Introduction
The dimorphic fungus Sporothrix schenckii
(S. schenckii) is the etiological agent of sporotrichosis,
an important cutaneous mycosis with a worldwide distribution
(1). S. schenckii grows at
room temperature (25°C) as a mold phase, while in vitro
incubation of mold cultures at body temperature (37°C) results in
the production of yeast cells (2). These in vitro forms are
virtually identical to the yeast cells of S. schenckii found
in diseased tissue. Therefore, the formation of yeast cells was
thought to be a requisite for the pathogenicity of S.
schenckii. The mechanisms that regulate the dimorphic switch,
however, remain unclear.
The mitogen-activated protein kinase (MAPK) cascade
and cyclic AMP (cAMP) signaling pathways are known to be involved
in fungal morphogenesis and pathogenic development. However, the
MAPK and cAMP pathways are both activated by an upstream branch,
two-component histidine kinase phospho-relay system. Nemecek et
al (3) recently uncovered a
long-sought regulator that controls the switch from a
non-pathogenic mold form to a pathogenic yeast form in dimorphic
fungi. They found that DRK1, a hybrid dimorphism-regulating
histidine kinase, functions as a global regulator of dimorphism and
virulence in Blastomyces dermatitidis (B. dermatitidis) and
Histoplasma capsulatum (H. capsulatum). DRK1 is
required for phase transition from mold to yeast, expression of
virulence genes, and pathogenicity in vivo. Disruption of
DRK1 locks B. dermatitidis in the mold form at temperatures
(37°C) that normally trigger phase transition to yeast. RNA
silencing of DRK1 expression in B. dermatitidis results in
impaired BAD1 expression, severe alterations in the cell wall, and
reduction in transcription of α-(1,3)-glucan synthase and the yeast-phase
specific gene BYS1. In H. capsulatum, DRK1 also regulates
expression of the yeast-phase specific genes CBP1, AGS1 and yps-3.
We previously reported differentially expressed genes between the
mycelial and the yeast phases of S. schenckii using 2DE. The
expressed sequence tag of spotC homologous to the DRK1 histidine
kinase from B. dermatitidis clearly increases in the yeast
form of S. schenckii (4).
We describe the molecular cloning of the DRK1 gene
from the yeast-form S. schenckii, designated SsDRK1.
We performed necessary function analysis of the SsDRK1 gene
as well as detection of the differential gene expression in the
dimorphic switch of S. schenckii. These findings establish
the primary foundation of understanding the function of
SsDRK1. The cloning and characterization of the DRK1 gene in
S. schenckii is reported for the first time.
Materials and methods
Fungal strain, media and growth
conditions
The strain of S. schenckii used, ATCC10268,
was maintained at the Research Center for Pathogenic Fungi, Dalian
Medical University, China. To obtain a mycelial culture, the
ATCC10268 isolate was inoculated on Sabouraud dextrose agar (SDA)
medium and incubated at 25°C. The mycelial colonies thus obtained
were inoculated in Sabouraud’s fluid medium and cultured with
shaking at 100 rpm at 25°C for 72 h. To achieve the switch of S.
schenckii from the mycelial to the yeast phase, mycelial
colonies were transferred to brain heart infusion (BHI) liquid
medium at 37°C and shaken at 100 rpm for 96 h. Mycelial and yeast
pellets were collected by centrifugation and stored at −80°C
immediately, or processed for total RNA isolation directly.
Total RNA, genomic DNA isolation and gene
cloning
Approximately 100 mg samples of S. schenckii
mycelia and yeast were separately pulverized under liquid nitrogen
with a mortar and pestle. Total RNA isolation was carried out
according to the manufacturer’s protocol using the Trizol Reagent
kit (Invitrogen, Carlsbad, CA, USA) and treated with the RNase-free
DNase I kit from Takara Bio, Inc. (Tokyo, Japan) to eliminate DNA
contamination. Genomic DNA was isolated from yeast phase colonies
following the manufacturer’s protocol using the InstaGene™ Matrix
kit (Bio-Rad, Hercules, CA, USA). cDNA was synthesized from 500 μg
of total RNA of ATCC10268 by murine leukemia virus reverse
transcriptase (MLV-RT) (Takara Bio, Inc.) primed with oligo(dT)
following the manufacturer’s instructions, and used as template for
PCR. Degenerate primers, SsDRK1-F1 and SsDRK1-R1,
were designed based on multiple alignments of the high conserved
DRK1 domains of Coccidioides immitis (C. immitis)
(EAS33695.2), Paracoccidioides brasiliensis (P.
brasiliensis) (EEH34763.1), B. dermatitidis (EGE84246.1)
and H. capsulatus H88 (EGC45940.1) amino acid sequences. PCR
product of expected size was cloned into pMD18 vector (Takara Bio,
Inc.) and sequenced. The degenerate primers yielded two fragments,
with the length of 161 and 160 bp, respectively. Primers HBB-F and
HBB-R were designed to amplify the cDNA sequence between the above
two fragments. To obtain the full-length cDNA sequence of the
SsDRK1 gene, 5′-RACE and 3′-RACE were performed with 5′-Full
RACE kit and 3′-Full RACE Core Set Ver.2.0 kit (Takara Bio, Inc.)
according to the manufacturer′s instructions. Nest-PCR was
performed. Briefly, five specific primers CTE869-F and CTE869-R of
3′-RACE and R132-1, R132-2 and R132-3 of 5′-RACE were synthesized
based on the cDNA sequence obtained by the degenerate primers. PCR
products of 5′- and 3′-RACE were both cloned into pMD18 vector
(Takara Bio, Inc.) and sequenced.
To determine the nucleotide sequence of the genomic
DNA corresponding to the SsDRK1, PCR was performed using the
primers SsDRK1-P1 and SsDRK1-B3 and genomic DNA as
template. The PCR products were then sequenced. The sequences of
all the primers used in this study are listed in Table I.
 | Table ISequence of primers in this study. |
Table I
Sequence of primers in this study.
| Primer | Sequence (5′-3′) |
|---|
| DRK1-F1 |
ACNGANAAYGTVAAYACYATGGC |
| DRK1-R1 |
CGRTCMACCATRTBRTTGATNGT |
| HBB-F |
TCACCAAAAAGATTGAGCGTCC |
| HBB-R |
TGTCACCGTTGGCGATGGCTT |
| CTE869-F |
GGCAACGCCATCAAGTTCACC |
| CTE869-R |
GCTCGCGCTCACGGTTTTTTTCGAGC |
| R132-1 (GSP1) |
ATTCCCTTCACGCCCT |
| R132-2 (GSP2) |
TGGTTTGTTGCAGTTGCAGGAT |
| R132-3 (GSP3) |
TGAGATCACCGAACGCGACAGC |
| P1 |
ATGACCGTTGTACCGACGAC |
| B3 |
ATGTGAGGGCCTCTCTTAGC |
| 8F |
GAATCTGCACGGTATTCTGA |
| 58R |
CTCAACCTCCACATCCTCAA |
| 24T |
FAM-CGTCGAGTCTGGTTACTAC-TAMRA |
Bioinformatics and phylogenetic analysis
of SsDRK1
Nucleotide sequences and deduced amino acid
sequences of the cloned SsDRK1 gene were analyzed. The
nucleotide sequences were analyzed using Sequencer software
(Sequencer, USA) and BLAST Network service of the National Center
for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/blast). The open reading
frame (ORF) was found by the ORF finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). For the
exact localization of the exon/intron boundaries the
mRNA-to-genomic alignment program Spidey (http://www.ncbi.nlm.nih.gov/IEB/Research/Ostell/Spidey/index.html)
was used. The deduced amino acid sequence was analyzed with the
Expert Protein Analysis System (http://www.expasy.org/) and the protein domain
features of SsDRK1 were determined by using Simple Modular
Architecture Research Tool (http://hits.isb-sib.ch/cgi-bin/PFSCAN). Isoelectric
point and molecular weight prediction were carried out at
(http://cn.expasy.org/tools/pi_tool.html). Multiple
alignments of SsDRK1 were performed with the ClustalW
Multiple Alignment Program (http://www.ebi.ac.uk/clustalw/).
Differential expression of SsDRK1 in two
stages during dimorphic switch
The expression of SsDRK1 transcript in
different stages (mycelial, yeast) was measured by real-time
RT-PCR. Primers and a TaqMan probe for target genes were designed
with primer select in DNASTAR software (Lasergene) and are listed
in Table I (24T, 8F, 58R). Fifty
nanograms of total RNA were assayed from two stages of S.
schenckii in triplicate using the PrimeScript RT-PCR kit
(Takara Bio, Inc.). The minus-reverse transcriptase control was
also performed in triplicate. The amplification conditions were
optimized for the ABI PRISM-7500 instrument (Applied Biosystems).
The cycling conditions using TaqMan probe detection were 95°C for 2
min followed by 40 cycles at 95°C for 10 sec, 61°C for 10 sec, 72°C
for 40 sec. 18srDNA was selected as the endogenous control.
Relative quantification of target gene expression was evaluated
using the comparative cycle threshold (CT) method as
previously described by Livak and Schmittgen (5). The ΔCT value was
determined by subtracting the target CT of each sample
from its respective 18srDNA CT value. Calculation of
ΔΔCT involved using the mycelial sample ΔCT
value as an arbitrary constant to subtract from yeast sample
ΔCT values. Differences in expression of target genes
were determined by 2−ΔΔCT. Data are expressed as
arithmetic means ± SD unless otherwise indicated. Comparison
between mycelial and yeast samples was performed using the
Student’s t-test. Differences with a P-value of <0.05 were
considered to be statistically significant.
Results
Cloning and genomic structure of
SsDRK1
First, 828 bp cDNA fragment which had a high
sequence similarity to the DRK1 of P. brasiliensis Pb01 was
obtained from the total RNA of ATCC10268. Following RACE PCR, a
full-length SsDRK1 cDNA 4743 including an ORF of 4071 bp,
encoding 1356 amino residues, was flanked by a 31 bp
5′-untranslated region (5′-UTR) and a 641 bp 3′-UTR. The most
probable CAAT box is located at -2, which is critical for
eukaryotic transcription initiation (6). As in other PKC genes, no TATA box
was identified within this sequence (7). Sequencing results showed that there
is a poly (A) tails in 3′-UTR. The SsDRK1 genomic DNA is
4065 bp in length. The aligned results revealed that there are no
introns between the sequences of the genomic DNA and the cDNA.
Based on the sequence of cDNA, the molecular weight of the
predicted amino acid is approximately 147.3 kDa, the theoretical pI
is 5.46. Suggested models for transmembrane topology indicated that
the amino acid sequence may be a soluble histidine kinase that
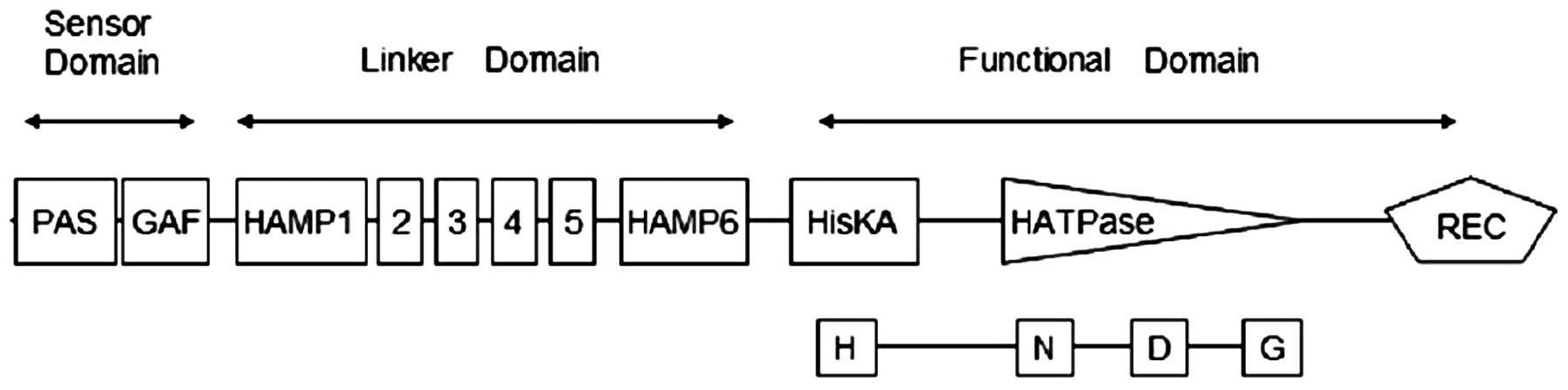
lacks transmembrane segments. Fig.
1 shows that SsDRK1 contains three parts: sensor domain,
linker domain and functional domain. The PAS and GAF domains, two
structural families of cytoplasmic sensor domains, are found at
positions 12–83 and 33–212 in the amino acid sequence. HAMP, which
is an approximately 50-amino acid α-helical region, begins at
position 231 in the linker domain part. It has been suggested that
the HAMP domain possesses a role of regulating the phosphorylation
of homodimeric receptors by transmitting the conformational changes
in periplasmic ligand-binding domains to cytoplasmic signaling
kinase domains. The functional domain of SsDRK1 is predicted
to have the necessary elements for histidine kinase function,
including the histidine-containing H-box and aspartate containing
D-box involved in phosphorelay. The sequence also contains the
N-and G-boxes used in ATP-binding and catalytic function, and an
aspartate-containing receiver domain (Fig. 4). SsDRK1 is homologous to
the hybrid histidine kinase SLN1 in Saccharomyces
cerevisiae (S. cerevisiae), DRK1 in B.
dermatitidis and to sequences in the genomes of H.
capsulatum and C. immitis, dimorphic fungi for which
extensive genome sequence is available.
Homology and phylogenetic analysis of
SsDRK1
Multi-alignment analysis by ClustalW indicated that
SsDRK1 has a high identity to DRK1 reported in other
species, sharing a similarity of 66% identity to P.
brasiliensis (EEH34763.1), 65% identity to B.
dermatitidis (EGE84246.1), 65% identity to C. immitis
(EAS33695.2), 67% identity to H. capsulatus (EGC45940.1)
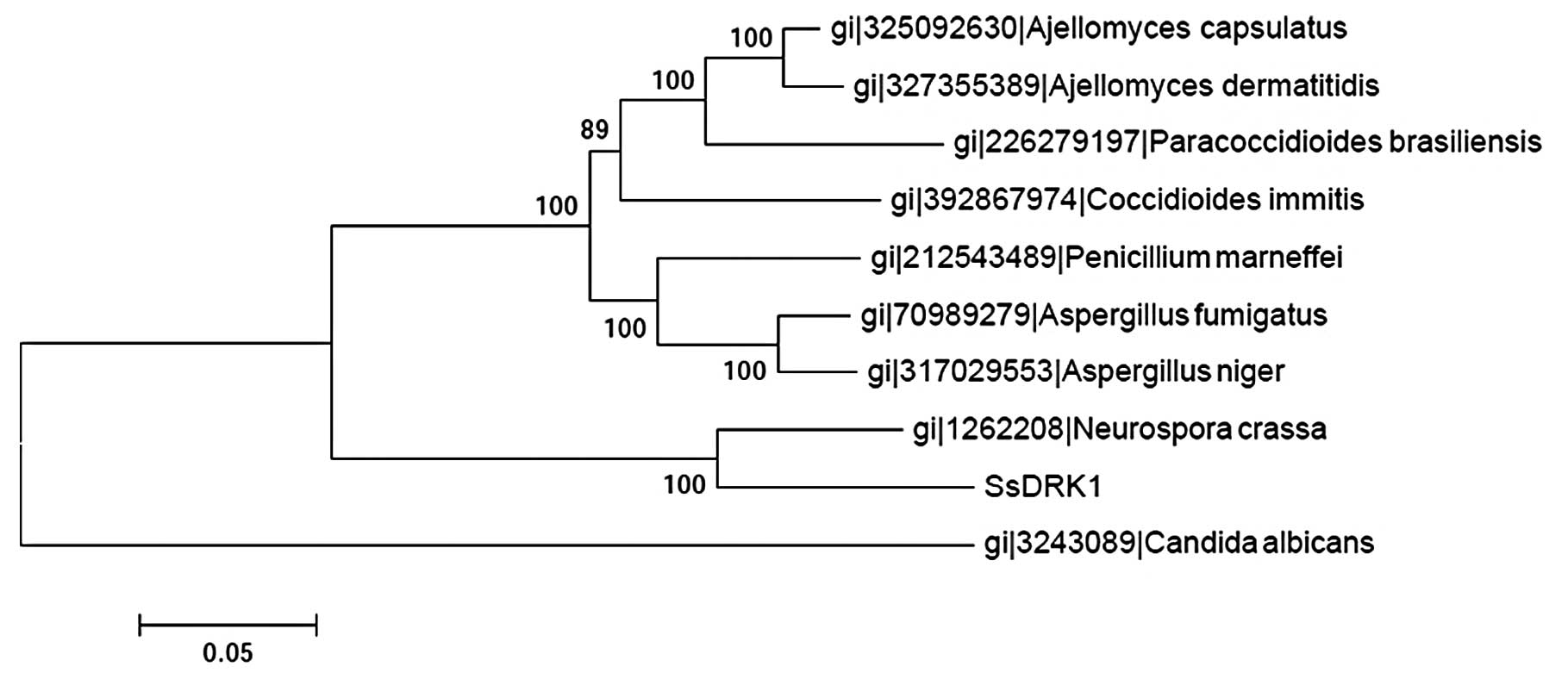
(Fig. 2). Based on the results of
the alignment of DRK1 sequences of the former and some common
fungi, the phylogenetic trees were constructed using the ClustalW
software (Fig. 3). Three groups
were clearly generated in the phylogenetic tree. The SsDRK1
identified in this study appeared most closely related to sequences
from Neurospora crassa (N. crassa), a member of the
ascomycetous class pyrenomycetes. The results also suggested that
the evolutionary relationship of SsDRK1 might be different
from that in Candida albicans (C. albicans).
Expression of SsDRK1 in two stages of
Sporothrix schenckii
The mRNA expression of SsDRK1 in different
stages was analyzed by real-time RT-PCR normalized against 18SrDNA
levels. Following amplification, Ct, ΔCt and ΔΔCt values were
calculated. Expression was determined as fold increased
2−ΔΔCt levels relative to the stage with lowest
expression (mycelia) set to 1. The SsDRK1 gene was expressed
in two stages of S. schenckii, with higher mRNA levels
observed in yeast (24.42-fold). There were significant differences
between the mycelial and the yeast form (Table II).
 | Table IIRelative abundance of differential
expression gene as determined by real-time RT/PCR (mean ± SD)
(P<0.01). |
Table II
Relative abundance of differential
expression gene as determined by real-time RT/PCR (mean ± SD)
(P<0.01).
| cDNA name | Phase | Target
CT | 18srDNA
CT | ΔCT |
ΔΔCT |
2−ΔΔCT |
|---|
| DRK1 histidine
kinase | Mycelial | 27.08±0.52 | 20.66±0.27 | 6.42±0.68 | 0 | 1 |
| Yeast | 23.90±0.26 | 22.09±0.64 | 1.81±0.87 | −4.61±0.23 | 24.42 |
Accession number
The full length of cDNA sequence and genomic DNA
sequence of the SsDRK1 gene were submitted to the GenBank
database under the accession number JX312331 and JX416706,
respectively.
Discussion
Histidine protein kinases (HPKs) are a large family
signal-transduction enzymes that autophosphorylate on a conserved
histidine residue. HPKs form two-component signaling systems
together with their downstream target proteins, the response
regulators, which have a conserved aspartate in a ‘receiver domain’
that is phosphorylated by the HPK. The dimorphism regulating kinase
DRK1 was recently proved to mediate the thermally induced
transition to the pathogenic yeast-phase program in both B.
dermatitidis and H. capsulatum (3). In this study, based on the conserved
structures of the DRK1 in four types of fungi cells, the degenerate
primers were designed to obtain the homologs of DRK1 in S.
schenckii. The production of PCR has a very high identity to
the DRK1 of P. brasiliensis Pb01. The ORF of SsDRK1
encoded protein was mostly similar in identity to the DRK1 of N.
crassa, similar with previous molecular phylogenetic analyses
both on a pertussis toxin-sensitive G protein α subunit (8) and three chitin synthase genes
(9). Aligned to the other fungal
DRK1, the identities were 64 to 74%. However, SsDRK1 shares
limited sequence similarity with histidine kinases that regulate
filamentation in the more distantly related fungus C.
albicans.
The amino sequence of SsDRK1 is predicted to
have the necessary elements for histidine kinase function including
H-box, D-box, N- and G-boxes. This indicates that the SsDRK1
has similar functions to other fungi histidine kinases. The typical
HPK is a transmembrane receptor with an aminoterminal extracellular
sensing domain and a carboxy-terminal cytosolic signaling domain;
however, a type of soluble histidine kinase that lacks
transmembrane segments was also identified. The cytoplasmic sensor
domain including GAF, PAS and PCD may reside N-terminal to the
C-terminal transmitter domain in the soluble histidine kinase
(10). SsDRK1 in the
present study was proved to be lacking transmembrane segments and
carrying GAF and PAS domains in the sensor part, which suggested
that SsDRK1 is a soluble histidine kinase.
Histidine kinase two component signaling systems
have recently been shown to play the role in environmental sensing
and all development in eukaryotes. In C. albicans, they
regulate filamentation whereas in B. dermatitidis and H.
capsulatum, they may control phase transition and virulence
gene expression as well as cell development and sporulation in the
other systemic dimorphic fungi. Does SsDRK1 have the same
functions during the process of dimorphic switch in S.
schenckii? In this study, the mRNA expression of SsDRK1
in yeast cells was higher than in mycelial cells, which suggested
that SsDRK1 is involved in regulating phase transition.
What is the environmental signal that SsDRK1
senses to regulate phase transition and virulence gene expression?
In S. cerevisiae (11),
histidine kinase Sln1p detects osmotic stress, whereas in
Schizosaccharomyces pombe (12), the histidine kinase-regulated SPC1
MAPK cascade senses osmotic, oxidative, heat stress and nutrient
deprivation. Potential signals for histidine kinase sensing in
dimorphic fungi include temperature, osmotic or oxidative stress,
nutrient deprivation, redox potential, and host-derived factors
including hormones such as 17-β-estradiol, which induces germ tubes
in C. albicans (13) and
block mold-to-yeast transition of P. brasiliensis (14). In this study, the mycelial cells
of S. schenckii switched to yeast cells when they were
incubated in BHI liquid medium at 37°C, which suggests
SsDRK1 can detect the change of temperature and nutrient
deprivation in the environment.
The detailed functions of the SsDRK1 and its
up- and down-stream proteins as well as their interactions require
further investigation. If the formation mechanism of the yeast
cells (the parasitic form) of S. schenckii is elucidated,
this may lead to a therapy strategy for sporotrichosis.
Acknowledgements
This study was partly supported by a grant from the
National Natural Science Foundation of China (grant no. 81000069).
The authors thank Zhang Zhenying and Yu Zhen for their assistance
with image and statistical analysis.
References
|
1
|
Travassos LR and Lloyd KO: Sporothrix
schenckii and related species of Ceratocystis. Microbiol Rev.
44:683–721. 1980.
|
|
2
|
Guarro J, Gené J and Stchigel AM:
Developments in fungal taxonomy. Clin Microbiol Rev. 12:454–500.
1999.
|
|
3
|
Nemecek JC, Wuthrich M and Klein BS:
Global control of dimorphism and virulence in fungi. Science.
312:583–588. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang ZY, Hou BB, Xin Y and Liu XM:
Protein profiling of the dimorphic pathogenic fungus, Sporothrix
schenckii. Mycopathologia. 173:1–11. 2012. View Article : Google Scholar
|
|
5
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Feng P, Xie Z, Sun J, Zhang J, Li X, Lu C
and Xi L: Molecular cloning, characterization and expression of
PmRsr1, a Ras-related gene from yeast form of Penicillium
marneffei. Mol Biol Rep. 37:3533–3540. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Aquino-Piñero E and Rodríguez-del Valle N:
Characterization of a protein kinase C gene in Sporothrix
schenckii and its expression during the yeast-to-mycelium
transition. Med Mycol. 40:185–199. 2002.PubMed/NCBI
|
|
8
|
Delgado N and Rodríguez-del VN: Presence
of a pertussis toxin-sensitive G protein alpha subunit in
Sporothrix schenckii. Med Mycol. 38:109–121. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chua SS, Momany M, Mendoza L and Szaniszlo
PJ: Identification of three chitin synthase genes in the dimorphic
fungal pathogen Sporothrix schenckii. Curr Microbiol.
29:151–156. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kimura S, Shiraiwa Y and Suzuki I:
Function of the N-terminal region of the phosphate-sensing
histidine kinase, SphS, in Synechocystis sp PCC 6803. Microbiology.
155:2256–2564. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Van Wuytswinkel O, Reiser V, Siderius M,
Kelders MC, Ammerer G, Ruis H and Mager WH: Response of
Saccharomyces cerevisiae to severe osmotic stress: evidence
for a novel activation mechanism of the HOG MAP kinase pathway. Mol
Microbiol. 37:382–397. 2000.
|
|
12
|
Pöhlmann J and Fleig U: Asp1, a conserved
1/3 inositol polyphosphate kinase, regulates the dimorphic switch
in Schizosaccharomyces pombe. Mol Cell Biol. 30:4535–4547.
2010.PubMed/NCBI
|
|
13
|
Cheng G, Yeater KM and Hoyer LL: Cellular
and molecular biology of Candida albicans estrogen response.
Eukaryot Cell. 5:180–191. 2006.
|
|
14
|
Salazar ME, Restrepo A and Stevens DA:
Inhibition by estrogens of conidium-to-yeast conversion in the
fungus Paracoccidioides brasiliensis. Infect Immun.
56:711–713. 1988.PubMed/NCBI
|