Introduction
Differentiated embryo-chondrocyte expressed gene 1
(DEC1, also known as BHLHE40/Stra13/Sharp2) is a basic
helix-loop-helix (bHLH) transcription factor ubiquitously expressed
in both embryonic and adult tissues (1,2).
DEC1 exerts multiple biological functions, such as neurogenesis,
apoptosis, cell proliferation, cell differentiation and circadian
rhythms (3–8). Recent studies have revealed that the
expression of DEC1 is correlated with the malignancy of a number of
cancer types (9). The expression
patterns of DEC1 and their impact on tumor development are largely
tumor-type specific (1,9). Low DEC1 expression was found to be
associated with poor histological differentiation and malignancy
progression in hepatocellular carcinoma (10), whereas, overexpression of DEC1 was
significantly correlated with poor survival of patients with
esophageal squamous cell carcinoma following surgery (11). In breast cancer, the expression of
DEC1 was found to be increased upon progression from normal tissues
to invasive carcinomas (12).
However, the mechanisms by which DEC1 modulates cancer progression
are largely unclear.
Claudin-1 is a key component of the tight junction
complex and is thereby important for maintaining tight junction
barrier integrity. Downregulation of claudin-1 is intimately
associated with tumorigenesis in breast (13,14), prostate (15) and melanocytic neoplasia (16). It is postulated that the impact of
claudin-1 on cancer invasion and metastasis is through its
regulation of certain invasion/metastasis suppressors or enhancers
(17,18). As significant loss of claudin-1
expression and overexpression of DEC1 are commonly observed in the
progression of various types of cancers including breast cancer, we
aimed to ascertain whether DEC1 modulates breast cancer invasion
through its regulation of claudin-1. Our study revealed that DEC1
modulates breast cancer invasion through regulation of
claudin-1.
Materials and methods
Patients and tissue samples
A total of 147 invasive ductal breast carcinoma
tissue samples were obtained from patients who underwent surgery at
the First Affiliated Hospital of China Medical University, China,
between 2005 and 2009. Informed consent was obtained prior to
surgery from all enrolled patients. Formalin-fixed
paraffin-embedded sections of tissues obtained from surgical
samples were stained routinely with hematoxylin and eosin (H&E
staining), and reviewed by two senior pathologists in order to
determine the histological type according to World Health
Organization breast carcinoma histological classification criteria
(2003). Accordingly, the grade of ductal carcinomas was classified
into three groups: grade 1 (low grade), grade 2 (moderate grade)
and grade 3 (high grade). Patients included in the study ranged
from 31 to 79 years of age (mean, 51 years). All patients with
tumors had axillary node status confirmed histologically. Details
of the clinical data regarding age, nodal status, tumor size,
grade, estrogen and progesterone receptor status of these patients
are listed in Table I. The study
was conducted according to the regulations stipulated by the
Institution Review Board of the China Medical University.
 | Table IThe expression of DEC1 and claudin-1
in invasive breast ductal carcinomas. |
Table I
The expression of DEC1 and claudin-1
in invasive breast ductal carcinomas.
| DEC1 expression | Claudin-1
expression |
|---|
|
|
|
|---|
| Negative | Positive | P-value | Negative | Positive | P-value |
|---|
| No. of patients | 39 | 108 | | 106 | 41 | |
| Age (years) | | | 0.873 | | | 0.847 |
| <50 | 20 | 57 | | 55 | 22 | |
| ≥50 | 19 | 51 | | 51 | 19 | |
| Nodal status | | | 0.825 | | | 0.028 |
| Negative | 31 | 84 | | 78 | 37 | |
| Positive | 8 | 24 | | 28 | 4 | |
| Tumor size
(cm) | | | 0.451 | | | 0.934 |
| ≤2 | 16 | 37 | | 38 | 15 | |
| >2 | 23 | 71 | | 68 | 26 | |
| Grade | | | 0.023 | | | 0.069 |
| I | 9 | 7 | | 15 | 1 | |
| II | 25 | 79 | | 72 | 32 | |
| III | 5 | 22 | | 19 | 8 | |
| ER status | | | 0.660 | | | 0.042 |
| Negative | 16 | 40 | | 35 | 21 | |
| Positive | 23 | 68 | | 71 | 20 | |
| PR status | | | 0.428 | | | 0.298 |
| Negative | 15 | 34 | | 38 | 11 | |
| Positive | 24 | 74 | | 68 | 30 | |
Immunohistochemistry
All resected specimens were fixed with 10% neutral
buffered formalin and embedded in paraffin blocks. Tissue blocks
were sliced into 4-μm sections. The sections were then
deparaffinized, rehydrated and immunostained with polyclonal rabbit
anti-DEC1 antibody (1:200; Novus Biologicals, Littleton, CO, USA)
and polyclonal rabbit anti-claudin-1 antibody (1:100; Zymed
Laboratories, Inc., South San Francisco, CA, USA) at 4°C overnight.
Antibodies were detected by the streptavidin-peroxidase method.
Evaluation of immunostaining
All the immunostained sections were evaluated by two
senior pathologists who were blinded to the clinical data.
According to the previous evaluation methods, only nuclear
expression of DEC1 indicates positivity (19) and ≥10% cytoplasm and/or membrane
expression of claudin-1 indicates positivity (20).
Cell culture and treatment
The human breast cancer cell lines MCF-7 and
MDA-MB-231 were obtained from the American Type Culture Collection
(ATCC, Manassas, VA, USA). The cells were cultured in Dulbecco’s
modified Eagle’s medium (DMEM)-high glucose (Sigma Chemical Co.,
St. Louis, MO, USA) supplemented with 10% fetal bovine serum at
37°C in a humidified atmosphere of 95% air and 5%
CO2.
Reverse transcription-polymerase chain
reaction (RT-PCR)
Total RNA was isolated using an RNeasy RNA isolation
kit (Qiagen, Hilden, Germany). First-strand cDNA was synthesized
from 1 μg of total RNA using ReverTra Ace (Toyobo, Osaka, Japan).
PCR was performed using an aliquot of first-strand cDNA as a
template under standard conditions with TaqDNA polymerase (Takara,
Shiga, Japan). The cDNAs for human DEC1, claudin-1 and
glyceraldehyde 3-phosphate dehydrogenase (GAPDH) were amplified for
up to 28, 25 and 22 cycles respectively. The primers used are
listed as follows: DEC1 forward, 5′-GTGAGTCACTCTCCAGTTT-3′ and
reverse, 5′-ATCCGTGTCTAGCTGTGCAAT-3′; claudin-1 forward,
5′-CAGCTGTTGGGCTTCATTCTC-3′ and reverse, 5′-ATC
ACTCCCAGGAGGATGCC-3′; GAPDH forward, 5′-CCA
CCCATGGCAAATTCCATGGCA-3′ and reverse, 5′-AGA
CCACCTGGTGCTCAGTGTAGC-3′. The predicted sizes of the amplified
products for DEC1, claudin-1 and GAPDH were 534, 277 and 696 bp,
respectively. The PCR products were separated on 1.5% (w/v) agarose
gels.
Small interfering RNA (siRNA)
siRNA against DEC1 and the negative control siRNA
(scrambled siRNA) were purchased from Santa Cruz Biotechnology,
Inc. (catalog no. of DEC1 siRNA (h): sc-106769; catalog no. of
control siRNA-A: sc-37007). For the siRNA transfection, MCF-7 and
MDA-MB-231 cells were seeded at 5×104 cells/35-mm well.
Then 24 h later, the siRNA was transfected into the cells using
Lipofectamine™ RNAiMAX reagent (Invitrogen, Carlsbad, CA, USA).
After transfection, the cells were incubated for 48 h and subjected
to various analyses.
Western blotting
The cells transfected with siRNA or plasmid DNA were
lysed using M-PER lysis buffer (Pierce, Rockford, IL, USA). Protein
concentrations were determined by a bicinchoninic acid assay. The
obtained lysates (20 μg protein) were subjected to SDS-PAGE, and
the acquired proteins were transferred to PVDF membranes
(Millipore, Billerica, MA, USA). The membranes were incubated with
antibodies specific for DEC1 (1:20,000), claudin-1 (1:1,000) and
actin (1:30,000; Sigma Chemical Co.), followed by horseradish
peroxidase-conjugated secondary antibody (Immuno-Biological
Laboratories Co., Ltd., Gunma, Japan). The ECL, ECL-Plus or
ECL-Advance Western Blotting Detection System (Amersham, Uppsala,
Sweden) was used for detection.
Matrigel invasion assay
The Matrigen invasion assay was performed according
to the manufacturer’s instructions. In each upper chamber,
5×105 cells were grown in serum-free medium on 8-μm
porous polycarbonate membranes (Corning, Acton, MA, USA), which
were coated with Matrigel basement membrane matrix (BD Biosciences,
San Jose, CA, USA). The lower chambers were filled with DMEM-high
glucose supplemented with 10% fetal bovine serum. After incubation
for 24 h at 37°C in a humid atmosphere with 5% CO2, the
cells that had migrated through the pores were fixed with methanol
for 30 min and stained with hematoxylin. For each filter, the
numbers of cells in five different fields under ×200 magnification
were visualized and counted using a Nikon E200 microscope. Each
experiment was performed in triplicate.
Statistical analysis
The statistical package SPSS version 13.0 for
Windows (SPSS, Chicago, IL, USA) was used for all data analysis.
The Chi-square test was used to assay whether the expression levels
of DEC1 or claudin-1 were related to the clinicopathological
characteristics of the breast cancers. The Spearman correlation
test was used to examine the correlations between DEC1 and
claudin-1 expression. A P-value <0.05 was considered to indicate
a statistically significant result.
Results
Relationship between DEC1, claudin-1
expression and clinicopathological factors
We examined the expression patterns of DEC1 and
claudin-1 using immunohistochemistry in 147 invasive breast ductal
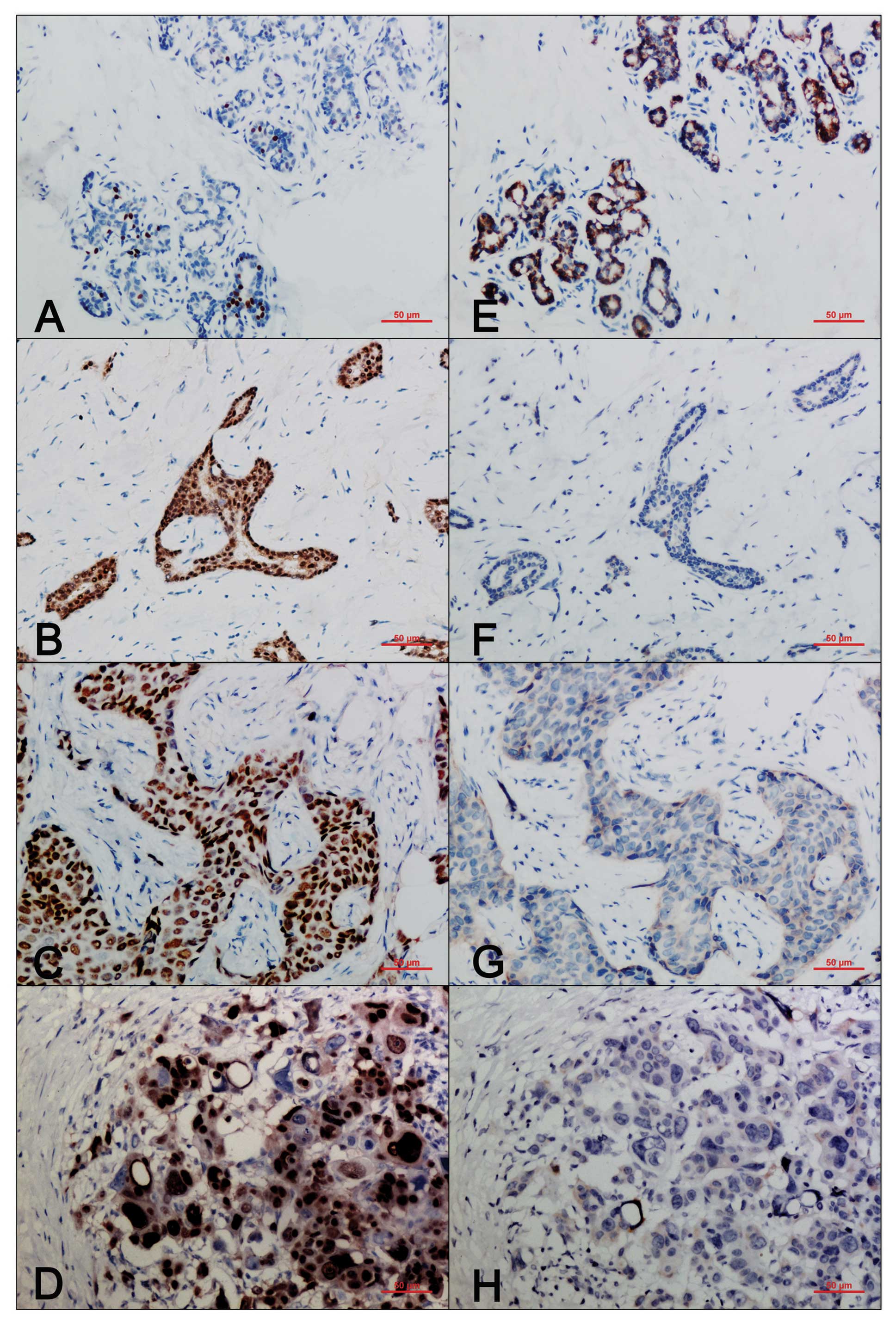
carcinomas. In normal human breast tissues, the expression of DEC1
was weak and patchy, and was mostly presented in the nucleus
(Fig. 1A). However, in the
invasive ductal breast carcinomas, the expression of DEC1 was
detected in both the nucleus and the cytoplasm, and the nuclear
staining was much stronger (Fig.
1B–D). The percentage of tumors positive for DEC1 was elevated
as tumor grade increased; 43.75% (7/16) for grade 1, 75.96%
(75/104) for grade 2, and 81.48% (22/27) for grade 3, respectively.
In contrast, in the normal breast tissues, claudin-1 was expressed
mainly in the cytoplasm and/or on the cell membrane (Fig. 1E). Loss or attenuation of
claudin-1 expression was noted in the invasive ductal breast
carcinomas (Fig. 1F–H). By
Chi-square analysis, we found that DEC1 expression was correlated
with tumor grade (P=0.023). However, no significant associations
were observed in regards to patient age (P=0.873), lymph node
status (P=0.825), tumor size (P=0.451), estrogen receptor (ER)
(P=0.660) or progesterone receptor (PR) (P=0.428). Loss of
claudin-1 expression was correlated with lymph node status
(P=0.028) and ER status (P=0.042), but not with patient age
(P=0.847), tumor size (P=0.934) or PR status (P=0.298).
Importantly, our correlation analysis provided the evidence that
there is a strong inverse correlation between DEC1 and claudin-1
expression in breast cancers (P=0.003, correlation coefficient
=−0.245) (Table II).
 | Table IICorrelations between the expression
of DEC1 and claudin-1. |
Table II
Correlations between the expression
of DEC1 and claudin-1.
| Nuclear expression
of DEC1 | | |
|---|
|
| | |
|---|
| Cytoplasmic and/or
membrane expression of claudin-1 | Negative | Positive | P-value | Correlation
coefficient |
|---|
| Negative | 21 | 85 | 0.003 | −0.245 |
| Positive | 18 | 23 | | |
DEC1 knockdown upregulates the expression
of claudin-1 in breast cancer cell lines
MCF-7 and MDA-MB-231 breast cancer cell lines were
employed to further illustrate the impact of DEC1 on claudin-1
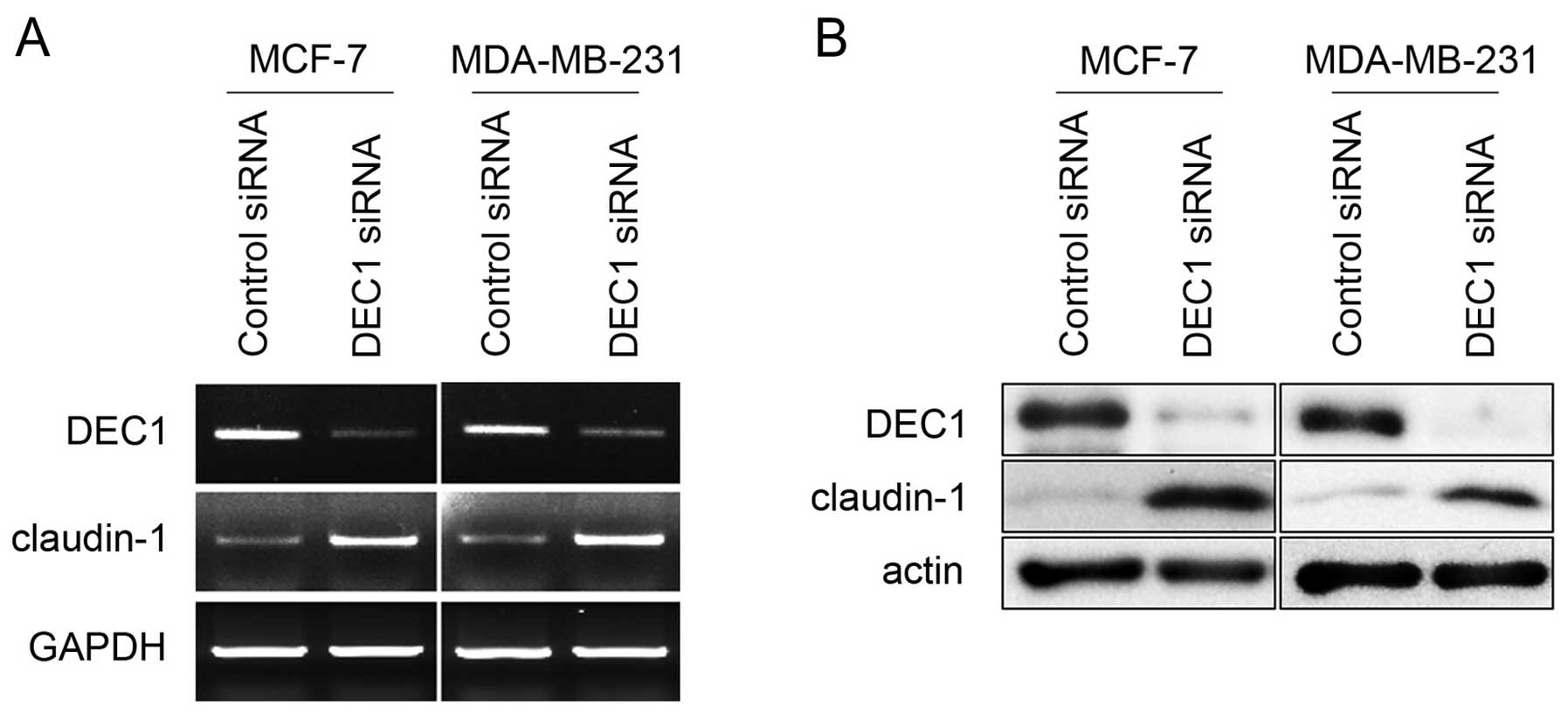
expression. As shown in Fig. 2A,
DEC1 siRNA considerably reduced the mRNA level of DEC1, whereas,
scrambled siRNA had no effect on the expression of DEC1. Notably,
ablation of endogenous DEC1 by siRNA resulted in enhanced claudin-1
expression at the mRNA level in both MCF-7 and MDA-MB-231 cells.
Consistently, the protein expression of claudin-1 was also
upregulated upon DEC1 knockdown in both cell lines (Fig. 2B). Taken together, these data
imply that DEC1 downregulates claudin-1 expression in breast cancer
cell lines. These data confirm that DEC1 downregulates claudin-1
expression and thereby results in the progressive invasiveness of
breast cancer.
Role of DEC1 in the invasiveness of
breast cancer cells
Breakdown of cell-cell interactions and
downregulation of junctional proteins are key steps in tumor
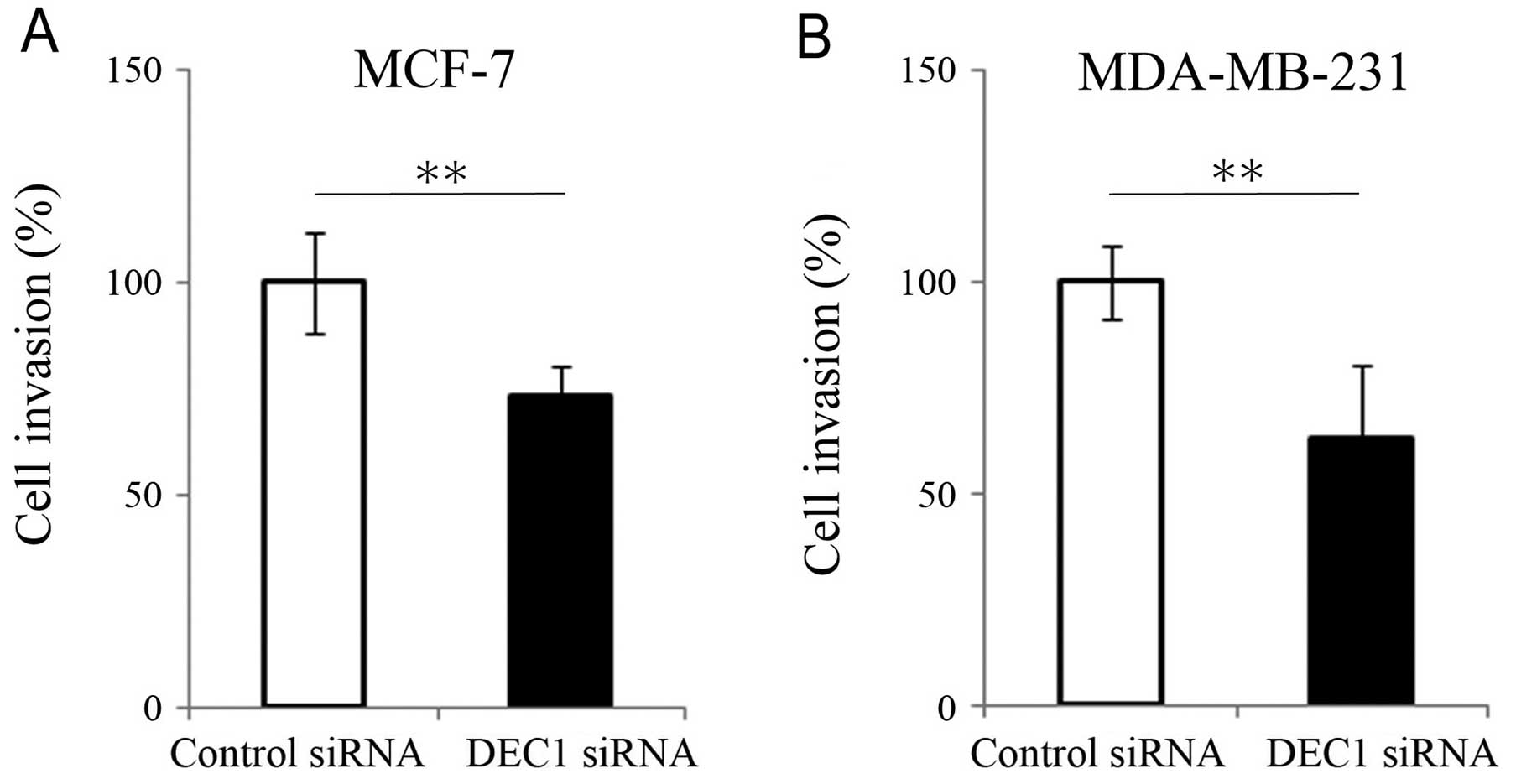
invasion. Therefore, we employed the Matrigel invasion assay to
test whether the expression level of DEC1 had any impact on cell
invasive capability. In both MCF-7 and MDA-MB-231 cells,
downregulation of DEC1 by siRNA knockdown resulted in a dramatic
reduction in the cell invasive capability compared to the control
group (Fig. 3). These data
further support the postulate that DEC1 is closely linked to the
invasive capability of breast cancer. Together with the
observations that DEC1 and claudin-1 are inversely correlated in
invasive ductal carcinomas and that DEC1 modulates the claudin-1
expression level, our data suggest that DEC1 may promote breast
cancer invasion through its regulation of claudin-1.
Discussion
DEC1 is involved in the control of proliferation,
differentiation and apoptosis of various types of cells (2). Overexpression of DEC1 was reportedly
correlated with a wide array of cancers (7,12,19,21,22). Chakrabarti et al (12) found that the expression of DEC1
increased upon progression from normal tissues to invasive breast
carcinomas. Agreeably, in this study, we also observed increased
DEC1 expression in breast invasive ductal adenocarcinoma. In
addition, the results of our Matrigel invasion assay revealed that
DEC1 may be involved in breast cancer invasion. Since the breakdown
of cell-cell interaction and downregulated expression of junctional
proteins are key steps in tumor invasion, it is possible that the
upregulation of DEC1 may have an inhibitory effect on the
expression of junctional proteins.
Tight junctions are the most apical of intercellular
junctions and appear as a network of continuous filaments on the
protoplasmic face of the plasma membrane (23). They contribute to the
transepithelial barrier that controls the transport of ions and
small molecules through the paracellular pathway (24,25). Claudins are the major component of
the tight junction, and to date, 24 claudin family members have
been identified (26). Loss of
claudin-1, a member of the tight junction, is associated with
cancer invasion and the acquisition of the metastatic phenotype
(13,14). Functional studies suggest that
claudin-1 may be a key player in the progression of breast cancer
(18). According to our results,
loss of claudin-1 was correlated with lymph nodal metastasis and
ER-positive status. Regarding the correlation between ER status and
claudin-1, our result is consistent with the findings reported by
Blanchard et al (27).
Their study also showed that, in ER-negative invasive breast
cancers, a higher number of cases were claudin-4 positive, whereas
fewer cases were claudin-3 positive. Taken together, this suggests
that different claudins have unique expression patterns in
different subtypes of breast cancers.
Our clinical data provide the evidence that there is
a strong negative correlation between DEC1 and claudin-1 expression
in invasive ductal breast carcinomas. We also found that knockdown
of endogenous DEC1 enhanced the expression of claudin-1 at both the
mRNA and protein levels. DEC1, a transcriptional factor, has been
shown to regulate the transcription of target genes by binding to
the E-box elements (28,29). There are two E-box motifs in the
promoter of claudin-1 (30).
Thus, it is likely that DEC1 may bind to the E-boxes in the
promoter of claudin-1 and regulate its transcription. Further
investigation by us will be directed towards further addressing the
mechanism.
Kon et al (31) along with our previous study
demonstrated that DEC1, as a downstream target gene, was induced by
transforming growth factor-β (TGF-β) and tumor necrosis factor-α
(TNF-α) (32). TNF-α and TGF-β
were widely reported as regulators in epithelial-mesenchymal
transition (EMT) through activation of NF-κB and TGF-β signal
pathway, respectively (11,33). Notably, TNF-α can also induce the
expression of claudin-1 (34),
while TGF-β increases tumor-initiating cells in ‘claudin-low’
breast cancer cell lines (35).
Based on these results, it is possible that the regulation of
claudin-1 by DEC1 may be a bridge which links TNF-α/TGF-β and the
induction of EMT.
In conclusion, both DEC1 and claudin-1 are closely
related with tumorigenesis of breast cancer. DEC1 is positively
associated with tumor grade in invasive breast cancer and is
negatively correlated with the expression of claudin-1. DEC1 may
influence the progression of invasive breast cancer through its
regulation of claudin-1.
Acknowledgements
We are grateful to Dr Xiaoman Li for the help in
preparing the manuscript.
References
|
1
|
Ivanova A, Liao SY, Lerman MI, Ivanov S
and Stanbridge EJ: STRA13 expression and subcellular localisation
in normal and tumour tissues: implications for use as a diagnostic
and differentiation marker. J Med Genet. 42:565–576. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Shen M, Kawamoto T, Yan W, et al:
Molecular characterization of the novel basic helix-loop-helix
protein DEC1 expressed in differentiated human embryo chondrocytes.
Biochem Biophys Res Commun. 236:294–298. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Nakashima A, Kawamoto T, Honda KK, et al:
DEC1 modulates the circadian phase of clock gene expression. Mol
Cell Biol. 28:4080–4092. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sato F, Bhawal UK, Kawamoto T, et al:
Basic-helix-loop-helix (bHLH) transcription factor DEC2 negatively
regulates vascular endothelial growth factor expression. Genes
Cells. 13:131–144. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liu Y, Sato F, Kawamoto T, et al:
Anti-apoptotic effect of the basic helix-loop-helix (bHLH)
transcription factor DEC2 in human breast cancer cells. Genes
Cells. 15:315–325. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bhawal UK, Sato F, Arakawa Y, et al: Basic
helix-loop-helix transcription factor DEC1 negatively regulates
cyclin D1. J Pathol. 224:420–429. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li Y, Zhang H, Xie M, et al: Abundant
expression of Dec1/stra13/sharp2 in colon carcinoma: its
antagonizing role in serum deprivation-induced apoptosis and
selective inhibition of procaspase activation. Biochem J.
367:413–422. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Boudjelal M, Taneja R, Matsubara S,
Bouillet P, Dolle P and Chambon P: Overexpression of Stra13, a
novel retinoic acid-inducible gene of the basic helix-loop-helix
family, inhibits mesodermal and promotes neuronal differentiation
of P19 cells. Genes Dev. 11:2052–2065. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Turley H, Wykoff CC, Troup S, Watson PH,
Gatter KC and Harris AL: The hypoxia-regulated transcription factor
DEC1 (Stra13, SHARP-2) and its expression in human tissues and
tumours. J Pathol. 203:808–813. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shi XH, Zheng Y, Sun Q, et al: DEC1
nuclear expression: a marker of differentiation grade in
hepatocellular carcinoma. World J Gastroenterol. 17:2037–2043.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xu Q, Ma P, Hu C, et al: Overexpression of
the DEC1 protein induces senescence in vitro and is related to
better survival in esophageal squamous cell carcinoma. PLoS One.
7:e418622012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chakrabarti J, Turley H, Campo L, et al:
The transcription factor DEC1 (stra13, SHARP2) is associated with
the hypoxic response and high tumour grade in human breast cancers.
Br J Cancer. 91:954–958. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tokés AM, Kulka J, Paku S, et al:
Claudin-1, -3 and -4 proteins and mRNA expression in benign and
malignant breast lesions: a research study. Breast Cancer Res.
7:R296–R305. 2005.PubMed/NCBI
|
|
14
|
Swisshelm K, Macek R and Kubbies M: Role
of claudins in tumorigenesis. Adv Drug Deliv Rev. 57:919–928. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sheehan GM, Kallakury BV, Sheehan CE,
Fisher HA, Kaufman RP Jr and Ross JS: Loss of claudins-1 and -7 and
expression of claudins-3 and -4 correlate with prognostic variables
in prostatic adenocarcinomas. Hum Pathol. 38:564–569. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cohn ML, Goncharuk VN, Diwan AH, Zhang PS,
Shen SS and Prieto VG: Loss of claudin-1 expression in
tumor-associated vessels correlates with acquisition of metastatic
phenotype in melanocytic neoplasms. J Cutan Pathol. 32:533–536.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chao YC, Pan SH, Yang SC, et al: Claudin-1
is a metastasis suppressor and correlates with clinical outcome in
lung adenocarcinoma. Am J Respir Crit Care Med. 179:123–133. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Myal Y, Leygue E and Blanchard AA: Claudin
1 in breast tumorigenesis: revelation of a possible novel ‘claudin
high’ subset of breast cancers. J Biomed Biotechnol.
2010:9568972010.PubMed/NCBI
|
|
19
|
Giatromanolaki A, Koukourakis MI, Sivridis
E, et al: DEC1 (STRA13) protein expression relates to
hypoxia-inducible factor 1-alpha and carbonic anhydrase-9
overexpression in non-small cell lung cancer. J Pathol.
200:222–228. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Morohashi S, Kusumi T, Sato F, et al:
Decreased expression of claudin-1 correlates with recurrence status
in breast cancer. Int J Mol Med. 20:139–143. 2007.PubMed/NCBI
|
|
21
|
Ivanova AV, Ivanov SV,
Danilkovitch-Miagkova A and Lerman MI: Regulation of STRA13 by the
von Hippel-Lindau tumor suppressor protein, hypoxia, and the
UBC9/ubiquitin proteasome degradation pathway. J Biol Chem.
276:15306–15315. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li Y, Xie M, Yang D, et al: The expression
of antiapoptotic protein survivin is transcriptionally upregulated
by DEC1 primarily through multiple sp1 binding sites in the
proximal promoter. Oncogene. 25:3296–3306. 2006. View Article : Google Scholar
|
|
23
|
Anderson JM and Van Itallie CM: Physiology
and function of the tight junction. Cold Spring Harb Perspect Biol.
1:a0025842009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Diamond JM: Twenty-first Bowditch lecture.
The epithelial junction: bridge, gate, and fence. Physiologist.
20:10–18. 1977.PubMed/NCBI
|
|
25
|
Tobioka H, Isomura H, Kokai Y, Tokunaga Y,
Yamaguchi J and Sawada N: Occludin expression decreases with the
progression of human endometrial carcinoma. Hum Pathol. 35:159–164.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tsukita S, Furuse M and Itoh M:
Multifunctional strands in tight junctions. Nat Rev Mol Cell Biol.
2:285–293. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Blanchard AA, Watson PH, Shiu RP, et al:
Differential expression of claudin 1, 3, and 4 during normal
mammary gland development in the mouse. DNA Cell Biol. 25:79–86.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li Y, Xie M, Song X, et al: DEC1
negatively regulates the expression of DEC2 through binding to the
E-box in the proximal promoter. J Biol Chem. 278:16899–16907. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
St-Pierre B, Flock G, Zacksenhaus E and
Egan SE: Stra13 homodimers repress transcription through class B
E-box elements. J Biol Chem. 277:46544–46551. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Martinez-Estrada OM, Cullerés A, Soriano
FX, et al: The transcription factors Slug and Snail act as
repressors of Claudin-1 expression in epithelial cells. Biochem J.
394:449–457. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kon N, Hirota T, Kawamoto T, Kato Y,
Tsubota T and Fukada Y: Activation of TGF-beta/activin signalling
resets the circadian clock through rapid induction of Dec1
transcripts. Nat Cell Biol. 10:1463–1469. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liu Y, Wang L, Lin XY, et al:
Anti-apoptotic effect of claudin-1 on TNF-α-induced apoptosis in
human breast cancer MCF-7 cells. Tumour Biol. 33:2307–2315.
2012.PubMed/NCBI
|
|
33
|
Wu Y, Sato F, Yamada T, et al: The BHLH
transcription factor DEC1 plays an important role in the
epithelial-mesenchymal transition of pancreatic cancer. Int J
Oncol. 41:1337–1346. 2012.PubMed/NCBI
|
|
34
|
Kondo J, Sato F, Kusumi T, et al:
Claudin-1 expression is induced by tumor necrosis factor-α in human
pancreatic cancer cells. Int J Mol Med. 22:645–649. 2008.
|
|
35
|
Bruna A, Greenwood W, Le Quesne J, et al:
TGFβ induces the formation of tumour-initiating cells in
claudinlow breast cancer. Nat Commun. 3:10552012.
|