthase | −0.402 | −0.115 |
| SCD | Stearoyl-CoA
desaturase | −0.399 | −0.021 |
| MET |
| EBP | Emopamil binding
protein | −0.441 | 0.245 |
| ELOVL2 | Elongation of very
long chain fatty acids (fen1/elo2, sur4/elo3, yeast)-like 2 | −0.427 | −0.146 |
| FADS1 | Fatty acid
desaturase 1 | −0.411 | 0.115 |
| FADS2 | Fatty acid
desaturase 2 | −0.596 | −0.099 |
| FASN | Fatty acid
synthase | −0.526 | 0.108 |
| IDI1 |
Isopentenyl-diphosphate δ isomerase 1 | −0.463 | 0.301 |
| LSS | Lanosterol
synthase | −0.390 | 0.117 |
| MVK | Mevalonate
kinase | −0.380 | −0.391 |
| PGS1 |
Phosphatidylglycerophosphate synthase
1 | −0.421 | −0.061 |
Functional gene categories associated
with lipid metabolism genes anti-correlated to EGFR
Next, associations of the ‘anti-EGFR/MET’ lipid
metabolism genes with other genes were evaluated by calculation of
the intergene Pearson correlation coefficients in lung cancer
samples. Table II shows the
number of genes demonstrating significant positive or negative
associations with mevalonate pathway genes (FDFT1, FDPS, HMGCS1,
IDI1, LSS, EBP and MVK).
 | Table IINumber of genes significantly
correlated with lipid metabolism genes. |
Table II
Number of genes significantly
correlated with lipid metabolism genes.
| Symbol | Positive | Negative |
|---|
| Mevalonate pathway
genes |
| FDFT1 | 68 | 162 |
| FDPS | 438 | 480 |
| HMGCS1 | 372 | 381 |
| IDI1 | 505 | 573 |
| LSS | 166 | 192 |
| EBP | 294 | 431 |
| MVK | 3012 | 1462 |
| Fatty acid
synthesis pathways genes |
| ELOVL2 | 1428 | 459 |
| FA2H | 87 | 93 |
| FADS1 | 182 | 214 |
| FADS2 | 130 | 200 |
| FASN | 67 | 325 |
| PGS1 | 147 | 127 |
| SCD | 152 | 188 |
Among these seven genes, FDPS, HMGCS1, IDI1 and MVK,
all of which mediate farnesyl pyrophosphate synthesis from
mevalonate, showed particularly large numbers of correlated genes.
In addition, 166 genes in the microarray dataset displayed
significant positive associations with three or more of the
mevalonate pathway genes. According to DAVID, gene functional
categories were dominated by GO Biological Processes related to the
cell cycle, DNA replication, response to DNA damage, and lipid
metabolism, suggesting close links between the regulation of cell
division and cholesterol biosynthesis (Table III). On the other hand, 235
genes had significant negative associations with three or more of
the mevalonate pathway genes. The functional categories were
principally related to cell adhesion, cell migration, blood vessel
development, extracellular matrix synthesis, and defense responses
(Table III). This gene set also
included regulators of cell proliferation, including endothelin
receptor type A (EDNRA), platelet-derived growth factor receptor, α
polypeptide (PDGFRA), protein kinase Cα (PRKCA), ras-related C3
botulinum toxin substrate 2 (RAC2), transforming growth factor β,
receptor II (TGFBR2), and vitamin D receptor (VDR). These data may
suggest that mevalonate pathway genes were negatively associated
with processes mediating signal transduction from the extracellular
space, but positively associated with pathways involving the
nucleus. Similarly, anti-EGFR/MET lipid metabolism genes involved
in fatty acid synthesis (FADS1, FADS2, FASN, SCD, ELOVL2, PGS1 and
FA2H) were evaluated (Table II).
Most of these genes showed smaller numbers of correlations than
genes of the mevalonate pathway. Only 18 and 35 genes displayed
significant positive and negative correlations, respectively, with
three or more of the fatty acid synthesis genes. The positively
correlated genes belonged to sets of functional categories similar
to those positively correlated with mevalonate pathway genes
(Table III); these were genes
of the cell cycle, cell division and lipid metabolism. No
functional category was significantly enriched in negatively
correlated genes.
 | Table IIIEnriched GO biological process terms
showing significant correlations with mevalonate and fatty acid
synthesis pathway genes. |
Table III
Enriched GO biological process terms
showing significant correlations with mevalonate and fatty acid
synthesis pathway genes.
| Term | Count | Benjamini |
|---|
| Enriched GO
biological process terms in genes positively correlated with the
mevalonate pathway genes |
| GO:0007049, cell
cycle | 67 | 3.56E-37 |
| GO:0000278, mitotic
cell cycle | 46 | 4.44E-36 |
| GO:0022403, cell
cycle phase | 45 | 1.01E-32 |
| GO:0007067,
mitosis | 38 | 1.21E-31 |
| GO:0000279, M
phase | 41 | 1.34E-31 |
| GO:0000087, M phase
of mitotic cell cycle | 38 | 1.42E-31 |
| GO:0022402, cell
cycle process | 55 | 2.26E-29 |
| GO:0051301, cell
division | 37 | 3.18E-29 |
| GO:0006259, DNA
metabolic process | 43 | 1.37E-15 |
| GO:0006260, DNA
replication | 24 | 2.92E-13 |
| GO:0000075, cell
cycle checkpoint | 15 | 5.30E-13 |
| GO:0007051, spindle
organization and biogenesis | 11 | 5.83E-13 |
| GO:0007017,
microtubule-based process | 22 | 2.24E-12 |
| GO:0000074,
regulation of progression through cell cycle | 31 | 2.71E-12 |
| GO:0051726,
regulation of cell cycle | 31 | 2.92E-12 |
| GO:0006996,
organelle organization and biogenesis | 44 | 2.00E-11 |
| GO:0000070, mitotic
sister chromatid segregation | 11 | 8.75E-11 |
| GO:0000819, sister
chromatid segregation | 11 | 1.19E-10 |
| GO:0007059,
chromosome segregation | 13 | 3.18E-10 |
| GO:0006974,
response to DNA damage stimulus | 23 | 3.46E-10 |
| GO:0000226,
microtubule cytoskeleton organization and biogenesis | 14 | 4.49E-10 |
| GO:0007088,
regulation of mitosis | 13 | 1.03E-09 |
| GO:0051276,
chromosome organization and biogenesis | 24 | 2.02E-09 |
| GO:0009719,
response to endogenous stimulus | 23 | 2.09E-08 |
| GO:0007093, mitotic
cell cycle checkpoint | 9 | 2.01E-07 |
| GO:0016126, sterol
biosynthetic process | 9 | 2.50E-07 |
| GO:0007010,
cytoskeleton organization and biogenesis | 24 | 5.16E-07 |
| GO:0051325,
interphase | 12 | 7.14E-07 |
| GO:0006281, DNA
repair | 17 | 2.01E-06 |
| GO:0006261,
DNA-dependent DNA replication | 12 | 2.29E-06 |
| GO:0016125, sterol
metabolic process | 11 | 2.68E-06 |
| GO:0051329,
interphase of mitotic cell cycle | 11 | 4.48E-06 |
| GO:0007052, mitotic
spindle organization and biogenesis | 6 | 1.13E-05 |
| GO:0006694, steroid
biosynthetic process | 10 | 2.43E-05 |
| GO:0006695,
cholesterol biosynthetic process | 7 | 2.70E-05 |
| GO:0006270, DNA
replication initiation | 7 | 8.05E-05 |
| GO:0009987,
cellular process | 150 | 1.10E-04 |
| GO:0008203,
cholesterol metabolic process | 9 | 1.25E-04 |
| GO:0044237,
cellular metabolic process | 112 | 3.23E-04 |
| GO:0016043,
cellular component organization and biogenesis | 52 | 3.59E-04 |
| GO:0006139,
nucleobase, nucleoside, nucleotide and nucleic acid metabolic
process | 68 | 3.63E-04 |
| GO:0008202, steroid
metabolic process | 12 | 3.69E-04 |
| GO:0044238, primary
metabolic process | 111 | 7.69E-04 |
| GO:0048015,
phosphoinositide-mediated signaling | 9 | 1.33E-03 |
| GO:0006268, DNA
unwinding during replication | 5 | 1.42E-03 |
| GO:0031570, DNA
integrity checkpoint | 6 | 1.75E-03 |
| GO:0007018,
microtubule-based movement | 9 | 1.87E-03 |
| GO:0000910,
cytokinesis | 6 | 1.96E-03 |
| GO:0032508, DNA
duplex unwinding | 5 | 2.19E-03 |
| GO:0032392, DNA
geometric change | 5 | 2.19E-03 |
| GO:0008610, lipid
biosynthetic process | 13 | 2.45E-03 |
| GO:0008152,
metabolic process | 117 | 3.33E-03 |
| GO:0008299,
isoprenoid biosynthetic process | 5 | 3.38E-03 |
| GO:0030705,
cytoskeleton-dependent intracellular transport | 9 | 5.04E-03 |
| GO:0031577, spindle
checkpoint | 4 | 5.08E-03 |
| GO:0006066, alcohol
metabolic process | 13 | 9.20E-03 |
| GO:0042770, DNA
damage response, signal transduction | 6 | 1.05E-02 |
| GO:0043283,
biopolymer metabolic process | 77 | 1.33E-02 |
| GO:0000077, DNA
damage checkpoint | 5 | 1.65E-02 |
| GO:0006950,
response to stress | 25 | 1.67E-02 |
| GO:0006720,
isoprenoid metabolic process | 5 | 1.85E-02 |
| Enriched GO
biological process terms in genes negatively correlated with the
mevalonate pathway genes |
| GO:0022610,
biological adhesion | 61 | 1.51E-25 |
| GO:0007155, cell
adhesion | 61 | 1.51E-25 |
| GO:0016337,
cell-cell adhesion | 33 | 8.98E-17 |
| GO:0007156,
homophilic cell adhesion | 25 | 4.41E-16 |
| GO:0009605,
response to external stimulus | 31 | 5.78E-06 |
| GO:0009611,
response to wounding | 25 | 6.36E-06 |
| GO:0048518,
positive regulation of biological process | 38 | 3.50E-04 |
| GO:0032501,
multicellular organismal process | 85 | 4.84E-04 |
| GO:0032502,
developmental process | 77 | 4.97E-04 |
| GO:0006954,
inflammatory response | 18 | 5.46E-04 |
| GO:0048731, system
development | 50 | 5.76E-04 |
| GO:0048856,
anatomical structure development | 57 | 6.46E-04 |
| GO:0048513, organ
development | 40 | 9.45E-04 |
| GO:0006950,
response to stress | 35 | 1.76E-03 |
| GO:0006952, defense
response | 24 | 1.79E-03 |
| GO:0007275,
multicellular organismal development | 59 | 1.80E-03 |
| GO:0008283, cell
proliferation | 29 | 1.90E-03 |
| GO:0007167, enzyme
linked receptor protein signaling pathway | 16 | 3.11E-03 |
| GO:0001944,
vasculature development | 13 | 4.01E-03 |
| GO:0048522,
positive regulation of cellular process | 32 | 4.99E-03 |
| GO:0048523,
negative regulation of cellular process | 34 | 9.29E-03 |
| GO:0009887, organ
morphogenesis | 18 | 1.50E-02 |
| GO:0001568, blood
vessel development | 12 | 1.51E-02 |
| GO:0006959, humoral
immune response | 8 | 1.52E-02 |
| GO:0048519,
negative regulation of biological process | 34 | 1.77E-02 |
| Enriched GO
biological process terms in genes positively correlated with fatty
acid synthesis pathways genes |
| GO:0008610, lipid
biosynthetic process | 7 | 1.83E-03 |
| GO:0000278, mitotic
cell cycle | 7 | 2.01E-03 |
| GO:0007049, cell
cycle | 9 | 2.03E-03 |
| GO:0007051, spindle
organization and biogenesis | 4 | 2.22E-03 |
| GO:0000226,
microtubule cytoskeleton organization and biogenesis | 5 | 2.35E-03 |
| GO:0000087, M phase
of mitotic cell cycle | 6 | 2.47E-03 |
| GO:0006695,
cholesterol biosynthetic process | 4 | 2.52E-03 |
| GO:0007067,
mitosis | 6 | 2.71E-03 |
| GO:0051301, cell
division | 6 | 3.06E-03 |
| GO:0016126, sterol
biosynthetic process | 4 | 3.46E-03 |
| GO:0000279, M
phase | 6 | 5.39E-03 |
| GO:0022403, cell
cycle phase | 6 | 1.39E-02 |
| GO:0044255,
cellular lipid metabolic process | 7 | 1.48E-02 |
Transcriptional regulatory mechanisms
associated with anti-EGFR lipid metabolism genes
Gene expression correlation analysis showed that
lipid metabolism genes were associated with specific biological
processes, particularly the cell cycle. To determine a possible
mechanism of correlated expression, enrichment of predicted
transcription factor binding sites was examined by DAVID. It was
found that genes positively associated with mevalonate pathway
genes were enriched in the NFY binding site, with a Benjamini score
of 3.4E-8. To examine the relationship between NFY and genes
positively correlated with mevalonate pathway genes, a search was
instituted for genes showing significant positive correlations with
NFY. As NFY is composed of subunits encoded by three genes, NFYA,
NFYB and NFYC, genes with positive correlations with at least one
subunit were selected. Respectively 202, 889 and 133 genes were
found to display a correlation with NFYA, NFYB and NFYC, and, in
total, 1,166 genes displayed significant positive correlations with
one or more of the NYF subunit genes. For each gene identified,
Pearson correlation coefficients were calculated with respect to
genes positively correlated with mevalonate pathway genes, and the
number of significant positive correlations was enumerated. This
disclosed that 53 genes showed positive correlations with 81 or
more of mevalonate pathway-associated genes. This threshold of 81
is the top 2.5th percentile of the number of mevalonate pathway
genes positively correlated with each gene in the microarray
dataset. These 53 genes will be simply termed ‘NFY-correlated
genes’ below.
A literature search found no reported direct
physical association between NFY and any of the 53 gene products.
However, according to DAVID, many of these genes were related to
DNA metabolic processes, DNA repair, or mRNA metabolism (Table IV). To account for the observed
associations between NFY and NFY-correlated genes, known protein
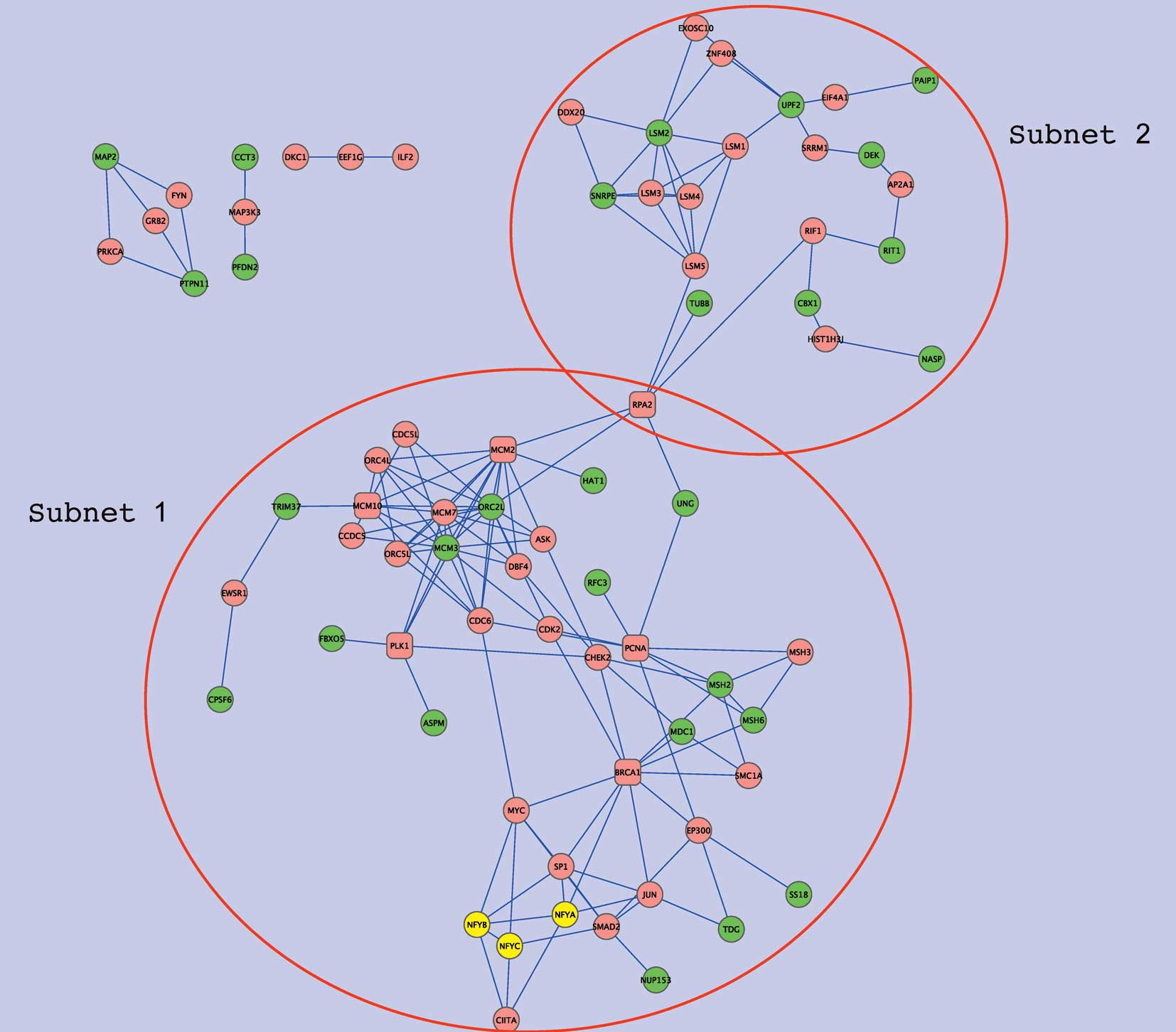
interactions were sought using Genes2Networks (25). Fig.
3 shows the overall network, formed by NFY genes,
NFY-correlated genes, and intermediate genes which connect these
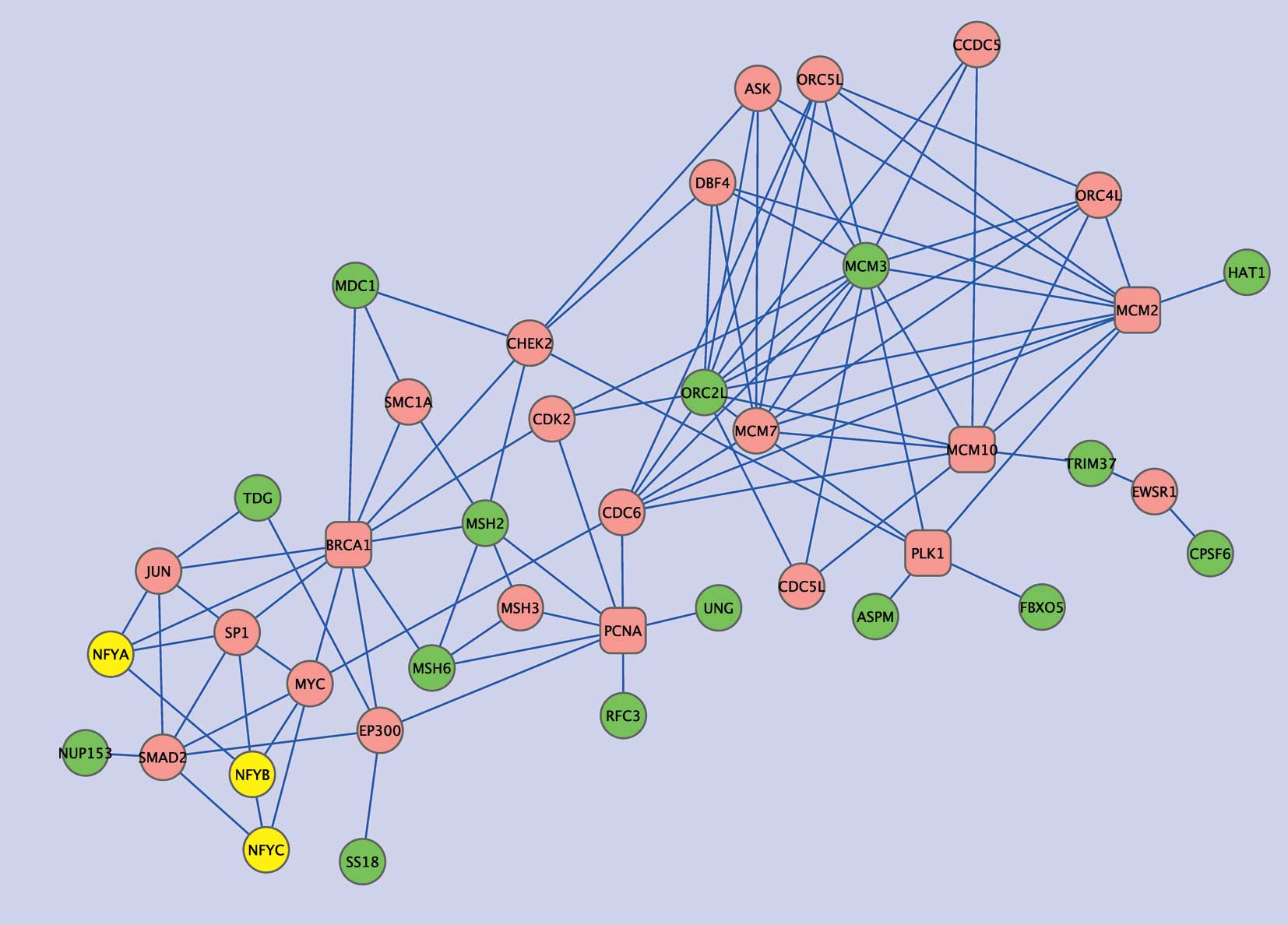
two gene sets. Extracts from the network, subnets 1 and 2, are
shown in Figs. 4 and 5, respectively. Subnet 1 has 15
NFY-correlated genes showing relatively close associations with NFY
genes in the interaction network (Table V). Six such genes are involved in
DNA repair and five are associated with either the cell cycle
(ASPM, FBXO5), DNA metabolic processes (ORC2L, HAT1), or both
(MCM3). In this subnetwork, several intermediate or ‘hub’ genes
were closely connected to the NFY-correlated genes. Namely, PCNA
and BRCA1 were connected to four of the NFY-correlated genes, and
each of MCM10, PLK1, MCM2 and RPA2 to three. In addition to these
hub genes, CHEK2, CDK2, MCM7, CDC6, EP300 and ORC4L were connected
to two of the NFY-correlated genes as well as to two hub genes. Of
these genes, PCNA, MCM2, CDK2 and MCM7 showed significantly
negative correlations with EGFR (Pearson coefficients, −0.446,
−0.399, −0.381 and −0.401, respectively), whereas PLK1, MCM2 and
CDK2 displayed significantly negative correlations with MET
(Pearson coefficients, −0.373, −0.486 and −0.495, respectively).
Moreover, the mean Pearson coefficients of all hub genes were
−0.252 for EGFR and −0.240 for MET, both of which were
significantly lower than the means for all genes in the dataset
(−0.0089 for EGFR and −0.0313 for MET; P=1.678E-4 and 0.0029 by
t-tests, respectively), demonstrating negative associations between
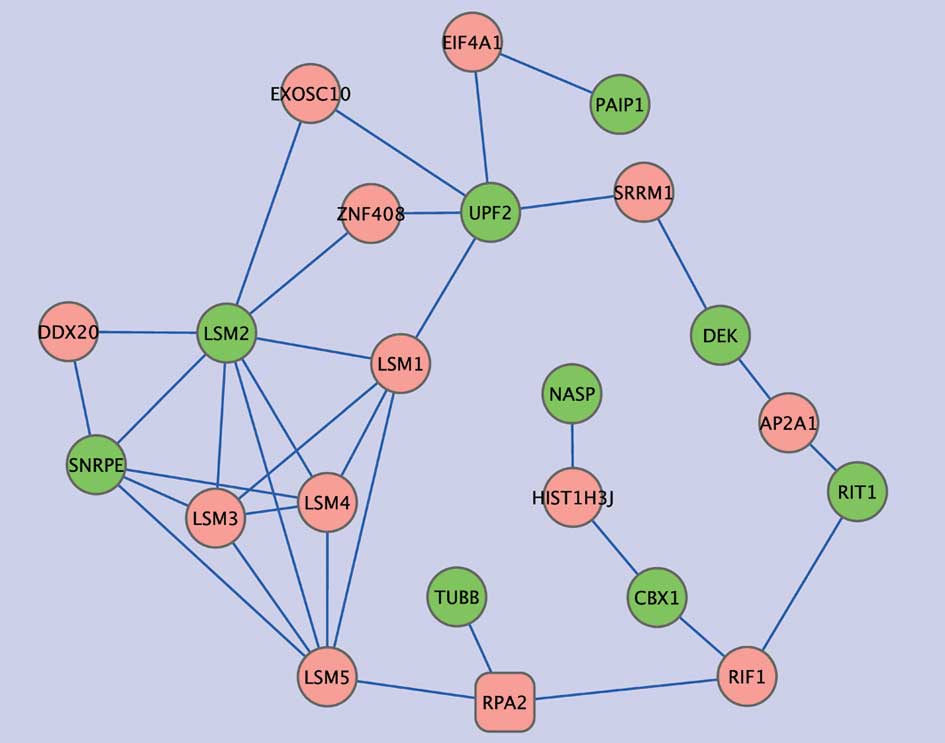
hub genes and growth signals. Subnet 2 includes nine of the
NFY-associated genes that were only distantly connected with NFY
genes in the protein-protein interaction network. Five of these
genes were related to RNA metabolic processes (PAIP1, SNRPE, DEK,
UPF and LSM2) and two genes encoded proteins with histone-binding
properties (NASP and CBX1). In this subnetwork, LSM1 showed high
connectivity, displaying two edges with the NFY-correlated genes,
and three with other intermediate genes. LSM1 is highly expressed
in lung cancer and mesothelioma, and LSM1 inhibition retards tumor
growth (26). Four other LMS
genes were present in the subnet but there was no evidence of
association with lung cancer.
 | Table IVEnriched GO biological process terms
with the NFY-correlated genes. |
Table IV
Enriched GO biological process terms
with the NFY-correlated genes.
| Term | Count | Benjamini |
|---|
| GO:0006259, DNA
metabolic process | 16 | 1.49E-04 |
| GO:0006139,
nucleobase, nucleoside, nucleotide and nucleic acid metabolic
process | 31 | 4.00E-04 |
| GO:0006974,
response to DNA damage stimulus | 10 | 1.05E-03 |
| GO:0043170,
macromolecule metabolic process | 40 | 1.12E-03 |
| GO:0006260, DNA
replication | 9 | 1.24E-03 |
| GO:0006281, DNA
repair | 9 | 1.36E-03 |
| GO:0009719,
response to endogenous stimulus | 10 | 2.74E-03 |
| GO:0044238, primary
metabolic process | 41 | 1.07E-02 |
| GO:0044237,
cellular metabolic process | 41 | 1.11E-02 |
| GO:0006261,
DNA-dependent DNA replication | 6 | 1.20E-02 |
| GO:0043283,
biopolymer metabolic process | 32 | 1.23E-02 |
| GO:0016071, mRNA
metabolic process | 8 | 2.00E-02 |
 | Table VList of genes in subnets 1 and 2 that
were positively associated with the NFY-correlated genes. |
Table V
List of genes in subnets 1 and 2 that
were positively associated with the NFY-correlated genes.
| Symbol | Name |
|---|
| Genes in subnet
1 |
| ASPM | ASP (abnormal
spindle) homolog, microcephaly associated (Drosophila) |
| CPSF6 | Cleavage and
polyadenylation specific factor 6, 68 kDa |
| FBXO5 | F-box protein
5 |
| HAT1 | Histone
acetyltransferase 1 |
| MCM2 | MCM2 minichromosome
maintenance deficient 2, mitotin (S. cerevisiae) |
| MDC1 | Mediator of DNA
damage checkpoint 1 |
| MSH2 | MutS homolog 2,
colon cancer, nonpolyposis type 1 (E. coli) |
| MSH6 | MutS homolog 6
(E. coli) |
| NUP153 | Nucleoporin 153
kDa |
| ORC2L | Origin recognition
complex, subunit 2-like (yeast) |
| RFC3 | Replication factor
c (activator 1) 3, 38 kDa |
| SS18 | Synovial sarcoma
translocation, chromosome 18 |
| TDG | Thymine-DNA
glycosylase |
| TRIM37 | Tripartite
motif-containing 37 |
| UNG | Uracil-DNA
glycosylase |
| Genes in subnet
2 |
| CBX1 | Chromobox homolog 1
(hp1 β homolog Drosophila) |
| DEK | Dek oncogene (DNA
binding) |
| LSM2 | LSM2 homolog, U6
small nuclear RNA associated (S. cerevisiae) |
| NASP | Nuclear
autoantigenic sperm protein (histone-binding) |
| PAIP1 | Poly(a) binding
protein interacting protein 1 |
| RIT1 | Ras-like without
caax 1 |
| SNRPE | Small nuclear
ribonucleoprotein polypeptide e |
| TUBB | Tubulin, β |
| UPF2 | UPF2 regulator of
nonsense transcripts homolog (yeast) |
Discussion
In the present study, gene expression correlation
patterns predicted that mevalonate metabolism and fatty acid
synthesis processes were negatively associated with expression of
EGFR and MET, but positively associated with cell division.
Promoter analysis suggested that the NFY transcription factor may
be involved in the regulation of genes involved in mevalonate
metabolism, and the processes positively associated with them.
Finally, gene expression correlation patterns and protein-protein
interaction data indicate that the transcriptional regulation by
NFY may be mediated by its interactions with other regulators of
DNA metabolic processes and cell cycle genes.
The negative correlations between growth factor
signaling and lipid metabolic pathways reported here seem to
indicate an inhibitory effect of cholesterol on EGFR pathways in
lung adenocarcinoma. Polyunsaturated fatty acids, such as oleic
acid, are also known to inhibit the EGFR pathway, although the
effects depend both on particular combinations of fatty acids and
the cell type (27–29). In lung adenocarcinoma, the
mevalonate pathway synthesizes more non-sterol and fewer sterol
products than seen in fibroblasts (30). This can result in a higher degree
of prenylation of small GTP-binding proteins, and reduced levels of
plasma membrane cholesterol, possibly leading to enhanced EGFR
activity. Mevalonate metabolites can also influence the expression
of metabolic genes through the intermediacy of the liver X receptor
(LXR). For example, LXR can activate FDPS synthesis (31), but LXR is inhibited by
geranylgeraniol (32), which is
produced from isopentenyl-PP and farnesyl-PP. Indeed, expression of
NR1H3 (LXR-α) showed a significant correlation with FDPS and EBP
synthesis in lung cancer samples but not in normal lung samples
(data not shown), suggesting a cancer-specific regulation of
mevalonate pathway genes by LXR-α.
The positive correlations seen between the lipid
metabolic pathway and cell division-related processes appear to be
consistent with previous experimental evidence. Pravastatin is
known to inhibit DNA synthesis, whereas addition of
geranylgeranylpyrophosphate restores such synthesis and promotes
the G1/S transition (33).
However, inhibition of farnesyl-protein transferase induces p21
expression and G1 blockade in a p53-dependent manner, suggesting
that regulation of the cell cycle by mevalonate metabolites occurs
at both the transcriptional and translational levels. In lung
carcinoma cell lines, farnesyl transferase inhibitors block
farnesylation of centromeric proteins and inhibit the association
of such proteins with microtubules (34). In retinoblastoma gene-deficient
thyroid tumors, FDPS is overexpressed, leading to increased
isoprenylation and activation of N-Ras and induction of the DNA
damage response (35). These
experimental findings seem to suggest that mevalonate metabolites
can directly regulate the expression of genes related to cell
division as well. Unsaturated fatty acids are also known to
increase cell proliferation (36)
(37), although the mechanism of
such action is not clear. One possibility is that increased
activity of intracellular signaling cascades, such as those
mediated by intracellular calcium (38) or AKT (39), may enhance the response of cells
to mitogenic signals. However, unsaturated fatty acids are
substrates for lipid peroxidation and may cause DNA damage in lung
cancer cells (40–42). This may lead, in turn, to apparent
(thus not real) correlated expression of unsaturated fatty acid
metabolism genes and DNA damage response genes.
Transcription factor binding sequence analysis
suggested that NFY may have a considerable influence on
associations of lipid metabolism genes. NFY is a ubiquitous
transcriptional factor which recognizes promoter CCAAT boxes
(43). NFY is known to be
involved in transcriptional regulation of a wide range of genes,
but the regulatory roles of NFY in lipogenesis, the cell cycle, DNA
repair, and DNA synthesis are of particular interest in the present
context. In lipogenic gene regulation, NFY often functions with
SREBPs and SP1 (44), and recent
genome-wide scanning of SREBP1, SP1 and NFY occupancy showed that
NFY shares about 20 and 40% of target genes with SREBP1 and SP1,
respectively, in HepG2 cells (45). In the lung adenocarcinoma dataset,
some mevalonate pathway genes displayed significant correlation
with SREBP1 and SREBP2, but not SP1 (data not shown), suggesting
possible coordinated regulation of such genes by NFY and SREBPs in
cancer cells.
The regulation of cell cycle and DNA metabolism
genes by NFY is also well documented. Expression of a
dominant-negative NFY subunit significantly decreased the number of
cells entering the S-phase and delayed the progress of this phase,
resulting in retarded cell growth (46). NFY seems be involved in induction
of S-phase-specific transcription, such as that resulting in
synthesis of ribonucleotide reductase R2 (47), histone H3 (48), and cyclin B1 (49). NFY also mediates genotoxic
stress-induced gene expression in a p53-independent manner
(50), and suppresses gene
expression in the presence of active p53 (51), suggesting a functional dependency
on co-regulators. Therefore, it was important to define proteins
interacting with NFY in the lung cancer cells of the present study.
Combined analysis of gene expression correlation and
protein-protein interaction identified several ‘hub’ genes which
displayed high connectivity with NFY-correlated genes and other hub
genes. Importantly, many of the hub genes have been associated with
lung cancer. These include BRCA1 (52,53), PCNA (54,55), PLK1 (56,57), MCM2 (58), CHEK2 (59,60), CDK2 (61) and MCM7 (62), suggesting that the network
discovered here is likely to be involved in progression of lung
cancer. As some such genes were also sensitive to inhibition of the
mevalonate pathway [BRCA1 (63),
PCNA (64), MCM2 (65), CDK2 (66) and MCM7 (67)], hub genes may also be involved in
the antitumor effects of pathway inhibitors in lung cancer. These
hub genes do not have direct links to NFY-correlated genes and,
although functional association with NFY has been experimentally
shown for BRCA1 (68), CDK2
(49,69) and EP300 (70), other hub genes likely interact
with NFY through intermediate genes, the expression of which was
found to be correlated with that of NFY.
Finally, the results presented in this article have
several important clinical implications for the treatment of lung
adenocarcinoma. First, the data support the importance of lipid
metabolic pathway inhibition in adenocarcinoma patients,
particularly in those insensitive to anti-EGFR therapy or patients
who have developed resistance to such therapy. The effects of
chemotherapy may be enhanced by downregulating genes related to
cell division. Some of the hub genes identified in this article are
already known as lung cancer markers, but exploration of the
activity of combinations of such genes should better indicate the
parts of the network that are active or inactive in cancer cells,
thus possibly increasing therapeutic predictive power. Finally,
drugs targeting NFY may be useful to improve the efficacy of other
chemotherapeutic agents, by blocking multiple pathways related to
lung carcinogenesis. The roles played by NFY in a variety of
cancers have been highlighted in recent reports (71,72), and I believe that a new
therapeutic strategy based on inhibition of NFY warrants further
research and development.
Acknowledgements
This study was funded by AstraZeneca, UK.
References
|
1
|
TE StinchcombeMA SocinskiCurrent
treatments for advanced stage non-small cell lung cancerProc Am
Thorac Soc6233241200910.1513/pats.200809-110LC19349493
|
|
2
|
T MitsudomiY YatabeMutations of the
epidermal growth factor receptor gene and related genes as
determinants of epidermal growth factor receptor tyrosine kinase
inhibitors sensitivity in lung cancerCancer
Sci9818171824200710.1111/j.1349-7006.2007.00607.x
|
|
3
|
J BeanC BrennanJY ShihG RielyA VialeL
WangMET amplification occurs with or without T790M mutations in
EGFR mutant lung tumors with acquired resistance to gefitinib or
erlotinibProc Natl Acad Sci
USA1042093220937200710.1073/pnas.071037010418093943
|
|
4
|
S YanoW WangQ LiK MatsumotoH SakuramaT
NakamuraHepatocyte growth factor induces gefitinib resistance of
lung adenocarcinoma with epidermal growth factor
receptor-activating mutationsCancer
Res6894799487200810.1158/0008-5472.CAN-08-164319010923
|
|
5
|
WR FarwellRE ScrantonEV LawlerRA LewMT
BrophyLD FioreThe association between statins and cancer incidence
in a veterans populationJ Natl Cancer
Inst100134139200810.1093/jnci/djm28618182618
|
|
6
|
V KhuranaHR BejjankiG CalditoMW
OwensStatins reduce the risk of lung cancer in humans: a large
case-control study of us
veteransChest13112821288200710.1378/chest.06-093117494779
|
|
7
|
ML TaylorBJ WellsMJ SmolakStatins and
cancer: a meta-analysis of case-control studiesEur J Cancer
Prev17259268200810.1097/CEJ.0b013e3282b721fe18414198
|
|
8
|
J HaukkaR SankilaT KlaukkaJ LonnqvistL
NiskanenA TanskanenIncidence of cancer and statin usage-record
linkage studyInt J Cancer126279284201010.1002/ijc.2453619739258
|
|
9
|
AJ ManthaKE McFeeN NiknejadG GossIA
LorimerJ DimitroulakosEpidermal growth factor receptor-targeted
therapy potentiates lovastatin-induced apoptosis in head and neck
squamous cell carcinoma cellsJ Cancer Res Clin
Oncol129631641200310.1007/s00432-003-0490-2
|
|
10
|
AJ ManthaJE HansonG GossAE LagardeIA
LorimerJ DimitroulakosTargeting the mevalonate pathway inhibits the
function of the epidermal growth factor receptorClin Cancer
Res1123982407200510.1158/1078-0432.CCR-04-195115788691
|
|
11
|
T KusamaM MukaiT IwasakiM TatsutaY
MatsumotoH AkedoInhibition of epidermal growth factor-induced RhoA
translocation and invasion of human pancreatic cancer cells by
3-hydroxy-3-methylglutaryl-coenzyme a reductase inhibitorsCancer
Res61488548912001
|
|
12
|
IH ParkJY KimJI JungJY HanLovastatin
overcomes gefitinib resistance in human non-small cell lung cancer
cells with K-Ras mutationsInvest New
Drugs28791799201010.1007/s10637-009-9319-419760159
|
|
13
|
I BuhaescuH IzzedineMevalonate pathway: a
review of clinical and therapeutical implicationsClin
Biochem40575584200710.1016/j.clinbiochem.2007.03.01617467679
|
|
14
|
LJ PikeL CaseyCholesterol levels modulate
EGF receptor-mediated signaling by altering receptor function and
traffickingBiochemistry411031510322200210.1021/bi025943i12162747
|
|
15
|
T RingerikeFD BlystadFO LevyIH MadshusE
StangCholesterol is important in control of EGF receptor kinase
activity but EGF receptors are not concentrated in caveolaeJ Cell
Sci11513311340200211884532
|
|
16
|
G ParmigianiE Garrett-MayerR AnbazhaganE
GabrielsonA cross-study comparison of gene expression studies for
the molecular classification of lung cancerClin Cancer
Res1029222927200410.1158/1078-0432.CCR-03-049015131026
|
|
17
|
DA WigleI JurisicaN RadulovichM PintilieJ
RossantN LiuMolecular profiling of non-small cell lung cancer and
correlation with disease-free survivalCancer
Res6230053008200212036904
|
|
18
|
DR RhodesJ YuK ShankerN DeshpandeR
VaramballyD GhoshLarge-scale meta-analysis of cancer microarray
data identifies common transcriptional profiles of neoplastic
transformation and progressionProc Natl Acad Sci
USA10193099314200410.1073/pnas.040199410115184677
|
|
19
|
MT LandiT DrachevaM RotunnoJD FigueroaH
LiuA DasguptaGene expression signature of cigarette smoking and its
role in lung adenocarcinoma development and survivalPLoS
One3e1651200810.1371/journal.pone.000165118297132
|
|
20
|
T BarrettDB TroupSE WilhiteP LedouxD
RudnevC EvangelistaNCBI GEO: archive for high-throughput functional
genomic dataNucleic Acids
Res37D885D890200910.1093/nar/gkn76418940857
|
|
21
|
G DennisBT ShermanDA HosackJ YangW GaoHC
LaneDAVID: Database for annotation, visualization, and integrated
discoveryGenome Biol4P3200310.1186/gb-2003-4-5-p312734009
|
|
22
|
DW HuangBT ShermanRA LempickiSystematic
and integrative analysis of large gene lists using DAVID
bioinformatics resourcesNat Protoc44457200919131956
|
|
23
|
SY RheeV WoodK DolinskiS DraghiciUse and
misuse of the gene ontology annotationsNat Rev
Genet9509515200810.1038/nrg236318475267
|
|
24
|
Y BenjaminiD DraiG ElmerN KafkafiI
GolaniControlling the false discovery rate in behavior genetics
researchBehav Brain
Res125279284200110.1016/S0166-4328(01)00297-211682119
|
|
25
|
SI BergerJM PosnerA Ma’ayanGenes2networks:
connecting lists of gene symbols using mammalian protein
interactions databasesBMC
Bioinformatics8372200710.1186/1471-2105-8-37217916244
|
|
26
|
PM WatsonSW MillerM FraigDJ ColeDK
WatsonAM BoylanCaSm (LSm-1) overexpression in lung cancer and
mesothelioma is required for transformed phenotypesAm J Respir Cell
Mol Biol38671678200810.1165/rcmb.2007-0205OC18218995
|
|
27
|
X CasabiellJL ZugazaCM PomboA PandiellaFF
CasanuevaOleic acid blocks epidermal growth factor-activated early
intracellular signals without altering the ensuing mitogenic
responseExp Cell Res205365373199310.1006/excr.1993.1099
|
|
28
|
KE McKenzieGK BandyopadhyayW ImagawaK SunS
NandiOmega-3 and omega-6 fatty acids and PGE2 stimulate the growth
of normal but not tumor mouse mammary epithelial cells: evidence
for alterations in the signaling pathways in tumor
cellsProstaglandins Leukot Essent Fatty
Acids51437443199410.1016/0952-3278(94)90062-0
|
|
29
|
S MollerupA HaugenDifferential effect of
polyunsaturated fatty acids on cell proliferation during human
epithelial in vitro carcinogenesis: involvement of epidermal growth
factor receptor tyrosine kinaseBr J
Cancer74613618199610.1038/bjc.1996.410
|
|
30
|
F BennisG FavreFL GaillardG
SoulaImportance of mevalonate-derived products in the control of
HMG-CoA reductase activity and growth of human lung adenocarcinoma
cell line A549Int J
Cancer55640645199310.1002/ijc.29105504218406993
|
|
31
|
J FukuchiC SongAL KoS LiaoTranscriptional
regulation of farnesyl pyrophosphate synthase by liver X
receptorsSteroids68685691200310.1016/S0039-128X(03)00100-412957674
|
|
32
|
BM FormanB RuanJ ChenGJ SchroepferRM
EvansThe orphan nuclear receptor LXRalpha is positively and
negatively regulated by distinct products of mevalonate
metabolismProc Natl Acad Sci
USA941058810593199710.1073/pnas.94.20.105889380679
|
|
33
|
T TeranoT ShiinaY NoguchiT TanakaI
TatsunoY SaitoGeranylgeranylpyrophosphate plays a key role for the
G1 to S transition in vascular smooth muscle cellsJ Atheroscler
Thromb516199810.5551/jat1994.5.110077451
|
|
34
|
HR AsharL JamesK GrayD CarrS BlackL
ArmstrongFarnesyl transferase inhibitors block the farnesylation of
CENP-E and CENP-F and alter the association of CENP-E with the
microtubulesJ Biol
Chem2753045130457200010.1074/jbc.M00346920010852915
|
|
35
|
A ShammaY TakegamiT MikiS KitajimaM NodaT
ObaraRb regulates DNA damage response and cellular senescence
through E2F-dependent suppression of N-ras isoprenylationCancer
Cell15255269200910.1016/j.ccr.2009.03.00119345325
|
|
36
|
S KasayamaM KogaH KouharaS SumitaniK WadaT
KishimotoUnsaturated fatty acids are required for continuous
proliferation of transformed androgen-dependent cells by fibroblast
growth factor family proteinsCancer Res54644164451994
|
|
37
|
CB RenardB AskariLA SuzukiF KramerKE
BornfeldtOleate, not ligands of the receptor for advanced glycation
end-products, promotes proliferation of human arterial smooth
muscle
cellsDiabetologia4616761687200310.1007/s00125-003-1247-914595542
|
|
38
|
MN GraberA AlfonsoDL GillRecovery of
Ca2+ pools and growth in Ca2+ pool-depleted
cells is mediated by specific epoxyeicosatrienoic acids derived
from arachidonic acidJ Biol Chem272295462955319979368016
|
|
39
|
MR YunJY LeeHS ParkHJ HeoJY ParkSS
BaeOleic acid enhances vascular smooth muscle cell proliferation
via phosphatidylinositol 3-kinase/AKT signaling pathwayPharmacol
Res5497102200610.1016/j.phrs.2006.03.00116621593
|
|
40
|
E NikiLipid peroxidation: physiological
levels and dual biological effectsFree Radic Biol
Med47469484200910.1016/j.freeradbiomed.2009.05.03219500666
|
|
41
|
A TrombettaM MaggioraG MartinassoP
CotogniRA CanutoG MuzioArachidonic and docosahexaenoic acids reduce
the growth of A549 human lung-tumor cells increasing lipid
peroxidation and PPARsChem Biol
Interact165239250200710.1016/j.cbi.2006.12.01417275799
|
|
42
|
L MaehleE LystadE EilertsenE EinarsdottrAT
HstmarkA HaugenGrowth of human lung adenocarcinoma in nude mice is
influenced by various types of dietary fat and vitamin EAnticancer
Res1916491655199910470096
|
|
43
|
K MatuokaKY ChenTranscriptional regulation
of cellular ageing by the CCAAT box-binding factor CBF/NF-YAgeing
Res Rev1639651200210.1016/S1568-1637(02)00026-012362892
|
|
44
|
SD ClarkePolyunsaturated fatty acid
regulation of gene transcription: a molecular mechanism to improve
the metabolic syndromeJ Nutr13111291132200111285313
|
|
45
|
BD ReedAE CharosAM SzekelySM WeissmanM
SnyderGenome-wide occupancy of SREBP1 and its partners NFY and SP1
reveals novel functional roles and combinatorial regulation of
distinct classes of genesPLoS
Genet4e1000133200810.1371/journal.pgen.100013318654640
|
|
46
|
Q HuSN MaityStable expression of a
dominant negative mutant of CCAAT binding factor/NF-Y in mouse
fibroblast cells resulting in retardation of cell growth and
inhibition of transcription of various cellular genesJ Biol
Chem27544354444200010.1074/jbc.275.6.4435
|
|
47
|
AL ChabesS BjrklundL ThelanderS
phase-specific transcription of the mouse ribonucleotide reductase
R2 gene requires both a proximal repressive E2F-binding site and an
upstream promoter activating regionJ Biol
Chem2791079610807200410.1074/jbc.M312482200
|
|
48
|
H KoesslerJ KahleC BodeD DoeneckeW
AlbigHuman replication-dependent histone H3 genes are activated by
a tandemly arranged pair of two CCAAT boxesBiochem
J384317326200410.1042/BJ2004050215320874
|
|
49
|
KS KatulaKL WrightH PaulDR SurmanFJ
NuckollsJW SmithCyclin-dependent kinase activation and S-phase
induction of the cyclin B1 gene are linked through the CCAAT
elementsCell Growth Differ881182019979218875
|
|
50
|
S JinF FanW FanH ZhaoT TongP
BlanckTranscription factors OCT-1 and NF-YA regulate the
p53-independent induction of the GADD45 following DNA
damageOncogene2026832690200110.1038/sj.onc.120439011420680
|
|
51
|
I ManniG MazzaroA GurtnerR MantovaniU
HaugwitzK KrauseNF-Y mediates the transcriptional inhibition of the
cyclin B1, cyclin B2, and cdc25C promoters upon induced G2 arrestJ
Biol Chem27655705576200110.1074/jbc.M00605220011096075
|
|
52
|
HT KimJE LeeES ShinYK YooJH ChoMH
YunEffect of BRCA1 haplotype on survival of non-small-cell lung
cancer patients treated with platinum-based chemotherapyJ Clin
Oncol2659725979200810.1200/JCO.2008.16.649619018088
|
|
53
|
I BoukovinasC PapadakiP MendezM TaronD
MavroudisA KoutsopoulosTumor BRCA1, RRM1 and RRM2 mRNA expression
levels and clinical response to first-line gemcitabine plus
docetaxel in non-small-cell lung cancer patientsPLoS
One3e3695200810.1371/journal.pone.000369519002265
|
|
54
|
M VolmR KoomgiRelevance of proliferative
and proapoptotic factors in non-small-cell lung cancer for patient
survivalBr J Cancer8217471754200010817513
|
|
55
|
T OyamaT OsakiN NoseY IchikiM InoueH
ImotoEvaluations of p53 immunoreactivity, nucleolar organizer
regions, and proliferating cell nuclear antigen in non-small cell
lung carcinomaAnticancer Res20505510200010769714
|
|
56
|
Q ZhouY SuM BaiEffect of antisense RNA
targeting polo-like kinase 1 on cell growth in A549 lung cancer
cellsJ Huazhong Univ Sci Technolog Med
Sci282226200810.1007/s11596-008-0106-918278450
|
|
57
|
B Spnkuch-SchmittG WolfC SolbachS LoiblR
KnechtM StegmllerDownregulation of human polo-like kinase activity
by antisense oligonucleotides induces growth inhibition in cancer
cellsOncogene2131623171200210.1038/sj.onc.120541212082631
|
|
58
|
DF TanJA HubermanA HylandGM LoewenJS
BrooksAF BeckMCM2-a promising marker for premalignant lesions of
the lung: a cohort studyBMC
Cancer16200110.1186/1471-2407-1-611472637
|
|
59
|
D ThompsonS SealM SchutteL McGuffogR
BarfootA RenwickA multicenter study of cancer incidence in CHEK2
1100delC mutation carriersCancer Epidemiol Biomarkers
Prev1525422545200610.1158/1055-9965.EPI-06-068717164383
|
|
60
|
C CybulskiB MasojcD OszutowskaE
JaworowskaT GrodzkiP WaloszczykConstitutional CHEK2 mutations are
associated with a decreased risk of lung and laryngeal
cancersCarcinogenesis29762765200810.1093/carcin/bgn04418281249
|
|
61
|
M VolmR KoomgiW RittgenClinical
implications of cyclins, cyclin-dependent kinases, RB and E2F1 in
squamous-cell lung carcinomaInt J
Cancer79294299199810.1002/(SICI)1097-0215(19980619)79:3%3C294::AID-IJC15%3E3.0.CO;2-89645354
|
|
62
|
S FujiokaK ShomoriK NishiharaK YamagaK
NosakaK ArakiExpression of minichromosome maintenance 7 (MCM7) in
small lung adenocarcinomas (pT1): prognostic implicationLung
Cancer65223229200910.1016/j.lungcan.2008.11.00719144445
|
|
63
|
HL Neville-WebbeCA EvansRE ColemanI
HolenMechanisms of the synergistic interaction between the
bisphosphonate zoledronic acid and the chemotherapy agent
paclitaxel in breast cancer cells in vitroTumour
Biol2792103200610.1159/000092489
|
|
64
|
WT GunningPM KramerRA LubetVE SteeleDW
EndW WoutersMA PereiraChemoprevention of benzo(a)pyrene-induced
lung tumors in mice by the farnesyltransferase inhibitor
R115777Clin Cancer Res919271930200312738751
|
|
65
|
C MorganPD LewisRM JonesG BertelliGA
ThomasRC LeonardThe in vitro anti-tumour activity of zoledronic
acid and docetaxel at clinically achievable concentrations in
prostate cancerActa
Oncol46669677200710.1080/0284186060099644717562444
|
|
66
|
SF Doisneau-SixouP CestacJC FayeG FavreRL
SutherlandAdditive effects of tamoxifen and the farnesyl
transferase inhibitor FTI-277 on inhibition of MCM-7 breast cancer
cell-cycle progressionInt J
Cancer106789798200310.1002/ijc.1126312866041
|
|
67
|
D BruemmerF YinJ LiuT KiyonoE FleckAV
HerleAtorvastatin inhibits expression of minichromosome maintenance
proteins in vascular smooth muscle cellsEur J
Pharmacol4621523200310.1016/S0014-2999(03)01317-712591091
|
|
68
|
W FanS JinT TongH ZhaoF FanMJ
AntinoreBRCA1 regulates GADD45 through its interactions with the
OCT-1 and CAAT motifsJ Biol
Chem27780618067200210.1074/jbc.M11022520011777930
|
|
69
|
HD ChaeJ YunYJ BangDY ShinCdk2-dependent
phosphorylation of the NF-Y transcription factor is essential for
the expression of the cell cycle-regulatory genes and cell cycle
G1/S and G2/M
transitionsOncogene2340844088200410.1038/sj.onc.120748215064732
|
|
70
|
V SalsiG CarettiM WasnerW ReinhardU
HaugwitzK EngelandInteractions between P300 and multiple NF-Y
trimers govern cyclin b2 promoter functionJ Biol
Chem27866426650200310.1074/jbc.M21006520012482752
|
|
71
|
H GoodarziO ElementoS TavazoieRevealing
global regulatory perturbations across human cancersMol
Cell36900911200910.1016/j.molcel.2009.11.01620005852
|
|
72
|
K YamanakaS MizuaraiT EguchiH ItadaniH
HiraiH KotaniExpression levels of NF-Y target genes changed by
CDKN1B correlate with clinical prognosis in multiple
cancersGenomics94219227200910.1016/j.ygeno.2009.06.00319559782
|