Introduction
Congenital heart defects (CHDs) are the most common
type of major birth defect, and account for the majority of
morbidity and mortality related to birth defects (1). Tetralogy of Fallot (TOF) is the most
commonly observed conotruncal congenital heart defect, accounting
for 10% of all CHDs, with an incidence of 3.6 per 10,000 live
births (2). The treatment of
these patients has evolved significantly over the last few decades
(3); however a genetic
explanation, particularly epigenetic factors, is lacking for the
failure of cardiac development in the majority of children with
TOF. Epigenetic changes refer to the heritable changes in genome
function that occur without a change in primary DNA sequence,
characterized by covalent modifications of cytosine bases and
histones, and changes in the positioning of nucleosomes (4). They are fundamental to the
regulation of many cellular processes, including gene and microRNA
expression, DNA-protein interactions, the suppression of
transposable element mobility, cellular differentiation,
embryogenesis, X-chromosome inactivation and genomic imprinting
(5).
Currently, the most widely studied epigenetic
modification in humans is DNA methylation. DNA methylation occurs
almost exclusively in the context of CpG dinucleotides, which
control the transcriptional activity of genes by various mechanisms
(6). The hypomethylation of
genomic DNA has been reported to contribute to chromosome
instability and to alter gene expression, cell differentiation and
apoptosis during embryogenesis (7). Long interspersed nuclear element-1
(LINE-1) constitutes 17–25% of the human genome (8). Since LINE-1 sequences are highly
repeated and widely interspersed human retrotransposons, their
methylation level can serve as a surrogate marker for global
genomic DNA methylation (9).
LINE-1 hypomethylation is frequently observed in certain diseases,
such as colon cancer (10),
neural tube defects (NTDs) (11)
and systemic lupus erythematosus (SLE) (12). LINE-1 hypomethylation have been
proven to be associated with the increased occurrence of
non-syndromic CHDs (9). We have
previously shown that lower LINE-1 methylation levels are
associated with an increased risk of TOF (13). However, the molecular mechanisms
leading to LINE-1 hypomethylation in patients with TOF are not yet
well understood.
DNA methylation is catalyzed and maintained by a
family of DNA methyltransferases (DNMTs), including DNMT1, DNMT3A
and DNMT3B. DNMT1 is known as a maintenance DNMT; it has a high
preference for hemimethylated DNA substrates and maintains
methylation patterns during DNA replication and accurately
replicates genomic DNA methylation patterns during cell division in
mammalian cells (14). DNMT3A and
DNMT3B are responsible for the de novo methylation of DNA
during early embryogenesis, thereby establishing a somatic
methylation pattern (15).
Methyl-CpG-binding domain protein 2 (MBD2) is one of the methyl-CpG
binding proteins (MBPs); it binds methylated cytosine and attracts
chromatin inactivation complexes containing histone deacetylase.
Moreover, MBD2 has also been shown to possess demethylase activity
(16). The aberrant expression of
DNMTs and MBD2 may influence the DNA methylation pattern and
consequently lead to diseases. The overexpression of DNMTs has been
found in several types of cancer, such as lung cancer (17), breast cancer (18) and hepatocellular carcinoma
(19). Moreover, lower mRNA
levels of DNMTs have been reported in patients with spermatogenic
arrest (20), systemic lupus
erythematosus (21) and atopic
dermatitis (22). Of note,
transcript levels of DNMTs, showing no differences between patients
and controls, have also been reported in patients with systemic
lupus erythematosus (23).
However, little is known about the variations in the mRNA levels of
DNMT1, DNMT3A, DNMT3B and MBD2 and their association with the
LINE-1 methylation status in patients with TOF.
In this study, to obtain a deeper understanding of
the role of epigenetic mechanisms in patients with TOF, we measured
and compared the mRNA levels of DNMTs and MBD2 and the methylation
pattern of LINE-1 in normal and TOF samples. The association
between the transcript levels of DNMTs, MBD2 and the LINE-1
methylation status was also investigated in normal and TOF
samples.
Materials and methods
Patients and controls
In this study, 48 patients with TOF were recruited
from the Children’s Hospital of Fudan University, Shanghai, China,
including 31 males (64.58%) and 17 females (35.42%), age ranging
from 1 to 48 months (14.88±11.27, mean ± SD). Patients were
diagnosed by an echocardiogram, and the diagnoses were confirmed by
surgery. Normal heart tissue samples were obtained from autopsy
cases at the Department of Forensic Medicine, Fudan University.
Samples from 16 healthy control subjects who had died by traffic
accidents were also recruited in this study, including samples from
10 males (62.50%) and 6 females (37.50%), age ranging from 0.5 to
38 years (20.91±14.22, mean ± SD). The clinical features of the
study subjects are summarized in Table I. To exclude the tissue
heterogeneity that may affect methylation levels, all the tissue
samples obtained from right ventricular outflow tracts were saved
in RNAlater® (Ambion, Inc., Austin, TX, USA) immediately
after surgical resection or autopsy and stored until use.
 | Table IClinical characteristics of TOF and
control samples. |
Table I
Clinical characteristics of TOF and
control samples.
| Characteristic | TOF (n=48) | Control (n=16) |
|---|
| Age (mean ± SD) | 14.88±11.27
(months) | 20.91±14.22
(years) |
| Gender, n (%) |
| Male | 31 (64.58) | 11 (68.75) |
| Female | 17 (35.42) | 5 (31.25) |
This study was approved by the local Ethics
Committee of Fudan University. Written informed consent was
obtained from the parents and relatives of all the study
participants.
RNA extraction and quantitative RT-PCR
(qRT-PCR)
Total RNA was extracted from the heart tissue
samples using TRIzol reagent (Invitrogen, Carlsbad, CA, USA)
according to the manufacturer’s instructions. RNA was
reverse-transcribed using the PrimeScript RT reagent kit with gDNA
Eraser (Perfect Real Time) (Takara Bio, Inc., Shiga, Japan) and the
integrity of the synthesized cDNA was confirmed using
glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as an endogenous
control. qRT-PCR was performed using SYBR Premix Ex Taq GC (Perfect
Real Time) (Takara Bio, Inc.) in a 10 μl reaction volume,
containing 5 μl SYBR Premix Ex Taq GC, 0.2 μM of each primer, 0.2
unit ROX1 Reference Dye and 100 ng cDNA. The reactions were
performed in triplicate and analyzed using an ABI 7900 Sequence
Detection System (Applied Biosystems, Carlsbad, CA, USA). Relative
expression levels were calculated according to the standard
2−ΔΔCt method using beta-2 microglobulin (B2M) and the
GAPDH gene as an endogenous control for normalization. The primer
sequences used in qRT-PCR analysis are listed in Table II.
 | Table IIPrimer sequences and product lengths
for qPT-PCR analysis. |
Table II
Primer sequences and product lengths
for qPT-PCR analysis.
| Genes | Forward primer
(5′→3′) | Reverse primer
(5′→3′) | Product length
(bp) |
|---|
| DNMT1 |
AAACCCCTTTCCAAACCTCG |
CTGGTGCTTTTCCTTGTAATCC | 101 |
| DNMT3A |
CCAAGTTCAGCAAAGTGAGGAC |
TGGACTGGGAAACCAAATACC | 145 |
| DNMT3B |
TCCCAGCTCTTACCTTACCATC |
ATCTCCACTGTCTGCCTCCA | 151 |
| MBD2 |
TTCAAGGAGTTGGTCCAGGTAG |
GCAGGGTTCTTTTCCACAGC | 121 |
| B2M |
TGCTGTCTCCATGTTTGATGTATCT |
TCTCTGCTCCCCACCTCTAAGT | 161 |
| GAPDH |
AGAAGGCTGGGGCTCATTTG |
AGGGGCCATCCACAGTCTTC | 220 |
DNA extraction and sodium bisulfite
conversion
Genomic DNA was extracted from the heart tissue
samples using a QIAamp DNA Micro kit (Qiagen, Hilden, Germany)
according to the manufacturer’s instructions. The concentration and
purity of the DNA were determined by absorbance at 260 and 280 nm
using a NanoDrop™ 1000 Spectrophotometer (Thermo Scientific,
Wilmington, MA, USA) on an agrose gel. Sodium bisulfite
modification for the extracted DNA was performed using an EZ DNA
Methylation kit™ (Zymo Research Corp., Orange, CA, USA) according
to the manufacturer’s instructions. The bisulfite-converted DNA was
resuspended in 10 μl elution buffer and stored at −80°C until the
samples were ready for use.
MassARRAY quantitative methylation
analysis
The Sequenom MassARRAY platform was used to perform
the quantitative methylation analysis of LINE-1 as previously
described (13). The primers used
in this study were designed using EpiDesigner (http://epidesigner.com). Based on the analyzed
information for the LINE-1 promoter region, we designed the primers
as follows: forward, cagtaatacgactcactatagggagaagg-TTTTATT
AGGGAGTGTTAGATAGTGGG and reverse,
aggaagagag-CCCCAAAAATAAAACCTACAAAAAC. The analyzed sequence
represents a 468 base pair fragment (positions 835–386) in the
5′-untranslated region (5′-UTR) of the LINE-1 element.
Briefly, bisulfite-treated DNA was amplified with
primers and the PCR products were spotted on a 384-pad SpectroChip
(Sequenom, Inc., San Diego, CA, USA) and followed by spectral
acquisition on a MassARRAY analyzer. The spectra and the
methylation values of matrix-associated laser desorption/ionization
time-of-flight mass spectrometry (Sequenom, Inc.) were collected
and analyzed using EpiTYPER software (version 1.0; Sequenom,
Inc.).
Statistical analysis
Data were analyzed using GraphPad Prism (version
5.0; GraphPad Software Inc., San Diego, CA, USA) and SPSS software
(version 13.0; SPSS Inc., Chicago, IL, USA). The Mann-Whitney U
test was performed to evaluate the significance of any differences
between the TOF and control groups. Spearman’s rank correlation was
used to examine the correlation between two continuous variables.
All statistical analyses were two-sided and a P-value <0.05 was
considered to indicate a statistically significant difference.
Results
mRNA levels of DNMT1, DNMT3A, DNMT3B and
MBD2 in patients with TOF and the control samples
We performed qRT-PCR to determine the expression
levels of DNMT1, DNMT3A, DNMT3B and MBD2 genes in the 48 TOF and 16
normal samples. The patients with TOF had statistically significant
lower mRNA levels of DNMTs (DNMT1, DNMT3A and DNMT3B) compared with
the controls (P<0.001). The MBD2 gene also showed a
significantly decreased mRNA level in the TOF samples (P<0.001)
(Table III). Of note, the DNMT1
and DNMT3B mRNA levels were significantly decreased in the TOF
samples compared with the normal controls (P<0.0001). Moreover,
in the control group, when the age was taken into account,
Spearman’s rho correlation coefficients were obtained with respect
to all the enzymes: r=0.046, P=0.876 (DNMT1); r=0.169, P=0.530
(DNMT3A); r=−0.221, P=0.411 (DNMT3); r=−0.013, P=0.961 (MBD2);
however, no significant correlations were observed. In the patient
groups, two significant negative correlations with age were
observed for DNMT1 (r=−0.327, P=0.023) and DNMT3A (r=−0.292,
P=0.044), while DNMT3B and MBD2 showed no significant correlation
(r=−0.069, P=0.640; r=−0.077, P=0.605; respectively). When
considering the gender difference, we found no significant
difference or correlation between DNMT1, DNMT3A, DNMT3B and MBD2 in
the control and TOF samples (P>0.05).
 | Table IIImRNA level of DNMT1, DNMT3A, DNMT3B
and MBD in the control samples and patients with TOF. |
Table III
mRNA level of DNMT1, DNMT3A, DNMT3B
and MBD in the control samples and patients with TOF.
| Genes | Control (mean ± SD,
n=16) | TOF (mean ± SD,
n=48) | Difference | P-valuea |
|---|
| DNMT1 | 4.186±2.77 | 1.245±0.791 | 2.941±1.78 | <0.0001 |
| DNMT3A | 2.797±1.78 | 1.638±0.692 | 1.159±1.24 | 0.0241 |
| DNMT3B | 17.31±21.21 | 1.678±3.67 | 15.63±12.45 | <0.0001 |
| MBD2 | 2.450±1.78 | 0.992±0.375 | 1.459±1.08 | 0.0007 |
Analysis of the correlation between the
mRNA levels of DNMT1, DNMT3A, DNMT3B and MBD2
To determine the correlation between the mRNA levels
of DNMT1, DNMT3A, DNMT3B and MBD2 in the control samples and
patients with TOF, we performed several Spearman’s correlation
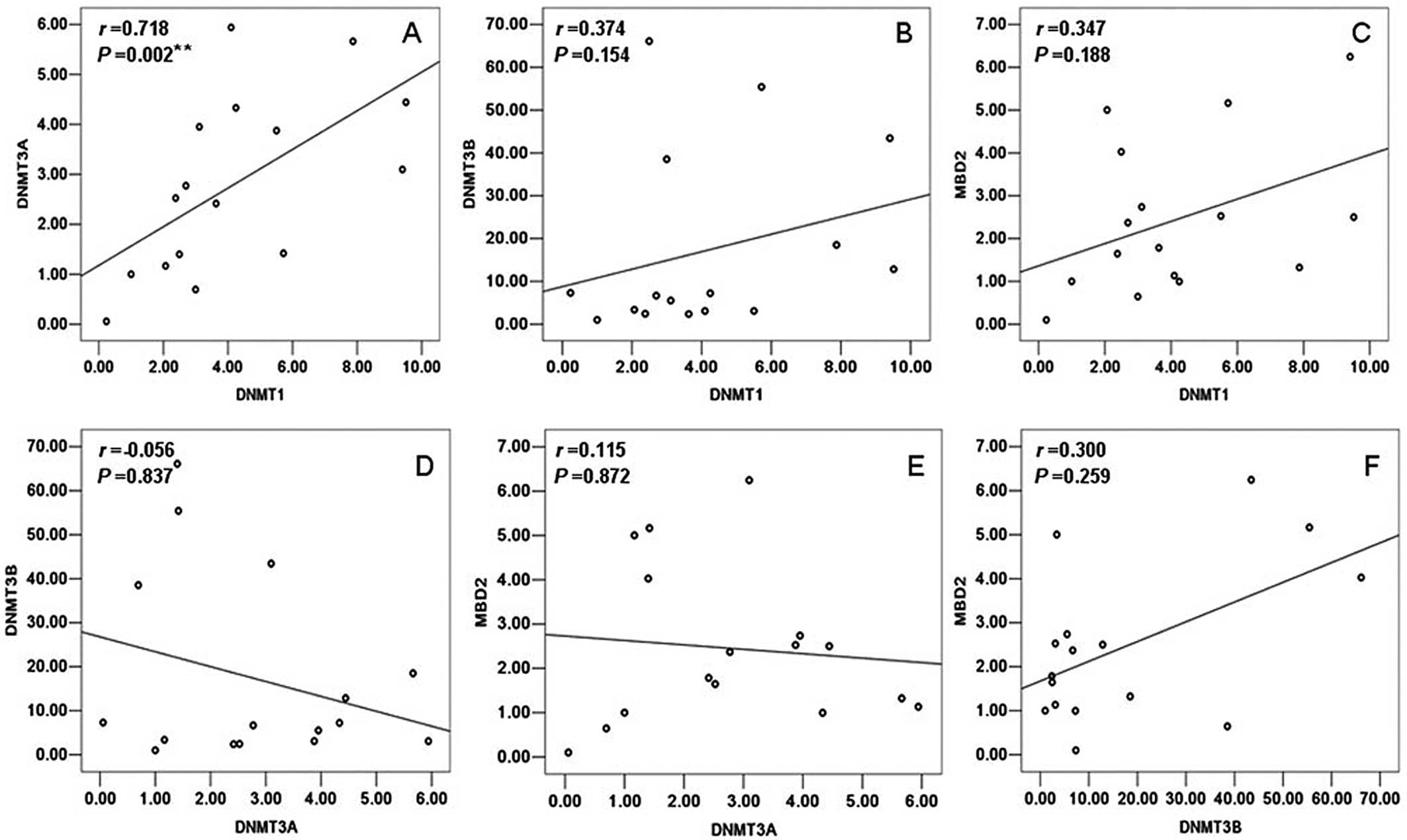
tests by SPASS 13.0. In the control samples, a statistically
significant positive correlation was observed only between DNMT1
and DNMT3A (r=0.718, P=0.002) (Fig.
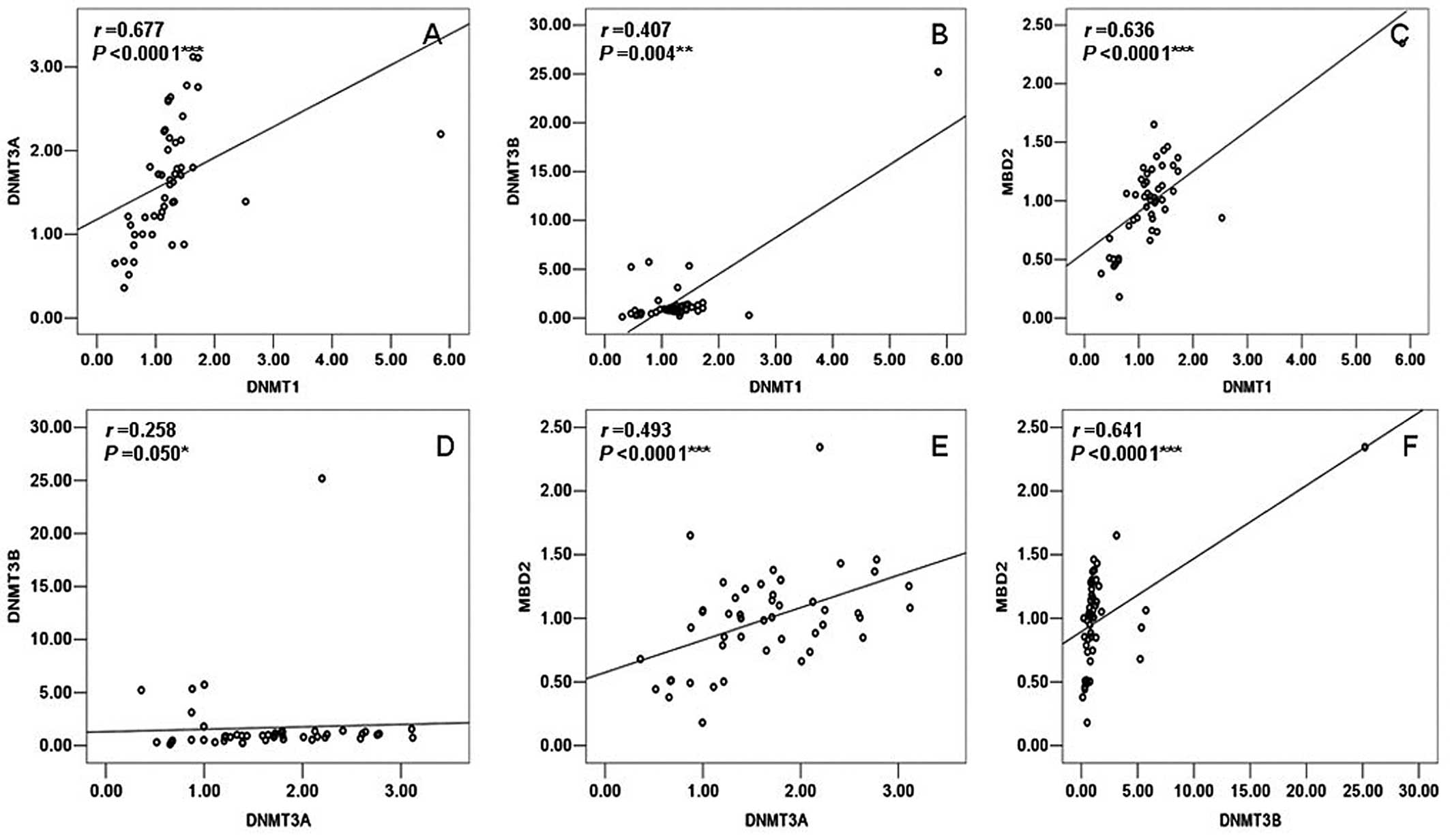
1). However, DNMT1 had a significant positive correlation with
DNMT3A (r=0.677, P<0.0001), DNMT3B (r=0.407, P=0.004) and MBD2
(r=0.636, P<0.0001) in the group of TOF patients. Moreover,
DNMT3A also showed a significant positive correlation with DNMT3B
(r=0.258, P=0.050) and MBD2 (r=0.493, P<0.0001); DNMT3B had a
significant positive correlation with MBD2 (r=0.641, P<0.0001)
in the patients with TOF (Fig.
2).
LINE-1 methylation levels in patients
with TOF and the control samples
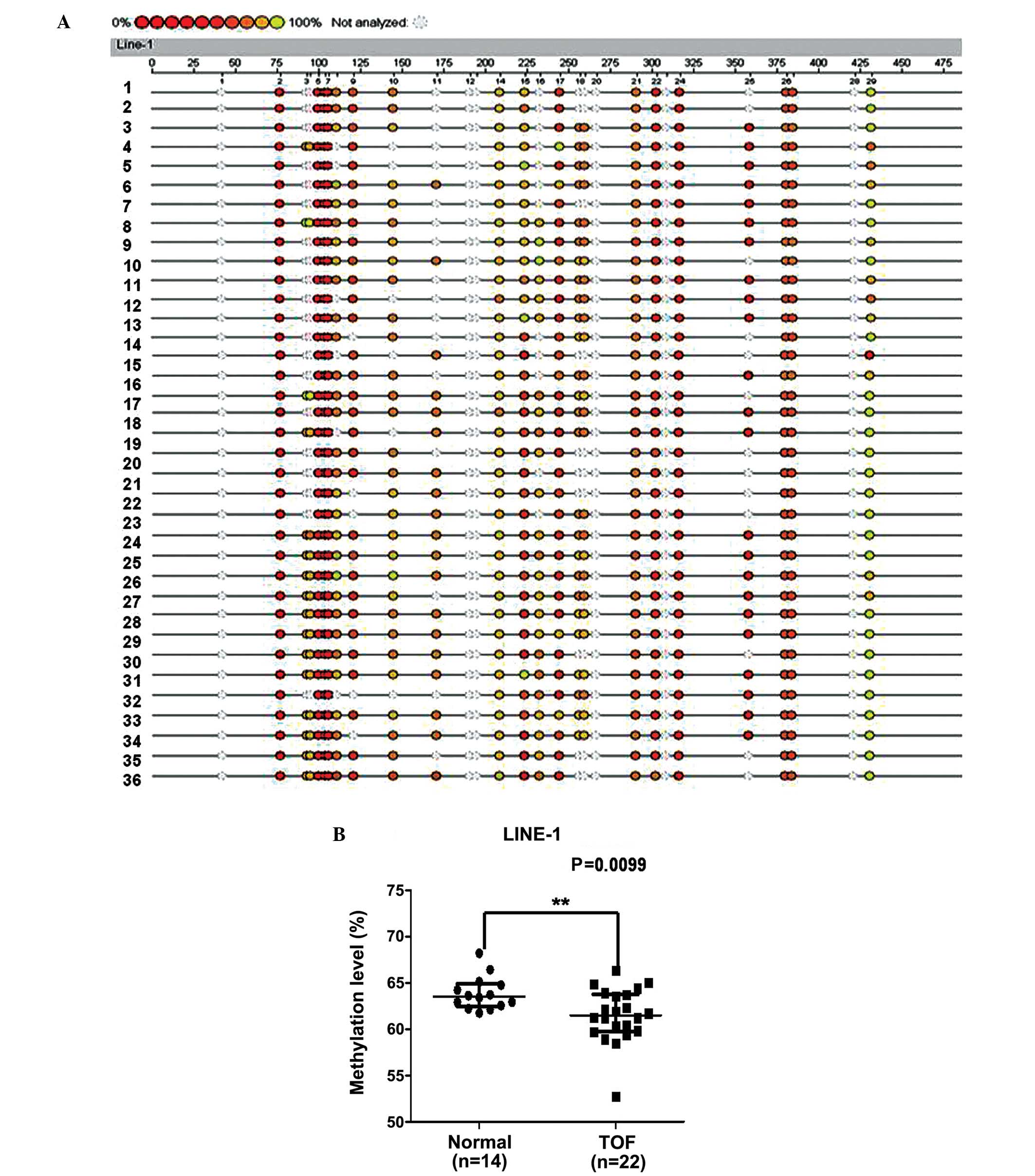
To explore the global DNA methylation status, we
analyzed the methylation status of LINE-1 in a similar region of
the cardiac samples obtained from 22 patients with TOF and 14
control samples. The methylation level of LINE-1 was significantly
lower in the patients with TOF with a median of 61.50%
[interquartile range (IQR), 59.78–63.77] compared with 63.54% (IQR,
62.49–64.88) among the controls (P=0.0099) (Fig. 3).
Moreover, taking into consideration the age or
gender differences in the individual samples, we analyzed the
correlation between age or gender and the LINE-1 methylation status
in the normal and TOF samples. In the normal group, no correlation
between age and the LINE-1 methylation level was observed
(r=0.2709, P=0.3488). The males and females had similar methylation
values (63.32 vs. 63.54%, median) and showed no significant
difference (P=0.9527). In the TOF patient panels, there was no
correlation observed between age and the methylation status of
LINE-1 (r=−0.3980, P=0.0666). Although the females showed a higher
methylation level than the males (62.04 vs. 61.25%, median), no
statistically significant difference was observed (P=0.7074).
Correlation between the mRNA levels of
DNMT1, DNMT3A, DNMT3B and MBD2 and LINE-1 methylation status
To ascertain whether a correlation exists between
the LINE-1 methylation status and the mRNA levels of DNMT1, DNMT3A,
DNMT3B and MBD2 and whether such a correlation exists in both the
control and TOF samples, we analyzed the data from the same
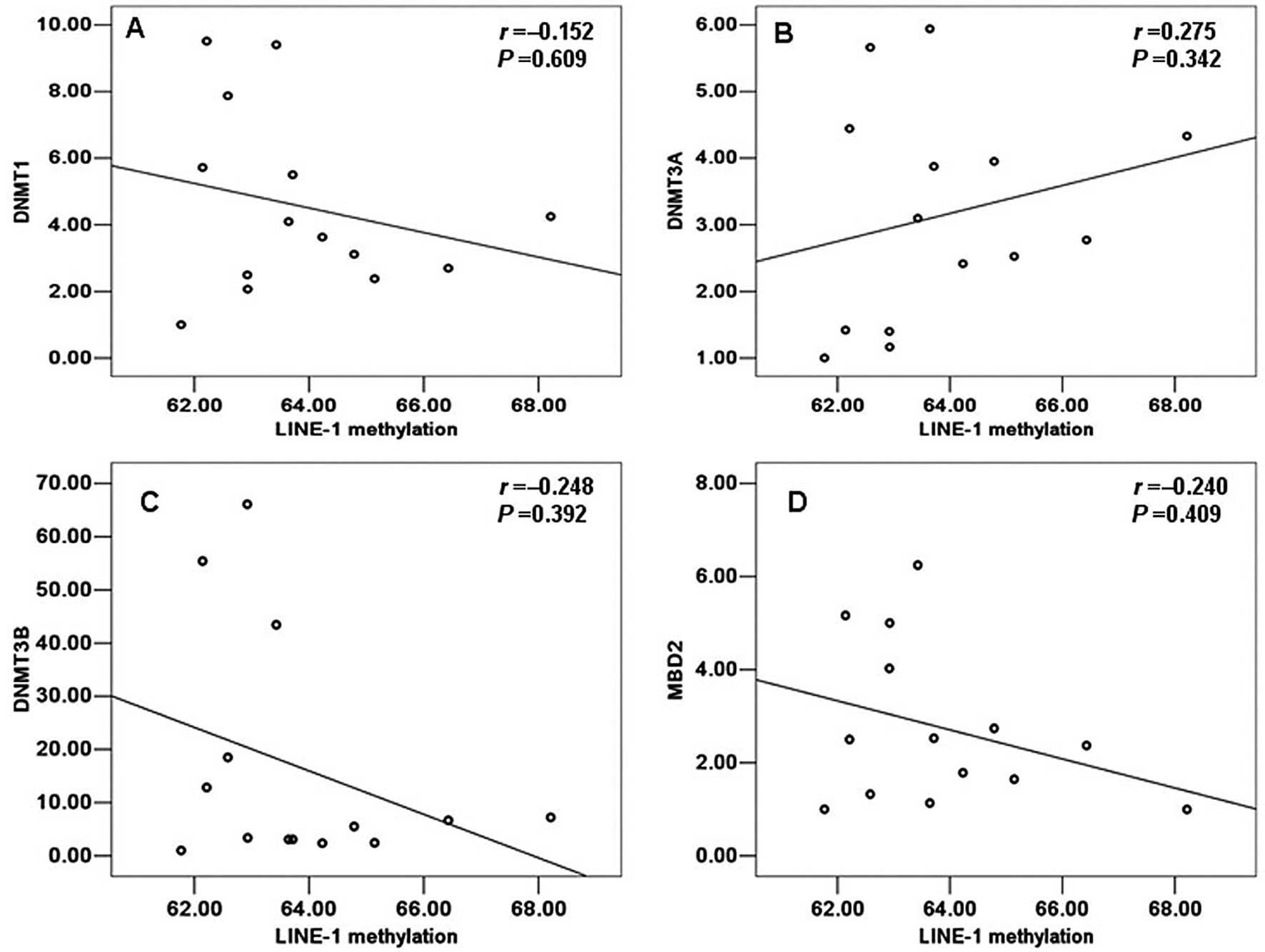
individual from 14 normal and 22 TOF samples by SPASS 13.0. A
positive correlation with the LINE-1 methylation status was
detected only for DNMT3A in the control group, but no statistically
significant value was observed (r=0.275, P=0.342) (Fig. 4). Moreover, in the control group,
DNMT1, DNMT3B and MBD2 all a showed negative correlation with the
LINE-1 methylation status and this was not statistically
significant (r=−0.152, P=0.609; r=−0.248, P=0.392; r=−0.240,
P=0.409; respectively). However, on the contrary, in the group of
TOF patients, DNMT3A showed a negative correlation with the LINE-1
methylation status but this was not statistically significant
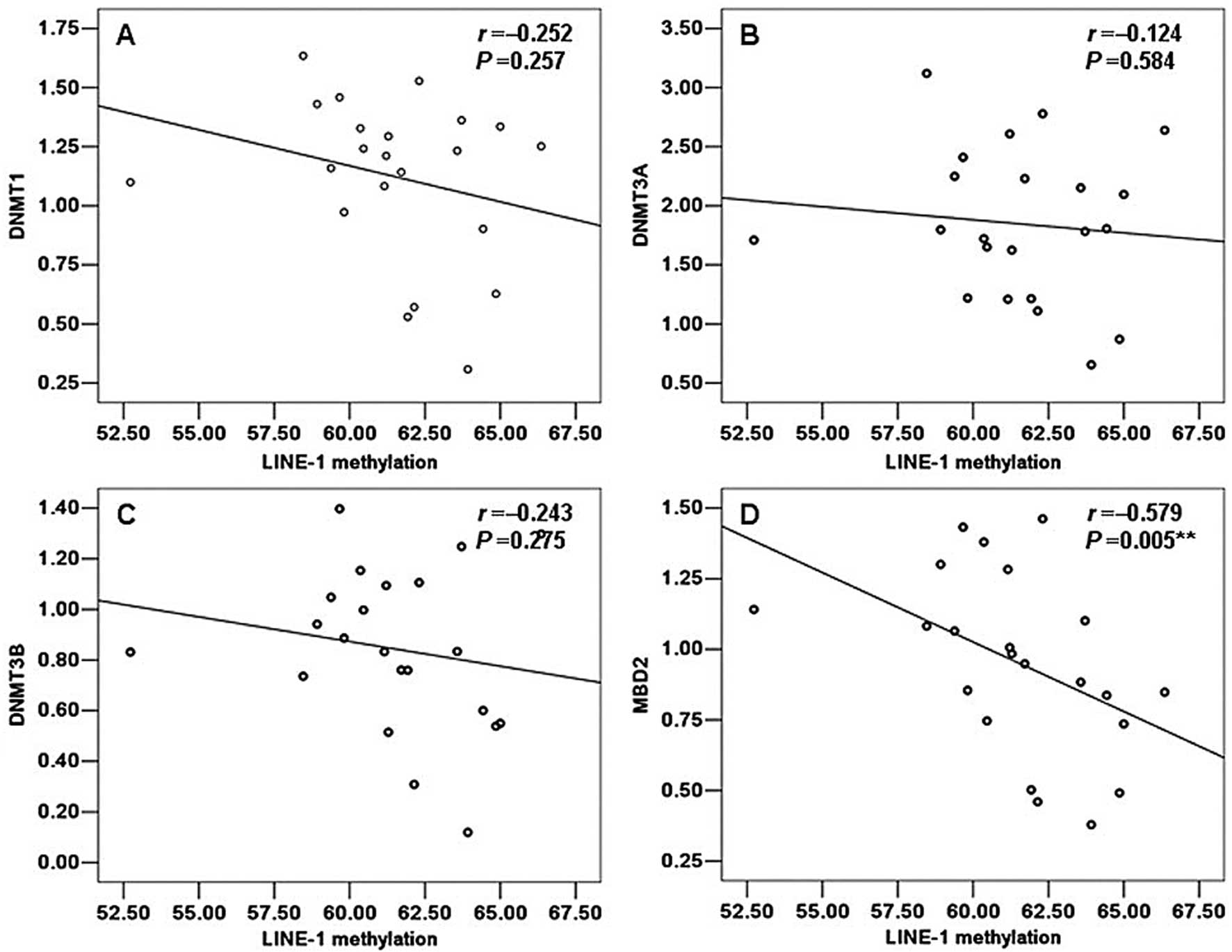
(r=−0.124, P=0.584) (Fig. 5B). Of
note, we also observed a negative correlation between the LINE-1
methylation status and the mRNA levels of DNMT1, DNMT3B and MBD2 in
the patients with TOF which was similar to that in the normal
samples. However, only MBD2 showed a statistically significant
correlation with the LINE-1 methylation status (r=−0.579, P=0.005)
(Fig. 5D). Moreover, DNMT1,
DNMT3B did not show a significant correlation with the LINE-1
methylation status (r=−0.252, P=0.257; r=−0.243, P=0.275;
respectively) (Fig. 5A and
C).
Discussion
In this study, we demonstrated that patients with
TOF had significantly lower global DNA methylation levels than
those of the controls, which may be associated with the complex
etiology of TOF. Moreover, the mRNA levels of the DNMTs (DNMT1,
DNMT3A and DNMT3B) and the MBD2 gene were significantly decreased
in the TOF samples; in particular, the decrease in the levels of
DNMT1 and DNMT3B was more significant. A significant negative
correlation between the LINE-1 methylation status and the MBD2 mRNA
levels was observed in the patients with TOF. These findings may
provide important insight into the development of pathologies from
an epigenetic viewpoint.
DNA methylation, the most investigated epigenetic
hallmark, is a reversible mechanism which modifies genome function
and chromosomal stability through the addition of methyl groups to
cytosine located in CpG dinucleotides to form 5 methylcytosine
(5mC). The CpG dinucleotides tend to cluster in regions termed CpG
islands (24). Unlike DNA
sequence mutations, the inheritance patterns of these epigenetic
events in humans are poorly understood. The hypomethylation of the
global genome largely promotes chromosomal instability,
translocations, gene disruption and reactivation of endoparasitic
sequences (25). LINE-1
methylation patterns may serve as a potential indicator of global
DNA methylation (11), which has
been proven to be frequently hypomethylated in several types of
cancer (26). Moreover, maternal
LINE-1 hypomethylation has also been found to be associated with an
increased occurrence of non-syndromic CHDs (9). Of note, higher methylation levels of
LINE-1 have also been reported in patients with Alzheimer’s disease
(AD) (27). These findings
suggest that changes in the LINE-1 methylation status may not be
restricted to cancers but may be present in other diseases and may
show hypo- or hype-methylation status under different conditions.
In the present study, we found that patients with TOF had
significantly lower levels of LINE-1 methylation compared with
samples from normal subjects, suggesting that reduced LINE-1
methylation levels may increase the chromosomal instability and
consequently alter the expression of genes associated with heart
development, eventually leading to TOF. However, the exact
molecular mechanisms of LINE-1 hypomethylation associated with the
etiology of TOF remain unclear and require further study.
DNA methylation is regulated mainly by four DNMTs
(DNMT1, DNMT2, DNMT3A and DNMT3B). MBD2 possesses demethylase
activity and participates in DNA methylation and demethylation
events (16). In the present
study, we found that the mRNA levels of DNMT1, DNMT3A, DNMT3B and
MBD2 were significantly decreased in patients with TOF; in
particular, the decrease in the levels of DNMT1 and DNMT3B was more
significant compared with the normal control samples. It is known
that lower mRNA levels of DNMT1 and DNMT3B eliminate
methyltransferase activity and thereby reduce DNA methylation by
>95% (28). DNA methylation
requires cooperative interactions of DNMT1 and DNMT3B. Thus, we
hypothesized that global DNA hypomethylation in patients with TOF
may be induced by the simultaneous decrease in the mRNA levels of
DNMT1 and DNMT3B. Moreover, due to the demethylase activity of
MBD2, the increased mRNA level of MBD2 is considered to contribute
to global DNA hypomethylation (29). By contrast, we observed the
decreased expression of MBD2 mRNA in patients with TOF; thus, we
hypothesized that the decreased expression of MBD2 mRNA may be due
to the global DNA hypomethylation (due to feedback mechanisms) in
patients with TOF.
In this study, we only observed a significant
positive correlation between the mRNA levels of DNMT1 and DNMT3A
and no correlation was observed among the other DNMTs and MBD2 in
the normal samples; this suggests that changes in the expression of
DNMT1 and DNMT3A may occur simultaneously. On the contrary, in the
patients with TOF, we found that the mRNA levels of DNMT1, DNMT3A,
DNMT3B and MBD2 positively correlated with each other and this was
statistically significant. Moreover, no correlation was observed
between age and the mRNA levels of the DNMTs and MBD2 in the normal
group; however, a significant negative correlation with age was
observed for DNMT1 and DNMT3A in the patient group; this result is
in accordance with the study by Zhang et al (30). No significant correlation with
gender was observed among DNMT1, DNMT3A, DNMT3B and MBD2 in the
control and TOF samples. Furthermore, we found no correlation
between LINE-1 hypomethylation and the mRNA levels of the DNMTs and
MBD2 in the normal group; however, a significant negative
correlation between the LINE-1 hypomethylation and the MBD2 mRNA
level was observed in the patient group. It has been reported that
the decrease in DNMT activity is due to the decrease in the mRNA
levels of DNMTs (31). However, a
previous study suggested that the DNMT1 protein levels were
elevated in human breast cancer cells due to increased stability,
while the DNMT1 mRNA levels were unaltered (32). Luo et al (33) found that the mRNA levels of other
enzymes (MBD1, MBD2, MBD3 and MBD4) involved in the DNA methylation
process were significantly higher in patients with lupus
erythematosus. Moreover, although patients with SLE have been shown
to have significantly lower levels of DNMT1 mRNA than the controls,
a correlation between the mRNA levels of DNMT1 and global DNA
methylation was not observed (21). A recent study also suggested that
there was no significant correlation between the global methylation
levels and DNMT1 and MBD2 mRNA expression in patients with active
and inactive SLE (29). However,
in our study, we found that the reduced mRNA level of MBD2 had a
significant correlation with global DNA hypomethylation in the
patient group. We concluded that MBD2 may influence the DNA
methylation pattern through an indirect method; the exact
mechanisms involved require further investigation in the
future.
As heat tissue samples are difficult to collect from
healthy controls and TOF patients, one limitation of this study was
that we were unable to obtain enough complete age-matched samples.
Thus, the findings presented in our study require further
investigation and validation in larger sample sizes. Moreover,
based only on these data from clinical samples, we cannot ascertain
whether methylation changes that are noted occur after the heart is
already formed or after heart defect already exists. At the same
time, we cannot determine whether these changes are reflective of
the disease physiology or related to disease etiology. All the
related research will be further explored using cell lines or
animal models in future studies.
In conclusion, global DNA hypomethylation is one of
the possible epigenetic variations associated with the complex
etiology of TOF and significantly correlates with the aberrant
expression of MBD2 mRNA in patients with TOF. The decreased
expression of DNMT1 and DNMT3B mRNA may play an important role in
the pathogenesis of TOF. The mechanisms of global DNA
hypomethylation in patients with TOF are complex. Enzymes that
participate in DNA methylation and demethylation events should be
investigated further in a larger number of samples, and may thus
provide important insight into the development of novel treatments
for TOF, as well as provide a deeper understanding of the etiology
of congenital heart disease.
Acknowledgements
This study was supported by grants from the National
Basic Research Program of China (973 Program; 2010CB529504 and
2009CB941704) and the Key Program of the National Natural Science
Foundation of China (30930096).
References
|
1
|
Bittel DC, Butler MG, Kibiryeva N, et al:
Gene expression in cardiac tissues from infants with idiopathic
conotruncal defects. BMC Med Genomics. 4:12011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bedard E, McCarthy KP, Dimopoulos K,
Giannakoulas G, Gatzoulis MA and Ho SY: Structural abnormalities of
the pulmonary trunk in tetralogy of fallot and potential clinical
implications: a morphological study. J Am Coll Cardiol.
54:1883–1890. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Di Felice V and Zummo G: Tetralogy of
fallot as a model to study cardiac progenitor cell migration and
differentiation during heart development. Trends Cardiovasc Med.
19:130–135. 2009.PubMed/NCBI
|
|
4
|
Rodenhiser D and Mann M: Epigenetics and
human disease: translating basic biology into clinical
applications. CMAJ. 174:341–348. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Portela A and Esteller M: Epigenetic
modifications and human disease. Nat Biotechnol. 28:1057–1068.
2010. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Goll MG and Bestor TH: Eukaryotic cytosine
methyltransferases. Annu Rev Biochem. 74:481–514. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lees-Murdock DJ, De Felici M and Walsh CP:
Methylation dynamics of repetitive DNA elements in the mouse germ
cell lineage. Genomics. 82:230–237. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Weisenberger DJ, Campan M, Long TI, et al:
Analysis of repetitive element DNA methylation by MethyLight.
Nucleic Acids Res. 33:6823–6836. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chowdhury S, Cleves MA, MacLeod SL, James
SJ, Zhao W and Hobbs CA: Maternal DNA hypomethylation and
congenital heart defects. Birth Defects Res A Clin Mol Teratol.
91:69–76. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sunami E, de Maat M, Vu A, Turner RR and
Hoon DS: LINE-1 hypomethylation during primary colon cancer
progression. PLoS One. 6:e188842011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang L, Wang F, Guan J, et al: Relation
between hypomethylation of long interspersed nucleotide elements
and risk of neural tube defects. Am J Clin Nutr. 91:1359–1367.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Nakkuntod J, Avihingsanon Y, Mutirangura A
and Hirankarn N: Hypomethylation of LINE-1 but not Alu in
lymphocyte subsets of systemic lupus erythematosus patients. Clin
Chim Acta. 412:1457–1461. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sheng W, Wang JH, Ma JX, et al: LINE-1
methylation status and its association with tetralogy of fallot in
infants. BMC Med Genomics. 5:202012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Rajendran G, Shanmuganandam K, Bendre A,
Muzumdar D, Goel A and Shiras A: Epigenetic regulation of DNA
methyltransferases: DNMT1 and DNMT3B in gliomas. J Neurooncol.
104:483–494. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Okano M, Bell DW, Haber DA and Li E: DNA
methyltransferases Dnmt3a and Dnmt3b are essential for de novo
methylation and mammalian development. Cell. 99:247–257. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Detich N, Theberge J and Szyf M:
Promoter-specific activation and demethylation by MBD2/demethylase.
J Biol Chem. 277:35791–35794. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lin RK, Hsu HS, Chang JW, Chen CY, Chen JT
and Wang YC: Alteration of DNA methyltransferases contributes to
5′CpG methylation and poor prognosis in lung cancer. Lung Cancer.
55:205–213. 2007.
|
|
18
|
Girault I, Tozlu S, Lidereau R and Bieche
I: Expression analysis of DNA methyltransferases 1, 3A, and 3B in
sporadic breast carcinomas. Clin Cancer Res. 9:4415–4422.
2003.PubMed/NCBI
|
|
19
|
Fan H, Zhao ZJ, Cheng J, Su XW, Wu QX and
Shan YF: Overexpression of DNA methyltransferase 1 and its
biological significance in primary hepatocellular carcinoma. World
J Gastroenterol. 15:2020–2026. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Adiga SK, Ehmcke J, Schlatt S, et al:
Reduced expression of DNMT3B in the germ cells of patients with
bilateral spermatogenic arrest does not lead to changes in the
global methylation status. Mol Hum Reprod. 17:545–549. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhu XH, Liang J, Li F, Yang YS, Xiang LH
and Xu JH: Analysis of associations between the patterns of global
DNA hypomethylation and expression of DNA methyltransferase in
patients with systemic lupus erythematosus. Int J Dermatol.
50:697–704. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Nakamura T, Sekigawa I, Ogasawara H, et
al: Expression of DNMT-1 in patients with atopic dermatitis. Arch
Dermatol Res. 298:253–256. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Balada E, Ordi-Ros J, Serrano-Acedo S,
Martinez-Lostao L, Rosa-Leyva M and Vilardell-Tarrés M: Transcript
levels of DNA methyltransferases DNMT1, DNMT3A and DNMT3B in
CD4+ T cells from patients with systemic lupus
erythematosus. Immunology. 124:339–347. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Crews D and McLachlan JA: Epigenetics,
evolution, endocrine disruption, health, and disease.
Endocrinology. 147(Suppl 6): S4–S10. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gaudet F, Hodgson JG, Eden A, et al:
Induction of tumors in mice by genomic hypomethylation. Science.
300:489–492. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wilson AS, Power BE and Molloy PL: DNA
hypomethylation and human diseases. Biochim Biophys Acta.
1775:138–162. 2007.PubMed/NCBI
|
|
27
|
Bollati V, Galimberti D, Pergoli L, et al:
DNA methylation in repetitive elements and Alzheimer disease. Brain
Behav Immun. 25:1078–1083. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Rhee I, Bachman KE, Park BH, et al: DNMT1
and DNMT3b cooperate to silence genes in human cancer cells.
Nature. 416:552–556. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu CC, Ou TT, Wu CC, et al: Global DNA
methylation, DNMT1, and MBD2 in patients with systemic lupus
erythematosus. Lupus. 20:131–136. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang Z, Deng C, Lu Q and Richardson B:
Age-dependent DNA methylation changes in the ITGAL (CD11a)
promoter. Mech Ageing Dev. 123:1257–1268. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Christman JK, Sheikhnejad G, Dizik M,
Abileah S and Wainfan E: Reversibility of changes in nucleic acid
methylation and gene expression induced in rat liver by severe
dietary methyl deficiency. Carcinogenesis. 14:551–557. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Agoston AT, Argani P, Yegnasubramanian S,
et al: Increased protein stability causes DNA methyltransferase 1
dysregulation in breast cancer. J Biol Chem. 280:18302–18310. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Luo Y, Li Y, Su Y, et al: Abnormal DNA
methylation in T cells from patients with subacute cutaneous lupus
erythematosus. Br J Dermatol. 159:827–833. 2008. View Article : Google Scholar : PubMed/NCBI
|