Introduction
The thymus is a pivotal central lymphoid organ that
has a unique role in the development of bone-marrow-derived
precursor cells into mature, functional T cells with a
self-restriction and self-tolerant mechanism. These functions of
the thymus are mediated by the thymic stroma which provides a
specialized microenvironment for the survival, proliferation,
differentiation, and maturation of immature T cells. In particular,
thymic epithelial cells (TECs), which form the major
sub-compartment of the stroma, provide the unique combination of
cellular interactions, cytokines, chemokines and various factors
including thymic hormones to induce thymocyte precursors to undergo
a differentiation program that leads to the generation of
functional T cells (1).
The thymus gland is most prominent during early
life. The thymus reaches its greatest relative weight at the time
of birth, although its absolute weight continues to increase until
the onset of puberty. Thereafter, the thymus begins to undergo a
physiological process of involution, whereby it progressively
decreases in size and the production of T cells progressively
declines during adult life. The thymus continues to play an
immunological role throughout life, although its function declines
with age. Furthermore, the gradual process of physiological
involution of the thymus can be acutely accelerated by acute
involution or accidental involution of the thymus due to various
stimuli including severe stress, ionizing radiation, adrenal and
sex steroids, adrenocorticotrophic hormones, toxins, diseases such
as tumors, and cytotoxic agents such as anti-neoplastic agents. In
an acute involution of the thymus, the massive death of thymocytes
can result in suppression of host immunity and increased
susceptibility to disease. Thus, it is critical to understand the
mechanisms of thymus regeneration and to thereby develop
therapeutic strategies for restoring or improving host immunity
when immune function is suppressed due to thymic involution.
We previously found the ultrastructural features on
the hyperfunctional activities, including the high synthetic
activity, in the TECs among various thymic cell types during rat
thymus regeneration, thereby providing the first direct evidence
that TECs, via production of several factors, are involved in the
process of proliferation, maturation and differentiation of the T
cell precursors during recovery from acute thymic involution
(2). These facts led us to
perform the present study for the identification of gene expression
profiles in the thymus during its regeneration. Although some
progress has been made in understanding the mechanisms of
TEC-mediated thymus regeneration, they remain poorly understood
(3). Most of the molecules
involved in the enhancement of thymus regeneration and T cell
reconstitution remain to be identified. Thus, identification of
genes expressed during thymus regeneration is an essential step in
understanding the mechanisms of thymus regeneration and in
discovering new thymotrophic factors that promote thymus
regeneration and T cell reconstitution.
A number of techniques, such as differential
display, subtractive hybridization, SAGE (serial analysis of gene
expression) and gene arrays, have been developed for the
identification of genes that are specifically expressed in a
particular cell, tissue, organ or organism, or are differentially
expressed under varying conditions such as development,
differentiation, activation, stimulation, environmental changes and
disease, over the last several decades. The expressed sequence tag
(EST) is short single-pass sequence reads of randomly selected
clones from cDNA libraries (4).
The EST approach is also an effective technology for characterizing
genes expressed during various biological processes since ESTs
provide a reliable method for gene discovery as well as a resource
for the large-scale analysis of gene expression of known and
unknown genes. A cDNA library is constructed from total RNA or
poly(A) RNA derived from specific tissues or cells, and represents
genes expressed in the original cellular population. Therefore,
ESTs have been shown to be a powerful tool in identifying
homologues of reported genes and gene expression in tissues or
cells (5,6). The discovery of novel or known genes
expressed in thymus regeneration and the determination of their
expression patterns in the thymus during regeneration will help
elucidate their roles in thymus regeneration and will provide the
basis for the development of effective therapeutic strategies for
promoting thymus regeneration. Thus, in this study, EST analysis
was performed following generation of the regenerating thymus cDNA
library and the expressed genes were compiled to identify genes
expressed during thymus regeneration.
Materials and methods
Experimental acute thymic involution and
regeneration model
Adult male specific pathogen-free Sprague-Dawley
rats were purchased from Dae Han Bio Link (Seoul, Korea). All rats
were housed 3–4/cage and maintained under a 12 h light/dark cycle
at 24°C in a specific pathogen-free and humidity-controlled
facility. Animals were provided with standard sterile food and
water ad libitum, and were allowed to adjust to their
environment for one week. Animals were used at 8–10 weeks of age,
by administering them with a single intraperitoneal dose of
cyclophosphamide (150 mg/kg body weight; Sigma, St. Louis, MO, USA)
in normal saline, and were sacrificed in groups of six at 3 and 7
days, for the construction of the cDNA library, or in groups of
four at 3, 7 and 14 days after injection, for RT-PCR analysis. Rats
given the same amount of normal saline were used as controls.
Animal care and all experimental procedures were conducted in
accordance with the ‘Guide for Animal Experiments’ edited by the
Korean Academy of Medical Sciences.
cDNA library construction and EST
sequencing
Total RNA was extracted from regenerating thymus 3
and 7 days after cyclophosphamide treatment using total RNA
isolation kit (CPG, Inc., Lincoln Park, NJ, USA), following the
manufacturer’s instructions. Poly(A) RNA was purified from the
total RNA using mRNA extraction kit (Stratagene, La Jolla, CA,
USA). Briefly, 5 mg of total RNA was mixed with 5 ml of elution
buffer and was then hybridized to 0.2 g of oligo(dT) cellulose at
room temperature with gentle agitation, followed by high-salt
(low-stringency) and low-salt (high-stringency) washes. These
washes removed unwanted components of the crude lysate, such as
proteins, carbohydrates, lipids, DNA, transfer RNA (tRNA) and a
significant amount of ribosomal RNA (rRNA) from poly(A) mRNA. The
oligo(dT) cellulose was loaded onto a push column, and mRNA was
eluted with 65°C elution buffer. The mRNA was concentrated by
ethanol precipitation. The mRNA concentration was determined by
absorbance at 260 nm.
Directionally cloned (EcoRI/XhoI) cDNA
libraries were generated from poly(A) RNA using a λ-Uni-Zap-XR cDNA
synthesis kit (Stratagene) and ZAP-cDNA Gigapack III Gold cloning
kit (Stratagene) according to the manufacturer’s instructions. For
all libraries, individual white colonies were inoculated in LB
broth containing ampicillin (100 μg/ml) and were incubated
for 12 h. Double-stranded plasmid DNA from individual cDNA clones
was isolated with a Qiagen plasmid DNA preparation kit (Qiagen,
Valencia, CA, USA). The titer of the unamplified library was
1.2×106 pfu/ml and average insert size was ∼0.8–2.5 kb.
Individual clones were randomly selected for plasmid DNA
purification and were sequenced by Macrogen Inc. (Seoul,
Korea).
Thymocytes and thymic stromal cell
isolation
Two to three thymi were dissected from freshly
sacrificed rats and trimmed of fat and connective tissue. Small
cuts (2–3 mm) were made into the capsules with a pair of razors,
and thymi were gently agitated in 30 ml of RPMI-1640 using a
magnetic stirrer at 4°C for 40 min. The resulting thymic fragments
and supernatants were transferred into separate tubes. For the
isolation of thymocytes, the supernatant was passed three times
through 70 μm mesh and centrifuged. The cell pellet was
resuspended in 20 ml of ACK lysis solution (0.15 M
NH4Cl, 1 mM KHCO3 and 0.1 mM
Na2EDTA) to remove red blood cells. The thymocyte
suspension was washed three times with HBSS buffer. The thymocytes
were then resuspended in HBSS buffer, and viable cells were counted
using the hemocytometer following trypan blue staining.
For the isolation of thymic stromal cells, the
thymic fragments were transferred into 5 ml of RPMI-1640 containing
0.125% (w/v) collagenase D and 0.1% (w/v) DNase I (both from Roche
Diagnostics GmbH, Mannheim, Germany) and were then incubated for 15
min with gentle shaking in a water bath at 37°C. The thymic
fragments in the enzyme mixtures were carefully dispersed with a
Pasteur pipette several times, and the supernatant was removed
after fragments had settled, and replaced with fresh enzyme
mixture. Gentle mechanical agitation was provided using a 5 ml
syringe and 18G needle, then a 21G needle followed by a 23G needle.
Tissue fragments were allowed to settle and the supernatant was
discarded. This digestion process was repeated four more times
until the tissue was fully digested. Cells liberated by the fourth,
fifth and sixth digests were saved, filtered through a 100
μm mesh to remove undigested particles, and washed three
times with HBSS buffer. They were then resuspended in HBSS buffer,
and viable cells were counted using the hemocytometer following
trypan blue staining.
Reverse transcriptase-polymerase chain
reaction (RT-PCR)
Total RNA was isolated from Easy-Blue RNA Extraction
Reagent (Intron, Seoul, Korea) following the manufacturer’s
protocol. First strand cDNA was obtained by reverse transcription
(RT) using 2 μg of total RNA. The reaction was conducted in
20 μl of buffer containing 0.5 μg of
Oligo(dT)12–18 primer, 50 mM Tris-HCl (pH 8.3), 75 mM
KCl, 3 mM MgCl2, 40 mM DTT, 0.5 mM deoxynucleotide
triphosphate (dNTP) mixture, 10 units of RNAse inhibitor, and 200
units of M-MLV reverse transcriptase (Gibco-BRL). Following
incubation at 37°C for 60 min, the reaction was stopped by heating
at 70°C for 15 min. To remove the remaining RNA, 1 μl of
E. coli RNase H (4 mg/ml) was added to the reaction mixture
and incubated at 37°C for 30 min. The cDNA was used as a template
for PCR amplification using gene-specific primers (Table I). PCR amplification of the cDNA
was performed in an automated thermal cycler (Techne, Teddington,
UK) in a final volume of 25 μl containing 4 μl of
cDNA solution, 20 mM Tris-HCl (pH 8.4), 50 mM KCl, 1.5 mM
MgCl2, 0.1% Triton X-100, 0.2 mM dNTP mixture
(Gibco-BRL), 0.5 pmol of each primer and 5 units of TaqDNA
polymerase (Promega, Madison, WI, USA). The amplification included
an initial denaturation at 94°C for 5 min and then 25 or 30 cycles
of denaturation at 94°C for 30 sec, primer annealing at 55°C for 30
sec, and extension at 72°C for 30 sec, followed by a final
extension at 72°C for 10 min, ending with a 4°C hold cycle. After
the PCR, the amplified products were analyzed by electrophoresis in
1.5% agarose gel and visualized by ethidium bromide staining under
UV light illumination. Band intensities of the PCR products were
measured and plotted using an image analysis program (MetaMorph;
Universal Imaging Corporation, Downingtown, PA, USA).
 | Table IGenes, oligonucleotide primers and
size of their RT-PCR products. |
Table I
Genes, oligonucleotide primers and
size of their RT-PCR products.
| Symbol | Gene name | GenBank accession
no. | Primer
sequence | Size (bp) |
|---|
| Crip3 | Thymus LIM protein
(cysteine-rich protein 3) | AF367972 | F:
5′-AAGTGTCCCAAGTGTGACAAGA-3′ | 234 |
| R:
5′-CGGAGGCTTCTCGTAGATGTAG-3′ | |
| CHCHD8 | E2IG2
(coiled-coil-helix-coiled-coil-helix domain containing 8) | AAF60345 | F:
5′-TGTCAACCTCAGTCCCACAA-3′ | 264 |
| R:
5′-GGCTTGCTCTTCCCTCCTCT-3′ | |
| Ehd4 | Pincher (EH-domain
containing 4) | AAM09109 | F:
5′-AACCCACAGATTCCTTCATTGCT-3′ | 337 |
| R:
5′-GATCGCCTCTGAGAACTCATCT-3′ | |
| Paip2 | Poly(A)-binding
protein-interacting protein 2 (Paip2) | NM_001014148 | F:
5′-AAGATCCAAGTCGCAGCAGTA-3′ | 318 |
| R:
5′-TCAGCACAAAATCTTCCAGAGA-3′ | |
| Tgfb1 | Transforming growth
factor β1 (Tgfb1) | NM_021578 | F:
5′-GCCCTGGATACCAACTACTGCT-3′ | 161 |
| R:
5′-AGGCTCCAAATGTAGGGGCAGG-3′ | |
| Tnfrsf9 | Tumor necrosis
factor receptor superfamily member 9 (Tnfrsf9) | NM_001025773 | F:
5′-CTATCTTCATCCGCATCCTACC-3′ | 339 |
| R:
5′-TGTTTATGGACGTTGGACTGAG-3′ | |
| Lama3 | Laminin-5 α3C chain
(Laminin α3) | CAA70073 | F: 5′-
GGACTCCTTCCTGGCTCTTTAT-3′ | 205 |
| R:
5′-GGTAGAATTTCCAGGCAAACTG-3′ | |
| GAPDH |
Glyceraldehyde-3-phosphate
dehydrogenase | AF106860 | F:
5′-CAACTCCCTCAAGATTGTCAGC-3′ | 449 |
| R:
5′-GGGAGTTGCTGTTGAAGTCACA-3′ | |
Statistical analysis
Data are expressed as the means ± SD for each
condition. For comparison of multiple groups, a one-way analysis of
variance (ANOVA) followed by a Scheffe’s post hoc test was
performed. P<0.05 was considered to indicate a statistically
significant difference.
Results
The regenerating thymus cDNA library was generated
by unidirectional insertion of oligo-dT-primed cDNAs into
λ-ZAP-Express. The phage library was converted through mass
excision to a plasmid library in the vector pBlue-script II. The
regenerating thymus cDNA library contained ∼2.18×1010
pfu. The identities of cloned sequences were determined by a BLAST
search of publicly available databases. The insert size was
distributed in a range from 0.8 to 2.5 kb. A total of 1,080
randomly selected clones from the regenerating thymus library were
sequenced. Each sequence was then aligned with all other EST
sequences in non-redundant nucleotide (NR) and dbEST databases of
NCBI using the BLASTN (BLAST nucleotide) program (www.ncbi.nlm.nih.gov/BLAST). In this study, E-value
scores <1.0 e−5 were considered to be
significant.
The average length of readable sequence was ∼400
nucleotides. Of the 1,080 clones, 1,000 were sequenced clones and
80 were read-fail clones. Of the 1,000 ESTs, 770 displayed
similarity to previously described genes with a BLASTN E-value of
≤1.0 e−5, representing known genes. A total of 1,000
ESTs were analyzed, of which 770 (77%) matched to known genes, 178
matched to unknown genes (17.8%) and 52 (5.2%) did not match any
known sequences (Table II).
 | Table IIResults of EST sequences for
regenerating rat thymus cDNA library. |
Table II
Results of EST sequences for
regenerating rat thymus cDNA library.
| Library | No. of clones | Percentage (%) |
|---|
| Total number of
gene cluster | 1,000 | 100 |
| Known genes | 770 | 77 |
| Unknown genes | 178 | 17.8 |
| Unidentified
genes | 52 | 5.2 |
The library complexity (i.e., the total number of
different sequences in the library) was estimated by NCBI search on
the basis of the 770 ESTs from the regenerating thymus cDNA
library. Among the 770 ESTs, 611 were found only once (87.3%)
without counting similar sequences in collected ESTs. The other 159
revealed more than two overlapping sequences.
Most abundant clones in the regenerating thymus cDNA
library were identified and listed in Table III. Highly abundant ESTs were 107
(17.5%) and sequenced more than four times. The EST which showed
the highest frequency was asparaginase-like 1 (L-asparaginase),
which occurred 10 times.
 | Table IIIMost abundant cDNA clones in the
regenerating rat thymus cDNA library. |
Table III
Most abundant cDNA clones in the
regenerating rat thymus cDNA library.
| Putative
identification
| | |
|---|
| Rank | Symbol | Gene name | GenBank accession
no. | No. of ESTs |
|---|
| 1 | Asrgl1 | Asparaginase like 1
(Asrgl1) | NM_145089 | 10 |
| 2 | Eef1a1 | Eukaryotic
translation elongation factor 1α1 (Eef1a1) | NM_175838 | 9 |
| 3 | Slc25a4 | Adenine nucleotide
translocator (Solute carrier family 25) | CAA43842 | 8 |
| 4 | Bcas1 | Breast
carcinoma-amplified sequence 1 homolog isoform 1 (breast carcinoma
amplified sequence 1) | NP_084091 | 6 |
| 5 | Tmsb4x | Thymosin β4 | NP_112398 | 6 |
| 6 | Hspa9 | Stress-70 protein,
mitochondrial [heat shock 70 kDa protein 9 (mortalin)] | NP_034611 | 5 |
| 7 | Ccng1 | Cyclin G | CAA50219 | 5 |
| 7 | Arpp21 | Cyclic
AMP-regulated phosphoprotein (cAMP-regulated phosphoprotein, 21
kDa) | BC089974 | 5 |
| 10 | Cd74 | Cd74 molecule,
major histocompatibility complex, class II invariant chain | BC059152 | 5 |
| 10 | Apoe | Apolipoprotein
E | BC060313 | 4 |
| 10 | Rnf111 | Arkadia (ring
finger protein 111) | AAK38272 | 4 |
| 10 | Naip2 | Baculoviral IAP
repeat-containing 1b | AAN77613 | 4 |
| 10 | Tm2d2 | TM2 domain
containing 2 | BC093382 | 4 |
| 10 | mt-COI | Cytochrome-c
oxidase I | CAA32956 | 4 |
| 10 | Dhfr | Dihydrofolate
reductase | NP_034179 | 4 |
| 10 | Pira6 | Immunoglobulin-like
receptor PIRA6 (12M1) | U96687 | 4 |
| 10 | Usp33 | Ubiquitin specific
peptidase 33 | BC092624 | 4 |
| 10 | Rps3a | Ribosomal protein
S3a | AAH58483 | 4 |
| 10 | Sf3b2 | Unnamed protein
product (splicing factor 3b, subunit 2, 145 kDa) | BAC32755 | 4 |
| 10 | Mtf2 | Metal-response
element-binding transcription factor 2 isoform 1 (Metal-response
element-binding transcription factor 2) | NP_038855 | 4 |
| 10 | Tob2 | Tob2 protein
(transducer of ERBB2, 2) | AAH11163 | 4 |
The 770 ESTs matched to known genes were grouped
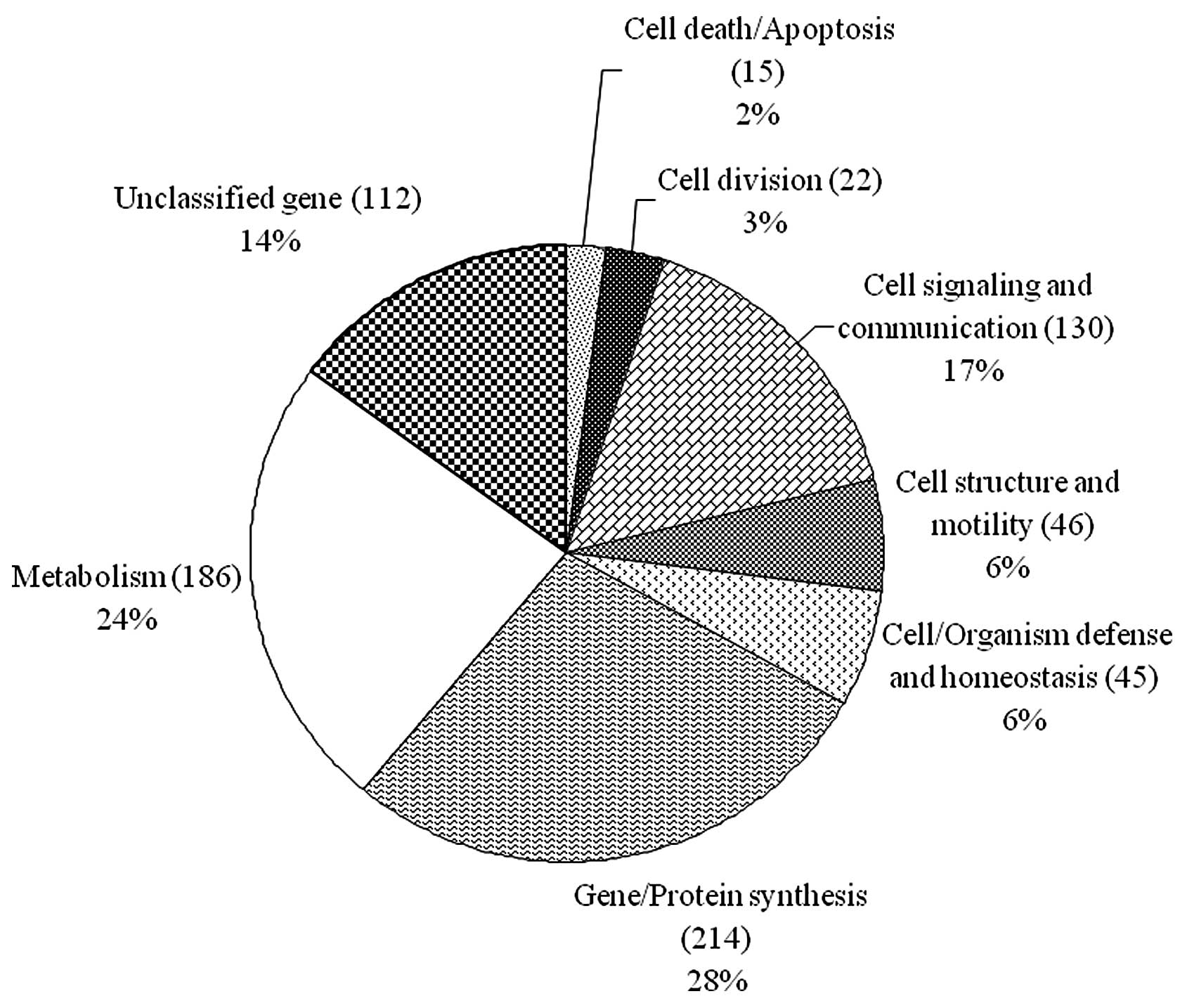
into eight functional categories based on the TIGR gene cellular
directory (http://www.tigr.org) (Fig. 1). Among the 770 ESTs, 214 (28%)
were gene or protein synthesis related genes, 186 (24%) were
metabolism related genes, 130 (17%) were cell signaling and
communication related genes, 46 (6%) were cell structure and
motility related genes, 45 (6%) were cell/organism defense and
homeostasis related genes, 22 (3%) were cell division related
genes, 15 (2%) were cell death/apoptosis related genes, and,
finally, 112 (14%) were unclassified genes (Fig. 1).
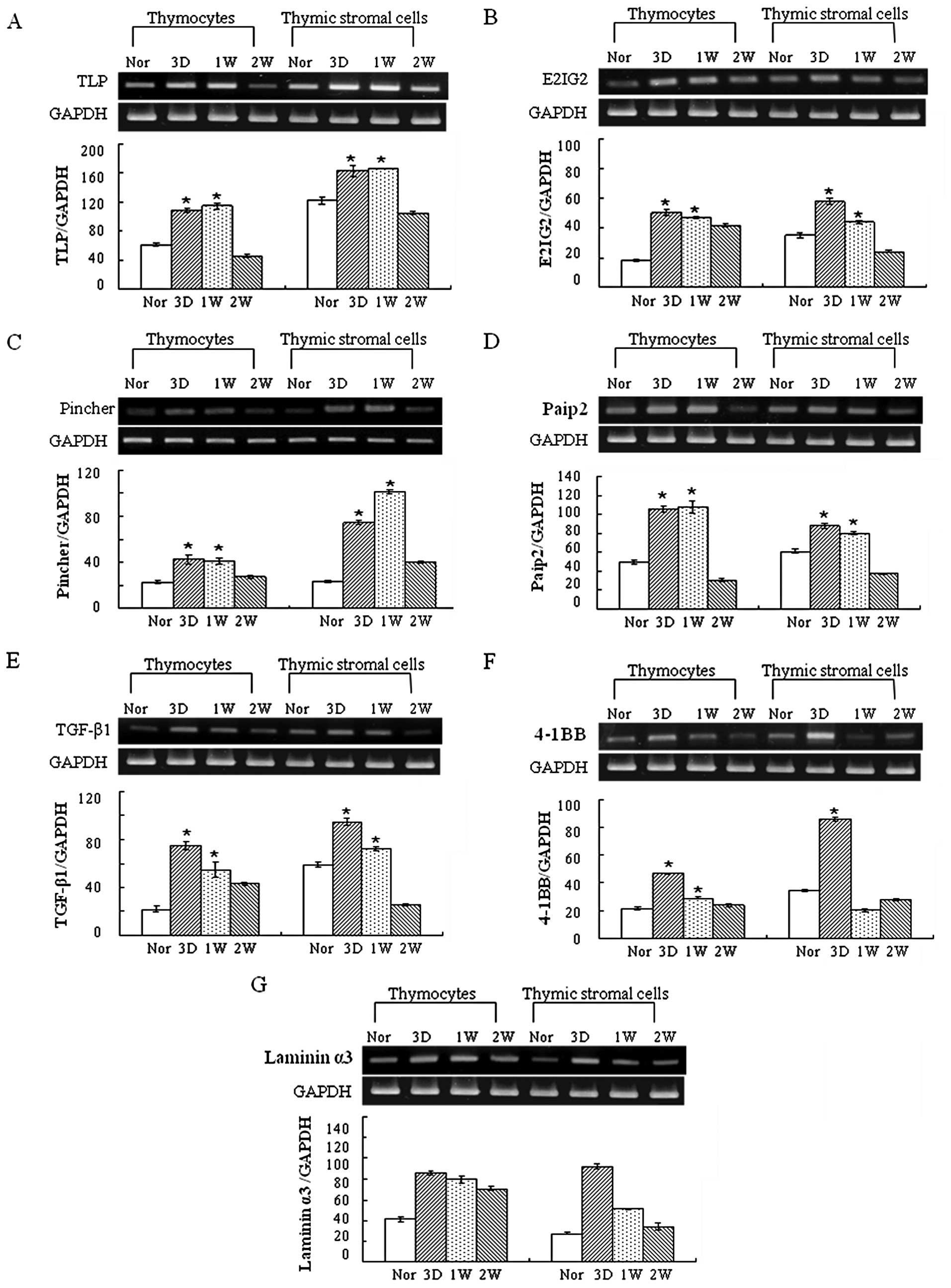
Seven genes that may be involved in thymus
regeneration were selected based on their biological function
rather than the frequency, and the gene expression was analyzed in
thymocytes and thymic stromal cells isolated from normal and
regenerating rat thymus using RT-PCR analysis. The seven genes
were: thymus LIM protein (TLP, cysteine-rich protein 3),
estradiol-induced gene 2 (E2IG2), pincher (pinocytic chaperone),
poly(A) binding protein interacting protein 2
(polyadenylate-binding protein-interacting protein 2, Paip2),
transforming growth factor-β1 (TGF-β1), tumor necrosis factor
receptor superfamily member 9 (4-1BB, Tnfrsf9) and laminin α3
(laminin-5 α3). Based on the data of RT-PCR analysis, the
expression of TLP, E2IG2, pincher, Paip2, TGF-β1, 4-1BB and laminin
α3 was increased in both the thymocytes and thymic stromal cells
during thymic regeneration, particularly 3 and 7 days after
cyclophosphamide treatment (Fig.
2A–G).
Discussion
In our previous studies, we established an
experimental thymus regeneration model to investigate the
mechanisms underlying T cell regeneration (2,7–10).
Using this model, in the present study, we analyzed several genes
that may be related to T cell regeneration from a regenerating rat
thymus cDNA library. Of note, our data showed an upregulated
expression of genes related to a translational machinery, as
inferred by the expression of protein synthesis-related genes
(28%), indicating the effectiveness of our experiment, since the
regulation of gene expression at the translational level plays a
key role in the induction of quiescent T cells to re-enter the cell
cycle and proliferate, which is a necessary step of cellular
regeneration.
Of particular interest, among genes expressed in the
regenerating rat thymus, we identified the seven genes which may be
involved in thymus regeneration. LIM domain proteins are known to
play a number of important roles in the development and
differentiation of normal as well as of cancer cells (11,12). In particular, it has been reported
that LIM domain proteins, in association with TAL1 (T-cell acute
leukaemia 1)/SCL (stem cell leukaemia), cause arrest of thymocyte
differentiation at the CD4−CD8− DN to
CD4+CD8+ DP transition (13). A novel LIM gene encoding TLP
expressed specifically in the thymus in a subset of cortical
epithelial cells near the corticomedullary junction (14). It was shown that TLP is
upregulated in a thymus in which selection of T cells is occurring
(Rag−/− OT-1 mice) compared to its expression in a
thymus in which selection is blocked at the
CD4+CD8+ DP stage of T cell development
(Rag−/−Tap−/− OT-1 mice) (14). They also suggested that TLP may
have a role in thymus development by demonstrating that targeted
disruption of TLP results in a reduction of thymus cellularity by
approximately 30%, with a proportional reduction in each
subpopulation (14). Furthermore,
accumulating evidence suggests that LIM domain proteins play a
crucial role in hematopoiesis (15–18). These facts, together with our
experimental findings, indicate a novel role of TLP in thymus
regeneration.
It has been established that sex hormones influence
lymphocyte development and, in particular, high levels of estrogen,
as in the case of pregnancy or estrogen administration, can induce
profound thymic atrophy and result in immunosuppressive effects on
the peripheral immune systems, whereas estrogen deficiency, as in
the case of ovariectomy, induces significant enlargement of the
thymus (19). In line with this
action of estrogen on the thymus, it is noteworthy that the gene
expression of E2IG2, a member of novel genes upregulated by
estrogen treatment and first identified by serial analysis of gene
expression in estrogen-responsive breast cancer cells following
exposure to this hormone, was upregulated during thymus
regeneration in the present study, suggesting its potential role in
the process of postnatal T cell development and production
(20). However, the function of
E2IG2 remains unclear.
It was observed that thymic cells are able to
produce nerve growth factor (NGF) protein and mRNA (8). In particular, we have shown that the
expression of NGF and its high affinity receptor TrkA are
upregulated in the rat TECs during thymus regeneration following
acute thymic involution (7,8).
Furthermore, we have shown that NGF induces upregulation of a
unique potent angiogenic factor, vascular endothelial growth factor
(VEGF) in TECs, which may play a key role in the reparative
angiogenesis during thymus regeneration (9). NGF also stimulates TEC activities
including cell proliferation, thymocyte adhesion to TECs, the
expression of critical cell adhesion molecules such as
intercellular adhesion molecule-1 (ICAM-1) and vascular cell
adhesion molecule-1 (VCAM-1), which mediate thymocyte-TEC
interactions and the production of major thymopoietic factors
including interleukin (IL)-7, granulocyte-macrophage
colony-stimulating factor (GM-CSF), stromal cell-derived factor-1
(SDF-1), thymus and activation-regulated chemokine (TARC) and
thymus-expressed chemokine (TECK) (21). These data collectively suggest
that NGF, particularly on TECs, may play a role in the T
lymphopoiesis associated with thymus regeneration during recovery
from acute thymic involution.
Pincher, an NGF-induced protein, functions in
endocytosis and trafficking of NGF and its receptor TrkA functions
as a pinocytic chaperone for vesicles containing these molecules
(22). Pincher function is
necessary for both the NGF-induced internalization of TrkA by a
pinocytic process, and the sorting of long-lived endosomal vesicles
with NGF signaling capability (22). Pincher-mediated macroendocytosis
for NGF generates Trk endosomes that are refractory to lysosomal
processing, thus ensuring sustained endosomal signaling after
retrograde transport, which is crucial for regulating neuronal
phenotype and survival (23,24). Thus, these results indicate that
pincher mediates pinocytic endocytosis of NGF and TrkA which play
an important role in the processes of thymus regeneration.
The cap structure and the poly(A) tail of eukaryotic
mRNAs act synergistically to enhance translation. This effect is
mediated by a direct interaction of eukaryotic initiation factor 4G
(eIF4G) and poly(A) binding protein (Pabp), which
results in circularization of the mRNA. Of the two identified
Pabp-interacting proteins (Paip), one, Paip1, stimulates
translation through its interaction with eukaryotic initiation
factor 3 (eIF3) (25),
and the other, Paip2, which competes with Paip1 for binding to
Pabp, represses translation by competing with eIF4G for
Pabp binding and by decreasing the affinity of Pabp for the poly(A)
tail (26,27). Two isoforms of Paip2, Paip2a and
Paip2b, have been defined in mammals and are expressed
differentially in tissues, indicating tissue-specific functions and
responses to distinct stimuli (28,29). In a previous study, we showed that
the expression of VEGF is upregulated during thymus regeneration
following thymic involution induced by cyclophosphamide treatment
(9). Thus, it is hypothesized
that Paip2 could modulate thymus regeneration by upregulating VEGF
expression. Further studies on Paip2 in thymus regeneration could
provide important information about its role in the postnatal T
cell development.
TGF-β1 may have a broad function not only for T cell
selection and death in immune responses and in the generation of
tolerance, but also for defining the mechanisms of programmed cell
death in general (30). It has
been demonstrated that TGF-β1 regulates the gene expression of
atrial natriuretic peptide (ANP) receptors in the thymic stromal
cells and that ANP and TGF-β1 may affect the thymic stromal cell
functions (31). Markedly, it was
demonstrated that TGF-β1 signaling in TECs accelerates the process
of age-related thymic involution and transiently hinders the early
phase of thymic reconstitution after myeloablative conditioning and
hematopoietic stem cell transplantation, suggesting that the
pharmacological inhibition of TGF-β1 activity could constitute a
novel component of a conditioning regimen to hasten thymopoietic
activity following hematopoietic stem cell transplantation
(32).
4-1BB, a member of the TNFR superfamily, is a major
costimulatory receptor that is rapidly expressed on the surface of
CD4+ and CD8+ T cells after antigen- or
mitogen-induced activation. Costimulation through 4-1BB by either
4-1BBL or agonistic monoclonal antibodies (mAbs) enhances T cell
activity (33) and promotes
CD8+ T cell survival (34). We have demonstrated that the
expression of 4-1BB and 4-1BBL is preferentially expressed in the
regenerating thymus, mainly in CD4+CD8+
double-positive (DP) thymocytes (10). The 4-1BB and 4-1BBL positive cells
among the CD4+CD8+ DP thymocytes present
during thymus regeneration were TCRhi and
CD69+, unlike the corresponding controls, indicating
that 4-1BB and 4-1BBL are predominantly expressed by the positively
selected population of the CD4+CD8+ DP during
thymus regeneration (10). We
also found that the interaction of 4-1BB with 4-1BBL promoted
thymocyte adhesion to TECs and suggested that 4-1BB and 4-1BBL are
involved in T lymphopoiesis associated with positive selection
during recovery from acute thymic involution (10).
Laminin α3 regulates various cellular functions,
including cell adhesion and migration (35). It has been found that laminin α3
chain plays a role in T cell development (36). However, much remains to be
clarified regarding the role of laminin α3 chain in thymus
function.
Among the most frequently expressed genes during
thymus regeneration, L-asparaginase, adenine nucleotide
translocator and thymosin β4 were striking. L-asparaginase, first
cloned as asparaginase-like sperm autoantigen from rat and human
testis cDNA libraries on the basis of reactivity with antibodies
produced after vasectomy, was found to be expressed not only in the
testis but also in the heart, brain, liver, skeletal muscle and
kidney (37). It has been shown
that loss of the eukaryotic initiation factor 2 (eIF2)
kinase, general control nonderepressible kinase 2 (GCN2) function
renders the immune system more sensitive to the cytotoxic effects
of asparaginase-mediated amino acid deprivation, leading to
enhanced cell death of lymphocytes (38,39). This finding provides insight into
the role of GCN2 in the immunosuppression caused by asparaginase.
Thus, it is conceivable that L-asparaginase that exists in high
frequency in the ESTs of regenerating rat thymus may be involved in
thymic involution by potentiating cyclophosphamide-induced
thymocyte depletion possibly via its action on GCN2.
Adenine nucleotide translocator serves as a
component of the mitochondrial permeability transition pore
complex, and plays a major role in promoting mitochondria-mediated
apoptosis (40). It has been
demonstrated that mitochondrial adenine nucleotide translocator 3
is regulated by IL-4 and IFN-γ via STAT-dependent pathways, and
regulation of mitochondrial adenine nucleotide translocator 3 by
IL-4 may have a functional implication in cytokine-mediated T cell
survival (41,42); however, the role of adenine
nucleotide translocator during thymus regeneration remains to be
investigated.
Thymosin β4, a small 43-amino acid polypeptide that
was first isolated from calf thymus, enhances cell migration and
adhesion, promotes angiogenesis and cell differentiation, prevents
apoptosis, promotes cell survival and tissue regeneration, and
enhances wound healing and repair (43). In particular, thymosin β4 acts on
lymphoid stem cells and controls the early stages of the maturation
process of thymus-dependent lymphocytes (44). Abundant expression of this gene in
the ESTs of regenerating rat thymus suggests that it may be
significantly involved in the thymus during regeneration.
Collectively, we have compiled genes expressed in
the regenerating thymus and identified specific genes which may be
involved in the processes of rat thymus regeneration, thereby
providing new comprehensive molecular information on thymus
regeneration for the first time. The regenerating thymus cDNA
library constructed in this study may be a useful source for
identifying various genes expressed during thymus regeneration.
Therefore, the present study provides further insight into the
development of novel therapeutic strategies for effective thymus
repopulation in the design of therapies for numerous clinical
conditions in which immune reconstitution is required.
Acknowledgements
This study was supported by the
Medical Research Institute Grant (2004–37), Pusan National
University Hospital, and the National Research Foundation of Korea
(NRF) Grant funded by the Korean government (MEST) (no.
2010-0014194).
References
|
1
|
Ritter MA and Palmer DB: The human thymic
microenvironment: new approaches to functional analysis. Semin
Immunol. 11:13–21. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yoon S, Yoo YH, Kim BS and Kim JJ:
Ultrastructural alterations of the cortical epithelial cells of the
rat thymus after cyclophosphamide treatment. Histol Histopathol.
12:401–413. 1997.PubMed/NCBI
|
|
3
|
Toubert A, Glauzy S, Douay C and Clave E:
Thymus and immune reconstitution after allogeneic hematopoietic
stem cell transplantation in humans: never say never again. Tissue
Antigens. 79:83–89. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gutierrez JA: Genomics: from novel genes
to new therapeutics in parasitology. Int J Parasitol. 30:247–252.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shiue YL, Wang LH, Chao TY, Lin CH and
Tsai CL: EST-based identification of genes expressed in the
hypothalamus of adult tilapia, Oreochromis mossambicus.
Biochem Biophys Res Commun. 316:523–527. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Biswas MK, Chai L, Qiang X and Deng X:
Generation, functional analysis and utility of Citrus
grandis EST from a flower-derived cDNA library. Mol Biol Rep.
39:7221–7235. 2012.PubMed/NCBI
|
|
7
|
Yoon S, Lee HW, Baek SY, Kim BS, Kim JJ
and Lee SA: Upregulation of TrkA neurotrophin receptor expression
in the thymic subcapsular, paraseptal, perivascular, and cortical
epithelial cells during thymus regeneration. Histochem Cell Biol.
119:55–68. 2003.
|
|
8
|
Lee HW, Kim SM, Shim NR, Bae SK, Jung IG,
Kwak JY, Kim BS, Kim JB, Moon JO, Chung JS and Yoon S: Expression
of nerve growth factor is upregulated in the rat thymic epithelial
cells during thymus regeneration following acute thymic involution.
Regul Pept. 141:86–95. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Park HJ, Kim MN, Kim JG, Bae YH, Bae MK,
Wee HJ, Kim TW, Kim BS, Kim JB, Bae SK and Yoon S: Up-regulation of
VEGF expression by NGF that enhances reparative angiogenesis during
thymic regeneration in adult rat. Biochim Biophys Acta.
1773:1462–1472. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kim YM, Kim HK, Kim HJ, Lee HW, Ju SA,
Choi BK, Kwon BS, Kim BS, Kim JB, Lim YT and Yoon S: Expression of
4-1BB and 4-1BBL in thymocytes during thymus regeneration. Exp Mol
Med. 41:896–911. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li A, Ponten F and dos Remedios CG: The
interactome of LIM domain proteins: the contributions of LIM domain
proteins to heart failure and heart development. Proteomics.
12:203–225. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Nouët Y, Dahan J, Labalette C, Levillayer
F, Julien B, Jouvion G, Cairo S, Vives FL, Ribeiro A, Huerre M,
Colnot S, Perret C, Van Nhieu JT, Tordjmann T, Buendia MA and Wei
Y: The four and a half LIM-only protein 2 regulates liver
homeostasis and contributes to carcinogenesis. J Hepatol.
57:1029–1036. 2012.PubMed/NCBI
|
|
13
|
Herblot S, Steff AM, Hugo P, Aplan PD and
Hoang T: SCL and LMO1 alter thymocyte differentiation: inhibition
of E2A-HEB function and pre-T alpha chain expression. Nat Immunol.
1:138–144. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kirchner J, Forbush KA and Bevan MJ:
Identification and characterization of thymus LIM protein: targeted
disruption reduces thymus cellularity. Mol Cell Biol. 21:8592–8604.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pinto do OP, Kolterud A and Carlsson L:
Expression of the LIM-homeobox gene LH2 generates immortalized
steel factor-dependent multipotent hematopoietic precursors. EMBO
J. 17:5744–5756. 1998.PubMed/NCBI
|
|
16
|
Yamada Y, Warren AJ, Dobson C, Forster A,
Pannell R and Rabbitts TH: The T cell leukemia LIM protein Lmo2 is
necessary for adult mouse hematopoiesis. Proc Natl Acad Sci USA.
95:3890–3895. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Terano T, Zhong Y, Toyokuni S, Hiai H and
Yamada Y: Transcriptional control of fetal liver hematopoiesis:
dominant negative effect of the overexpression of the LIM domain
mutants of LMO2. Exp Hematol. 33:641–651. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tao Y, Wang J, Tokusumi T, Gajewski K and
Schulz RA: Requirement of the LIM homeodomain transcription factor
tailup for normal heart and hematopoietic organ formation in
Drosophila melanogaster. Mol Cell Biol. 27:3962–3969. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zoller Al and Kersh GJ: Estrogen induces
thymic atrophy by eliminating early thymic progenitors and
inhibiting proliferation of beta-selected thymocytes. J Immunol.
176:7371–7378. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Charpentier AH, Bednarek AK, Daniel RL,
Hawkins KA, Laflin KJ, Gaddis S, MacLeod MC and Aldaz CM: Effects
of estrogen on global gene expression: identification of novel
targets of estrogen action. Cancer Res. 60:5977–5983.
2000.PubMed/NCBI
|
|
21
|
Lee HW, Na YJ, Jung PK, Kim MN, Kim SM,
Chung JS, Kim BS, Kim JB, Moon JO and Yoon S: Nerve growth factor
stimulates proliferation, adhesion and thymopoietic cytokine
expression in mouse thymic epithelial cells in vitro. Regul Pept.
147:72–81. 2008. View Article : Google Scholar
|
|
22
|
Shao Y, Akmentin W, Toledo-Aral JJ,
Rosenbaum J, Valdez G, Cabot JB, Hilbush BS and Halegoua S:
Pincher, a pinocytic chaperone for nerve growth factor/TrkA
signaling endosomes. J Cell Biol. 157:679–691. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Valdez G, Akmentin W, Philippidou P,
Kuruvilla R, Ginty DD and Halegoua S: Pincher-mediated
macroendocytosis underlies retrograde signaling by neurotrophin
receptors. J Neurosci. 25:5236–5247. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Philippidou P, Valdez G, Akmentin W,
Bowers WJ, Federoff HJ and Halegoua S: Trk retrograde signaling
requires persistent, Pincher-directed endosomes. Proc Natl Acad Sci
USA. 108:852–857. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Martineau Y, Martineau Y, Derry MC, Wang
X, Yanagiya A, Berlanga JJ, Shyu AB, Imataka H, Gehring K and
Sonenberg N: Poly(A)-binding protein-interacting protein 1 binds to
eukaryotic translation initiation factor 3 to stimulate
translation. Mol Cell Biol. 28:6658–6667. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Khaleghpour K, Svitkin YV, Craig AW,
DeMaria CT, Deo RC, Burley SK and Sonenberg N: Translational
repression by a novel partner of human poly(A) binding protein,
Paip2. Mol Cell. 7:205–216. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Karim MM, Svitkin YV, Kahvejian A, De
Crescenzo G, Costa-Mattioli M and Sonenberg N: A mechanism of
translational repression by competition of Paip2 with eIF4G for
poly(A) binding protein (PABP) binding. Proc Natl Acad Sci USA.
103:9494–9499. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Berlanga JJ, Baass A and Sonenberg N:
Regulation of poly(A) binding protein function in translation:
characterization of the Paip2 homolog, Paip2B. RNA. 12:1556–1568.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yanagiya A, Delbes G, Svitkin YV, Robaire
B and Sonenberg N: The poly(A)-binding protein partner Paip2a
controls translation during late spermiogenesis in mice. J Clin
Invest. 120:3389–3400. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen W, Jin W, Tian H, Sicurello P, Frank
M, Orenstein JM and Wahl SM: Requirement for transforming growth
factor beta1 in controlling T cell apoptosis. J Exp Med.
194:439–453. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Agui T, Xin X, Cai Y, Shim G, Muramatsu Y,
Yamada T, Fujiwara H and Matsumoto K: Opposite actions of
transforming growth factor-beta 1 on the gene expression of atrial
natriuretic peptide biological and clearance receptors in a murine
thymic stromal cell line. J Biochem. 118:500–507. 1995.PubMed/NCBI
|
|
32
|
Hauri-Hohl MM, Zuklys S, Keller MP, Jeker
LT, Barthlott T, Moon AM, Roes J and Holländer GA: TGF-beta
signaling in thymic epithelial cells regulates thymic involution
and postir-radiation reconstitution. Blood. 112:626–634. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Pollok KE, Kim YJ, Zhou Z, Hurtado J, Kim
KK, Pickard RT and Kwon BS: Inducible T cell antigen 4-1BB.
Analysis of expression and function. J Immunol. 150:771–781.
1993.PubMed/NCBI
|
|
34
|
Takahashi C, Mittler RS and Vella AT:
4-1BB is a bona fide CD8 T cell survival signal. J Immunol.
162:5037–5040. 1999.PubMed/NCBI
|
|
35
|
Tsubota Y, Mizushima H, Hirosaki T,
Higashi S, Yasumitsu H and Miyazaki K: Isolation and activity of
proteolytic fragment of laminin-5 alpha3 chain. Biochem Biophys Res
Commun. 278:614–620. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kim JM, Park WH and Min BM: The
PPFLMLLKGSTR motif in globular domain 3 of the human laminin-5
alpha3 chain is crucial for integrin alpha3beta1 binding and cell
adhesion. Exp Cell Res. 304:317–327. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Bush LA, Herr JC, Wolkowicz M, Sherman NE,
Shore A and Flickinger CJ: A novel asparaginase-like protein is a
sperm autoantigen in rats. Mol Reprod Dev. 62:233–247. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Bunpo P, Dudley A, Cundiff JK, Cavener DR,
Wek RC and Anthony TG: GCN2 protein kinase is required to activate
amino acid deprivation responses in mice treated with the
anti-cancer agent L-asparaginase. J Biol Chem. 284:32742–32749.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Bunpo P, Cundiff JK, Reinert RB, Wek RC,
Aldrich CJ and Anthony TG: The eIF2 kinase GCN2 is essential for
the murine immune system to adapt to amino acid deprivation by
asparaginase. J Nutr. 140:2020–2027. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kim EH, Koh EH, Park JY and Lee KU:
Adenine nucleotide translocator as a regulator of mitochondrial
function: implication in the pathogenesis of metabolic syndrome.
Korean Diabetes J. 34:146–153. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Jang JY and Lee CE: Mitochondrial adenine
nucleotide translocator 3 is regulated by IL-4 and IFN-gamma via
STAT-dependent pathways. Cell Immunol. 226:11–19. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Jang JY and Lee CE: IL-4-induced
upregulation of adenine nucleotide translocase 3 and its role in Th
cell survival from apoptosis. Cell Immunol. 241:14–25. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Crockford D, Turjman N, Allan C and Angel
J: Thymosin β4: structure, function, and biological properties
supporting current and future clinical applications. Ann NY Acad
Sci. 1194:179–189. 2010.
|
|
44
|
Low TL, Hu SK and Goldstein AL: Complete
amino acid sequence of bovine thymosin beta 4: a thymic hormone
that induces terminal deoxynucleotidyl transferase activity in
thymocyte populations. Proc Natl Acad Sci USA. 78:1162–1166. 1981.
View Article : Google Scholar
|