Introduction
The human ether-a-go-go-related gene (HERG)
gene encodes the α subunit of the rapid delayed rectifier potassium
current channel (Ikr), which is important for
cardiac repolarization. Mutations of the HERG gene that cause the
dysfunction of Ikr may lead to type 2 long QT
syndrome (LQTS). Approximately 10% of mutations are nonsense
mutations which introduce the premature termination codons into
HERG gene (PTC) (1,2).
Aminoglycosides showed the read-through effect on
nonsense mutations to produce full-lengh functional proteins in
genetic disease models, such as cystic fibrosis and Duchenne
muscular dystrophy (DMD) (3–6).
It was previously demonstrated that gentamicin partially restored
the HERG-like currents in HEK293 cells expressing R1014X mutant
(7). However, whether other
aminoglycosides had a similar effect, the most effective
concentration as well as whether mutation sites affected the rescue
effect remain to be determined.
PTC124
(3-[5-(2-fluorophenyl)-[1,2,4]oxadiazol-3-yl]-benzoic acid;
C15H9FN2O3) is a new
option for numerous genetic diseases related to nonsense mutations
(8–11). The chemical structure of PTC124 is
different from aminoglycosides. It can act as a PTC suppressor to
overcome the PTC and allow synthesis of a mature protein similar to
gentamicin but without serious toxic side-effects (11).
The aim of this study was to evaluate the ability of
three aminoglycosides (G418, gentamicin and tobramycin), which
share a 4,6-disubstituted deoxystreptamine ring, and PTC124 to
rescue nonsense mutations of HERG gene.
Materials and methods
Cell culture and transfection
Human embryonic kidney cells (HEK293) were grown in
Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10%
fetal bovine serum, 1% glutamine, 100 IU/ml penicillin, and 100
μg/ml streptomycin at 37°C in humidified 5% CO2, 95% air
incubator. The wild-type (WT) HERG cDNA cloned into pcDNA3.1 vector
was a kind gift from Dr Zhengfeng Zhou (Oregon Health and Science
University, Portland, OR, USA). Four HERG nonsense mutations,
R1014X, W927X, R863X and E698X were generated in the pcDNA3.1 WT
HERG plasmid using polymerase chain reaction-based mutagenesis
strategy and the sequence was then verified. Transfections were
carried out with Effectene transfection reagent (Qiagen, Hilden,
Germany) according to the manufacturer’s instructions. HEK293 cells
were transiently transfected with 0.4 μg WT or equal amounts of
mutant cDNA, respectively. Green fluorescent protein (GFP, 0.2 μg)
gene was cotransfected as an indicator during patch clamp
recording. Pharmacological rescue was examined by adding G418,
gentamicin, tobramycin or PTC124 into DMEM for 24 h.
Western blot analysis
After washing three times with PBS, cells were
disrupted by adding lysis buffer containing PMSF and a phosphatase
inhibitor mixture, followed by centrifugation at 12,000 × g for 10
min at 4°C. The supernatant was obtained. Protein concentration was
determined by using an Enhanced BCA protein assay kit (Beyotime,
Nantong, China). Protein samples were then separated by 10%
SDS-PAGE and transferred by semi-dry blotting to a nitrocellulose
membrane and incubated with rabbit anti-N or anti-C terminal HERG
antibodies (Alomone Laboratories Ltd., Jerusalem, Israel) overnight
at 4°C. The membranes were washed three times in TBST buffer,
incubated with 1:2,500 dilutions of anti-rabbit IgG at room
temperature for 1 h, and again washed three times in TBST.
Visualization was carried out using an enhanced chemiluminescence
kit (Thermo Fisher Scientific Inc., Rockford, IL, USA). Band
density on western blots was quantified by densitometry analysis of
the scanned blots, using β-actin as the internal control.
qPCR
Cells were collected and total RNA was extracted by
homogenization in TRIzol reagent according to the manufacturer’s
instructions. RNA was reverse transcribed. Quantitative polymerase
chain reaction (qPCR) was prepared with PCR reagents and primers
designed according to their recommendations, including HERG and
GAPDH. The primer sequences for the genes were: HERG: (forward)
5′-GCTTGCTCAACT CCACCTC-3′ and (reverse) 5′-TTTGGGGAATCTTGC
TAATG-3′; GAPDH: (forward) 5′-CCCTTCATTGACCTC AACTACATG-3′ and
(reverse) 5′-CTTCTCCATGGTGG TGAAGAC-3′. qPCR was performed using 1
μl of target cDNAs at 94°C for 2 min, followed by 40 cycles at 94°C
for 15 sec and at 58°C for 30 sec and 72°C for 15 sec with the use
of a SYBR-Green Master mix Kit. For all samples, GAPDH RNA was
amplified using an internal control. Relative gene expression was
calculated using the 2−ΔΔCt method.
Patch clamp recording
The whole-cell patch clamp method was applied in our
experiment to record the HERG current. The bath solution contained
the following (in mM): 135 NaCl, 5 KCl, 2 CaCl2, 1
MgCl2·6H2O, 10 glucose, 10 HEPES. The
internal pipette solution contained the following (in mM): 140 KCl,
0.5 MgCl2·6H2O, 5 HEPES, 2 EGTA, 4
K2ATP. The HERG current was activated by a depolarizing
pulse (4 sec) to 50 mV from the holding potential of −80 mV. The
tail current of the HERG channel was recorded following
repolarization to −50 mV. Voltage dependence of channel activation
was obtained by fitting normalized curves with the Boltzmann
function: I/Imax = 1/(1+exp[(V1/2 −V)/k]): I = measured tail
current, Imax = maximal tail current, V = applied membrane voltage,
V1/2 = voltage at which half of the channels are activated, and k =
slope factor. All the patch clamp experiments were performed at
room temperature (22±1°C).
Statistical analysis
Data are expressed as the means ± SE and were
analyzed by using SPSS v. 13.0 software. For statistical analysis,
the Student’s t-test was performed to compare between two data
groups. For comparisons of data among groups, analysis of variance
(ANOVA) was used. P<0.05 was considered to indicate statistical
significance.
Results
Nonsense mutations affected the protein
expression but not transcription
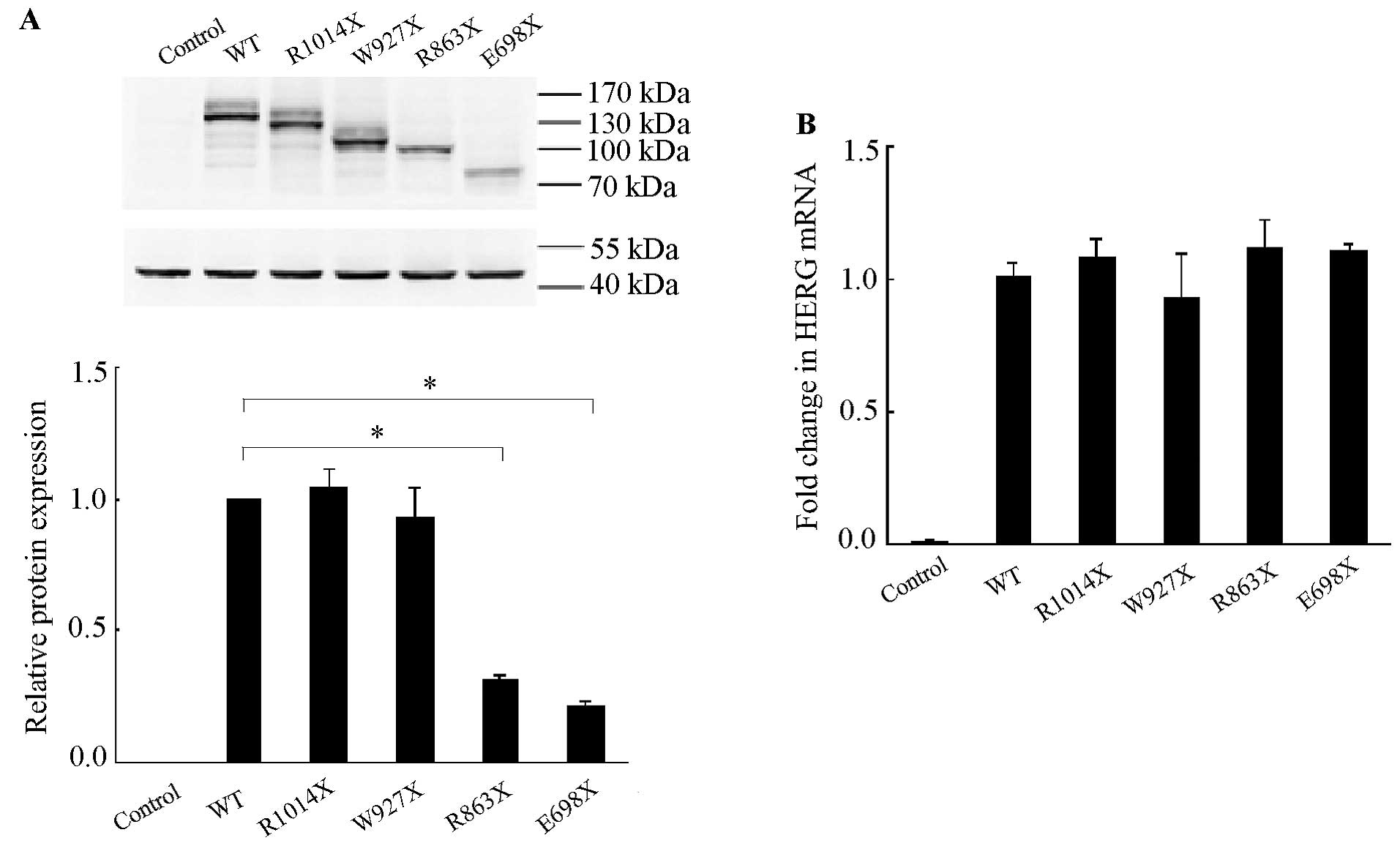
By using antibody against N-terminus of the HERG
protein, two protein bands were detected in the cells transfected
with WT cDNA: the upper band at 155 kDa and the lower band at 135
kDa representing the complex glycosylated mature and core
glycosylated immature form, respectively, of HERG proteins. There
were also two bands in cells expressing R1014X and W927X mutants
with molecular weights less than those of WT channels, representing
their mature and immature form. The expression levels did not
differ from those of WT. By contrast, the cells expressing R863X or
E698X mutants, where mutation sites approached the N-terminal, only
showed a single band with a molecular weight of <100 kDa and the
protein expression levels were decreased significantly compared
with those in cells expressing WT cDNA (31.7±0.4% for R863X,
21.7±0.5% for E698X vs. 100% for WT, n=3, both P<0.05) (Fig. 1A).
The transcription of WT and four mutants (R1014X,
W927X, R863X and E698X) were also examined. The mRNA levels were
not different among cells transfected with either WT or mutants of
HERG cDNA. The mRNA level was barely detectable in untransfected
cells (Fig. 1B). The results
above suggested that R863X and E698X mutations may have caused
trafficking defect that inhibited the glycosylation process.
Nonsense mutations inhibited the HERG
currents with different degrees
Ionic currents were recorded from HEK293 cells
transiently transfected with WT cDNA and four mutants (Fig. 2A). The mean peak tail current
densities of R1014X (4.75±0.88 pA/pF, n=7) and W927X (10.23±1.22
pA/pF, n=7) were lower than those of WT channels (52.83±4.01 pA/pF,
n=10, both P<0.05), while there were no tail currents observed
in cells transfected with R863X or E698X mutants (Fig. 2B). The voltage dependence of
activation was negatively shifted in R1014X (V1/2 =
0.20±1.13 mV) and W927X (V1/2 = 3.42±0.86 mV) compared
with that of the WT channel (V1/2 = 10.67±0.68 mV, both
P<0.05), while the slope factors were similar in WT and mutant
channels (Fig. 2C).
Aminoglycosides and PTC124 partially
restored the production of full-length HERG protein from R1014X
mutant
To examine the rescuing effect, three
aminoglycosides and PTC124 were used. The 135- and 155-kDa bands
were detected using antibodies against C-terminal of the HERG
protein when the R1014X mutant was rescued, suggesting the full
HERG protein was expressed. G418 promoted a significant effect of
readthrough in a dose-dependent manner (0, 14.3±1.1, 25.4±1.1 and
39.1±2.4% of the WT protein level, n=3, P<0.05) under the
concentrations of 0, 50, 100 and 400 μg/ml (Fig. 3A and B). Gentamicin showed less
maximal effect than G418, under the concentrations of 0, 200, 400
and 800 μg/ml, the WT protein levels were 0, 11.8±0.7, 18.6±0.3 and
14.7±1.1% (n=3, P<0.05) (Fig. 3A
and B). By contrast, tobramycin did not show any significant
effect on rescuing R1014X mutant (Fig. 3A). PTC124, a known compound for
rescuing nonsense mutation, also showed read-through ability with
lower efficiency (0, 2.9±0.6, 7.6±0.5 and 10.3±1.0% of the WT at
concentrations of 0, 10, 50, 200 μmol/l) (Fig. 3A and B).
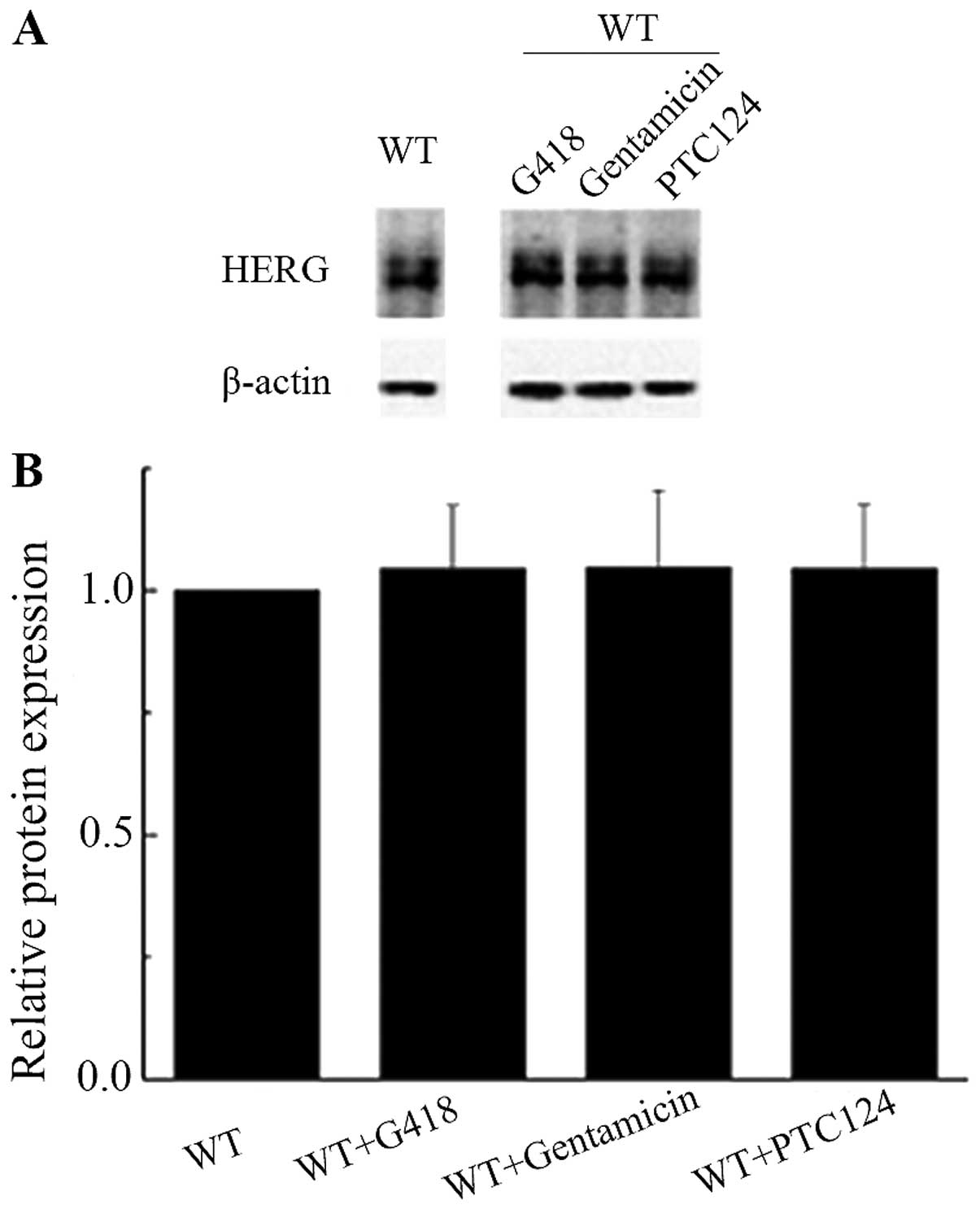
Then, we tested the effect of drugs on protein
expression in cells transfected with WT-HERG gene by adding G418
(400 μg/ml), gentamicin (400 μg/ml) and PTC124 (200 μmol/l) into
culture medium. The results showed there was no difference in
protein expression levels in cells cultured with or without drugs
(Fig. 4A and B), indicating that
these drugs did not affect the WT-HERG expression.
Aminoglycosides and PTC124 restored
current density and activation relation of R1014X mutant
channel
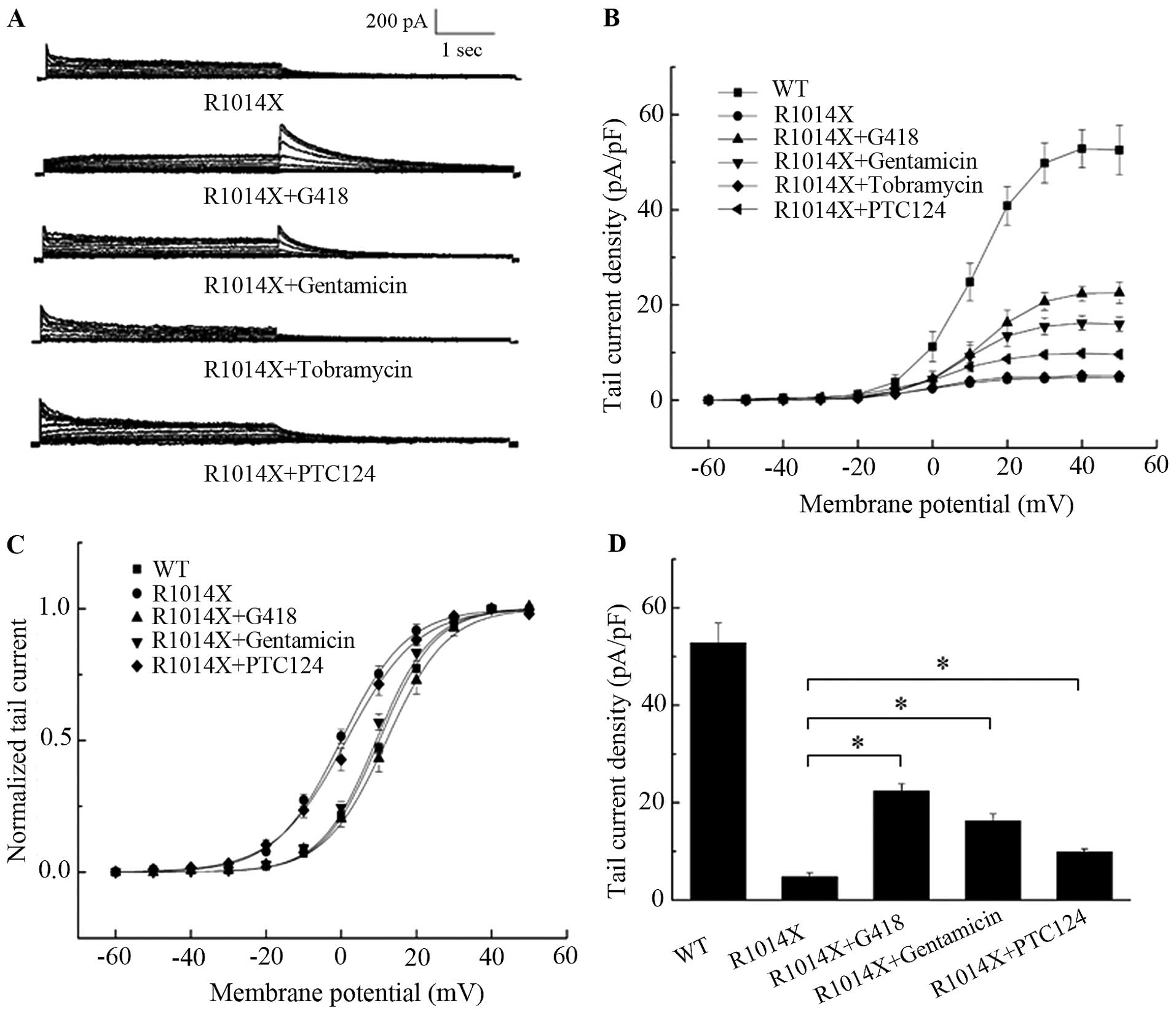
The functional expression of R1014X mutant with and
without drug treatment was tested by patch clamp recording
(Fig. 5A). As shown in Fig. 5D, peak tail current density was
increased in cells treated with 400 μg/ml G418 (22.57±2.26 pA/pF,
n=6, P<0.05) and gentamicin 400 μg/ml (16.21±1.49 pA/pF, n=5,
P<0.05) compared to that of untreated cells (4.75±0.88 pA/pF,
n=7), although this increase was lower than that of the WT channels
(52.83±4.01 pA/pF, n=10). The leftward shift of the activation
curve was corrected by the two drugs, i.e., V1/2 =
12.05±1.93 and 9.31±1.07 for G418 and gentamicin, respectively, vs.
and 0.20±1.13 mV for the untreated cells, all P<0.05 (Fig. 5C). PTC124 (200 μmol/l) treatment,
however, only slightly increased the peak tail current (from
4.75±0.88 pA/pF to 9.62±0.73 pA/pF, n=7, P<0.05; Fig. 5B and D) and did not correct the
shift of the activation curve (V1/2 = 0.20±1.13 mV vs.
V1/2 = 1.28±1.65 mV, P>0.05; Fig. 5C). Tobramycin treatment, however,
did not restore the current of R1014X mutant (Fig. 5A and B).
Aminoglycosides and PTC124 restored the
protein expression of W927X but not R863X and E698X mutants
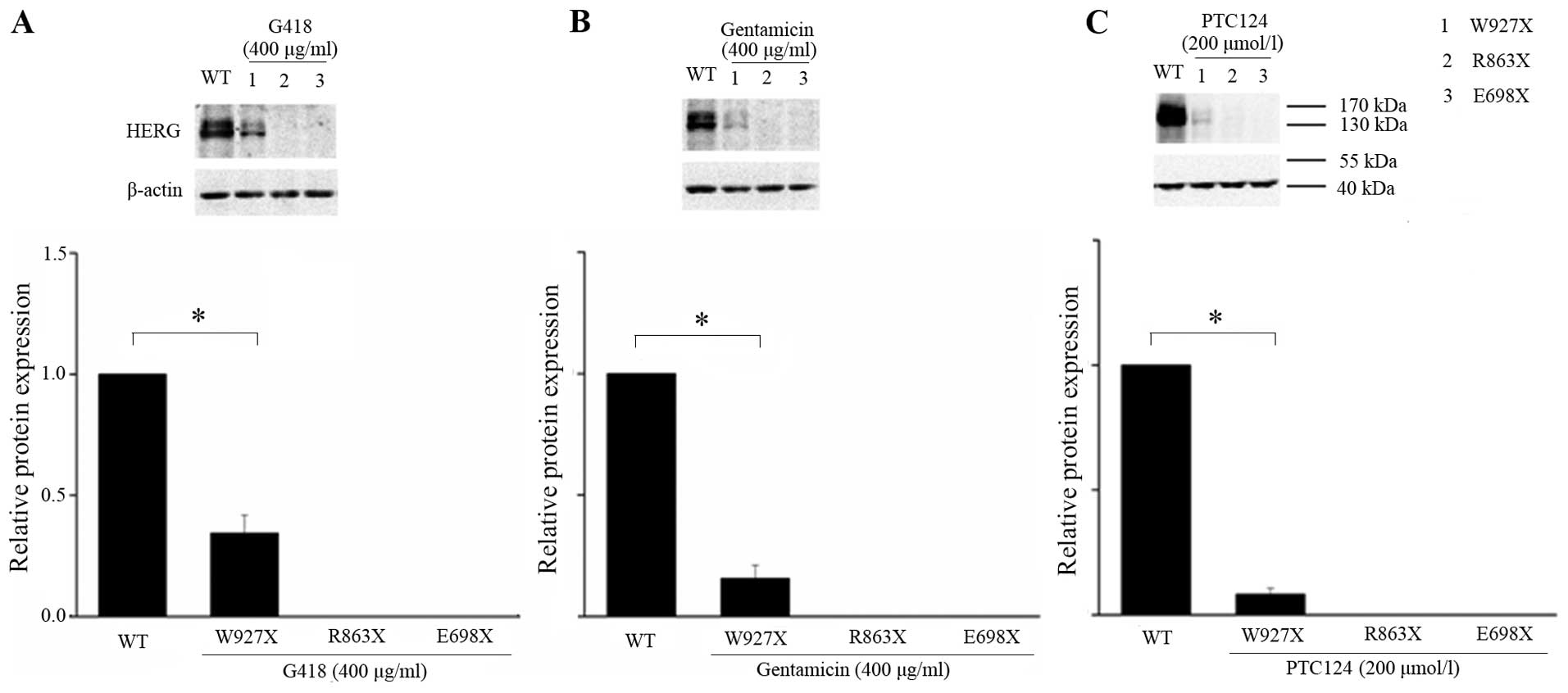
The read-through effects of G418, gentamicin and
PTC124 on the expression of W927X, R863X and E698X mutants were
tested. As shown in Fig. 6, all
three compounds partially induced the expression of full-length
protein in cells transfected with W927X mutant. The read-through
efficiency of G418 was higher than that of gentamicin and PTC124
(34.4±4.3% for G418 compared to 15.6±3.2 and 8.3±1.3% for
gentamicin and PTC124, respectively, n=3, both P<0.05, Fig. 6). For R863X and E698X mutants, the
read-through effect was not observed when the cells were treated by
G418, gentamicin or PTC124, suggesting that as the mutation site
approached the N-terminal, the pharmacological rescue efficiency
was decreased.
Discussion
In this study, we investigated the pharmacologic
rescue effect on nonsense mutations of the HERG gene. The results
showed that G418, gentamicin and PTC124 partially restored the
expression of full-length protein in cells transfected with R1014X
or W927X mutant. The leftward shift of the R1014X channel
activation curve was also corrected subsequent to G418 and
gentamicin treatment. Additionally, the mutation site was a factor
affecting the rescuing efficiency.
LQTS is an inherited cardiac arrhythmia syndrome
related to syncope and sudden death (12). It has been found that mutations in
13 encoding ion channels or structural protein genes are associated
with LQTs. Mutations of the HERG gene lead to type 2 LQTs (13,14).
In this study, we first observed the protein and
mRNA levels of the WT-HERG gene and four nonsense mutants that
produce C-terminus truncations. Results of the western blot
analysis revealed that the untransfected HEK293 cells did not react
with the antibody against the N-terminus of the HERG protein.
Following transfection of cells with WT or mutant HERG cDNAs,
different lengths of HERG proteins were detected according to
mutation sites. As the mutation site approached the N-terminal, the
protein became shorter. The protein level was decreased in cells
transfected with R863X or E698X. Nonsense-mediated mRNA decay (NMD)
is an effective mRNA surveillance mechanism which can detect and
degrade mRNAs with premature termination condons (PTCs) and
protects cells from the potentially deleterious effects of
truncated proteins. However, the results of qPCR suggested an
unchanged mRNA level, suggesting the mutants were not subjected to
nonsense-mediated decay (NMD) mechanism in our expression system. A
possible reason for this observation was that the cDNA did not
contain the full configuration of exons and introns (15).
Residues 750–870 of the HERG protein comprised the
cyclic nucleotide binding domain (cNBD) which was responsible for
protein trafficking (16).
Disruption of this region may result in defect of protein
trafficking. Residues 860–899 of the HERG protein were also
reported to be essential for endoplasmic reticulum exit and for the
stability of the channel protein (17). Nonsense mutation in this region
may result in defective trafficking as well as rapid degradation of
mutant channels in endoplasmic reticulum. Thus R863X and E698X only
produced single bands and the protein level was significantly
decreased compared to that of WT channels.
Aminoglycosides are classical antibiotics that bind
to the A site of bacterial ribosomal 16S RNA to reduce translation
fidelity, thus similar aminoacyl-tRNA enters the A site, inducing
synthesis of error protein or preventing the release of protein and
ultimately causing bacterial death (18). The crystal structure analysis
revealed that aminoglycosides combined with A1408 and G1491 16S RNA
by forming hydrogen bonds to cause conformational changes, reducing
its ability to identify aminoacyl-tRNA. The structure of the
eukaryotic ribosome 18S RNA was a little different from the
prokaryotes 16S RNA, with A1408→G and G1491→A substitution.
Previous studies have demonstrated that aminoglycosides also bind
to the 18S rRNA subunit, reducing discrimination of near-cognate
tRNAs, although this interaction was less stable than that in
bacteria (19–21). Due to the lack of upstream and
downstream normal stop codon sequence and polyA tail structure, the
premature stop codon (PTC) could be suppressed more readily by
aminoglycosides (22).
PTC suppression to restore the function of mutant
proteins in mammalian cells was first reported by Howard et
al (6). Their results showed
that G418 suppressed PTCs to restore the function of mutant protein
in transiently expressed cDNAs which contained mutations in the
cystic fibrosis transmembrane conductance regulator (CFTR) gene.
The potency of aminoglycosides for inducing the production of
full-length proteins from other genes carrying a PTC, such as P53,
APC and DMD genes has also been demonstrated (4,23,24). In a previous study we have
demonstrated that gentamicin suppressed PTC in R1014X and W927X
mutants of the HERG gene to produce full-length channel protein
(7).
Aminoglycosides were mainly divided into two
categories: one with a 4,6-disubstituted deoxystreptamine ring and
the other with a 4,5-disubstituted deoxystreptamine ring. In this
experiment, we mainly explored the read-through effect of G418,
gentamicin and tobramycin, which belong to the 4,6-disubstituted
type. Our results suggest that G418 and gentamincin effectively
rescued the R1014X mutant to produce full-length and functional
protein. However, tobramycin was unable to show a significant
read-through effect, consistent with the results of Manuvakhova
et al (25). The authors
of that study found that four compounds in the 4,6-disubstituted
class (kanamycin A, kanamycin B, tobramycin, and amikacin) did not
rescue the nonsense mutants constructed for that experiment. As the
four compounds shared a common ring 3 that was not found on G418
and gentamicin, they suggested that certain characteristics of the
ring 3 may have prevented the induction of the readthrough
effect.
The severe side-effects such as nephrotoxicity and
ototoxicity of aminoglycosides may limit their clinical
application. Recently it has been reported that PTC124, which has
low toxicity, may also act as a nonsense codon suppressor (10) in the studies using cell lines and
mouse models with Duchenne muscular dystrophy and cystic fibrosis
(11,26). The effect of PTC124 on clinical
trial in CF patients with nonsense mutations has also been
demonstrated (27). In this study
it has been demonstrated that PTC124 promoted the production of
full-length protein in cells expressing R1014X mutant with lower
efficiency, in contrast to previous studies which showed that
PTC124 was more effective than gentamicin. By contrast, results of
in vitro studies demonstrated readthrough induced by
aminoglycosides but not PTC124 (28–30), suggesting specificity of PTC124
for particular genes.
In this experiment, we also tested the rescuing
ability of G418, gentamicin and PTC124 on W927X, R863X and E698X
mutants. As the mutation site approached the N-terminal, the rescue
ability was decreased. These results were different from studies in
other genetic diseases (23,31). The exact mechanism accounting for
the phenomenon was unclear. Earlier studies indicated that
tryptophan or glutamine may be inserted into the premature stop
codon site (32,33). We proposed that the amino acid
that was inserted into the premature termination codon site induced
by the read-through mechanism may be different from the original
one in WT protein. Thus, it may result in the dysfunction of cNBD
domain or residues 860–899, leading to trafficking defect.
Other aminoglycosides, as well as aminoglycoside
derivatives with lower toxicity such as NB54 and NB84 have not been
studied in our experiments (25,34). Additionally, our study was limited
to channels expressed in the homologous cell system. Additionaly
studies are required to obtain an improved expression system or
animal models to confirm our findings.
In conclusion, aminoglycosides and PTC124 induced
different read-through effects on the nonsense mutations of HERG
gene. G418 and gentamicin had a higher ability to rescue the
nonsense mutants. The mutation site was a significant factor
determining the rescue efficiency. As the mutation site approached
the N-terminal, the rescue efficiency was decreased. These findings
suggest complexity in the pharmacological treatment of diseases
caused by nonsense mutations in the clinical setting.
Acknowledgements
This study was funded by the National Natural
Science Foundation of China (project no., 81070150, to J. Pu).
References
|
1
|
Splawski I, Shen J, Timothy KW, et al:
Spectrum of mutations in long-QT syndrome genes. KVLQT1, HERG,
SCN5A, KCNE1, and KCNE2. Circulation. 102:1178–1185. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Anderson CL, Delisle BP, Anson BD, et al:
Most LQT2 mutations reduce Kv11.1 (hERG) current by a class 2
(trafficking-deficient) mechanism. Circulation. 113:365–373. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lee HL and Dougherty JP: Pharmaceutical
therapies to recode nonsense mutations in inherited diseases.
Pharmacol Ther. 136:227–266. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zilberberg A, Lahav L and Rosin-Arbesfeld
R: Restoration of APC gene function in colorectal cancer cells by
aminoglycoside- and macrolide-induced read-through of premature
termination codons. Gut. 59:496–507. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Malik V, Rodino-Klapac LR, Viollet L, et
al: Gentamicin-induced readthrough of stop codons in Duchenne
muscular dystrophy. Ann Neurol. 67:771–780. 2010.PubMed/NCBI
|
|
6
|
Howard M, Frizzell RA and Bedwell DM:
Aminoglycoside antibiotics restore CFTR function by overcoming
premature stop mutations. Nat Med. 2:467–469. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yao Y, Teng S, Li N, Zhang Y, Boyden PA
and Pu J: Aminoglycoside antibiotics restore functional expression
of truncated HERG channels produced by nonsense mutations. Heart
Rhythm. 6:553–560. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wilschanski M, Miller LL, Shoseyov D, et
al: Chronic ataluren (PTC124) treatment of nonsense mutation cystic
fibrosis. Eur Respir J. 38:59–69. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Goldmann T, Overlack N, Wolfrum U and
Nagel-Wolfrum K: PTC124-mediated translational readthrough of a
nonsense mutation causing Usher syndrome type 1C. Hum Gene Ther.
22:537–547. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tan L, Narayan SB, Chen J, Meyers GD and
Bennett MJ: PTC124 improves readthrough and increases enzymatic
activity of the CPT1A R160X nonsense mutation. J Inherit Metab Dis.
34:443–447. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Welch EM, Barton ER, Zhuo J, et al: PTC124
targets genetic disorders caused by nonsense mutations. Nature.
447:87–91. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Crotti L, Celano G, Dagradi F and Schwartz
PJ: Congenital long QT syndrome. Orphanet J Rare Dis. 3:182008.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Curran ME, Splawski I, Timothy KW, Vincent
GM, Green ED and Keating MT: A molecular basis for cardiac
arrhythmia: HERG mutations cause long QT syndrome. Cell.
80:795–803. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Alders M and Mannens M: Romano-Ward
Syndrome. Pagon RA, Adam MP, Bird TD, et al: GeneReviews™
(internet). University of Washington; Seattle, WA: 2003, http://www.ncbi.nlm.nih.gov/books/NBK1129/.
Last update: May 31, 2012.
|
|
15
|
Schweingruber C, Rufener SC, Zünd D,
Yamashita A and Mühlemann O: Nonsense-mediated mRNA decay -
mechanisms of substrate mRNA recognition and degradation in
mammalian cells. Biochim Biophys Acta. 1829:612–623. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Akhavan A, Atanasiu R, Noguchi T, Han W,
Holder N and Shrier A: Identification of the
cyclic-nucleotide-binding domain as a conserved determinant of
ion-channel cell-surface localization. J Cell Sci. 118:2803–2812.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Akhavan A, Atanasiu R and Shrier A:
Identification of a COOH-terminal segment involved in maturation
and stability of human ether-a-go-go-related gene potassium
channels. J Biol Chem. 278:40105–40112. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Moazed D and Noller HF: Interaction of
antibiotics with functional sites in 16S ribosomal RNA. Nature.
327:389–394. 1987. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kondo J, Urzhumtsev A and Westhof E: Two
conformational states in the crystal structure of the Homo sapiens
cytoplasmic ribosomal decoding A site. Nucleic Acids Res.
34:676–685. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kondo J, Francois B, Urzhumtsev A and
Westhof E: Crystal structure of the Homo sapiens cytoplasmic
ribosomal decoding site complexed with apramycin. Angew Chem Int Ed
Engl. 45:3310–3314. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hermann T, Tereshko V, Skripkin E and
Patel DJ: Apramycin recognition by the human ribosomal decoding
site. Blood Cells Mol Dis. 38:193–198. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Amrani N, Ganesan R, Kervestin S, Mangus
DA, Ghosh S and Jacobson A: A faux 3′-UTR promotes aberrant
termination and triggers nonsense-mediated mRNA decay. Nature.
432:112–118. 2004.
|
|
23
|
Floquet C, Deforges J, Rousset JP and
Bidou L: Rescue of non-sense mutated p53 tumor suppressor gene by
aminoglycosides. Nucleic Acids Res. 39:3350–3362. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
De Luca A, Nico B, Rolland JF, et al:
Gentamicin treatment in exercised mdx mice: identification of
dystrophin-sensitive pathways and evaluation of efficacy in
work-loaded dystrophic muscle. Neurobiol Dis. 32:243–253.
2008.PubMed/NCBI
|
|
25
|
Manuvakhova M, Keeling K and Bedwell DM:
Aminoglycoside antibiotics mediate context-dependent suppression of
termination codons in a mammalian translation system. RNA.
6:1044–1055. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Du M, Liu X, Welch EM, Hirawat S, Peltz SW
and Bedwell DM: PTC124 is an orally bioavailable compound that
promotes suppression of the human CFTR-G542X nonsense allele in a
CF mouse model. Proc Natl Acad Sci USA. 105:2064–2069. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kerem E, Hirawat S, Armoni S, et al:
Effectiveness of PTC124 treatment of cystic fibrosis caused by
nonsense mutations: a prospective phase II trial. Lancet.
372:719–727. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ho G, Reichardt J and Christodoulou J: In
vitro read-through of phenylalanine hydroxylase (PAH) nonsense
mutations using aminoglycosides: a potential therapy for
phenylketonuria. J Inherit Metab Dis. Mar 27–2013.(Epub ahead of
print).
|
|
29
|
Brumm H, Mühlhaus J, Bolze F, et al:
Rescue of melanocortin 4 receptor (MC4R) nonsense mutations by
aminoglycoside-mediated read-through. Obesity (Silver Spring).
20:1074–1081. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Dranchak PK, Di Pietro E, Snowden A, et
al: Nonsense suppressor therapies rescue peroxisome lipid
metabolism and assembly in cells from patients with specific PEX
gene mutations. J Cell Biochem. 112:1250–1258. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bartolomeo R, Polishchuk EV, Volpi N,
Polishchuk RS and Auricchio A: Pharmacological read-through of
nonsense ARSB mutations as a potential therapeutic approach for
mucopolysaccharidosis VI. J Inherit Metab Dis. 36:363–371. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Engelberg-Kulka H: UGA suppression by
normal tRNA Trp in Escherichia coli: codon context effects.
Nucleic Acids Res. 9:983–991. 1981. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Nilsson M and Ryden-Aulin M: Glutamine is
incorporated at the nonsense codons UAG and UAA in a
suppressor-free Escherichia coli strain. Biochim Biophys
Acta. 1627:1–6. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Brendel C, Belakhov V, Werner H, et al:
Readthrough of nonsense mutations in Rett syndrome: evaluation of
novel aminoglycosides and generation of a new mouse model. J Mol
Med (Berl). 89:389–398. 2011. View Article : Google Scholar : PubMed/NCBI
|