Introduction
Macrophage migration inhibitory factor (MIF) was
identified in 1966 as the first T-cell cytokine inhibiting random
movement of macrophages (1). The
biological and physiological activities of human MIF were
elucidated from studies on recombinant human MIF generated by
molecular techniques. Structural analysis demonstrated that MIF is
a homotrimeric molecule with similar characteristics and activities
to the human D-dopachrome tautomerase (D-DT) enzyme
(2). MIF is considered a
multifunctional protein constitutively expressed and stored in
preformed cytoplasmic pools in several immune cells including
monocytes, macrophages, T and B lymphocytes, eosinophils,
neutrophils, and dendritic cells and is rapidly released in
response to stimuli. In addition, MIF has been identified in
various tissues and organs such as lung, the epithelial lining of
the skin, gastrointestinal and genitourinary tracts (reviewed in
ref. 3).
MIF plays a pivotal role in immune inflammatory
responses by acting as a major mediator in the counter-regulation
of the immunosuppressive effects of glucocorticoids. As it is an
upstream regulator, MIF exerts its effects in innate and adaptive
immune systems by the activation of leukocyte recruitment and
initiation of the inflammatory cascade for the release of other
pro-inflammatory cytokines and mediators such as interleukin
(IL)-1β, IL-2, IL-6, IL-8, TNF-α, nitric oxide and prostaglandin E2
(3–5). Furthermore, MIF inhibits
p53-mediated apoptosis of immune cells during inflammatory
activation (6). MIF biological
activity correlates with several catalytic activities such as D-DT
(7), phenylpyruvate tautomerase
(8), and thiol-protein
oxidoreductase (9). However, its
physiological substrate has yet to be identified.
Clinical studies have identified the correlation
between MIF levels in plasma or affected tissue and disease
severity in a number of inflammation-associated diseases. Elevated
levels of MIF have been detected in sepsis (10) as well as numerous infectious
diseases with exaggerated immune response such as dengue
hemorrhagic fever (11,12) and influenza virus infection
(13). An increase of secreted
MIF has also been observed in a number of inflammatory-mediated
autoimmune and also metabolic diseases (14–19). In addition, MIF has a direct
relationship with cancer growth and progression (20). Therefore, MIF has become a
promising biomarker for diagnosis and an attractive therapeutic
target for the treatment of a wide variety of diseases.
Inhibition of MIF activities by small molecules or
neutralizing antibodies has become crucial in identifying
therapeutic strategies to alleviate immuno-inflammatory disorders
in humans. It has been well documented that interference of MIF
tautomerase activity readily reduces inflammatory-mediated
pathogenesis in various disease models such as sepsis, diabetes and
allergic neuritis (21–24). Similar to MIF inhibitors,
neutralizing anti-MIF antibodies have been proven to be
therapeutically effective in several models of autoimmune and
inflammatory diseases (25–29). Recently, antibodies specific to
β-sheet structure containing the oxidoreductase motif of MIF have
been demonstrated for their protective effects in sepsis model or
contact hypersensitivity (30).
In the present study, fully human single-chain variable fragment
(HuScFv) antibody, which is five-fold smaller than the normal
antibody molecule, specific to MIF has been generated. HuScFv
selected from a human antibody phage display library, not only
recognizes human MIF, but also neutralizes its tautomerase
activity. The results of several biological experiments indicate
the potential development of MIF-specific HuScFv for therapeutic or
diagnostic applications.
Materials and methods
Cell culture and native protein
preparation
Human monoblastic leukemia (U937) cells were
cultured in RPMI-1640 medium (Gibco, Grand Island, NY, USA)
supplemented with 10% FBS (vol/vol), 2 mM L-glutamine and 100 U/ml
antibiotics at 37°C in a 5% CO2 atmosphere. Native MIF
in U937 cell lysate was used as an antigen in the western blot
analysis and experiments regarding tautomerase activity.
Production and purification of
recombinant human MIF (rMIF)
Full-length human MIF was amplified from
kidney Matchmaker cDNA library (Clontech, Mountain View, CA, USA)
by using MIF-specific primers (forward, 5′-CGG GAT CCA TGC CGA TGT TCA TCG
TAA ACA CC-3′ and reverse, 5′-CCG CTC GAG GGC GAA GGT GGA GTT GTT
C-3′). BamHI and XhoI endonuclease restriction sites
(underlined) were incorporated for DNA cloning purposes. The
amplicon was digested and ligated into pET21a(+) vector (Novagen,
Darmstadt, Germany) and introduced into E. coli BL21(DE3). A
colony of transformant E. coli carrying MIF was
induced by IPTG for rMIF production. Polyhistidine-tagged-rMIF was
purified by TALON™ Metal Affinity Resin (Clontech) under native
conditions. The purity of rMIF was determined by 15% SDS-PAGE and
Coomassie Brilliant Blue G-250 (Sigma, St. Louis, MO, USA)
staining.
Phage bio-panning
Phage clones carrying MIF-specific HuScFv were
selected from the human antibody phage display library by
bio-panning procedure (31).
Purified rMIF (1 μg) was coated into microtiter wells and phage
library (100 μl containing ~1011 pfu) was added. Phages
exhibiting HuScFv that bound to rMIF were rescued by E. coli
HB2151 infection and selected on selective agar plates (LB
containing 100 μg/ml ampicillin and 2% glucose). Individual
phagemid-transformed E. coli clones were screened for the
presence of huscfv in a phagemid vector by colony PCR using
phagemid-specific primers induced for monoclonal HuScFv production,
as previously described (31).
Bacterial lysates were detected for E-tagged HuScFv by western blot
analysis using anti-E-tag polyclonal antibody (Abcam, Cambridge,
UK) followed by HRP-conjugated swine anti-rabbit Ig (Dako,
Glostrup, Denmark) and DAB substrate.
Screening of MIF-specific HuScFv by
indirect enzyme-linked immunosorbent assay (ELISA)
Indirect ELISA was performed to determine the
binding of monoclonal HuScFv to rMIF. The wells of ELISA plate were
coated with 1 μg purified rMIF or BSA (negative antigen control) at
37°C overnight. After washing and blocking the wells,
HuScFv-containing preparations (1 mg in 100 μl) were added
individually to both rMIF and BSA wells and incubated at 37°C for 2
h. HuScFv binding to rMIF was detected by rabbit anti-E-tag
polyclonal antibody followed by HRP-conjugated swine anti-rabbit
IgG. Enzymatic reaction was developed following the addition of TMB
substrate (Invitrogen, Camarillo, CA, USA) and 1 N HCl. Color of
the content in the wells was measured at OD450nm using
ELISA reader (Multiskan EX; Thermo Scientific, Waltham, MA,
USA).
Preparation of purified 6xHis-tagged
HuScFv
The Huscfv sequence in the phagemid vector of the
selected E. coli clone was subcloned into modified pET23b(+)
vector and introduced into E. coli BL21(DE3) by
transformation (32). Bacterial
transformants containing pET23b(+)-huscfv were induced with
IPTG for the production of monoclonal 6xHis-tagged HuScFv. The
HuScFv in the bacterial lysate was purified using TALON Metal
Affinity Resin and prepared in 1X PBS (pH 7.4) by dropwise dialysis
prior to use.
Determination of the binding activity of
HuScFv to native MIF
Western blot analysis and immunofluorescence assay
were performed to determine the binding activity of HuScFv to
native MIF in human U937 cells. U937 whole cell lysate (40 μg) was
separated on SDS-PAGE and transferred onto nitrocellulose membrane.
Polyhistidine-tagged HuScFv was added to the membrane and
subsequently detected by mouse anti-His antibody. The reactive band
of HuScFv-MIF immune complexes was revealed by adding AP-conjugated
goat anti-mouse Ig and BCIP/NBT colorimetric substrate,
respectively. The irrelevant HuScFv (dengue virus capsid
protein-specific HuScFv) and mouse anti-MIF polyclonal antibody
were used as negative and positive antibody controls,
respectively.
Immunofluorescence assay was used to demonstrate and
localize the interaction of HuScFv to cellular MIF in U937 cells.
The cells were fixed with 4% paraformaldehyde and permeabilized
with 0.2% Triton X-100. After blocking, the cells were incubated
with purified HuScFv (1 μM) at 37°C for 2 h in a humidified
chamber. The HuScFv-MIF interaction was revealed by adding a
mixture of mouse anti-His antibody and rabbit anti-MIF polyclonal
antibody. The cells were then incubated with a mixture of Alexa
Flour 488-conjugated goat anti-mouse Ig (Molecular Probes,
Carlsbad, CA, USA), Cy™3-conjugated AffiniPure donkey anti-rabbit
Ig (Jackson ImmunoResearch Laboratories, West Grove, PA, USA), and
anti-nuclear staining reagent (Hoechst; Molecular Probes) to
localize HuScFv, endogenous MIF, and nuclear DNA, respectively.
Fluorescence images were visualized by using a laser scanning
confocal microscope (LSM 510 META; Carl Zeiss, Jena, Germany).
Neutralization of MIF tautomerase
activity by HuScFv
Recombinant MIF and native MIF in U937 cell lysates
were analyzed for its inherent tautomerase activity as previously
described with slight modifications (33). The enzymatic reaction was
initiated at 25°C by adding 20 μl of the dopachrome methyl ester
substrate (2 mM L-3,4-dihydroxyphenylalanine methyl ester and 4 mM
sodium periodate) to a cuvette containing 200 μl of either rMIF (30
μM) or native MIF in U937 cell lysate (6 μg) prepared in
tautomerase assay buffer (50 mM potassium phosphate, 1 mM EDTA, pH
6.0). The activity was determined by the semi-continuous reduction
of OD475nm at 60-sec interval for 300 sec using a
spectrophotometer (Shimadzu, Kyoto, Japan). The neutralization of
tautomerase activity by MIF-specific HuScFv was determined by
pre-incubation of HuScFv with rMIF or native MIF in U937 cell
lysate at 37°C for 1 h prior to the enzymatic reaction was
initiated. Irrelevant HuScFv was used as a negative antibody
control. Heat-denatured rMIF and U937 cell lysate were used as
tautomerase activity-negative controls. Results were presented as
the mean values from three independent experiments.
HuScFv mimotope searching and in silico
analysis of HuScFv-MIF interaction
Mimotope of MIF-specific HuScFv was determined by
using Ph.D.-12™ Phage Display Peptide Library (New England Biolabs;
Ipswich, MA, USA) as previously described (34). Purified HuScFv was used as the
target for selection of phage clones displaying 12-mer peptide
(mimotope). After three rounds of bio-panning, genomic DNA from a
number of phage clones displaying peptides bound to MIF-specific
HuScFv was sequenced and the 12-mer peptide sequences were deduced.
Consensus mimotope was obtained by multiple alignments of the
deduced peptide sequences. The mimotope was aligned to the MIF
protein sequence and residues on the MIF molecule matching with the
obtained mimotope were considered as the HuScFv epitope.
Molecular modeling of the antibody was initiated via
BLAST search analysis. The highest identity sequence with 3D
structure was used as a template for homology modeling by using the
modeler module embedded in the Discovery Studio 2.5 program
(Accelrys Software Inc., San Diego, CA, USA). The three-dimensional
structure of MIF was obtained from the PDB database (PDB entry
1GD0) and the crystal structure of a human anti-SARS spike protein
antibody, 80R (PDB entry 2GHW) was used as a template for modeling
of the HuScFv molecule. The constructed models were accessed for
the quality of structure via the Ramachandran plot. The plot was
generated using PROCHECK v3.4 program (University College London,
London, UK) (35). Subsequently,
the docked poses were subjected to structural refinements by using
RDOCK module embedded in Discovery Studio 2.5 program.
Statistical analysis
Data were analyzed using GraphPad Prism Software 4.0
(GraphPad Software, Inc., San Diego, CA, USA) and presented as the
mean ± SEM. Significant differences were determined by one-way
ANOVA (P<0.05) and Tukey’s HSD test.
Results
Production of rMIF with inherent
enzymatic activity
A full-length cDNA encoding MIF was amplified from
human cDNA library (345 bp). Following DNA sequencing, the sequence
was deposited into the GenBank database (accession no. JQ846015).
It showed 100% homology to the human MIF-coding sequence previously
reported in the database (NM_002415.1). Recombinant MIF was
abundantly expressed after induction of the bacterial clone
carrying recombinant plasmid. Polyhistidine-tagged rMIF was
purified as a soluble protein at 12.5 kDa. Tautomerase enzymatic
activity, a distinct pathogenesis-related property of MIF, was
verified as the reduction of dopachrome methyl ester substrate at
OD475nm (33).
Purified rMIF exhibited in vitro tautomerase activity,
suggesting that rMIF presented inherent physiological function, and
retained enzymatic epitope similar to the native molecule (data not
shown).
Selection and screening of MIF-specific
HuScFv
Purified rMIF was used as the antigen for selection
of phages carrying MIF-specific HuScFv by bio-panning of the human
antibody phage display library. A number of E. coli clones
carrying huscfv were obtained and randomly tested for the
production of monoclonal soluble HuScFv. Among
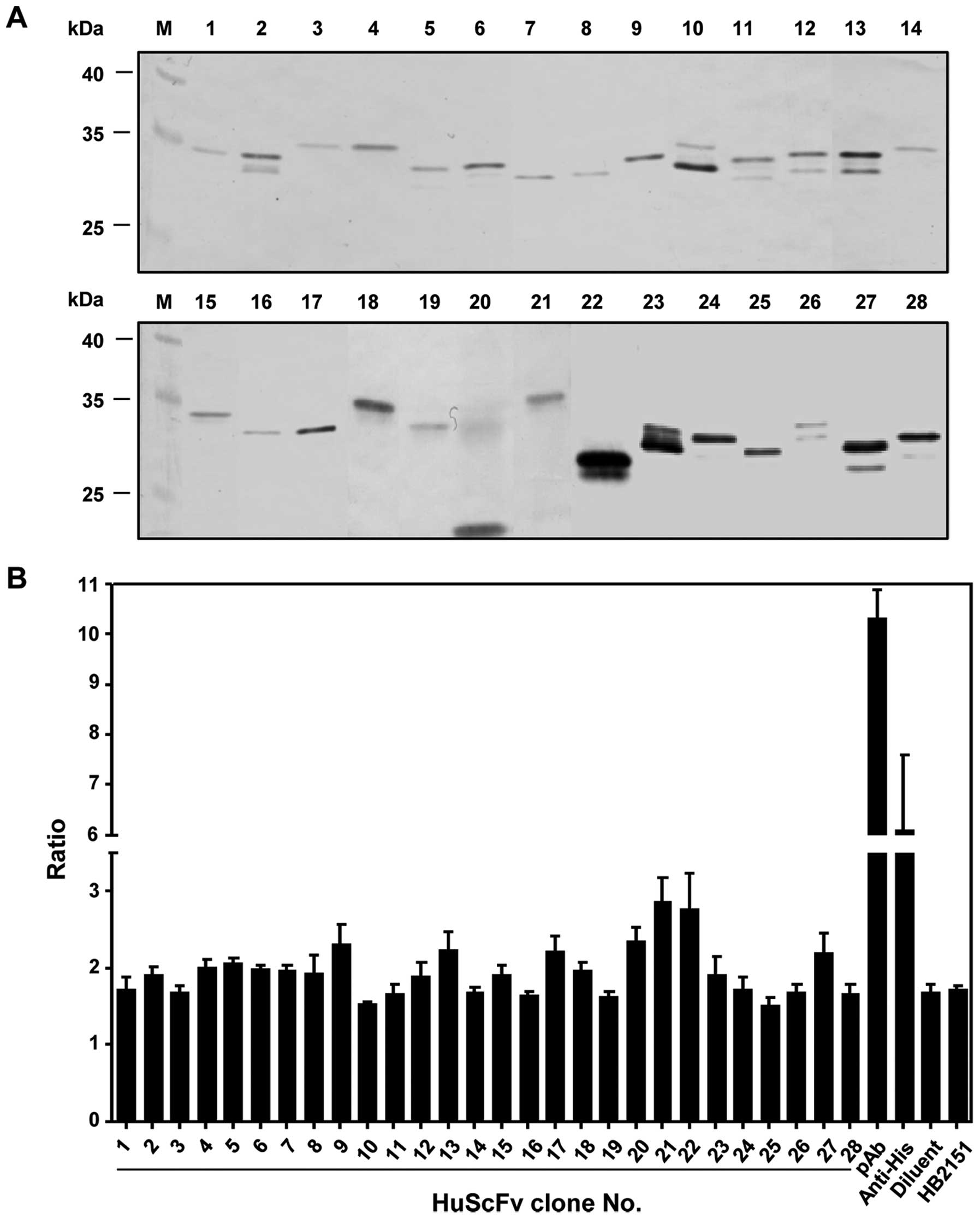
huscfv-positive transformants, 28 clones (35%) produced
HuScFv (20–35 kDa) as detected by western blot analysis using
anti-E-tag antibody (Fig. 1A).
These clones were individually screened for their binding
activities to rMIF by indirect ELISA. Seventeen clones exhibiting
ELISA signal ratios (OD450nm of HuScFv-rMIF
binding/OD450nm of HuScFv-BSA binding) were higher than
the ELISA signal ratio identified in the negative control (Fig. 1B). Huscfv of E. coli
clones exhibiting high binding activity were subcloned into a
modified pET23b(+) vector for benefits in HuScFv purification and
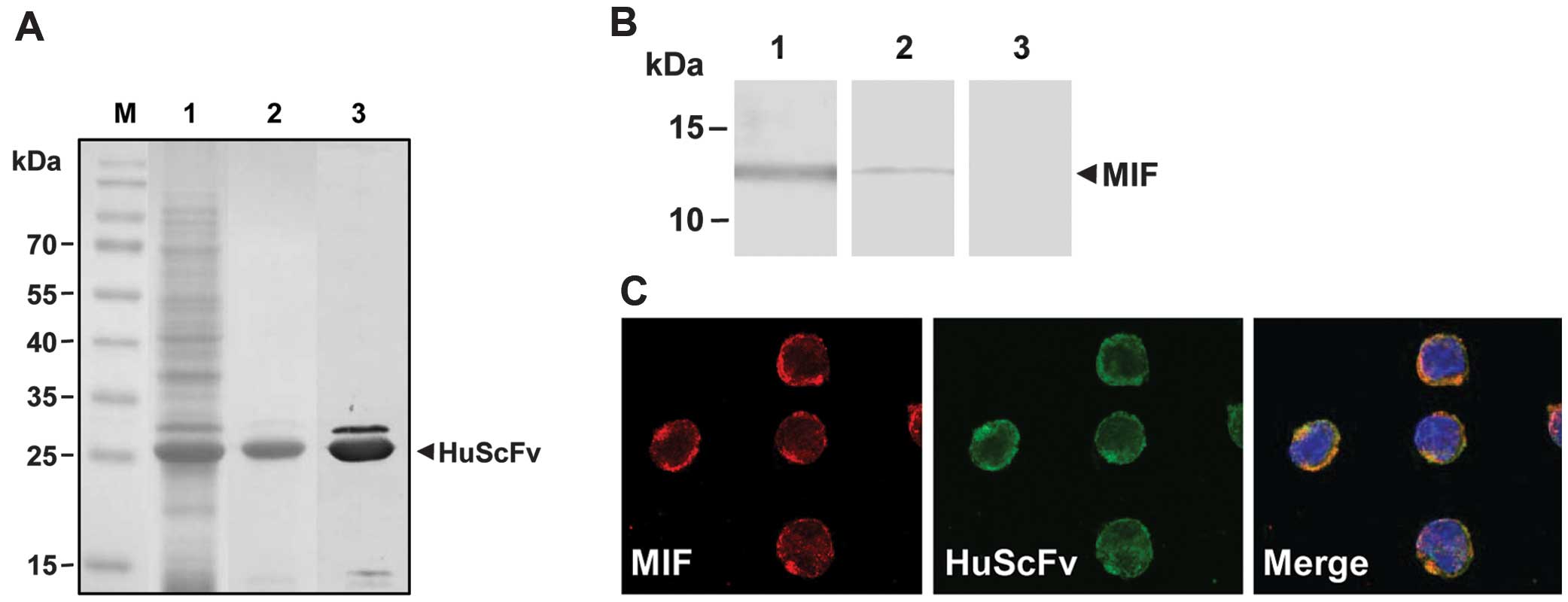
detection in subsequent experiments. Only the HuScFv of clone no.
22 (~25 kDa) was produced in an sufficient amount and appropriate
purity for subsequent experiments as revealed by SDS-PAGE and
western blot analysis detected with anti-6xHis antibody (Fig. 2A).
Characterization of MIF-specific
HuScFv
The HuScFv no. 22 was demonstrated for its binding
activity to native MIF by western blot analysis and
immunofluorescence staining. Western blot analysis revealed a
reactive band of native MIF from U937 cell lysate bound to HuScFv
at the same size as that of the native human MIF protein (12.5
kDa), similar to the positive antibody control (mouse anti-MIF
pAb), while the irrelevant HuScFv showed no reactive band (Fig. 2B). Immunofluorescence staining
confirmed the binding activity of HuScFv to intracellular MIF in
U937 cells. Co-localization of MIF and HuScFv throughout the
cytoplasm of U937 cells was clearly observed under confocal
microscopy (Fig. 2C).
Inhibition of MIF tautomerase activity by
HuScFv
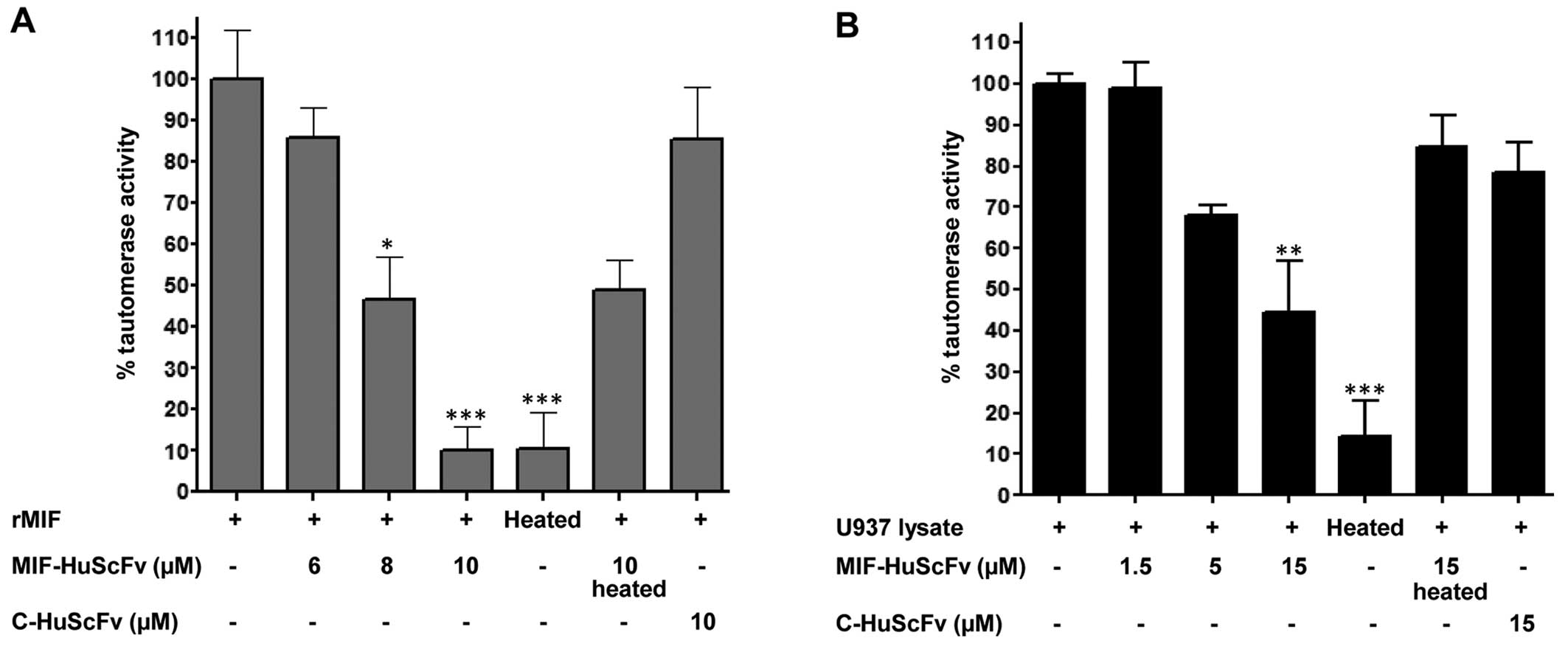
Inhibitory effect of HuScFv on MIF dopachrome
tautomerase activity was determined. Following pre-incubation with
HuScFv, tautomerization reaction indicated by substrate
decolorization of both rMIF and native MIF was reduced compared to
that of the non-HuScFv control reaction. Increased inhibition was
evident with the increasing amount of HuScFv present in the
reaction. At the end-point of rMIF-mediated enzymatic reaction,
HuScFv levels at concentrations of 6, 8 and 10 μM markedly reduced
tautomerase activity to 86, 46 and 10%, respectively, compared to
the non-HuScFv control reaction (Fig.
3A). On the other hand, heat-denatured and irrelevant HuScFv
did not significantly affect the enzymatic activity of rMIF.
The HuScFv selected from rMIF was also tested for
its inhibition of tautomerase activity mediated by native MIF in
human monoblastic leukemia (U937) cells. Tautomerase activity of
MIF in U937 cells was reduced as MIF-specific HuScFv was added
prior to initiation of enzymatic reaction. As the HuScFv
concentration increased (1.5, 5 and 15 μM) the average percentages
of tautomerase activity were reduced to 99, 68 and 44%,
respectively (Fig. 3B). By
contrast, denatured MIF-specific and irrelevant HuScFv failed to
hinder tautomerase activity.
Mimotope searching and in silico analysis
of HuScFv-MIF interaction
Human MIF-HuScFv interaction was analyzed by means
of mimotope searching and molecular docking. In mimotope searching,
20 phage clones carrying peptide mimotopes of HuScFv were obtained
and the 12-mer consensus mimotope sequence was ‘MSTPLGQYTGTK’.
HuScFv mimotope was aligned on the MIF sequence and it was found to
locate at the residues 22–33 of the human MIF-residing tautomerase
active site.
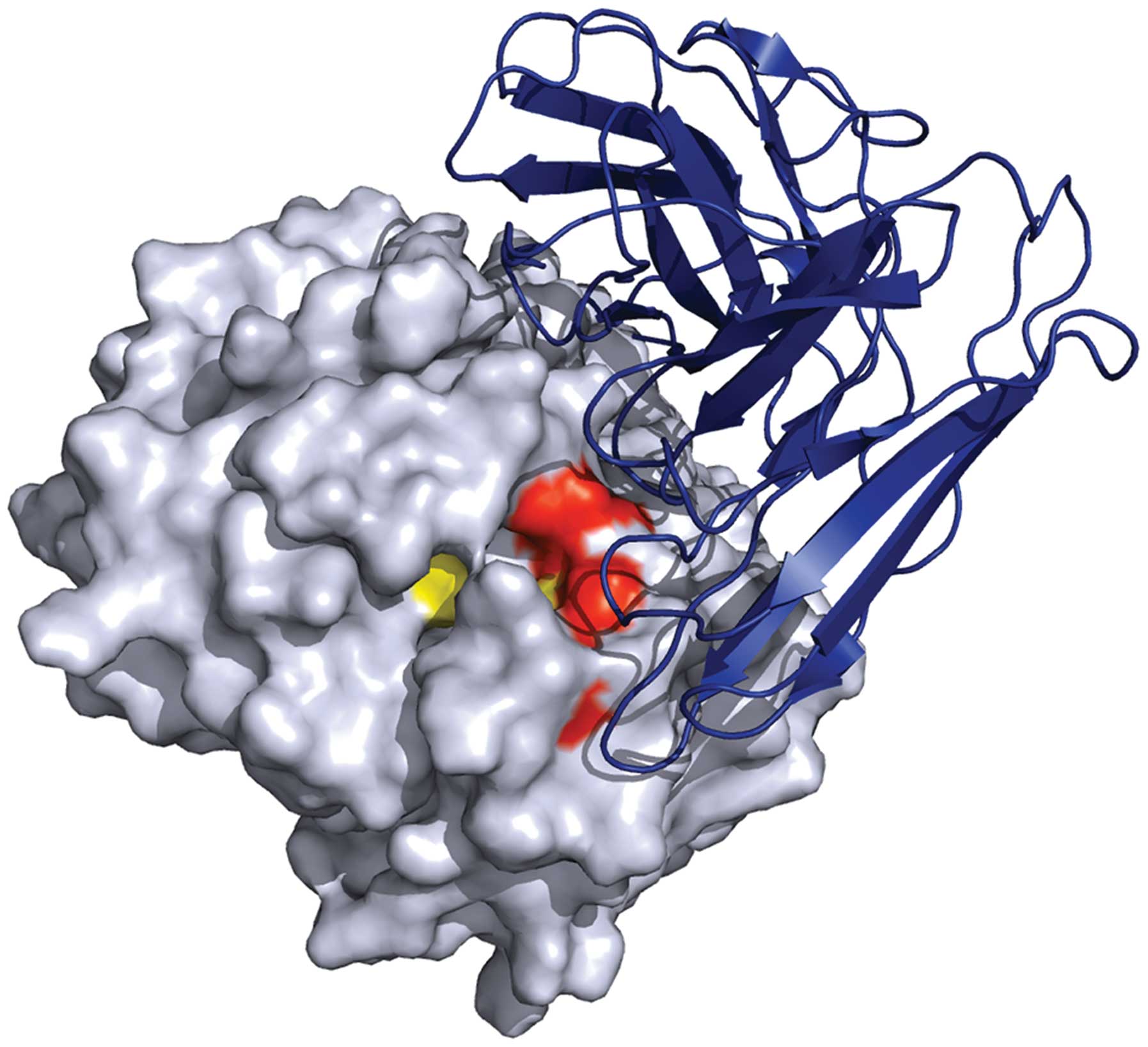
In molecular docking, HuScFv was modeled using a
human anti-SARS spike protein antibody with 75% sequence similarity
to the template. Ramachandran plot of the modeled HuScFv revealed
the acceptable disallowed region (1.0%), indicating that the
modeled structure is appropriate for the molecular docking
analysis. The docked pose calculated by ZDOCK and RDOCK shown in
Fig. 4, exhibited low-binding
energy of HuScFv-MIF binding (−17.493 kcal/mol) at residues Lys32
and Ile64 of the MIF tautomerase active site.
Discussion
The correlation of increased MIF levels in plasma
with severity of inflammatory diseases has been previously
reported. Thus, MIF is considered a promising biomarker (36) and therapeutic target for these
diseases. It has been shown that interference or abrogation of
enzymatic activity of MIF rescued individuals that succumbed to
inflammation-mediated pathogenesis (21,37). A number of inhibitors and
neutralizing antibodies have been introduced for the inhibition of
MIF activity. Recently, Kerschbaumer et al (30) generated fully human anti-MIF
antibodies by selection from a phage display library and
extensively analyzed these antibody molecules in vitro and
in vivo. Results of that study showed that only the
antibody-binding epitopes within amino acids 50–68 or 86–102 of the
MIF molecule, the regions forming a β-sheet structure in the MIF
oxidoreductase motif, exerted protective effects in models of
sepsis or contact hypersensitivity. Findings of this study have
demonstrated that fully human antibody fragments specific to MIF
inhibited the enzymatic activity of MIF and presented therapeutic
potentials. Identification of HuScFv specific to MIF is also
crucial. Unlike the previous study (30), we identified and reported HuScFv
that inhibits tautomerase activity of MIF.
In this study, we initially amplified the coding
sequence of human MIF from the human cDNA library, which showed
100% homology to the human MIF-coding sequence previously reported
in the NCBI database (NM_002415.1). The rMIF generated from the
human MIF cDNA construct exhibited tautomerase activity, suggesting
that it carried a native structure and active enzymatic sites;
thus, it was suitable for additional use as the target antigen for
selection of MIF-specific HuScFv from the human antibody phage
display library. This library, constructed in our laboratory, has
previously been used for the selection of a number of neutralizing
HuScFv specific to different pathogenic antigens (31,32,38,39). However, it has not been previously
used for the selection of HuScFv specific to a human protein.
Naturally, immature B lymphocytes carrying
antibodies specific to human autologous proteins are eliminated
during B-lymphocyte maturation; consequently, they are absent from
the pool of circulating lymphocytes. However, results of this study
have demonstrated that the human antibody phage display library
prepared from human B lymphocytes is composed of HuScFv specific to
human MIF that should be deleted from the matured B lymphocytes.
This finding is attributable to the fact that in the generation
step of human antibody phage display library the heavy and light
chains (VH and VL) of antibody genes were
individually amplified and allowed to randomly fuse to form
HuScFv. Thus, the combination of VH and VL that did not
exist in nature was available in the in vitro preparation of
synthetic antibody phage display library. This explains the fact
that a smaller number of huscfv-positive
phagemid-transformed E. coli from MIF-bio-panning (29%) was
obtained when they were compared to those from the selection of
HuScFv specific to pathogenic proteins reported in previous studies
(59–100%) (31,34,38).
Some of the HuScFv clones obtained exhibited strong
binding activity to rMIF as determined by indirect ELISA. The
HuScFv with high binding signals were produced and purified. Only
HuScFv of clone no. 22 yielded sufficient amount with appropriate
purity, and it was subsequently characterized by results of
different experiments. Western blot analysis revealed that HuScFv
no. 22 bound to native MIF protein prepared from the monocytic cell
line, U937. Moreover, immunofluorescence staining revealed
co-localization of HuScFv and native MIF in the cytoplasm of U937
cells. These results readily indicate that although it was selected
by rMIF, the HuScFv molecules interacted with its target MIF
antigen in the native form.
The tautomerase-active site is essential for the
pro-inflammatory activity of MIF; thus, attempts have been made to
develop tautomerase-neutralizing inhibitors (21,40). A number of these inhibitors have
also been proven beneficial for therapeutic efficacy in
MIF-associated diseases (24,40–42). In the present study, inhibition of
dopachrome tautomerization was performed to assess inhibition of
MIF tautomerase activity mediated by HuScFv. Subsequent to
pre-incubation with MIF-specific HuScFv, the tautomerization
activity of rMIF and native MIF, as detected by substrate
decolorization, was reduced in a dose-dependent manner, when
compared to that of the non-HuScFv control reaction, indicating its
specific inhibition to MIF enzymatic activity. The finding that
HuScFv inhibited the tautomerase activity of rMIF to a greater
extent than that of native MIF may indicate the presence of other
proteins containing the tautomerase activities in U937 cell lysate
(2). In vitro
neutralization of MIF-mediated enzymatic activity suggested the
possibility of using HuScFv to reduce MIF-induced pro-inflammatory
reactions, similar to the inhibitors or specific monoclonal
antibodies as demonstrated in previous studies (21,43). Experiments on in vivo
neutralization tests are required to evaluate anti-inflammatory
activities of MIF-specific HuScFv (30).
The HuScFv and MIF interaction was elaborated by the
target epitope searching and molecular docking approaches. The
12-mer consensus mimotope sequence of MIF-specific HuScFv is
located at the amino acid residues 22–33 of human MIF residing
within the tautomerase active site. Molecular docking also revealed
that the interaction between HuScFv and MIF spontaneously occurred
with low-binding energy at residues Lys32 and Ile64 of the MIF
tautomerase active site. This finding strongly supports the
previous description of tautomerase catalytic residues including
Pro1, Lys32, Ile64, Tyr95 and Asn97 (40). The phenomenon of the
HuScFv-mediated inhibition of MIF tautomerase activity is similar
to that exerted by the ISO1 chemical inhibitor (40).
In conclusion, HuScFv selected by using recombinant
human MIF readily recognized native MIF present in human
monoblastic leukemia (U937) cells. The antibody also neutralized
tautomerase activity mediated by both rMIF and native MIF as it
interacted with catalytic residues residing in the tautomerase
active site of the MIF molecule. The inhibitory effect was
elaborated by mimotope searching and the molecular docking
approaches. MIF-specific HuScFv, with its fully-human format and
small size, should be investigated in subsequent experiments to
demonstrate its anti-inflammatory activity and its ability to
develop into a therapeutic molecule for the treatment of human
inflammatory diseases.
Acknowledgements
MT and OP are TRF young research scholars
(supporting grant nos. TRG5680057 and TRG5480006, respectively). WC
is supported by the research grant under the National Research
University (NRU) Project, Office of Higher Education Commission
(OHEC). PY is a TRF-Senior Research Scholar and also supported by
Chalermphrakiat Grant, Faculty of Medicine Siriraj Hospital,
Mahidol University. The authors would like to thank the National
Nanotechnology Center (NANOTEC), the National Science and
Technology Development Agency (NSTDA) for providing the Discovery
Studio Package 2.5. The human antibody phage display library used
in this study is the property of the National Research Council of
Thailand (NRCT).
References
|
1
|
David JR: Delayed hypersensitivity in
vitro: its mediation by cell-free substances formed by lymphoid
cell-antigen interaction. Proc Natl Acad Sci USA. 56:72–77. 1966.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Merk M, Zierow S, Leng L, et al: The
D-dopachrome tautomerase (DDT) gene product is a cytokine
and functional homolog of macrophage migration inhibitory factor
(MIF). Proc Natl Acad Sci USA. 108:E577–E585. 2011.
|
|
3
|
Calandra T and Roger T: Macrophage
migration inhibitory factor: a regulator of innate immunity. Nat
Rev Immunol. 3:791–800. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bernhagen J, Mitchell RA, Calandra T,
Voelter W, Cerami A and Bucala R: Purification, bioactivity, and
secondary structure analysis of mouse and human macrophage
migration inhibitory factor (MIF). Biochemistry. 33:14144–14155.
1994. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mitchell RA, Metz CN, Peng T and Bucala R:
Sustained mitogen-activated protein kinase (MAPK) and cytoplasmic
phospholipase A2 activation by macrophage migration inhibitory
factor (MIF). Regulatory role in cell proliferation and
glucocorticoid action. J Biol Chem. 274:18100–18106. 1999.
View Article : Google Scholar
|
|
6
|
Mitchell RA, Liao H, Chesney J,
Fingerle-Rowson G, Baugh J, David J and Bucala R: Macrophage
migration inhibitory factor (MIF) sustains macrophage
proinflammatory function by inhibiting p53: regulatory role in the
innate immune response. Proc Natl Acad Sci USA. 99:345–350. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sugimoto H, Taniguchi M, Nakagawa A,
Tanaka I, Suzuki M and Nishihira J: Crystal structure of human
D-dopachrome tautomerase, a homologue of macrophage
migration inhibitory factor, at 1.54 A resolution. Biochemistry.
38:3268–3279. 1999.
|
|
8
|
Rosengren E, Aman P, Thelin S, et al: The
macrophage migration inhibitory factor MIF is a phenylpyruvate
tautomerase. FEBS Lett. 417:85–88. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kleemann R, Kapurniotu A, Frank RW, et al:
Disulfide analysis reveals a role for macrophage migration
inhibitory factor (MIF) as thiol-protein oxidoreductase. J Mol
Biol. 280:85–102. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bozza FA, Gomes RN, Japiassu AM, Soares M,
Castro-Faria-Neto HC, Bozza PT and Bozza MT: Macrophage migration
inhibitory factor levels correlate with fatal outcome in sepsis.
Shock. 22:309–313. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen LC, Lei HY, Liu CC, et al:
Correlation of serum levels of macrophage migration inhibitory
factor with disease severity and clinical outcome in dengue
patients. Am J Trop Med Hyg. 74:142–147. 2006.PubMed/NCBI
|
|
12
|
Assunção-Miranda I, Amaral FA, Bozza FA,
et al: Contribution of macrophage migration inhibitory factor to
the pathogenesis of dengue virus infection. FASEB J. 24:218–228.
2010.PubMed/NCBI
|
|
13
|
Hou XQ, Gao YW, Yang ST, Wang CY, Ma ZY
and Xia XZ: Role of macrophage migration inhibitory factor in
influenza H5N1 virus pneumonia. Acta Virol. 53:225–231. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kim HR, Park MK, Cho ML, et al: Macrophage
migration inhibitory factor upregulates angiogenic factors and
correlates with clinical measures in rheumatoid arthritis. J
Rheumatol. 34:927–936. 2007.PubMed/NCBI
|
|
15
|
Foote A, Briganti EM, Kipen Y, Santos L,
Leech M and Morand EF: Macrophage migration inhibitory factor in
systemic lupus erythematosus. J Rheumatol. 31:268–273.
2004.PubMed/NCBI
|
|
16
|
Lan HY, Yang N, Nikolic-Paterson DJ, et
al: Expression of macrophage migration inhibitory factor in human
glomerulonephritis. Kidney Int. 57:499–509. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Niino M, Ogata A, Kikuchi S, Tashiro K and
Nishihira J: Macrophage migration inhibitory factor in the
cerebrospinal fluid of patients with conventional and optic-spinal
forms of multiple sclerosis and neuro-Behçet’s disease. J Neurol
Sci. 179:127–131. 2000.PubMed/NCBI
|
|
18
|
Burger-Kentischer A, Goebel H, Seiler R,
et al: Expression of macrophage migration inhibitory factor in
different stages of human atherosclerosis. Circulation.
105:1561–1566. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Herder C, Kolb H, Koenig W, et al:
Association of systemic concentrations of macrophage migration
inhibitory factor with impaired glucose tolerance and type 2
diabetes: results from the Cooperative Health Research in the
Region of Augsburg, Survey 4 (KORA S4). Diabetes Care. 29:368–371.
2006. View Article : Google Scholar
|
|
20
|
Bucala R and Donnelly SC: Macrophage
migration inhibitory factor: a probable link between inflammation
and cancer. Immunity. 26:281–285. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Al-Abed Y, Dabideen D, Aljabari B, et al:
ISO-1 binding to the tautomerase active site of MIF inhibits its
pro-inflammatory activity and increases survival in severe sepsis.
J Biol Chem. 280:36541–36544. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cvetkovic I, Al-Abed Y, Miljkovic D, et
al: Critical role of macrophage migration inhibitory factor
activity in experimental autoimmune diabetes. Endocrinology.
146:2942–2951. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Nicoletti F, Créange A, Orlikowski D, et
al: Macrophage migration inhibitory factor (MIF) seems crucially
involved in Guillain-Barré syndrome and experimental allergic
neuritis. J Neuroimmunol. 168:168–174. 2005.PubMed/NCBI
|
|
24
|
Alam A, Haldar S, Thulasiram HV, et al:
Novel anti-inflammatory activity of epoxyazadiradione against
macrophage migration inhibitory factor: inhibition of tautomerase
and proinflammatory activities of macrophage migration inhibitory
factor. J Biol Chem. 287:24844–24861. 2012. View Article : Google Scholar
|
|
25
|
Calandra T, Echtenacher B, Roy DL, et al:
Protection from septic shock by neutralization of macrophage
migration inhibitory factor. Nat Med. 6:164–170. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bernhagen J, Bacher M, Calandra T, Metz
CN, Doty SB, Donnelly T and Bucala R: An essential role for
macrophage migration inhibitory factor in the tuberculin
delayed-type hypersensitivity reaction. J Exp Med. 183:277–282.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Mikulowska A, Metz CN, Bucala R and
Holmdahl R: Macrophage migration inhibitory factor is involved in
the pathogenesis of collagen type II-induced arthritis in mice. J
Immunol. 158:5514–5517. 1997.PubMed/NCBI
|
|
28
|
Cvetkovic I and Stosic-Grujicic S:
Neutralization of macrophage migration inhibitory factor-novel
approach for the treatment of immunoinflammatory disorders. Int
Immunopharmacol. 6:1527–1534. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Greven D, Leng L and Bucala R: Autoimmune
diseases: MIF as a therapeutic target. Expert Opin Ther Targets.
14:253–264. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kerschbaumer RJ, Rieger M, Völkel D, et
al: Neutralization of macrophage migration inhibitory factor (MIF)
by fully human antibodies correlates with their specificity for the
β-sheet structure of MIF. J Biol Chem. 287:7446–7455.
2012.PubMed/NCBI
|
|
31
|
Kulkeaw K, Sakolvaree Y, Srimanote P, et
al: Human monoclonal ScFv neutralize lethal Thai cobra, Naja
kaouthia, neurotoxin. J Proteomics. 72:270–282. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Poungpair O, Pootong A, Maneewatch S, et
al: A human single chain transbody specific to matrix protein (M1)
interferes with the replication of influenza A virus. Bioconjug
Chem. 21:1134–1141. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Dios A, Mitchell RA, Aljabari B, et al:
Inhibition of MIF bioactivity by rational design of pharmacological
inhibitors of MIF tautomerase activity. J Med Chem. 45:2410–2416.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Thanongsaksrikul J, Srimanote P,
Maneewatch S, et al: A VHH that neutralizes the zinc
metalloproteinase activity of botulinum neurotoxin type A. J Biol
Chem. 285:9657–9666. 2010.
|
|
35
|
Laskowski RA, MacArthur MW, Moss DS and
Thornton JM: PROCHECK: a program to check the stereochemical
quality of protein structures. J Appl Cryst. 26:283–291. 1993.
View Article : Google Scholar
|
|
36
|
Grieb G, Merk M, Bernhagen J and Bucala R:
Macrophage migration inhibitory factor (MIF): a promising
biomarker. Drug News Perspect. 23:257–264. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Dagia NM, Kamath DV, Bhatt P, et al: A
fluorinated analog of ISO-1 blocks the recognition and biological
function of MIF and is orally efficacious in a murine model of
colitis. Eur J Pharmacol. 607:201–212. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Thathaisong U, Maneewatch S, Kulkeaw K, et
al: Human monoclonal single chain antibodies (HuScFv) that bind to
the polymerase proteins of influenza A virus. Asian Pac J Allergy
Immunol. 26:23–35. 2008.PubMed/NCBI
|
|
39
|
Maneewatch S, Thanongsaksrikul J, Songserm
T, et al: Human single-chain antibodies that neutralize homologous
and heterologous stains and clades of influenza A virus subtype
H5N1. Antivir Ther. 14:221–230. 2009.PubMed/NCBI
|
|
40
|
Lubetsky JB, Dios A, Han J, et al: The
tautomerase active site of macrophage migration inhibitory factor
is a potential target for discovery of novel anti-inflammatory
agents. J Biol Chem. 277:24976–24982. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Al-Abed Y and Van Patten S: MIF as a
disease target: ISO-1 as a proof-of-concept therapeutic. Future Med
Chem. 3:45–63. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kithcart AP, Cox GM, Sielecki T, et al: A
small-molecule inhibitor of macrophage migration inhibitory factor
for the treatment of inflammatory disease. FASEB J. 24:4459–4466.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhang Y, Zeng X, Chen S, et al:
Characterization, epitope identification and mechanisms of the
anti-septic capacity of monoclonal antibodies against macrophage
migration inhibitory factor. Int Immunopharmacol. 11:1333–1340.
2011. View Article : Google Scholar
|