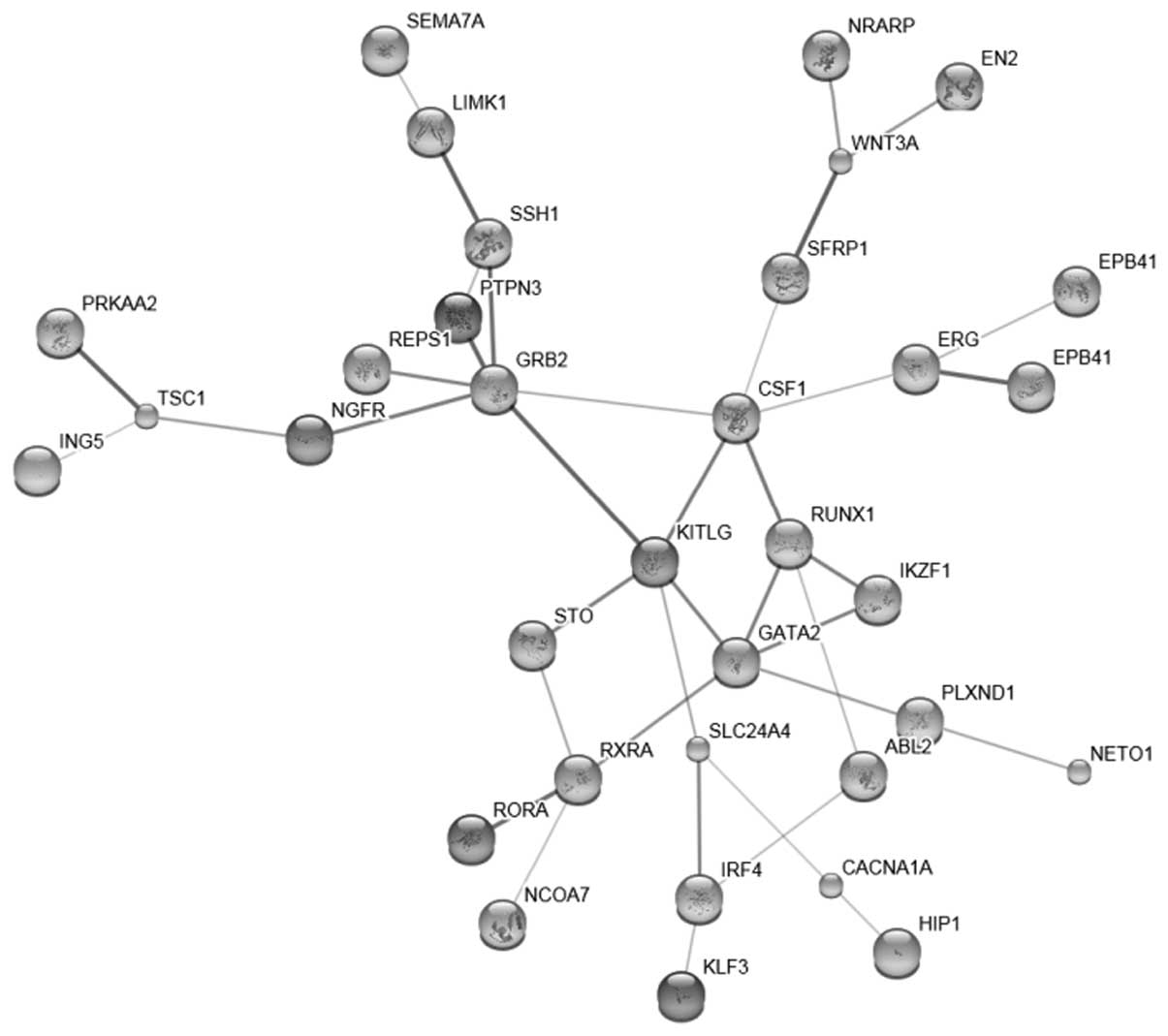
|
1
|
Hernandez BY, Green MD, Cassel KD,
Pobutsky AM, Vu V and Wilkens LR: Preview of Hawaii cancer facts
and figures 2010. Hawaii Med J. 69:223–224. 2010.PubMed/NCBI
|
|
2
|
Gray-Schopfer V, Wellbrock C and Marais R:
Melanoma biology and new targeted therapy. Nature. 445:851–857.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hatfield S and Ruohola-Baker H: microRNA
and stem cell function. Cell Tissue Res. 331:57–66. 2008.
View Article : Google Scholar
|
|
4
|
Agarwala SS: Current systemic therapy for
metastatic melanoma. Expert Rev Anticancer Ther. 9:587–595. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Calin GA and Croce CM: MicroRNA-cancer
connection: the beginning of a new tale. Cancer Res. 66:7390–7394.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Iorio MV, Ferracin M, Liu CG, et al:
MicroRNA gene expression deregulation in human breast cancer.
Cancer Res. 65:7065–7070. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ma L, Teruya-Feldstein J and Weinberg RA:
Tumour invasion and metastasis initiated by microRNA-10b in breast
cancer. Nature. 449:682–688. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yanaihara N, Caplen N, Bowman E, et al:
Unique microRNA molecular profiles in lung cancer diagnosis and
prognosis. Cancer Cell. 9:189–198. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhou J, Yu L, Gao X, et al: Plasma
microRNA panel to diagnose hepatitis B virus-related hepatocellular
carcinoma. J Clin Oncol. 29:4781–4788. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen CZ: MicroRNAs as oncogenes and tumor
suppressors. N Engl J Med. 353:1768–1771. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hatfield SD, Shcherbata HR, Fischer KA,
Nakahara K, Carthew RW and Ruohola-Baker H: Stem cell division is
regulated by the microRNA pathway. Nature. 435:974–978. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Giles KM, Barker A, Zhang PM, Epis MR and
Leedman PJ: MicroRNA regulation of growth factor receptor signaling
in human cancer cells. Methods Mol Biol. 676:147–163. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jiang S, Zhang HW, Lu MH, et al:
MicroRNA-155 functions as an OncomiR in breast cancer by targeting
the suppressor of cytokine signaling 1 gene. Cancer Res.
70:3119–3127. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shumway M, Cochrane G and Sugawara H:
Archiving next generation sequencing data. Nucleic Acids Res.
38:D870–D871. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cock PJ, Fields CJ, Goto N, Heuer ML and
Rice PM: The Sanger FASTQ file format for sequences with quality
scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res.
38:1767–1771. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kaminuma E, Mashima J, Kodama Y, et al:
DDBJ launches a new archive database with analytical tools for
next-generation sequence data. Nucleic Acids Res. 38:D33–D38. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Langmead B, Trapnell C, Pop M and Salzberg
SL: Ultrafast and memory-efficient alignment of short DNA sequences
to the human genome. Genome Biol. 10:R252009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Trapnell C and Salzberg SL: How to map
billions of short reads onto genomes. Nat Biotechnol. 27:455–457.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Friedlander MR, Chen W, Adamidi C, et al:
Discovering microRNAs from deep sequencing data using miRDeep. Nat
Biotechnol. 26:407–415. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Buermans HP, Ariyurek Y, van Ommen G, den
Dunnen JT and ‘t Hoen PA: New methods for next generation
sequencing based microRNA expression profiling. BMC Genomics.
11:7162010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Smyth GK: Limma: linear models for
microarray data. Bioinformatics and Computational Biology Solutions
using R and Bioconductor. Gentleman R, Carey V, Dudoit S, Irizarry
R and Huber W: Springer; New York: pp. 397–420. 2005, View Article : Google Scholar
|
|
22
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Friedman RC, Farh KK, Burge CB and Bartel
DP: Most mammalian mRNAs are conserved targets of microRNAs. Genome
Res. 19:92–105. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tong AH, Drees B, Nardelli G, et al: A
combined experimental and computational strategy to define protein
interaction networks for peptide recognition modules. Science.
295:321–324. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Szklarczyk D, Franceschini A, Kuhn M, et
al: The STRING database in 2011: functional interaction networks of
proteins, globally integrated and scored. Nucleic Acids Res.
39:D561–D568. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hosack DA, Dennis G Jr, Sherman BT, Lane
HC and Lempicki RA: Identifying biological themes within lists of
genes with EASE. Genome Biol. 4:R702003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Steeg PS: Tumor metastasis: mechanistic
insights and clinical challenges. Nat Med. 12:895–904. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Rabinowits G, Gercel-Taylor C, Day JM,
Taylor DD and Kloecker GH: Exosomal microRNA: a diagnostic marker
for lung cancer. Clin Lung Cancer. 10:42–46. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Katakowski M, Zheng X, Jiang F, Rogers T,
Szalad A and Chopp M: MiR-146b-5p suppresses EGFR expression and
reduces in vitro migration and invasion of glioma. Cancer Invest.
28:1024–1030. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mertens-Talcott SU, Chintharlapalli S, Li
X and Safe S: The oncogenic microRNA-27a targets genes that
regulate specificity protein transcription factors and the G2-M
checkpoint in MDA-MB-231 breast cancer cells. Cancer Res.
67:11001–11011. 2007. View Article : Google Scholar
|
|
31
|
Scott GK, Mattie MD, Berger CE, Benz SC
and Benz CC: Rapid alteration of microRNA levels by histone
deacetylase inhibition. Cancer Res. 66:1277–1281. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gottardo F, Liu CG, Ferracin M, et al:
Micro-RNA profiling in kidney and bladder cancers. Urol Oncol.
25:387–392. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liu T, Tang H, Lang Y, Liu M and Li X:
MicroRNA-27a functions as an oncogene in gastric adenocarcinoma by
targeting prohibitin. Cancer Lett. 273:233–242. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Guttilla IK and White BA: Coordinate
regulation of FOXO1 by miR-27a, miR-96, and miR-182 in breast
cancer cells. J Biol Chem. 284:23204–23216. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hiroki E, Akahira J, Suzuki F, et al:
Changes in microRNA expression levels correlate with
clinicopathological features and prognoses in endometrial serous
adenocarcinomas. Cancer Sci. 101:241–249. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Rhodes LV, Tilghman SL, Boue SM, et al:
Glyceollins as novel targeted therapeutic for the treatment of
triple-negative breast cancer. Oncol Lett. 3:163–171.
2012.PubMed/NCBI
|
|
37
|
Baffa R, Fassan M, Volinia S, et al:
MicroRNA expression profiling of human metastatic cancers
identifies cancer gene targets. J Pathol. 219:214–221. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Leidinger P, Keller A, Borries A, et al:
High-throughput miRNA profiling of human melanoma blood samples.
BMC Cancer. 10:2622010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Li X, Nishida T, Noguchi A, et al:
Decreased nuclear expression and increased cytoplasmic expression
of ING5 may be linked to tumorigenesis and progression in human
head and neck squamous cell carcinoma. J Cancer Res Clin Oncol.
136:1573–1583. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Bohm M, Locke WJ, Sutherland RL, Kench JG
and Henshall SM: A role for GATA-2 in transition to an aggressive
phenotype in prostate cancer through modulation of key
androgen-regulated genes. Oncogene. 28:3847–3856. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wang J, Cai Y, Ren C and Ittmann M:
Expression of variant TMPRSS2/ERG fusion messenger RNAs is
associated with aggressive prostate cancer. Cancer Res.
66:8347–8351. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Carver BS, Tran J, Gopalan A, et al:
Aberrant ERG expression cooperates with loss of PTEN to promote
cancer progression in the prostate. Nat Genet. 41:619–624. 2009.
View Article : Google Scholar : PubMed/NCBI
|