Contents
Introduction
T cell subsets and antigen-presenting cells
regulated by various factors
Signal transduction in T cell subsets
Genetic changes after T cell differentiation
Perspective
Introduction
When naive T lymphocytes are primed by MHC class
II-expressing dendritic cells, which are specialized
antigen-presenting cells, CD4-positive T lymphocytes exhibit
unusual effector function for host defense, and they play a major
role in the regulation of adaptive immunity (1–3).
Conversely, they are also fundamental regulators of autoimmunity
when tolerance is lost. Several cytokines are then key mediators in
the development of T lymphocytes playing crucial roles in
controlling immunity (4,5). Naive T cells differentiate into
different functional subsets. These include classical T helper
cells, Th1 and Th2, which regulate immunity against intracellular
and extracellular pathogens, respectively. Th1 and Th2 lineages
also generate either cellular or humoral immune responses. Th1
cells produce interferon-γ (IFNγ), and Th2 cells express the
cytokines IL-4, −5 and −13 (6,7).
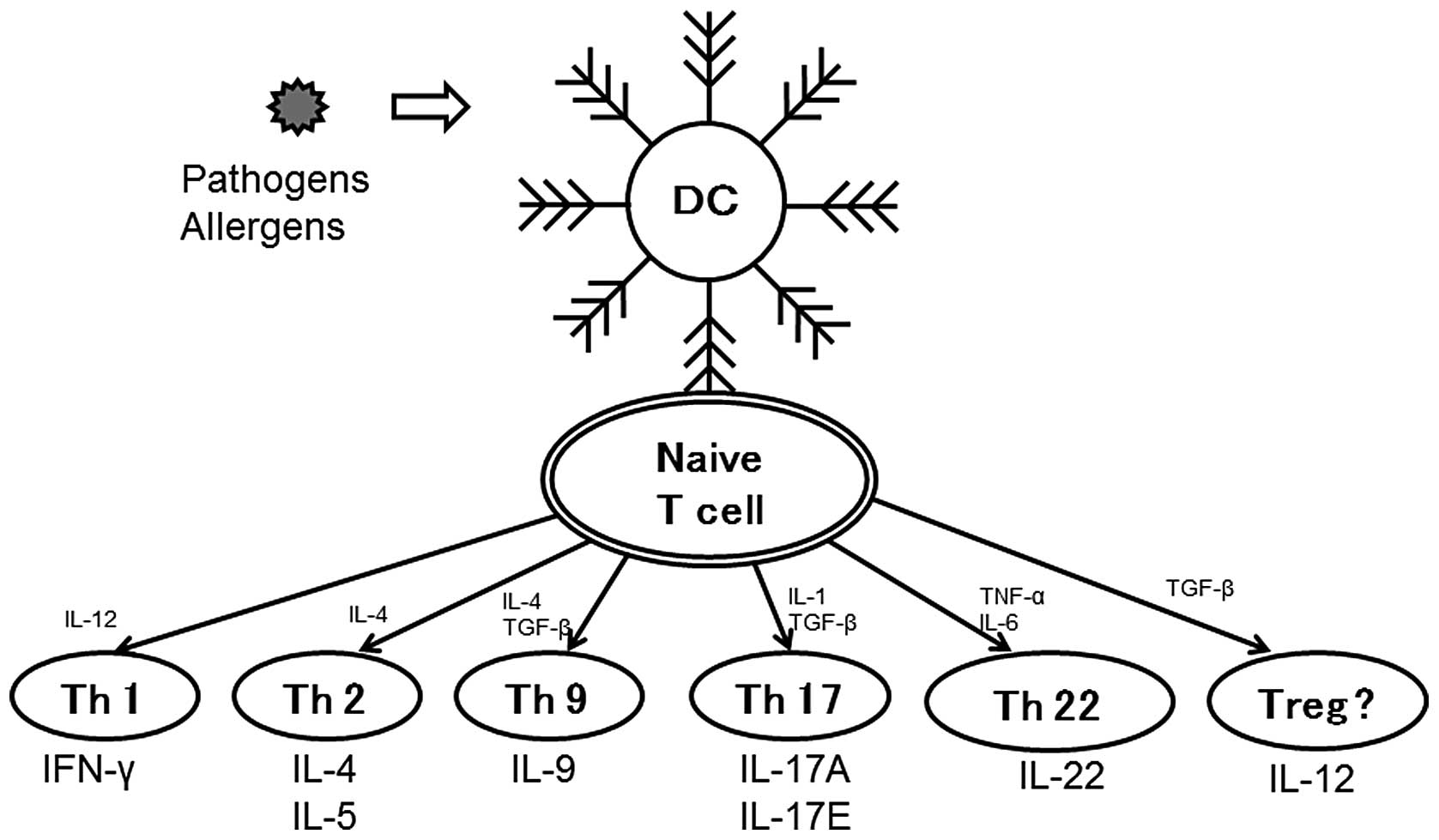
In addition, several effector T (Treg) cell subsets have been
identified. They include regulatory T cells, IL-17-producing Th17
cells, IL-9-producing Th9 cells and a subset of IL-22-producing
Th22 cells (8,9), which all determine the development
of different types of T cell immunity (Fig. 1). Proper regulation of Th
differentiation is critical for controlling immune responses and
for maintaining immunological homeostasis.
T cell lineage maturation is governed by the
expression of master regulator transcription factors that drive
differentiation. Actually, Th cell differentiation is regulated by
several distinct cytokines, which signal through ubiquitous
transcription factors including the STAT family (10,11). These factors upregulate the
expression of lineage-specific transcription factors, which
function not only to promote its own lineage differentiation but
also to inhibit alternative differentiation pathways. In addition,
epigenetic mechanisms are also important for regulating appropriate
gene expression in T cell differentiation (12–15). There are extensive
cross-regulations of lineage-determining transcription factors. In
addition, Th cell lineage commitment can be plastic in certain
circumstances. The T cell plasticity and lineage fate may also be
governed by cytokines and epigenetic regulations. There are many
examples of plasticity in Th cell subsets (16,17). Autoimmune diseases can be driven
by Th1, Th17 cells or their combinations. The inflammatory response
is supported by innate immune mechanisms that are relevant to
autoimmunity. Recent evidence on T cell subset reciprocal
regulation (18) and
counterbalance between Th1 and Th2 cells to Th17 and Treg has
influenced the peripheral tolerance (19). Many autoimmune diseases are driven
by cruel cycles of specialized T cells that are unable to be
suppressed by regulatory T cells. Here we summarize and speculate
on the current knowledge regarding the regulation and signaling for
T cell subsets. The understanding of these insights into the
mechanisms of autoimmune regulation may lead to novel therapeutic
opportunities.
T cell subsets and antigen-presenting cells
regulated by various factors
On antigen stimulation, naive CD4-positive T cells
can differentiate into Th1 and Th2 effector cells, which rapidly
produce IFN-γ and IL-4, respectively (1–4).
The Th1 cell phenotype is dominated by IL-2, IFN-γ and tumor
necrosis factor cytokine profiles. Th1 cells are involved in
cell-mediated defense against intracellular microorganisms, and
they also engage in the effector mechanisms of allergic disease.
Th1 cells not only are themselves prone to activation and
apoptosis, they also induce apoptosis of keratinocytes in atopic
dermatitis and of epithelium and bronchial smooth muscle cells in
asthma. Th1 cells differentiate after stimulation with IL-12 and
IL-17 (20). IL-12, which is
produced by macrophages, dendritic cells and B cells, is a
regulatory cytokine that has an important function in initiation
and regulation. Th1 cells thus respond to the release of IFN-γ by
IL-12 and suppression of Th2 cytokines (21). Th1 cells in addition to natural
killer cells and macrophages are the primary Th1 cytokine-producing
cells. The Th1 cytokines are also involved in immunoglobulin class
switching to the IgG2a isotype (22).
Th2 cells produce the highest amount of Th2
cytokines in addition to mast cells and basophils. The Th2
cytokines such as IL-4, IL-5, IL-10 and IL-13 are associated with
humoral immunity and immunoglobulin class switching to IgG1 and IgE
(23). The Th2 cytokines are also
involved in controlling immune responses against extracellular
parasites. In addition, IL-4 and IL-5 are implicated in atopic and
allergic disease because of the role in regulating IgE-mediated
immune responses via mast cells and eosinophils (24). Th2 cells predominantly mediate IgE
responses. The differentiation of naive T cells into Th2 cells is
induced in the presence of IL-4 (25). Cross-linking of IgE on effector
mast cells results in the release of vasoactive amines such as
histamine, leukotriene, chemokine and cytokines such as IL-4, IL-5
and IL-13, leading to the development of type 1 immediate
hypersensitivity reaction (26).
The cytokine expression patterns in Th1 and Th2 cells are
controlled by transcriptional activation and repression via each
subset differentiation.
Th9 is a distinct population of effector T cells
involved in tissue inflammation. This subset is characterized by
IL-9 and IL-10 secretion (27).
The cells differentiate from naive cells after IL-4 and TGF-β
stimulation. Th17 cells have been shown to induce host protection
against extracellular pathogens. IL-9 together with TGF-β
contribute to Th17 cell differentiation, and Th17 cells themselves
produce IL-9. Th9 and Th17 cells control tissue inflammation
through upregulation of inflammatory cytokines and chemokines
(28). In addition,
differentiation of Th17 cells is induced by IL-6, IL-21 and IL-23
(29). Th17 cells are also
implicated in the pathogenesis of autoimmune diseases. Tregs
suppress Th17 cells and autoimmunity. IL-1 also plays a crucial
role in early Th17 cell differentiation. In immune responses
against infection and autoimmune disease models, Th1 and Th17 cells
often develop simultaneously (30). Perturbation of one pathway may
result in augmentation of the other. They are involved in host
defence against extracellular pathogens such as bacteria and fungi,
but also in the pathogenesis of various autoimmune diseases.
Another T cell subset (Th22) (31) has been demonstrated in T cells
that independently express IL-22 with low expression levels of
IL-17 and play a role in atopic dermatitis. IL-22 can be protective
for colitis by induction of epithelial healing and mucus
production.
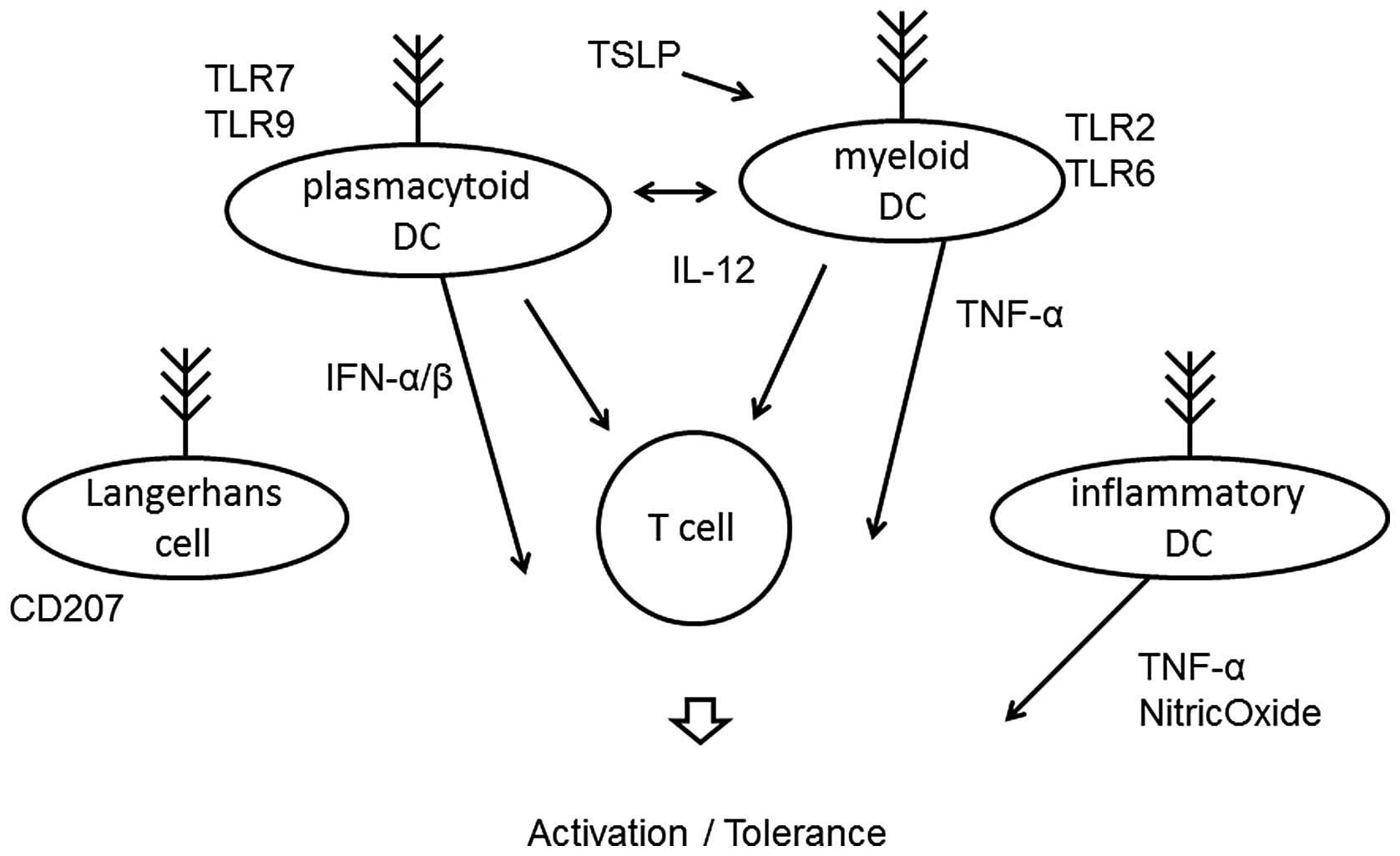
Dendritic cells (DCs) are a heterogeneous group of
antigen-presenting leukocytes (Fig.
2) that play an important role in the activation of both the
innate and acquired immune system. They are essential for the
differentiation of naive T cells via release of cytokines (32). Their role is to process antigens
and to migrate to local lymph nodes, where they present to
antigen-specific T cells. DCs loaded with allergen-derived peptides
reach the lymph nodes within 24 h, where they interact with naive
CD4-positive T cells to support the differentiation of Th1,
regulatory T cells (33) and Th2
cells within five days. These cells subsequently migrate into the
blood and back to mucosal tissues, resulting in allergen tolerance
or activation. Two distinct subsets of DCs have been identified.
Myeloid DCs express Toll-like receptor (TLR)2 and TLR6 and produce
IL-12 in response to bacterial and viral stimuli (34). Plasmacytoid DCs express TLR7 and
TLR9 (35), and release
interferon during the outcome of responses. Plasmacytoid DCs
directly suppress the ability of myeloid DCs to generate effector T
cells, and they are capable of stimulating the development of
regulatory T cells. The depletion of plasmacytoid DCs results in
lack of tolerance to certain antigens. In addition, the other two
DC populations that are present at inflammatory sites of the skin
are the classical Langerhans cells and the inflammatory dendritic
epidermal cells (36,37). The Langerhans cells are the
predominant DC population in the epidermis and are the first line
of defense against antigens. The inflammatory dendritic cells
activate Th1 subsets, whereas classical Langerhans cells induce Th2
subsets. Thymic stromal lymphopoietin seems to play an essential
role in allergic inflammation and activates myeloid DCs to induce
inflammatory Th2 responses.
Signal transductions in T cell subsets
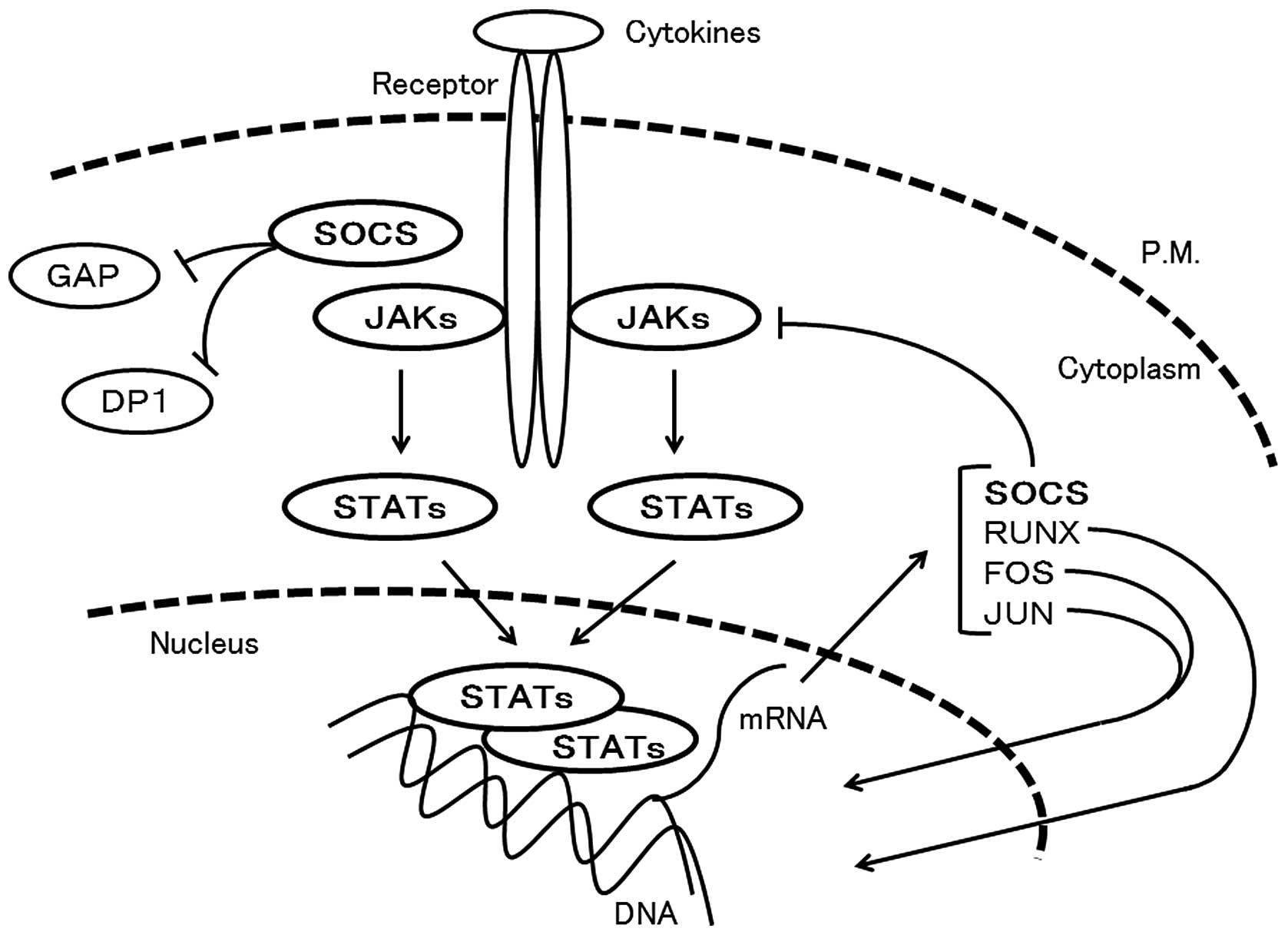
Cytokines play important roles in controlling
adaptive immunity. Upon cytokine-induced activation, Janus kinases
(JAKs) phosphorylate the cytoplasmic tail of the receptor, leading
to the recruitment of signal transducer and activator of
transcriptions (STATs), which also are phosphorylated by JAKs
(38) (Fig. 3). The JAKs play a critical role in
mediating immune responses, and their modulation represents a novel
approach to the therapies of inflammatory immune-mediated diseases.
Deficiency of JAK1 leads to nonresponsiveness to interferons (IFNs)
and cytokines, whereas JAK2-deficient cells fail to respond to
hormone-like cytokines such as erythropoietin, thrombopoietin and
GM-CSF. TYK2, which is one of the JAK family kinases (39), transmits the signals derived from
IFNs and the IL-12 receptor subunit, whereas JAK3 has a discrete
function (40) and is associated
only with the IL-2 receptor.
Activated STAT family proteins by JAKs dimerize and
translocate to the nucleus. They regulate the expression of a
multitude of genes including RUNX and SOCS. The STAT family
proteins have pivotal roles in transmitting cytokine-mediated
signals and specifying T cell differentiation. The STAT1 and STAT2
proteins have been discovered as a mediator of IFN action. STAT3
regulates the expression of Th17 cell-related cytokines and
transcription factors. For example, IL-21 is produced by Th17 cells
in a STAT3-dependent manner (41). Identified STAT3 target genes in T
cells include anti-apoptotic genes such as Bcl2, Fos and Jun. STAT3
is also activated throughout Th2 cell differentiation, and is
required for Th2 cytokine production and transcription factor
expression, and is required for Th2 cell-mediated allergic
inflammation. STAT4 is activated mainly by IL-12, IL-23 and IFNs,
and it predominantly functions in promoting Th1 cell
differentiation (42). STAT4 is
also the major regulator of IFN and IL-21 gene expression. STAT5
plays roles in Th2 cell differentiation by upregulating expression
of the IL-4 receptor (43). STAT5
competes with STAT3 for binding to IL-17 and inhibits the function
of STAT3 in activating IL-17 transcription, and consequently
inhibits Th17 cell differentiation. STAT6 mediates the expression
of the IL-4 regulated genes, and induces Th2 cell differentiation
(44). Another key role of STAT
family proteins includes shaping epigenetic patterns on target gene
loci to maintain cell lineage specificity.
RUNX transcription factors have a central role in
regulating Th cell differentiation. RUNX1, which is the direct
target of STAT6, inhibits Th2 cell differentiation by
downregulating GATA3 expression and binds to the IL4 silencer
region (45). In addition, RUNX1
forms a complex with FOXP3 and RORC, which is necessary for Treg
and Th17 cell function, respectively. Overexpression of RUNX1 is
sufficient to accelerate the effects of IL-17A production in Th17
cells. The RUNX3 transcription factor augments Th1 and
downmodulates Th2 phenotypes by interacting with GATA3. RUNX3 is
upregulated in CD4-positive T cells during Th1 cell differentiation
(46), and RUNX 3 functions with
the T-box family transcription factor, T-bet, which is a master
regulator of Th1 cell differentiation. BATF, which is also directly
regulated by STAT6, regulates both Th17 and Th2 cell
differentiation (47). STAT6
functions as a transcriptional activator, but it also functions as
a functional repressor for certain genes.
The suppressor of cytokine signaling (SOCS) family
of proteins is also a key regulator of cytokine responses, and
downregulates specific cytokine signals and consequently modifies
the immune response. SOCS1 and SOCS3 have been shown to affect the
Th1 and Th2 balance (48). SOCS2
expression regulates IL-2 and IL-3 signals (49), and plays an important role in
regulating Th2 cell expansion and development of type 2 allergic
responses. SOCS3 expression correlates with the severity of asthma
as well as serum IgE levels in patients with allergy (50). It is plausible that SOCS proteins
play a significant role in Th cell polarization. Constitutive
expression of SOCS3 facilitates Th2 expansion, whereas selective
deletion of SOCS3 facilitates STAT3 activation and elevated IL-17
production in T cells. SOCS3 may also play a role in controlling
Treg cell responses.
Genetic changes after T cell
differentiation
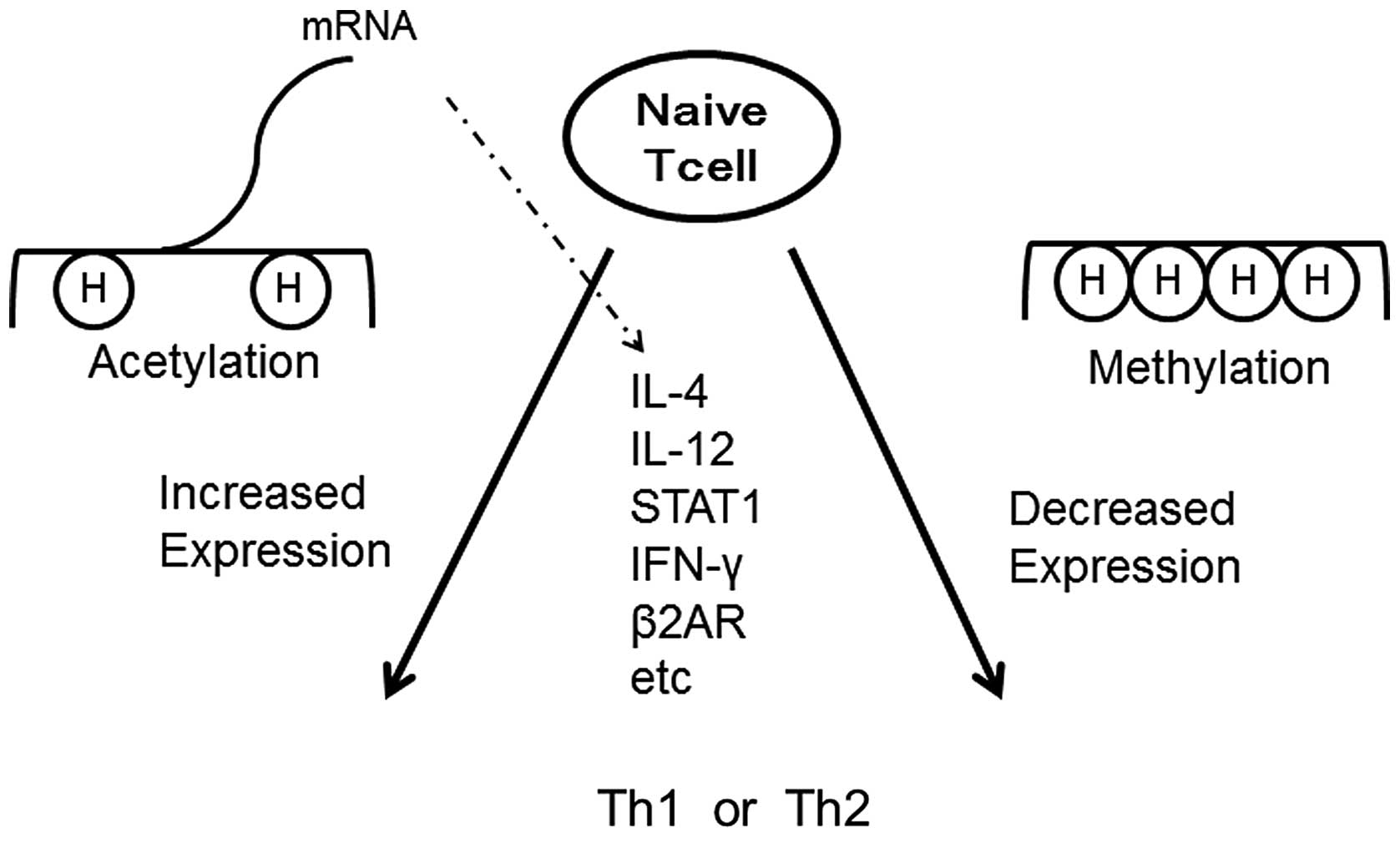
Gene silencing mediated by epigenetic mechanisms is
important for regulating proper gene expression in cell
differentiation. Histone modifications of genes encoding Th cell
subsets are of particular importance. In general, chromatin
modifications control the accessibility of transcriptional
activators and repressors (Fig.
4). Permissive marks on a particular cytokine gene are present
in the relevant lineage that expresses that cytokine (51). Conversely, repressive marks are
also present in other lineages that do not express the cytokine.
The presence of the DNase I hypersensitive site clearly suggests
functionally defined chromatin structures. For example, deficiency
of DNase I hypersensitive sites in the IL-4 gene decreases IL-4
production, as IL-4 is expressed in Th1 cells. Similarly, several
Th2-specific DNase I hypersensitive sites around IL-4 and IL-13
promoters in Th2 cells are found after differentiation. DNAs are
predominantly methylated in naive T cells. After each T cell
differentiation, CpG demethylation coincides at consensus GATA
binding sites and DNase I hypersensitive sites appear (52). It seems that CpG methylation in
certain genes could be a mechanism of suppression in permissive
lineages.
The chromatin remodeling that is associated with
both cytokine gene expression and repression in Th1 and Th2 cells
is promoted by the activation of the transcription factor GATA3 by
IL-4, STAT-4 by IL-12 and T-bet by IFN-γ. Chromatin structures in
the locus differ among naive, Th1 and Th2 cells. In these cells,
several GATA-3 consensus binding sites are present at DNase I
hypersensitive sites (53). The
GATA family members are associated with CREB-binding protein, which
is an acetyltransferase that acetylates not only histones but also
GATA proteins. As mentioned above, epigenetic mechanisms are
activated to promote the gene expression of cytokines resulting in
Th1 and Th2 cells in naive T cell differentiation. Similar changes
occur in the β2-adrenergic receptor (β2AR) promoter during
differentiation (54). It has
been shown that naive T cells and Th1 cell clones express the β2AR
(55), while Th2 cell clones do
not. Increased β2AR gene expression in Th1 cells is mediated by an
increase acetylation in histone 3 and histone 4. Genomic bisulfite
sequencing shows that the level of methylated CpG dinucleotides
within the promoter of the β2AR gene is increased in Th2 cells as
compared to Th1 cells. In contrast, Th1 cells show an increase in
pan acetylation and slight DNA methylation when compared to Th2
cells. β2AR gene expression is regulated in T cells as they
differentiate which implies chromatin remodeling in the β2AR gene
promoter. Catecholamine-mediated signal pathways may play a
fundamental role in acute stress-mediated immune alterations
(56).
Perspective
As T helper cells have emerged as an important
mediator of human immune diseases, many factors have been
identified as important in the Th cell differentiation process. How
these factors function individually and collectively requires
further elucidation. Another important issue involves the
plasticity of Th cells. What factors regulate and maintain this
plasticity require investigation. Furthermore, several other types
of chromatin modifications such as ribosylation and ubiquitination
may also occur as a mechanism controling expression of key genes.
Precise understanding of the regulation of T cell subsets and
development in this field will aid in the development of effective
therapies for immune diseases. It is possible that transcription
factors activated by cytokines may play roles in promoting
chromatin modifications that occur within the promoter of certain T
cell subsets. Understanding the epigenetic mechanisms may also lead
to the development of novel treatments for immune disease. Many
significant findings have emerged from this research field. It has
become clear that an important part of gene regulation depends on
epigenetic regulation. However, it is crucial to determine how STAT
proteins affect epigenetic mark functioning. Further epigenetic
analysis will provide evidence for both cytokine and regulator
genes in T cells. The extent to which T cell subsets behave as
flexible populations will be the focus of future research.
Acknowledgements
This study was supported by
grants-in-aid from the Ministry of Education, Culture, Sports,
Science and Technology in Japan and the Nara Women’s University
Intramural Grant for Project Research. In addition, this study was
supported in part by a grant from SHIN-EI Pharmaceutical Co.,
Ltd.
References
|
1.
|
VL CrotzerJS BlumAutophagy and adaptive
immunityImmunology1319172010
|
|
2.
|
A CorthayKB LorvikB BogenIs secretion of
tumour-specific antigen important for cancer eradication by CD4(+)
T cells? - Implications for cancer immunotherapy by adoptive T cell
transferScand J Immunol73527530201121388431
|
|
3.
|
O BoymanJ SprentThe role of interleukin-2
during homeostasis and activation of the immune systemNat Rev
Immunol12180190201222343569
|
|
4.
|
A AgrawalA SridharanS PrakashH
AgrawalDendritic cells and aging: consequences for
autoimmunityExpert Rev Clin
Immunol87380201210.1586/eci.11.7722149342
|
|
5.
|
SC JuvetL ZhangDouble negative regulatory
T cells in transplantation and autoimmunity: recent progress and
future directionsJ Mol Cell
Biol44858201210.1093/jmcb/mjr04322294241
|
|
6.
|
KA PackardMM KhanEffects of histamine on
Th1/Th2 cytokine balanceInt
Immunopharmacol3909920200310.1016/S1567-5769(02)00235-712810348
|
|
7.
|
W LiaoJX LinWJ LeonardIL-2 family
cytokines: new insights into the complex roles of IL-2 as a broad
regulator of T helper cell differentiationCurr Opin
Immunol23598604201110.1016/j.coi.2011.08.00321889323
|
|
8.
|
JA WisniewskiL BorishNovel cytokines and
cytokine-producing T cells in allergic disordersAllergy Asthma
Proc328394201110.2500/aap.2011.32.342821439160
|
|
9.
|
K HamzaouiTh17 cells in Behçet’s disease:
a new immunoregulatory axisClin Exp Rheumatol29Suppl
4S71S762011
|
|
10.
|
S RutzW OuyangRegulation of interleukin-10
and interleukin-22 expression in T helper cellsCurr Opin
Immunol23605612201110.1016/j.coi.2011.07.01821862302
|
|
11.
|
S CrottyRJ JohnstonSP
SchoenbergerEffectors and memories: Bcl-6 and Blimp-1 in T and B
lymphocyte differentiationNat
Immunol11114120201010.1038/ni.183720084069
|
|
12.
|
I TaniuchiW EllmeierTranscriptional and
epigenetic regulation of CD4/CD8 lineage choiceAdv
Immunol11071110201110.1016/B978-0-12-387663-8.00003-X21762816
|
|
13.
|
JP McAleerJK KollsMechanisms controlling
Th17 cytokine expression and host defenseJ Leukoc
Biol90263270201110.1189/jlb.021109921486905
|
|
14.
|
M GialitakisM SellarsDR LittmanThe
epigenetic landscape of lineage choice: lessons from the
heritability of CD4 and CD8 expressionCurr Top Microbiol
Immunol356165188201221989924
|
|
15.
|
T AkimovaUH BeierY LiuL WangWW
HancockHistone/protein deacetylases and T-cell immune
responsesBlood11924432451201210.1182/blood-2011-10-29200322246031
|
|
16.
|
K HiraharaG VahediK GhoreschiHelper T-cell
differentiation and plasticity: insights from
epigeneticsImmunology134235245201110.1111/j.1365-2567.2011.03483.x21977994
|
|
17.
|
TL GeigerS TauroNature and nurture in
Foxp3(+) regulatory T cell development, stability, and functionHum
Immunol73232239201222240298
|
|
18.
|
IV UstyugovaL ZhiMX WuReciprocal
regulation of the survival and apoptosis of Th17 and Th1 cells in
the colonInflamm Bowel Dis18333343201210.1002/ibd.2177221618360
|
|
19.
|
MR MaceyJL SturgillJK MoralesIL-4 and
TGF-beta 1 counterbalance one another while regulating mast cell
homeostasisJ
Immunol8446884695201010.4049/jimmunol.090347720304823
|
|
20.
|
L RovedattiT KudoP BiancheriDifferential
regulation of interleukin 17 and interferon gamma production in
inflammatory bowel
diseaseGut5816291636200910.1136/gut.2009.18217019740775
|
|
21.
|
A NeunkirchnerVM Leb-ReichlKG
SchmettererHuman TCR transgenic Bet v 1-specific Th1 cells suppress
the effector function of Bet v 1-specific Th2 cellsJ
Immunol18740774087201110.4049/jimmunol.100322021908735
|
|
22.
|
E MohrAF CunninghamKM ToellnerIFN-{gamma}
produced by CD8 T cells induces T-bet-dependent and -independent
class switching in B cells in responses to alum-precipitated
protein vaccineProc Natl Acad Sci USA10717292172972010
|
|
23.
|
RI NurievaY ChungUnderstanding the
development and function of T follicular helper cellsCell Mol
Immunol7190197201010.1038/cmi.2010.2420383172
|
|
24.
|
C PrussinY YinB UpadhyayaT(H)2
heterogeneity: Does function follow form?J Allergy Clin
Immunol12610941098201010.1016/j.jaci.2010.08.03120951419
|
|
25.
|
B MinMA BrownG LegrosUnderstanding the
roles of basophils: breaking
dawnImmunology135192197201210.1111/j.1365-2567.2011.03530.x22044049
|
|
26.
|
J SwedenborgMI MäyränpääPT KovanenMast
cells: important players in the orchestrated pathogenesis of
abdominal aortic aneurysmsArterioscler Thromb Vasc
Biol31734740201110.1161/ATVBAHA.110.21315721205988
|
|
27.
|
C TanI GeryThe unique features of Th9
cells and their productsCrit Rev
Immunol32110201210.1615/CritRevImmunol.v32.i1.1022428852
|
|
28.
|
M AkdisThe cellular orchestra in skin
allergy; are differences to lung and nose relevant?Curr Opin
Allergy Clin
Immunol10443451201010.1097/ACI.0b013e32833d7d4820736733
|
|
29.
|
T KornE BettelliM OukkaVK KuchrooIL-17 and
Th17 cellsAnnu Rev
Immunol27485517200910.1146/annurev.immunol.021908.13271019132915
|
|
30.
|
KH MillsTLR-dependent T cell activation in
autoimmunityNat Rev Immunol11807822201122094985
|
|
31.
|
N ZhangHF PanDQ YeTh22 in inflammatory and
autoimmune disease: prospects for therapeutic interventionMol Cell
Biochem3534146201110.1007/s11010-011-0772-y21384158
|
|
32.
|
K PaluckaJ BanchereauCancer immunotherapy
via dendritic cellsNat Rev
Cancer12265277201210.1038/nrc325822437871
|
|
33.
|
M BragaC QuecchiaE CavallucciT regulatory
cells in allergyInt J Immunopathol Pharmacol24Suppl 1S55S642011
|
|
34.
|
R SilvestreAM SilvaA Cordeiro-da-SilvaA
OuaissiThe contribution of Toll-like receptor 2 to the innate
recognition of a Leishmania infantum silent information
regulator 2
proteinImmunology128484499200910.1111/j.1365-2567.2009.03132.x19930041
|
|
35.
|
E EsashiM BaoYH WangW CaoYJ LiuPACSIN1
regulates the TLR7/9-mediated type I interferon response in
plasmacytoid dendritic cellsEur J
Immunol42573579201210.1002/eji.20114204522488361
|
|
36.
|
A WollenbergE KleinCurrent aspects of
innate and adaptive immunity in atopic dermatitisClin Rev Allergy
Immunol333544200710.1007/s12016-007-0032-918094945
|
|
37.
|
K SchäkelA HänselNews from dendritic cells
in atopic dermatitisCurr Opin Allergy Clin
Immunol11445450201121841470
|
|
38.
|
CA KnospJA JohnstonRegulation of
CD4+ T-cell polarization by suppressor of cytokine
signalling proteinsImmunology1351011112012
|
|
39.
|
J BustamanteS Boisson-DupuisE
JouanguyNovel primary immunodeficiencies revealed by the
investigation of paediatric infectious diseasesCurr Opin
Immunol203948200810.1016/j.coi.2007.10.00518083507
|
|
40.
|
Z XiongA MaH ChenJAK3 inhibitors in organ
transplantation and autoimmune diseaseRecent Pat Inflamm Allergy
Drug Discov47581201010.2174/18722131078989557719832695
|
|
41.
|
R SpolskiWJ LeonardInterleukin-21: basic
biology and implications for cancer and autoimmunityAnnu Rev
Immunol265779200810.1146/annurev.immunol.26.021607.09031617953510
|
|
42.
|
BD KormanDL KastnerPK GregersenEF
RemmersSTAT4: genetics, mechanisms, and implications for
autoimmunityCurr Allergy Asthma
Rep8398403200810.1007/s11882-008-0077-818682104
|
|
43.
|
L FainboimL ArruvitoMechanisms involved in
the expansion of Tregs during pregnancy: role of IL-2/STAT5
signallingJ Reprod
Immunol889398201110.1016/j.jri.2010.12.00721329987
|
|
44.
|
S GoenkaMH KaplanTranscriptional
regulation by STAT6Immunol
Res508796201110.1007/s12026-011-8205-2
|
|
45.
|
WF WongK KohuT ChibaT SatoM
SatakeInterplay of transcription factors in T-cell differentiation
and function: the role of
RunxImmunology132157164201110.1111/j.1365-2567.2010.03381.x21091910
|
|
46.
|
R YagiJ ZhuWE PaulAn updated view on
transcription factor GATA3-mediated regulation of Th1 and Th2 cell
differentiationInt
Immunol23415420201110.1093/intimm/dxr02921632975
|
|
47.
|
K HiraharaK GhoreschiA LaurenceSignal
transduction pathways and transcriptional regulation in Th17 cell
differentiationCytokine Growth Factor
Rev21425434201010.1016/j.cytogfr.2010.10.00621084214
|
|
48.
|
ID DimitriouL ClemenzaAJ ScotterPutting
out the fire: coordinated suppression of the innate and adaptive
immune systems by SOCS1 and SOCS3 proteinsImmunol
Rev224265283200810.1111/j.1600-065X.2008.00659.x18759933
|
|
49.
|
GM TannahillJ ElliottAC BarrySOCS2 can
enhance interleukin-2 (IL-2) and IL-3 signaling by accelerating
SOCS3 degradationMol Cell
Biol2591159126200510.1128/MCB.25.20.9115-9126.200516199887
|
|
50.
|
M KuboH InoueSuppressor of cytokine
signaling 3 (SOCS3) in Th2 cells evokes Th2 cytokines, IgE, and
eosinophiliaCurr Allergy Asthma
Rep63239200610.1007/s11882-006-0007-616476192
|
|
51.
|
PN CockerillMechanisms of transcriptional
regulation of the human IL-3/GM-CSF locus by inducible
tissue-specific promoters and enhancersCrit Rev
Immunol24385408200410.1615/CritRevImmunol.v24.i6.1015777160
|
|
52.
|
S KanajiT KanajiB JacquelinThrombopoietin
initiates demethylation-based transcription of GP6 during
megakaryocyte
differentiationBlood10538883892200510.1182/blood-2004-08-3109
|
|
53.
|
S Klein-HesslingT BoppMK JhaCyclic
AMP-induced chromatin changes support the NFATc-mediated
recruitment of GATA-3 to the interleukin 5 promoterJ Biol
Chem2833103031037200810.1074/jbc.M80592920018772129
|
|
54.
|
MJ LozaS FosterSP PetersRB
PennBeta-agonists modulate T-cell functions via direct actions on
type 1 and type 2
cellsBlood10720522060200610.1182/blood-2005-08-326516278302
|
|
55.
|
VM SandersRA BakerDS
Ramer-QuinnDifferential expression of the beta2-adrenergic receptor
by Th1 and Th2 clones: implications for cytokine production and B
cell helpJ Immunol1584200421019979126981
|
|
56.
|
L XiangKS Del BenKE RehmGD Marshall
JrEffects of acute stress-induced immunomodulation on TH1/TH2
cytokine and catecholamine receptor expression in human peripheral
blood
cellsNeuropsychobiology651219201210.1159/00032816022094268
|