Introduction
Although radiation therapy has been applied for
prostate cancer, it has been pointed out that improvement of
treatment strategies is necessary to manage the local recurrence of
prostate cancer after radiotherapy. In addition, the side effects
of radiation therapy are always a concern. Gene therapy is being
considered as one of the new procedures for prostate cancer
treatment. Prostate cancer is a particularly suitable malignancy to
study, due to its relatively accessible location and the
specificity of its gene products (1). However, there are still problems
that need to be solved before gene therapy can become a general
therapeutic procedure for prostate cancer.
It has been pointed out that combining gene therapy
with radiation may lead to a novel, more efficient therapy with
fewer side effects as the strategies can compensate for each
other’s defects (2). Thus, we
developed promoters that are activated in response to radiation,
wherein radiation stimulation may control the expression of
therapeutic genes. We demonstrated that it was possible to
efficiently obtain radiation-responsive promoters by linking the
TATA box signal to randomly combined DNA fragments containing
cis-elements of transcription factors activated by
radiation. However, the resulting promoters tended to be cell
type-specific (3). Therefore, we
constructed a radiation-responsive promoter for prostate cancer
cells with cis-elements of transcription factors activated
in prostate cancer cells by radiation. In addition, these were
successfully improved by randomly introducing point mutations. The
promoter that was designated clone 880-8 enhanced the expression of
the luciferase gene linked downstream more than 10-fold 12 h
following 10 Gy X-ray irradiation in vitro in prostate
cancer cells. In the case of malignant LNCaP cells implanted into a
mouse, the promoter also significantly enhanced the luciferase gene
expression after X-ray irradiation, although to a lesser degree.
However, in an in vitro simulation study of suicide gene
therapy, when a prodrug was applied at high concentrations,
significant cell death was observed even without X-ray irradiation
(4).
microRNAs (miRNAs) are low-molecular-weight RNAs
(19–24 nucleotides in length) that do not code for polypeptides and
control gene expression mainly by interfering with the translation
process after binding target sequences complimentary to the miRNA
seed sequence present in messenger RNAs. Over 1,000 miRNAs have
been identified in human cells and are reported to be involved in
many important biological processes, such as development,
differentiation, cell proliferation and cell death in a tissue
type-specific manner (5).
It was recently reported that expression of many
miRNAs is altered in response to stimuli. For instance, many cancer
cells have altered miRNA expression profiles in response to
radiation (6–8). In addition, the expression of the
let-7 family, known as antimirs, increases after stimulation with
radiation (9). These results
suggest that changes in miRNA expression may be involved in the
response of cells to radiation (10). Furthermore, it has been reported
that other stimuli, including anticancer drugs (11), ultrasound (10), hyperthermia (12) and hypoxia (13) also induce changes in miRNA
expression levels, suggesting that miRNAs may be involved in the
adaptive response of cells to various types of stimuli.
Several reports have indicated that changes in miRNA
expression may be useful for controlling gene expression for
tissue-specific or cancer-specific gene therapy. Research has shown
that inserting copies of a target sequence of an miRNA that is not
expressed in the target tissues into the 3′-untranslated region
(3′UTR) of a gene of interest resulted in gene expression in a
target tissue-specific manner (15–17).
In this study, we analyzed the miRNA expression
changes in a prostate cancer cell line after radiation stimulation.
We focused our attention on the miRNAs that significantly decreased
in response to radiation, and introduced copies of their
complementary target sequences into the 3′-UTR of our gene of
interest to see if it could lead to radiation-mediated gene
expression control. In addition, we combined target sequences with
a radiation-responsive clone (880-8) promoter, so that it could
lead to fine-tuned gene expression control, thus suppressing gene
expression in the absence of radiation, while increasing such
expression after radiation stimulation.
Materials and methods
Cells and bacteria
LNCaP cells from human prostate carcinoma were used
throughout this study. The cell line was purchased from Health
Science Research Resources Bank (Tokyo, Japan). Cells were grown
and maintained in RPMI-1640 medium supplemented with 10% fetal calf
serum and appropriate antibiotics at 37°C in a 5% CO2
atmosphere. The other cell line used in this study was AmphoPack293
(Takara Bio, Inc., Ohtsu, Japan), which is a genetically modified
cell line used for the generation of recombinant retrovirus
particles. It was derived from HEK293 cells, and the envelope gene
and gag gene of the moloney murine leukemia virus were inserted
into the genome to provide the viral proteins for packaging. A
retrovirus genome-like RNA containing the Ψ sequence transcribed
from a vector introduced into the cells is packaged in a virus-like
particle and buds into the culture medium. The particles carry
amphotropic envelope protein on the surface so that they can
recognize the ram 1 receptor. Therefore, they infect a broad range
of mammalian cells.
The DH5α strain of Escherichia coli (Nippon
Gene; Toyama, Japan) was used for the DNA manipulation experiments.
E. coli cells were grown in LB medium at 37°C. All medium
components were purchased from BD Diagnostics (Sparks, MD, USA).
DNA manipulation experiments with E. coli were performed
according to the methods described by Sambrook and Russell
(18).
RNA extraction and miRNA microarray
analysis
Total RNA was extracted from control cells as well
as cells exposed to radiation 6 h post-exposure using the miRNeasy
Extraction kit. Samples were treated with DNase I (RNase-free DNase
kit), (both from Qiagen Inc., Valencia, CA, USA) for 15 min at room
temperature during the procedure as directed in the instructions to
remove residual genomic DNA.
miRNA expression was analyzed using a GeneChip
system with the miRNA array ver. 1.0 or 2.0 (Affymetrix Inc., Santa
Clara, CA, USA). Samples for array hybridization were prepared
using the HRT FlashTag RNA Labeling kit for Affymetrix GeneChip
miRNA arrays (Genisphere Inc., Hatfield, PA, USA) following a
procedure described in the product manual. Briefly, 1 μg of
total RNA was labeled with biotin through ligation with 3DNA
dendrimer. The labeled RNA was hybridized to the GeneChip array at
48°C for 16 h. The arrays were washed, stained with
streptavidin-phycoerythrin and scanned using a probe array scanner.
The scanned chip data were converted to digitized information using
the miRNA QC Tool application (ver. 1.0.33.0 or 1.1.1.0).
Quantitative real-time PCR
To evaluate the miRNA expression, quantitative
real-time PCR was performed. Total RNA was collected from LNCaP
cells using the miRNeasy Mini kit (Qiagen Inc.) and treatment with
DNase I according to the manufacturer’s instructions, as described
above. cDNAs were synthesized using the extracted RNA as templates,
using a miScript Reverse Transcription kit (Qiagen Inc.) according
to the manufacturer’s instructions. The miRNA expression analysis
was performed using a M×3000P QPCR System (Agilent Technologies
Inc., Santa Clara, CA, USA) with the synthesized cDNA. Quantitative
PCR measurement by real-time monitoring of SYBR-Green integration
into the synthesized DNA was performed during the PCR process:
incubation at 95°C for 15 min, and then 40 thermal cycles for
reactions at 94°C for 15 sec, 55°C for 30 sec and 70°C for 30 sec,
followed by reactions at 55°C for 30 sec and 95°C for 30 sec with a
miScript SYBR-Green PCR Kit (Qiagen Inc.). After the PCR process,
the dissociation temperature of the synthesized DNA fragments was
also determined by monitoring the release of SYBR-Green from the
denatured DNA to confirm the integrity of the synthesized DNA
fragment. The primers used for the PCR reaction were selected from
a miRNA primer library provided in the miScript Primer assay
(Qiagen Inc.). We used a specific primer to detect human U1A small
nuclear RNA expression as an internal control. Relative standard
curves representing several 10-fold dilutions of cDNA from a
representative sample were used for the linear regression analysis
for other samples.
Vector construction
The initial assessment of the introduction of target
sequences into the 3′UTR of the luciferase gene was performed using
vectors constructed based on pmirGLO (Promega Corp., Madison, WI,
USA), in which the 3′UTR of the firefly luciferase gene carries a
multi-cloning site and is driven by the mouse PGK promoter, and the
Renilla luciferase gene was aligned in tandem with the
firefly luciferase gene and is driven by the SV40 promoter. We
inserted DNA fragments containing target sequences into the
multi-cloning site to see whether the target sequences affected the
luciferase expression after radiation.
We constructed two types of target sequences. One
was composed of randomly combined complimentary sequences of the 3
miRNAs that were chosen due to the decreases in their expression in
response to radiation, and the other was composed of 2 or 4 copies
of a single complimentary sequence of 1 of the 3 miRNAs. To
construct target DNA fragments composed of randomly combined
complimentary sequences, we synthesized DNA fragments with a sticky
end sequence that was not palindromic (5′-agtg-3′) so that the
complimentary sequences would have to be unidirectionally aligned.
The synthesized target sequences for miR-92-1-5p were as follows:
5′-aggttgggatcggttgcaatgctcact-3′ and 5′-agcattgcaaccga
tcccaacctagtg-3; for miR-19b, 5′-tgtgcaaatccatgcaaaactgac act-3′
and 5′-tcagttttgcatggatttgcacaagtg-3′; miR-503, 5′-tag
cagcgggaacagttctgcagcact-3′ and 5′-ctgcagaactgttcccgctgctaa gtg-3′.
These fragment pairs were annealed in annealing buffer (100 mM
NaCl, 50 mM Tris-Cl, pH 7.5) by incubating them at 95°C for 5 min
and then gradually decreasing the temperature to 4°C. The annealed
pairs were treated with polynucleotide kinase at 37°C for 60 min,
and then equimolar amounts were mixed. We also synthesized a pair
of DNA fragments containing a NheI recognition site
(5′-gtacgctagca gtg-3′ and 5′-gctagcgtaccact-3′) and a one
containing SalI and EcoRI recognition sites
(5′-gaattcgtcgacagtg-3′ and 5′-gtcgac gaattccact-3′) and similarly
treated them for annealing and kinase activity. They were also
mixed at equimolar concentrations.
The target mixture and the restriction site mixture
were further mixed at ratios of 20:1, 10:1 and 5:1 and then
separately ligated. Elongated DNA fragments were digested with
NheI and SalI, and purified using spin columns. They
were then inserted into the NheI and SalI sites of
pmirGLO. We judged that an insert carrying the EcoRI site
was cloned in the right direction. We examined 9 plasmid clones
designated as pmirGLO5-1, 5-2, 5-3, 5-4, 10-1, 10-2, 20-1, 20-2,
20-3, according to the mixing ratios.
We then constructed target fragments containing 2 or
4 copies of the complimentary sequences of the 3 miRNAs. For
miR-19b, 5′-ctagctcagttttgcatggatttgcacacagctcagttttgcatgg
atttgcacaaagcttg-3′ and 5′-tcgacaagctttgtgcaaatccatgcaaaactg
agctgtgtgcaaatccatgcaaaactgag-3′ were annealed and treated with
kinase, and then were inserted into the NheI and SalI
sites of pmirGLO to construct pmirGLO-19b-2. Furthermore,
5′-tcgactcagttttgcatggatttgcacacagctcagttttgcatggatttgcac
agaattcgc-3′ and 5′-ggccgcgaattctgtgcaaatccatgcaaaactgagctg
tgtgcaaatccatgcaaaactgag-3′ were annealed and treated with kinase,
and inserted into the SalI and NotI sites of
pmirGLO-19b-2 to construct pmirGLO-19b-4. As for miR-503,
5′-ctagcctgcagaactgttcccgctgctacagcctgcagaactgttcccgctgctaaa
gcttg-3′ and 5′-tcgacaagctttagcagcgggaacagttctgcaggctgta
gcagcgggaacagttctgcagg-3′ were annealed and treated with kinase,
and then were inserted into the NheI and SalI sites
of pmirGLO to construct pmirGLO-503-2. Furthermore,
5′-tcgacctgcagaactgttcccgctgctacagcctgcagaactgttcccgctg
ctagaattcgc-3′ and 5′-ggccgcgaattctagcagcgggaacagttctgcagg
ctgtagcagcgggaacagttctgcagg-3′ were annealed and treated with
kinase, and inserted into the SalI and NotI sites of
pmirGLO-503-2 to construct pmirGLO-503-4. For miR-92a-1-5p,
5′-ctagcagcattgcaaccgatcccaacctcagcagcattgcaacc
gatcccaacctaagcttg-3′ and 5′-tcgacaagcttaggttgggatcggttgcaa
tgctgctgaggttgggatcggttgcaatgctg-3′ were annealed and treated with
kinase, and inserted into the NheI and SalI sites of
pmirGLO to construct pmirGLO-92a-1-5p-2. In addition,
5′-tcgacagcattgcaaccgatcccaacctcagcagcattgcaaccgatcccaa
cctgaattcgc-3′ and 5′-ggccgcgaattcaggttgggatcggttgcaatgctgctg
aggttgggatcggttgcaatgctg-3′ were annealed and treated with kinase,
and inserted into the SalI and NotI sites of
pmirGLO-92a-1-5p-2 to construct pmirGLO-92a-1-5p-4.
Transient transfection and X-ray
irradiation
The constructed plasmid vectors were transfected
into cells using the Effecten reagent (Qiagen Inc.) according to
the manufacturer’s instructions. One and a half million cells were
washed once with pre-warmed RPMI-1640, and resuspended with 1.5 ml
of the medium. An Effecten complex containing 1.0 of a plasmid
vector, and in some cases 10 ng phRL-TK (Promega Corporation), was
added to each dish. The cells were harvested and resuspended with 5
ml of pre-warmed RPMI-1640 medium after incubation at 37°C for more
than 4 h, and then were re-incubated at 37°C overnight. A 35-mm
cell culture dish was seeded with 3.0×105 of the
incubated cells in 2 ml of pre-warmed RPMI-1640 and then the cells
were subjected to X-ray irradiation. Each cell culture dish was
placed on the turning table of an X-ray generator (MBR-1520-3;
Hitachi Medical Technology Corp., Tokyo, Japan) and irradiated with
5 to 15 Gy X-ray at 5 Gy/min.
Luciferase assay
At various times after X-ray irradiation, cells were
washed once with PBS, and 300 μl of passive lysis buffer
from the Dual Luciferase Assay kit (Promega Corp.) was added to
lyse the cells. Cells were incubated at room temperature for 15
min. A volume of 10 μl of cell lysate supernatant was mixed
with 50 μl of Luciferase Assay Reagent II from the kit to
measure the luminescence generated by the firefly luciferase.
Immediately following this, 50 μl of Stop & Glow reagent
from the kit was added to the mixture to measure the luminescence
generated by the Renilla luciferase expressed from phRL-TK
or pmrGLO. The amount of luciferase gene expression was determined
as relative luminescence units (RLU), where the value of
luminescence from the firefly luciferase was divided by that of the
Renilla luciferase expressed in the same lysate. The
increase or decrease in the luciferase expression was expressed as
the fold-activity, where the RLU value of a sample of treated cells
was divided by that of an identically prepared sample without
treatment.
In the case of a combination of stable transfection
to express firefly luciferase and transient transfection to express
Renilla luciferase, when X-ray irradiation was applied to
the cells, the cell proliferation would be modified, possibly
affecting the ratio of firefly and Renilla luciferase
activities, regardless of gene expression efficiency. We thus
employed a single luciferase assay for such cases, using the
protein concentration of the cell lysate as a reference for
standardization. At various times after X-ray irradiation, cells
were washed once in PBS, and 300 μl of passive lysis buffer
from the Dual Luciferase Assay kit was added to lyse the cells. The
cells were incubated at room temperature for 15 min. A volume of 10
μl of cell lysate supernatant was mixed with 50 μl of
Luciferase Assay Reagent II included in the kit to measure the
luminescence generated by the firefly luciferase. The protein
concentration was determined by the Bradford method with 5–20
μl of the cell lysate using a BioRad protein assay kit
(Bio-Rad, Hercules, CA, USA).
Recombinant retrovirus vector
construction and stable transfection
After the initial screening of pmirGLO derivative
vectors with miRNA target insertion, we excised 2 miRNA target
sequences out of agarose gels that had been subjected to
electrophoresis of pmirGLO-10-1 and pmirGLO-503-4 after digestion
with NheI and EcoRI. Each of these fragments was
inserted into the XbaI and EcoRI sites of
pRet-880-8-luc to generate pRet-880-8-luc-10-1 and
pRet-880-8-luc-503-4.
One million AmphoPack293 cells (Takara Bio, Inc.)
were seeded onto a 60-mm collagen-coated cell culture dish, and the
following day, they were transfected using a CalPhos™ Mammalian
Transfection kit (Takara Bio, Inc.) with 5 mg of each of the
constructed retrovirus-generating vectors. The virus-containing
conditioned medium was collected 48 h after transfection and passed
through a 0.45-mm filter to remove debris. Polybrene (Sigma-Aldrich
Inc., St. Louis, MO, USA) was added to the filtered medium at a
final concentration of 7.0 μg/ml. This prepared solution was
used as a virus source to infect 1×106 LNCaP cells. The
infected cells were concentrated by treatment with 0.5 μg/ml
puromycin, to which infected cells were resistant, in order to
establish a stably transfected cell line.
Statistical analysis
All values are expressed as the means ± standard
deviations. Differences were assessed with the Student’s unpaired
t-test. For comparisons of more than two groups, a one-way analysis
of variance (ANOVA) was used. Statistical significance was
established at a value of P<0.05.
Results
Changes in the miRNA expression profile
following X-ray irradiation
First, we investigated the miRNA profile in the
LNCaP cells using the miRNA array ver. 1.0 of the GeneChip system
developed by Affymetrix Inc. Total RNA was extracted from LNCaP
cells 4 h after X-ray irradiation at 5 or 10 Gy. The results showed
that the expression levels of many of the miRNAs were altered
following X-ray irradiation, suggesting that they may be involved
in the adaptive responses of cells to radiation, consistent with
reports showing that miRNAs are involved in the cellular response
to radiation (19).
We focused on the miRNAs downregulated after X-ray
irradiation, since we intended to determine whether such miRNAs can
be applied to increase X-ray irradiation-mediated gene expression.
We identified 15 miRNAs that were downregulated in the cells after
exposure to 5 Gy (Table I) or 10
Gy (Table II) irradiation. In
addition, we obtained another list of downregulated miRNAs by using
the miRNA array ver. 2.0 with the total RNA extracted from LNCaP
cells after 10 Gy X-ray irradiation (Table III).
 | Table I.Radiation-induced changes in
expression of miRNAs in LNCaP cells 4 h after X-ray irradiation at
5 Gy. |
Table I.
Radiation-induced changes in
expression of miRNAs in LNCaP cells 4 h after X-ray irradiation at
5 Gy.
| miRNAa | Change ratio | Relative expression
value (without radiation) |
|---|
| has-miR-126 | 0.80 | 27.60 |
| hsa-miR-20b | 0.80 | 106.11 |
| hsa-miR-203 | 0.80 | 50.80 |
| hsa-let-7g | 0.82 | 73.09 |
| hsa-miR-30b | 0.83 | 57.43 |
| hsa-miR-30a | 0.85 | 37.12 |
| hsa-miR-19b | 0.85 | 295.98 |
| hsa-miR-29a | 0.85 | 44.77 |
| hsa-miR-25 | 0.85 | 202.84 |
| hsa-miR-183 | 0.86 | 59.72 |
| hsa-miR-20a | 0.86 | 706.08 |
| hsa-miR-1826 | 0.87 | 2295.58 |
|
has-miR-1207-5p | 0.87 | 91.50 |
| has-miR-105 | 0.87 | 31.77 |
| hsa-miR-181a | 0.88 | 40.73 |
 | Table II.Radiation-induced changes in
expression of miRNAs in LNCaP cells 4 h after X-ray irradiation at
10 Gy. |
Table II.
Radiation-induced changes in
expression of miRNAs in LNCaP cells 4 h after X-ray irradiation at
10 Gy.
| miRNAa | Change ratio | Relative expression
value (without radiation) |
|---|
| has-miR-181a | 0.59 | 40.73 |
|
has-miR-1207-5p | 0.60 | 91.50 |
|
hsa-miR-193a-5p | 0.72 | 77.39 |
| hsa-mi-638 | 0.73 | 138.32 |
| hsa-miR-502-3p | 0.75 | 67.38 |
|
hsa-miR-138-1* | 0.77 | 25.87 |
| hsa-miR-181b | 0.78 | 44.83 |
| hsa-miR-500 | 0.78 | 57.41 |
| hsa-miR-20b | 0.78 | 106.11 |
| hsa-miR-19b | 0.79 | 295.98 |
| hsa-miR-767-5p | 0.80 | 44.19 |
| hsa-miR-149 | 0.80 | 50.79 |
| has-miR-30a | 0.81 | 37.12 |
|
has-miR-193b* | 0.81 | 23.68 |
| hsa-miR-720 | 0.81 | 131.87 |
 | Table III.Radiation-induced changes in
expression of miRNAs in LNCaP cells 6 h after X-ray irradiation at
10 Gy. |
Table III.
Radiation-induced changes in
expression of miRNAs in LNCaP cells 6 h after X-ray irradiation at
10 Gy.
| miRNAa | Change ratio | Relative expression
value (without radiation) |
|---|
| has-miR-3156 | 0.39 | 23.71 |
| hsa-miR-3175 | 0.50 | 77.52 |
| hsa-miR-139-3p | 0.59 | 20.19 |
|
hsa-lmiR-483-5p | 0.64 | 21.66 |
| hsa-miR-198 | 0.66 | 21.39 |
|
hsa-miR-92a-1* | 0.67 | 81.86 |
|
hsa-miR-30c-2* | 0.68 | 28.61 |
| hsa-miR-503 | 0.68 | 95.41 |
| hsa-miR-224 | 0.69 | 48.22 |
We chose miR-181a, -1207-5p, -20b, -19b, -3156,
-3175, -92a-1*, -503 and -224 for the subsequent
quantitative real-time PCR analyses for confirmation of the change
in expression and to assess the time course of the expression
decrease. We chose these miRNA since they exhibited larger
downregulation ratios after irradiation according to the microarray
results, and exhibited higher expression in cells without radiation
exposure. We also had to consider the availability of primers for
the real-time PCR analysis. Regarding Tables I and II, we gave a higher priority to Table II, but chose miRNAs listed in both
of the Tables. According to the results (Fig. 1), the data concerning miR-181a
obtained with the GeneChip and real-time PCR were conflicting.
Although the expression levels of miR-3156 and -3175 decreased
initially for a few hours, they increased rapidly by 12 h after
X-ray irradiation. Regarding miR-20b and -1207, their decreases in
expression reached a trough level 6 h after X-ray irradiation. The
expression levels of miR-92a-1* and miR-19b reached their greatest
decrease 12 h after X-ray irradiation, while miR-224 and miR-503
continued to decrease for up to 24 h after X-ray irradiation. Out
of these miRNAs, we chose miR-503, miR-92a-1* and
miR-19b to evaluate the potential of using their changes in
expression for gene expression control due to their large ratios of
decrease.
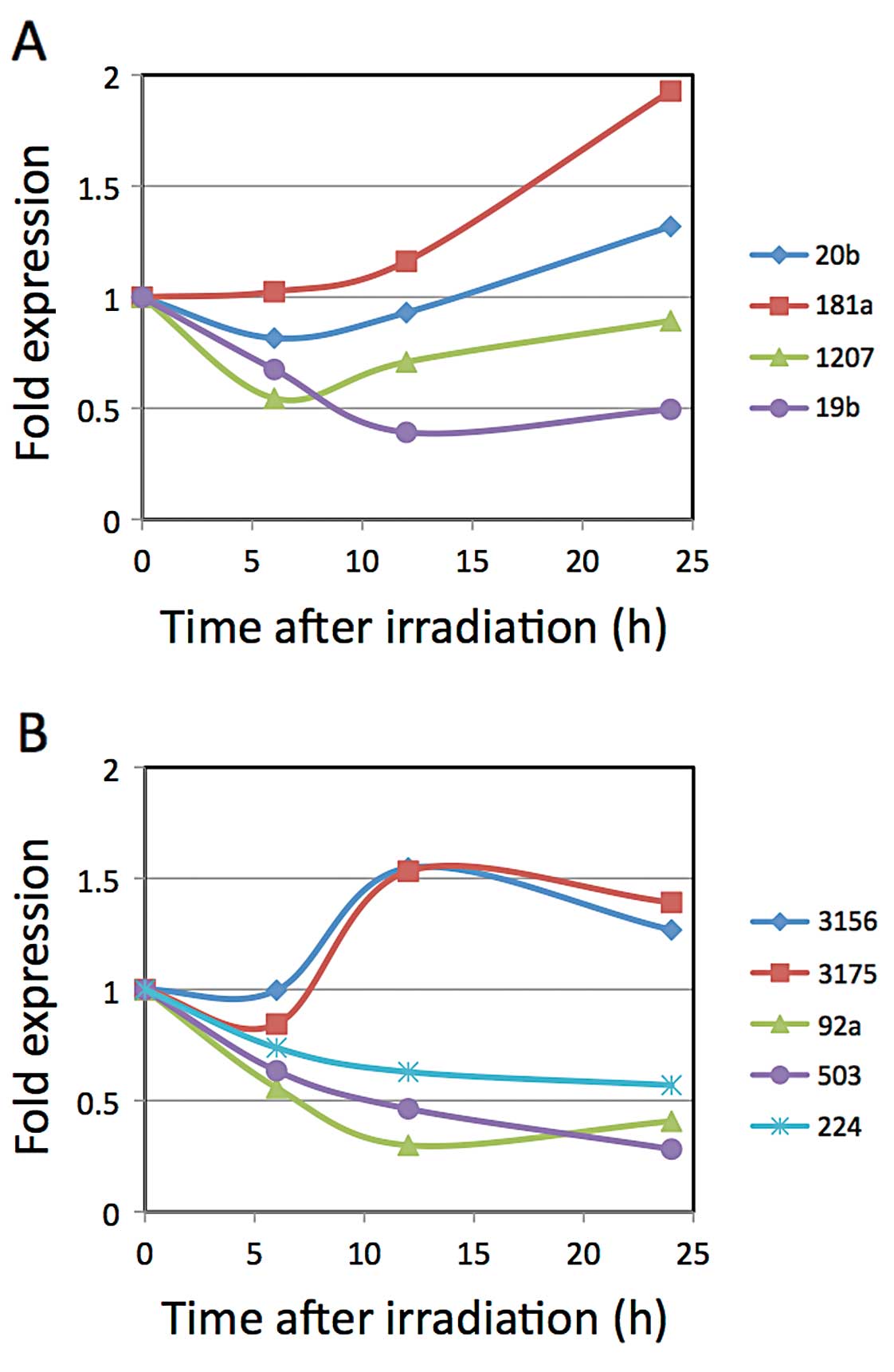 | Figure 1.Kinetics of miRNA expression after
X-ray irradiation. LNCaP cells were irradiated with 10 Gy X-rays.
Total RNAs were extracted at 0, 6, 12 and 24 h after irradiation,
and the cDNAs were synthesized for the real-time PCR analyses. (A)
Expression levels of miR-20b, -181a, -1207-5p and -19b, which were
chosen according to the results of the GeneChip miRNA array ver.
1.0, were analyzed by real-time PCR. (B) Expression levels of
miR-3156, -3175, -92a-1*, -503 and -224, which were
chosen according to the results of the GeneChip miRNA array ver.
2.0, were analyzed by real-time PCR. |
Effects of target sequences composed of
complimentary sequences on the luciferase expression levels
following X-ray irradiation
We constructed plasmid vectors containing the
firefly luciferase gene whose 3′-UTR carried a variety of target
sequences composed of complimentary sequences of the chosen
miRNA(s). As described in Materials and methods, we constructed two
types of target sequences. One was composed of randomly combined
complimentary sequences of the chosen miRNAs and the other
comprised multi-copies of tandemly-aligned complimentary sequence
of the miRNA. We designated vectors containing the former target
sequences as pmirGLO-miRT#5-1, 5-2, 5-3, 5-4, 10-1, 10-2, 20-1,
20-2 and 20-3, and designated the latter as pmirGLO-miRT#19b-2,
19b-4, 92a*-2, 92a*-4, 503-2 and 503-4. Each was transfected into
LNCaP cells and irradiated with 10 Gy X-rays. The cell lysates were
subjected to a dual luciferase assay 9 h after X-ray irradiation
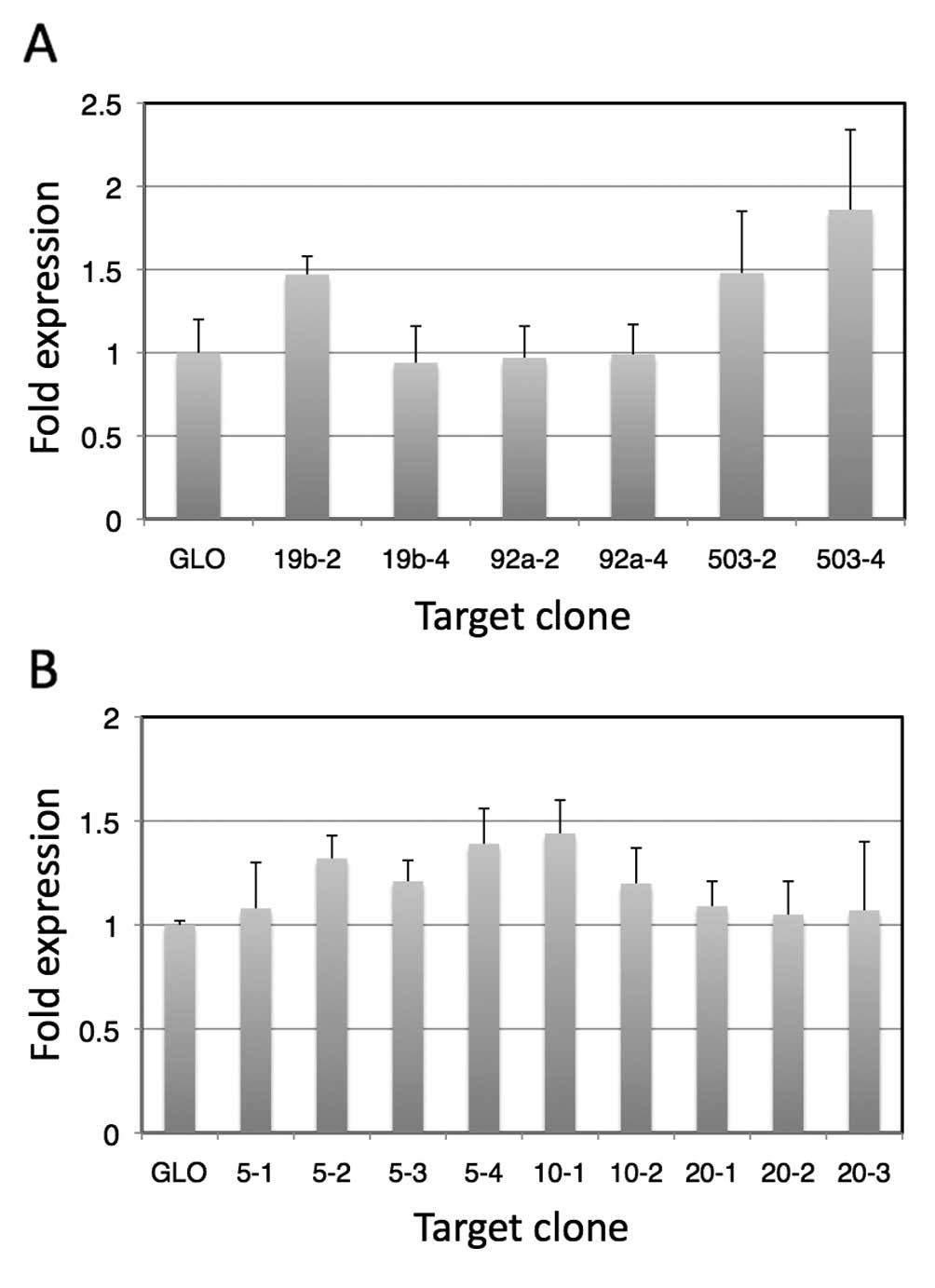
(Fig. 2).
We observed suppressed expression of the luciferase
gene in all of the plasmids containing target sequences. In
addition, when transfected cells were irradiated, the enhancement
ratios of luciferase expression increased compared to those of
cells transfected with pmirGLO. In the case of vectors containing
complementary sequences of a single miRNA, as expected, vectors
containing 4 copies of the complementary sequences showed higher
enhancement ratios for miR-92a* and miR-503, although
this was opposite in the case of the vectors containing
complementary sequences for miR-19b. These results suggest that the
mRNA conformation may be involved in the expression, presumably as
a result of sequestering the target sequences. Among the targets
composed of a single miRNA complementary sequence,
pmirGLO-miRT#503-4 showed the highest enhancement, which was
1.9-fold that without the target sequences. In addition, among the
randomly combined complementary sequences, pmirGLO-miRT#10-1 was
the highest, and showed 1.5-fold enhancement compared to that of
pmirGLO without a target sequence.
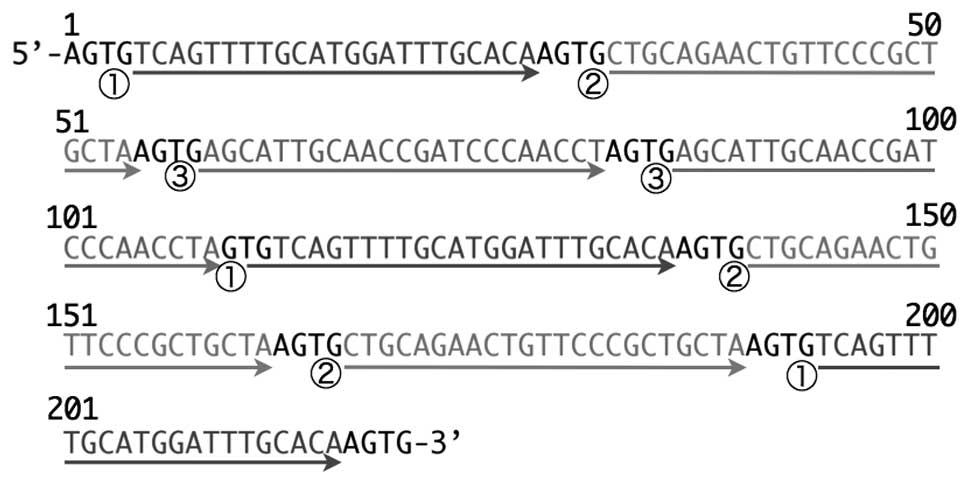
Nucleotide sequence analysis revealed that the
miRT#10-1 target sequence was 218 nucleotides long, and was
composed of 8 complementary sequences, and as shown in Fig. 3, 3 were complementary sequences for
miR-19b and miR-503, and the rest were composed of 2 copies of
miR-92a-1* complementary sequences.
Control of the luciferase gene expression
using the combination of a radiation-responsive promoter and miRNA
target sequence
We then combined the clone 880-8 promoter (a
previously constructed radiation-responsive promoter) and the
target sequences contained in pmirGLO-miRT#503-4 and
pmirGLO-miRT#10-1. The target sequences were PCR-amplified and
cloned downstream of the luciferase gene of pRet-880-8-luc, which
was a vector used to construct a recombinant retrovirus expressing
the luciferase gene under control of the clone 880-8 promoter. The
promoter is efficiently responsive to X-ray irradiation, but its
basal activity is a problem since it expresses genes linked
downstream even in the absence of radiation, and it killed cells at
a high concentration of prodrug in an in vitro suicide gene
therapy simulation study (4).
Thus, by combining these features, we wished to determine whether
the target sequences could suppress the basal activity while
retaining the peak activity after X-ray irradiation. We designated
a vector constructed by introducing the target sequence of
pmirGLO-miRT#503-4 into pRet-880-8-luc as
pRet-880-8-luc-miRT#503-4, and another vector with the target
sequence of pmirGLO-miRT#10-1 as pRet-880-8-luc-miRT#10-1.
Recombinant retroviruses were constructed and infected into LNCaP
cells to generate stably transfected cells. We then obtained stably
transfected LNCaP cells designated LNCaP-880-8-luc-miRT#10-1 using
pRet-880-8-luc-miRT#10-1. However, we could not obtain stably
transfected cells with pRet-880-8-luc-miRT#503-4, although the
reason remains unknown. It is possible that the target sequence may
contain some type of signal sequence affecting the stability of the
recombinant virus.
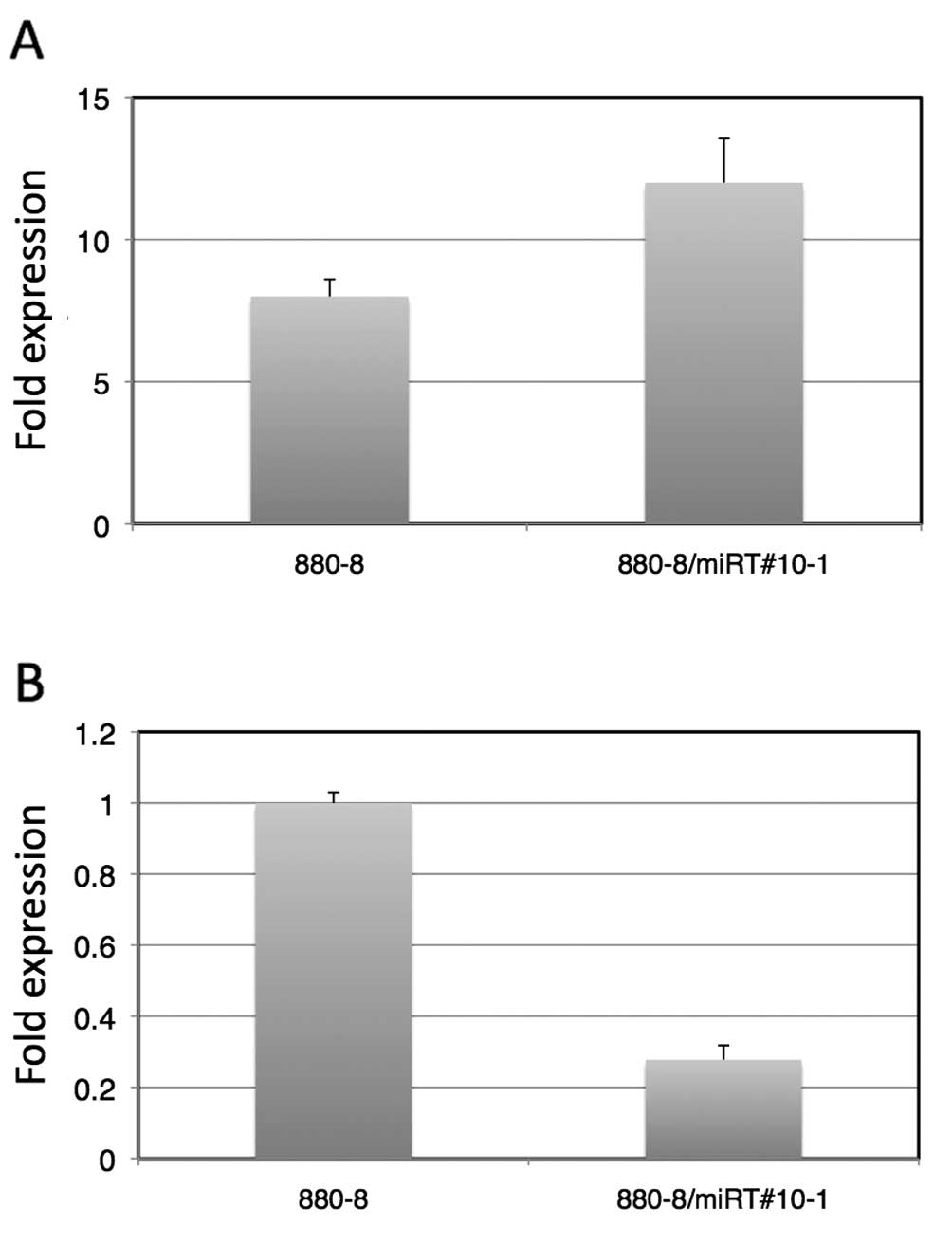
As shown in Fig.
4A, 9 h after the LNCaP-880-8-lucmiRT#10-1 cells were
irradiated with 10 Gy X-rays, the luciferase activity was enhanced
12-fold to that of the level of the LNCaP-880-8-luc-miRT#10-1 cells
without X-ray irradiation. This was an ~1.5-fold enhancement
compared to that of the LNCaP-880-8-luc without a target sequence.
Fig. 4B shows a comparison of the
luciferase activities of LNCaP-880-8-luc and
LNCaP-880-8-luc-miRT#10-1 without X-ray irradiation, indicating
that there was a 70% suppression of luciferase expression by the
target sequence contained in LNCaP-880-8-luc-miRT#10-1. As
expected, the target sequence of miRNAs was able to be used for
fine-tuning the gene expression control induced by another
mechanism.
Discussion
In the present study, we initially investigated
change in the miRNA profiles in LNCaP cells after radiation. We
confirmed that there were several miRNAs that exhibited changes in
their expression levels in the prostate cancer cell line after
X-ray irradiation as determined by the microarray and quantitative
real-time PCR analyses. In addition, reports have suggested that
expression levels of miRNAs are altered in response to stimuli
including ultrasound (10),
anticancer agents (11),
hyperthermia (12) and hypoxia
(13). As Simone et al
(20) demonstrated that the
oxidative stress induced after radiation alters the expression
levels of miRNAs, the changes in miRNA expression after radiation
that were observed in our study may have been caused by oxidative
stress. It was also suggested that such miRNA expression changes
may be involved in the responses of cells to stimulation, including
apoptosis (3). In our case,
several miRNAs may have played a role in the response of the cells
to radiation.
We employed two different versions of
GeneChip® micro-arrays for the miRNA profile analyses.
The lists of miRNAs shown in Tables
I and III were substantially
different from each other. The reason for the differences remains
unknown, but may have been derived from inter-experimental
differences in techniques or the differences between the two
versions of the arrays (e.g. sensitivity, sample numbers). We
consider that it may have been due to inter-experimental
differences, since most of the miRNAs (8 out of 9) listed as being
decreased after radiation were confirmed to be decreased after
X-ray irradiation by real-time PCR.
We focused on the miRNAs that exhibited decreased
expression levels after radiation to determine whether their target
sequences could be applied for gene expression regulation. Few
reports exist concerning the use of miRNAs to regulate gene
expression. For instance, Xie et al (17) found that the introduction of a
target sequence from a liver-specific miRNA into the 3′UTR of a
gene in an expression vector for the central nervous system
resulted in suppression of the expression in the liver after
systemic administration. In the present study, we attempted to use
radiation-responsive miRNAs rather than tissue-specific miRNAs. We
introduced the target sequences of miRNAs whose expression was
decreased following radiation into a gene of interest so that the
gene could be suppressed under normal conditions, but retain a
higher level of expression following radiation exposure. We
demonstrated that this was possible. However, the degree of miRNA
expression changes induced by radiation was limited. Thus, we
applied the system for fine-tuning another gene regulation
system.
The system was combined with a radiation-responsive
promoter that was previously developed by us. The promoter was
sensitively responsive to radiation in vitro, and in
vivo, and its transcriptional activity was activated after
radiation. However, the promoter was active to some extent even
without radiation. Thus, in a suicide gene therapy simulation study
in vitro, cells were killed in the presence of a high
concentration of prodrug even without radiation. We inserted two
target sequences that enhanced gene expression after radiation into
the 3′UTR of the luciferase gene under control of the
radiation-responsive promoter. The transfected cells successfully
showed gene regulation that was more strict than that by
LNCaP-880-8-luc with the promoter alone, with approximately
one-third of the basal activity (activity without radiation) and a
1.5-fold increase in the enhancement ratio after radiation.
We did not obtain stably transfected cells with the
luciferase gene containing a target sequence of miRT#503-4, which
exhibited the best results in the transient transfection experiment
among the various constructed target sequences, since all of the
cells were killed during the process of concentrating the
transfected cells by cultivating them in medium containing
puromycin. Although the reason for the cell death remains unknown,
we considered that the sequence may have affected the stability of
the recombinant retrovirus. We are still trying to obtain stably
transfected LNCaP cells with the luciferase gene carrying the
target sequence under the control of the clone 880-8 promoter.
Our present findings showed that this method
utilizing changes in the expression levels of miRNAs in response to
stimulation may be used for fine-tuning a gene regulation system,
even though it may be difficult to apply alone for the purpose of
obtaining sufficient gene regulation. The stimulation-responsive
promoter developed using our method was associated with some basal
activity. Thus, this method utilizing miRNA expression changes
should be useful for promoters developed according to our method.
At present, we are aiming to devise a more effective application of
miRNA expression change for developing new gene regulation
systems.
Acknowledgements
This study was supported, in part, by
Grants-in-Aid for Scientific Research (C) (21500403) and for Young
Scientists (B) (20377253 and 25861412) from the Japan Society for
the Promotion of Science, and, in part, by a Collaborative Research
Project of the Wakasa Wan Energy Research Center. The authors thank
Dr Loreto B. Feril Jr for his critical reading of the
manuscript.
References
|
1.
|
Steiner MS and Gungrich JR: Gene therapy
for prostate cancer: where are we now? J Urol. 164:1121–1136. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Mauceri HJ, Hanna NN, Staba MJ, Beckett
MA, Kufe DW and Weichselbaum RR: Radiation-inducible gene therapy.
CR Acad Sci III. 322:225–228. 1999. View Article : Google Scholar
|
|
3.
|
Ogawa R, Lee SI, Kagiya G, Hirano H,
Fukuda S, Kondo T and Kodaki T: Construction of X-ray-inducible
promoters through cis-acting element elongation and
error-prone polymerase chain reaction. J Gene Med. 10:316–324.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Morii A, Ogawa R, Watanabe A, Kakutani S,
Zhao QL, Kume K, et al: Regulation of gene expression in prostate
cancer cells with an artificially constructed promoter responsive
to radiation. Gene Ther. 19:219–227. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Esquela-Kerscher A and Slack FJ: Oncomirs
- microRNA with a role of cancer. Nat Rev Cancer. 6:259–269. 2006.
View Article : Google Scholar
|
|
6.
|
Josson S, Sung SY, Lao K, Cung LW and
Johnstone PA: Radiation modulation of microRNA in prostate cancer
cell lines. Prostate. 68:1599–1606. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Shin S, Cha HJ, Lee EM, Lee SJ, Seo SK,
Jin HO, Park IC, Jin YW and An S: Alteration of miRNA profiles by
ionizing radiation in A549 human small-cell lung cancer cells. Int
J Oncol. 35:81–86. 2009.PubMed/NCBI
|
|
8.
|
Chaudhry MA, Sachdeva H and Omaruddin RA:
Radiation-induced microRNA modulation in glioblastoma cells
differing in DNA-repair pathways. DNA Cell Biol. 29:553–561. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Oh JS, Kim JJ, Byun JY, et al: Lin28-let7
modulates radiosensitivity of human cancer cells with activation of
K-Ras. Int J Radiat Oncol Biol Phys. 76:5–8. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Ogawa R, Morii A and Watanabe A:
Ultrasound stimulation induces microRNA expression changes that
could be involved in sonication-induced apoptosis. J Med
Ultrasonics. 39:207–216. 2012. View Article : Google Scholar
|
|
11.
|
Fornari F, Gramantieri L, Giovannini C, et
al: miR-122/cyclin G1 interaction modulates p53 activity and
affects doxorubicin sensitivity of human hepatocarcinoma cells.
Cancer Res. 69:5761–5767. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Wilmink GJ, Roth CL, Ibey BL, et al:
Identification of microRNAs associated with hyperthermia-induced
cellular stress response. Cell Stress Chaperones. 15:1027–1038.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Kulshreshtha R, Ferracin M, Wojcik SE, et
al: A microRNA signature of hypoxia. Mol Cell Biol. 27:1859–1867.
2007. View Article : Google Scholar
|
|
14.
|
Brown BD, Venneri MA, Zingale A, Sergi
Sergi L and Naldini L: Endogenous microRNA regulation suppresses
transgene expression in hematopoietic lineages and enables stable
gene transfer. Nat Med. 12:585–591. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Brown BD, Gentner B, Cantore A, Colleoni
S, Amendola M, Zingale A, Baccarini A, Lazzari G, Galli C and
Naldini L: Endogenous microRNA can be broadly exploited to regulate
transgene expression according to tissue, lineage and
differentiation state. Nat Biotechnol. 25:1457–1467. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Wu C, Lin J, Hong M, Choudhury Y, Balani
P, Leung D, Dang LH, Zhao Y, Zeng J and Wang S: Combinatorial
control of suicide gene expression by tissue-specific promoter and
microRNA regulation for cancer therapy. Mol Ther. 17:2058–2066.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Xie J, Xie Q, Zhang H, Ameres SL, Hung JH,
Su Q, He R, Mu X, Ahmed S, Park S, Kato H, Li C, Mueller C, Weng Z,
Flotte TR, Zamore PD and Gao G: MicroRNA-regulated, systemically
delivered rAVV9: a step closer to CNS-restricted transgene
expression. Mol Ther. 19:526–535. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Sambrook J and Russell DW: Molecular
Cloning: A Laboratory Manual. 3rd edition. Cold Spring Harbor
Laboratory Press; New York: 2001
|
|
19.
|
Li B, Shi XB, Nori D, Chao CK, Chen AM,
Valicenti R and White Rde V: Down-regulation of microRNA106b is
involved in p21-mediated cell cycle arrest in response to radiation
in prostate cancer cells. Prostate. 71:567–574. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Simone NL, Soule BP, Ly D, et al: Ionizing
radiation-induced oxidative stress alters miRNA expression. PLoS
One. 4:e63772009. View Article : Google Scholar : PubMed/NCBI
|