Introduction
Mammary gland is a highly efficient organ that
produces milk during nursing and is composed of epithelial, adipose
and other stromal cells (1).
Breast cancer is a type of cancer originating from breast tissue
such as the inner lining of milk ducts or the lobules that supply
the ducts with milk (2–4). Breast cancer is 100 times more
common in women than in men and accounts for 22.9% of all cancer
types in women (5,6). Therefore, the primary risk factors
for breast cancer are female gender and older age (7,8).
Other potential risk factors include lack of childbearing or
breastfeeding, high hormone levels, diet and obesity (9,10).
However, the exact risk factors that contribute to breast cancer
development are largely unknown. Although mutations in BRCA
genes have been linked to breast cancer initiation, they only
account for a small percentage of breast cancers (11,12). These data suggest that the breast
microenvironment such as milk may also contribute to breast cancer
initiation and development.
MicroRNAs (miRNAs or miRs) are small non-coding
single-stranded RNAs transcribed from genomic DNA and processed to
mature miRNAs by Drosha in the nucleus and subsequently by Dicer in
the cytoplasm (13,14). A variety of miRNAs have been
identified to play a role in breast cancer carcinogenesis (15,16). Let-7 family and miR-200 family are
relatively well-characterized miRNAs that are crucial to breast
cancer initiation and breast cancer cell self-renewal (17–20). In addition, results of a recent
study showed that the combination of miR-145 and miR-451 in the
blood may discriminate breast cancer patients from healthy controls
(21). A notable and important
biological feature of miRNAs is their greater stability relative to
mRNAs, which makes them stable and powerful biomarkers for breast
cancer. Milk and the contained miRNAs act as an important breast
cancer microenvironment, however, their roles in breast cancer
development are largely undetermined.
It has previously been demonstrated that miRNAs are
present in bovine and human breast milk (22–26). miRNAs in breast milk are stable
even under extremely acidic conditions (pH 1) and the
freeze-thawing process, and are resistant to RNase treatment
(22,27). Milk is produced but instead of
discharging during feeding it remains in the breast, resulting in
‘milk stasis’ (28–30). The duration required for milk
stasis ranges from several years to ≥30 years. Milk stasis in
patients is usually found via fiberoptic ductoscopy or operation,
with most patients presenting non-spontaneous nipple discharge.
Milk stasis is the main cause of mastitis, however, its role in
breast cancer remains to be determined.
The aim of this study was to determine miRNA
profiling in milk stasis. Comparison of milk miRNA profiles between
patients with milk stasis only and patients with both milk stasis
and breast tumor showed differential miRNA expression in the two
cohorts, confirmed by quantitative PCR. Our results demonstrated a
potential role of milk stasis and the associated miRNAs in breast
cancer development.
Materials and methods
Subject cohorts
Two cohorts of women with milk stasis were enrolled
in this study. One cohort included 10 patients with milk stasis but
without tumor as evidenced by breast ultrasound and mammography.
The other cohort included 10 patients with milk stasis and breast
tumor. Pathological examination confirmed that one case was ductal
carcinoma in situ, one case was atypical ductal epithelial
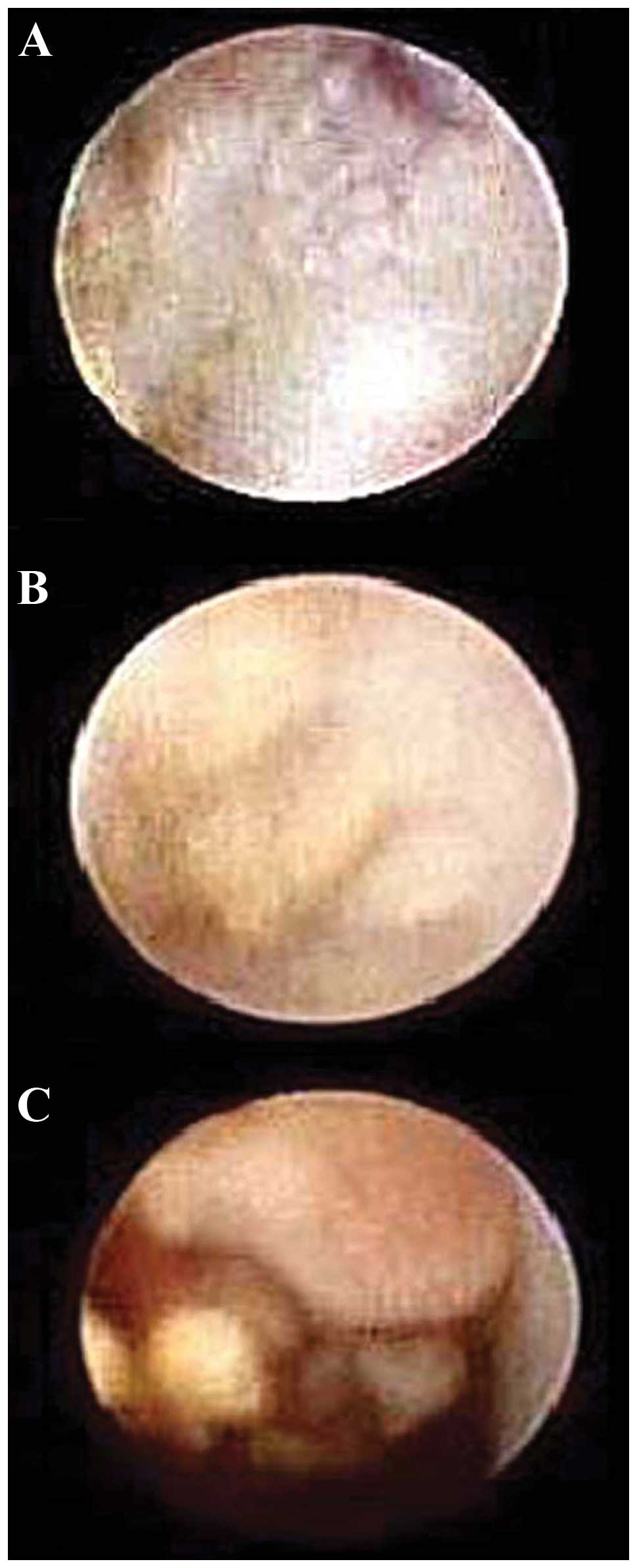
hyperplasia and eight cases were intraductal papilloma (Fig. 1). All the women involved in this
study provided signed informed consent. The study was approved by
the Ethics Committees of Jilin University (Changchun, China).
miRNA isolation
Samples were collected by lavaging the breast milk
ducts with 0.9% saline, and frozen until centrifugation in
RNAse-free tubes at 4°C. The substratum liquid layer containing
miRNAs was transferred to a new RNAse-free tube, and stored at
−80°C until RNA isolation. miRNA isolation was performed using a
mirVana miRNA Isolation kit (Ambion, Inc., Foster City, CA, USA).
The purity and quantity of isolated miRNA were determined by a
spectrophotometer (UV2800 ultraviolet spectrophotometer; Unico, New
York, NY, USA).
miRNA identification
Total RNA isolated from the lavage milk obtained
from each woman was used for library construction and subjected to
single-end sequencing in 36-bp reads using an Illumina Genome
Analyzer II (Illumina, San Diego, CA, USA). The raw reads that
passed through a series of filters (such as the length and sequence
comparison) were termed ‘map-pable reads’. The map-pable reads were
counted and the identical reads were combined into a single type,
and then mapped to the 1,527 human pre-miRNAs registered in the
miRBase 18.0 with no more than one mismatch. The most abundant
mature variant of a given miRNA was chosen as a reference sequence,
which provided the most robust approach for the evaluation of the
expression level of miRNA.
Quantitative PCR
cDNA was generated from 5 μl total RNA using. One
Step PrimeScript® miRNA cDNA Synthesis kit (Takara Bio,
Inc., Shiga, Japan) according to the manufacturer’s instructions.
Quantitative PCR was performed using a High-Specificity miRNA
qRT-PCR Detection kit (Takara) on the ABI PRISM® 7300
real-time PCR system. The ΔΔCt method was used to determine the
expression level of miRNAs in the surveyed samples.
Results
miRNAs profiling of milk from the two
groups
To profile miRNAs in human breast milk from patients
with milk stasis (all patients stopped breastfeeding for ≥3 years;
average stop breastfeeding years, 10; average age, 39.2 years), we
extracted total RNA from human saline lavage from breast ducts with
milk stasis. The concentration of miRNAs was 88 and 128.4 μg/ml in
milk from the milk stasis only group and milk stasis plus breast
neoplasm group, respectively.
A total of 821,967 unique sRNAs were detected in 10
milk stasis only samples and a total of 677,177 sRNAs were detected
in 10 milk stasis plus breast neoplasma samples. The two groups
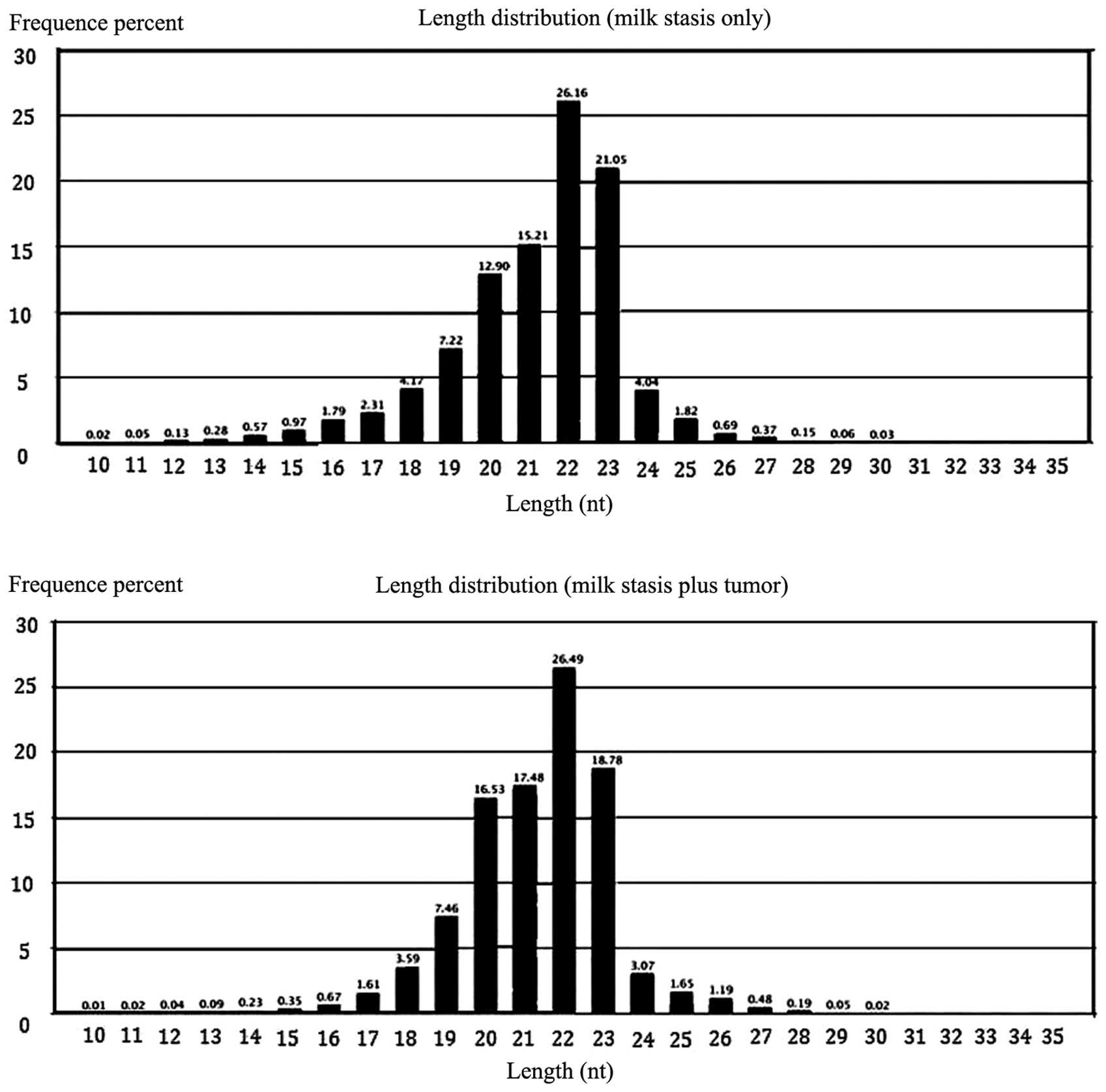
showed similar length distribution of sRNAs. The majority of sRNAs
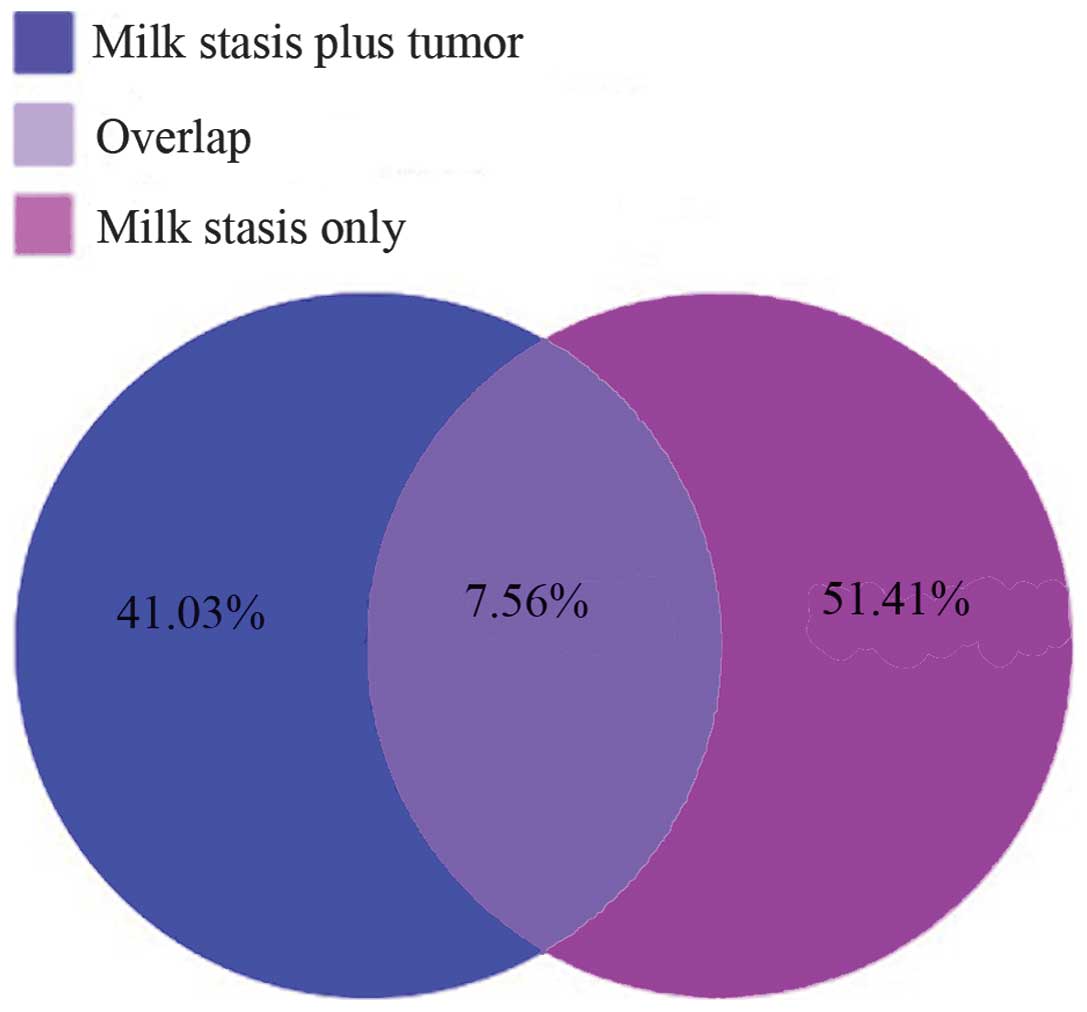
were at a length of 20, 21, 22 and 23 nt (Fig. 2). A total of 7.56% unique sRNAs
were identified in the two groups, and 41.03 and 51.41% were
specifically detected in the milk stasis plus breast neoplasm group
and milk stasis only group, respectively (Fig. 3). Among them, 266 known miRNAs
together with 271 novel miRNAs were detected in 10 milk stasis only
samples and 271 known miRNAs together with 140 novel miRNAs were
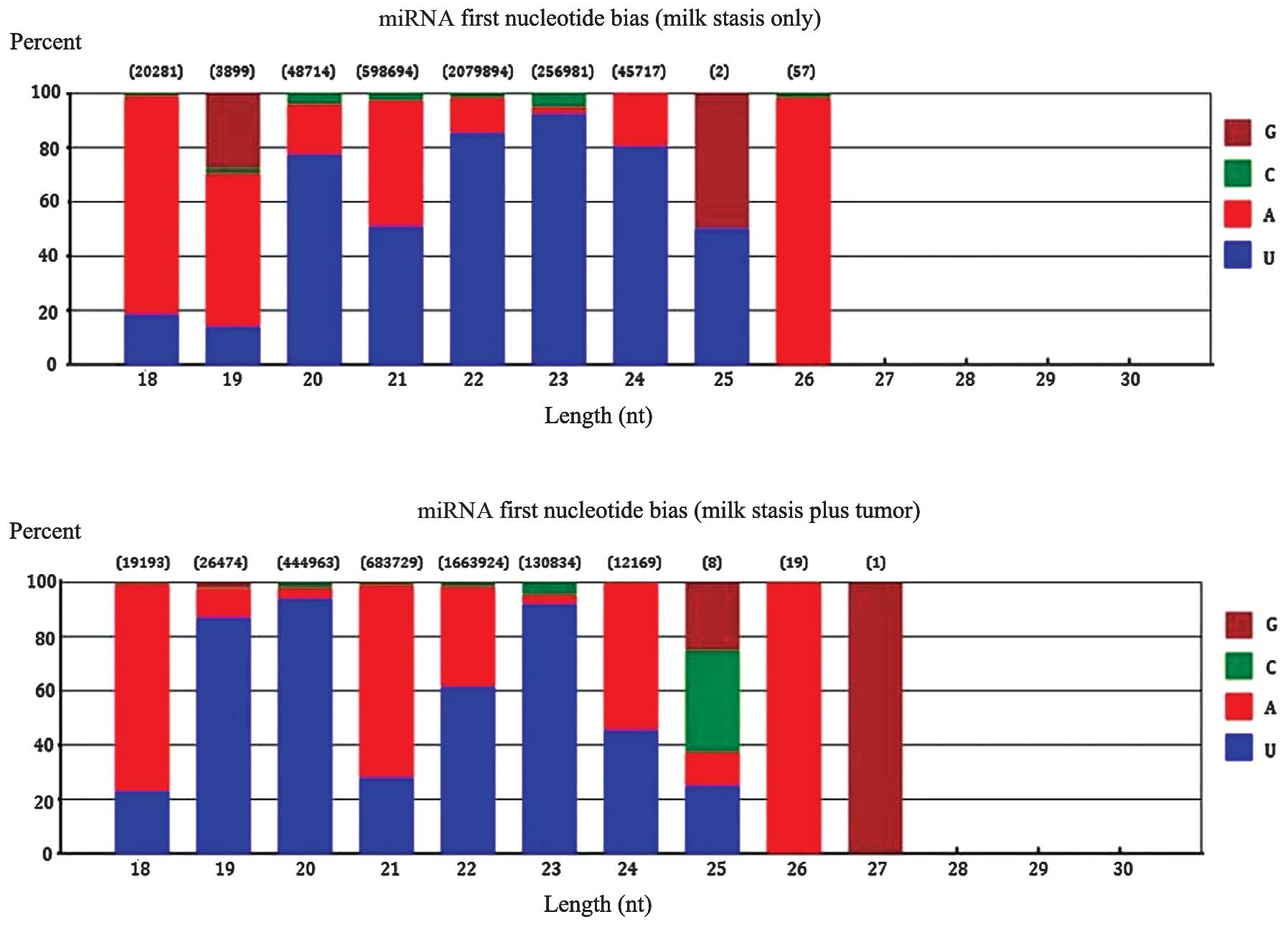
detected in 10 milk stasis plus breast neoplasm samples (Table I). The first nucleotide of miRNAs
at a length of 18–30 nt is shown in Fig. 4. Nucleotides A and U were the most
frequent first nucleotides.
 | Table ISummary of known and novel miRNAs in
the two groups. |
Table I
Summary of known and novel miRNAs in
the two groups.
| Variables | miRNA | miRNA-5p | miRNA-3p | miRNA precursors | Novel miRNA |
|---|
| Known miRNA in
miRbase 18 | 845 | 533 | 542 | 1527 | |
| Milk stasis | 266 | 292 | 292 | 736 | 271 |
| Milk stasis plus
breast tumor | 271 | 292 | 294 | 732 | 140 |
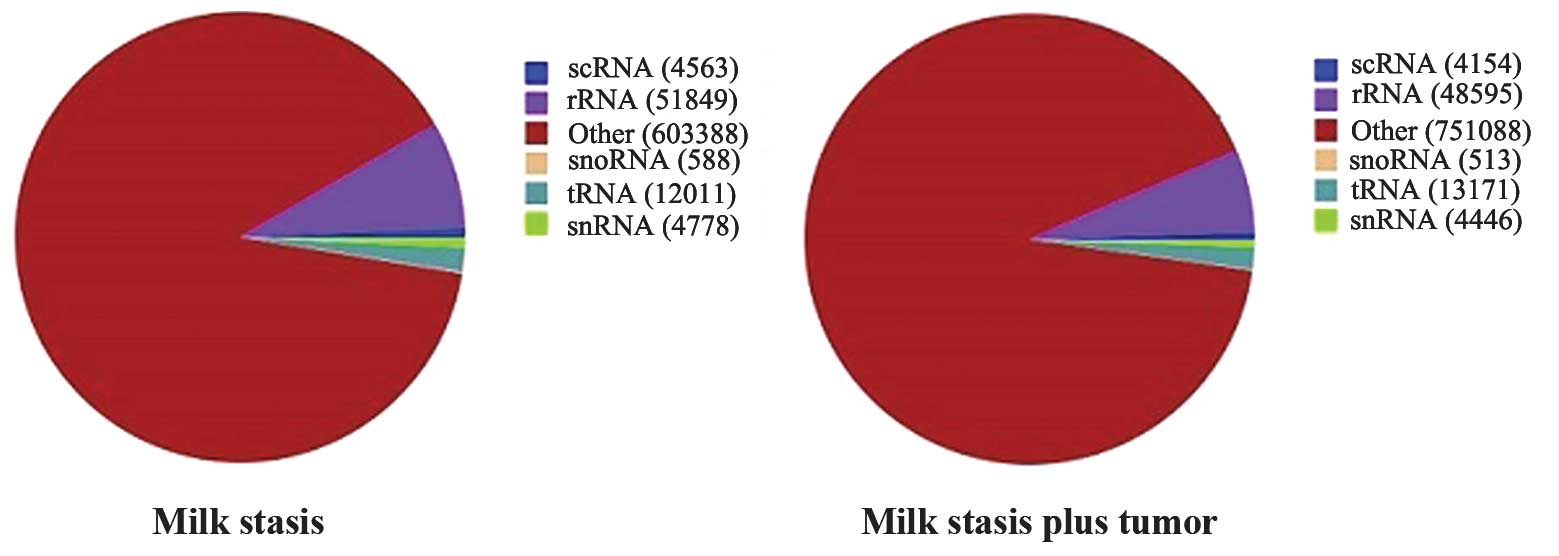
To understand the features of sRNAs in milk stasis,
subtypes of RNAs were identified in the two groups (Fig. 5) and the sequences were aligned to
the human genome. A total of 69.82% of unique sRNA in milk stasis
only patients and 67.70% of unique sRNA in milk stasis plus
neoplasm patients were mapped to the genome. Collectively, these
data clearly demonstrated that sRNAs are abundant in milk stasis
patients. The microenvironment of the epithelial cell was altered
during the years following breastfeeding.
Differential expression of miRNA in the
two groups
Milk is an important microenviroment for breast
cancer development, thus we hypothesized that miRNAs involved in
milk stasis may regulate breast cancer development. We compared the
expression of miRNAs between milk stasis only patients and milk
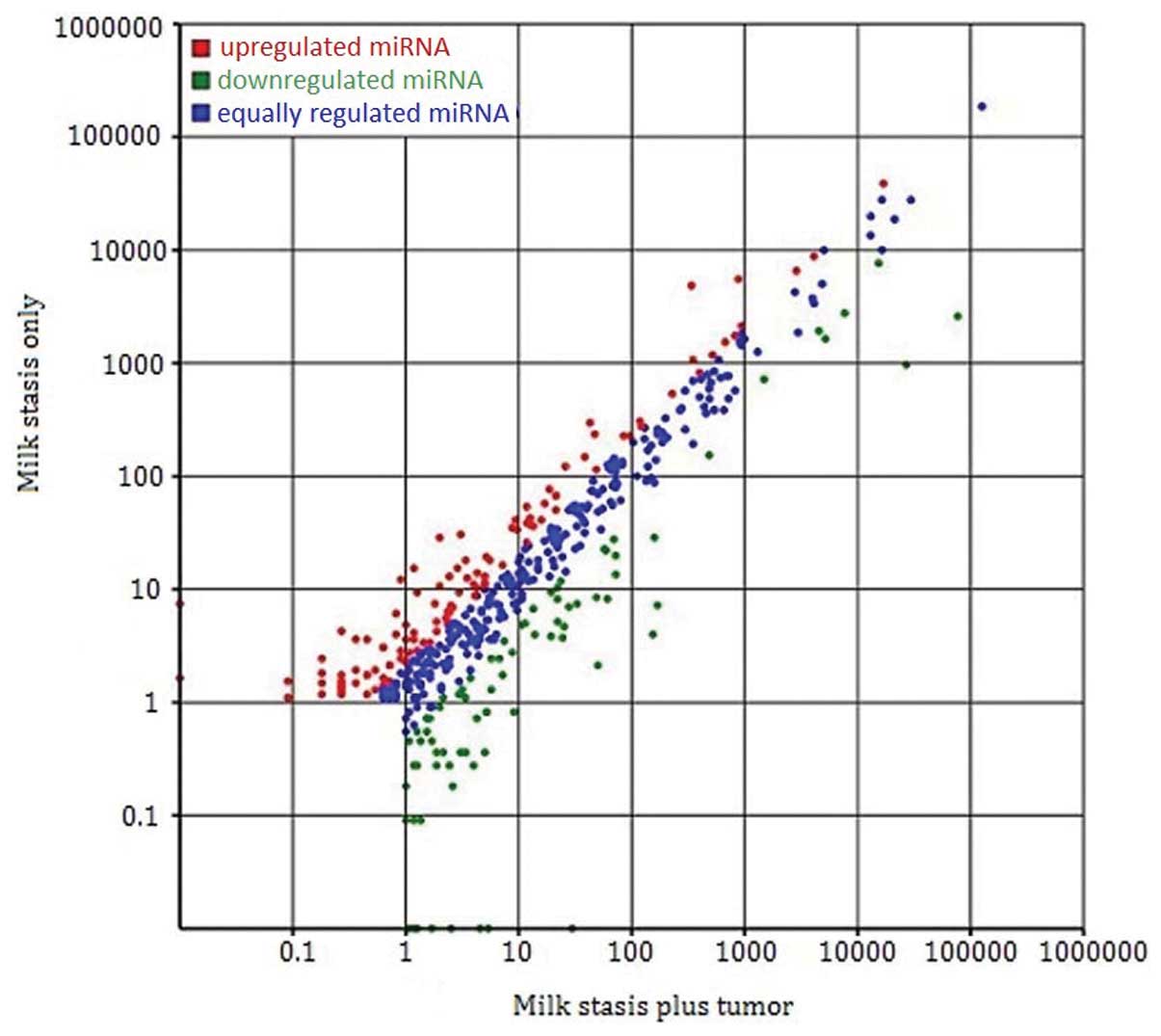
stasis plus breast neoplasm patients. Fig. 6 shows the differential expression
of miRNA in the two groups by plotting of Log2-ratio and scatter
plot. A total of 174 known miRNAs showed a differential expression
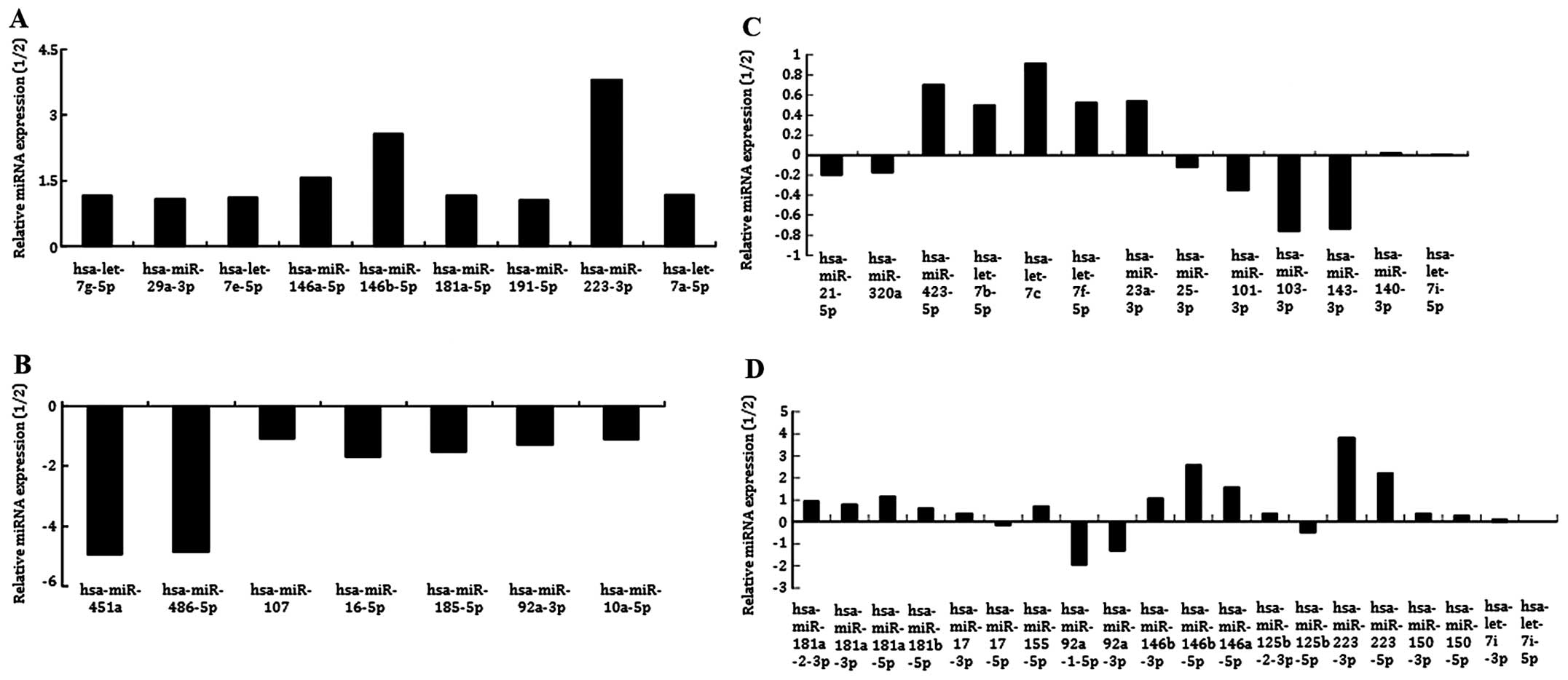
(p<0.01). Among them, nine miRNAs (hsa-let-7g-5p,
hsa-miR-29a-3p, hsa-let-7e-5p, hsa-miR-146a-5p, hsa-miR-146b-5p,
hsa-miR-181a-5p, hsa-miR-191-5p, hsa-miR-223-3p, hsa-let-7a-5p)
were significantly downregulated (p<0.01, fold-change >1)
compared to milk stasis only patients (Fig. 7A), six of which were tumor
suppressors (hsa-let-7g-5p, hsa-miR-29a-3p, hsa-let-7e-5p,
hsa-miR-146b-5p, hsa-miR-223-3p, hsa-let-7a-5p). By contrast, seven
miRNAs (hsa-miR-451a, hsa-miR-486-5p, hsa-miR-107, hsa-miR-16-5p,
hsa-miR-185-5p, hsa-miR-92a-3p, hsa-miR-10a-5p) were significantly
upregulated (p<0.01, fold-change >1) in milk stasis plus
breast neoplasm patients, compared to milk stasis only patients
(Fig. 7B), five of which have an
oncogenic function (hsa-miR-451a, hsa-miR-486-5p, hsa-miR-107,
hsa-miR-92a-3p, hsa-miR-10a-5p).
We also detected 13 highly expressed miRNAs in the
two groups (p>0.05, fold-change <1), nine of which were tumor
suppressors (hsa-miR-21-5p, hsa-miR-423-5p, hsa-let-7b-5p,
hsa-let-7c, hsa-let-7f-5p, hsa-miR-23a-3p, hsa-miR-101-3p,
hsa-miR-143-3p, hsa-let-7i-5p), while hsa-miR-140-3p had oncogenic
function (Fig. 7C).
Twenty immune-related miRNAs, which are reported in
the milk of <11 months of lactation were compared. Six of the 20
were significantly downregulated in the milk stasis plus breast
neoplasm group, however, mir-92-3p and mir-92-5p which have an
oncogenic function were significantly upregulated (Fig. 7D).
The differential miRNA profiling between the two
groups suggested that miRNA involved in milk stasis may play
important role in breast carcinogenesis.
Verification of the expression of miRNAs
in milk stasis
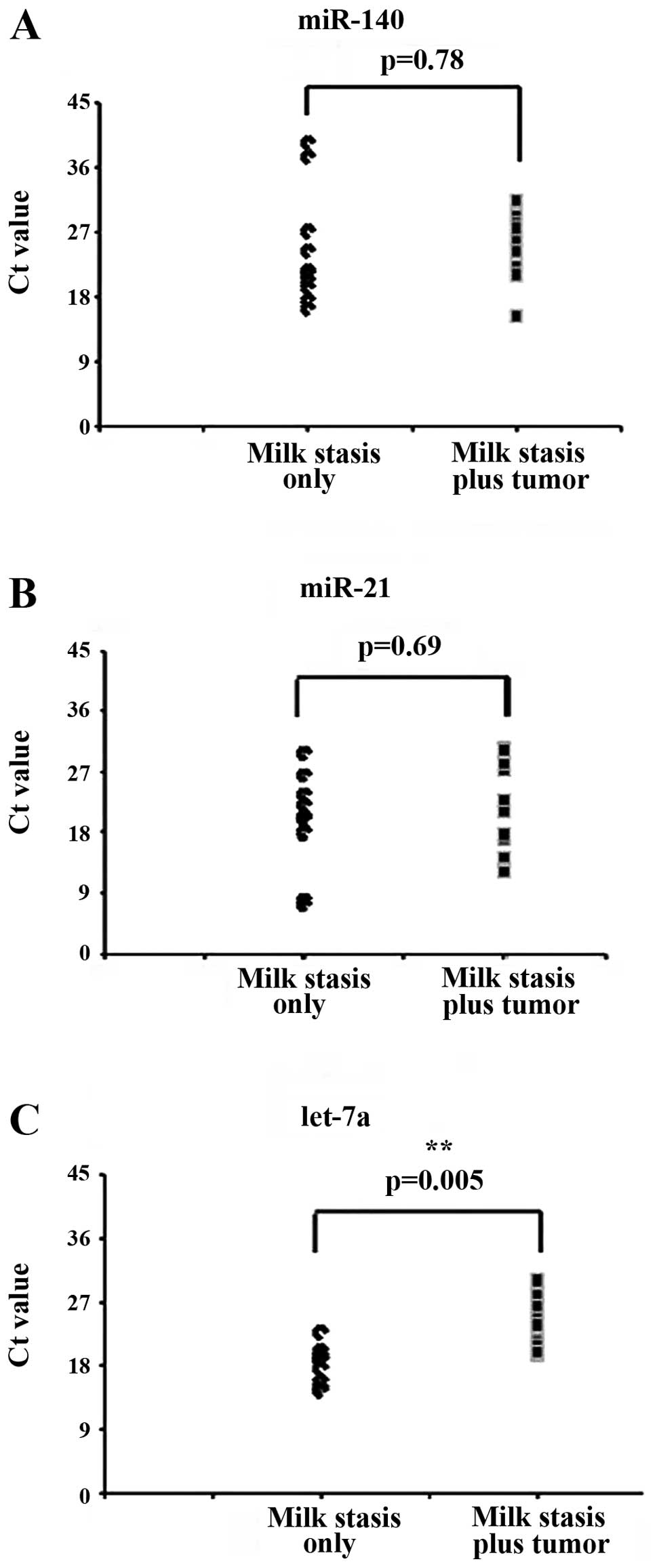
To confirm that miRNAs are abundant in milk stasis,
we used quantitative PCR to quantify the expression of three miRNAs
(miR-140, miR-21 and let-7a) in randomly selected 20 milk samples,
10 of which were milk stasis samples, and the remaining 10 milk
stasis plus tumor samples. Consistent with our sequencing data,
miR-140, miR-21 and let-7a were highly expressed in the selected
samples (Fig. 8). Thus, these
results confirm that miRNAs robustly exist in milk from milk stasis
patients.
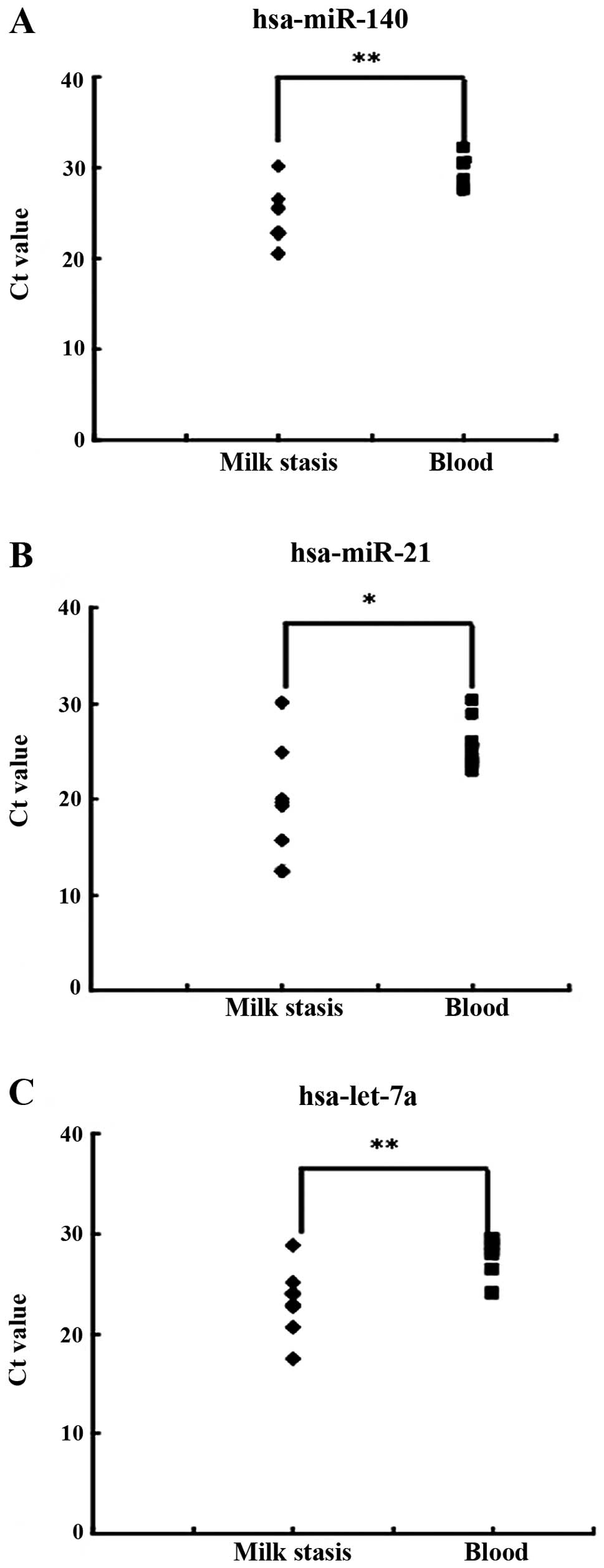
miRNAs are more abundant in milk stasis
than in the blood
Identification of biomarkers that predict or prevent
breast cancer is important for breast cancer patients. Due to the
abundance of miRNAs in milk stasis, we examined whether milk miRNAs
are better biomarkers for breast cancer than blood miRNAs. We
selected seven patients and determined miRNAs (miR-140, miR-21 and
let-7a) both in the milk and the blood. Results showed that
miR-140, miR-21 and let-7a were more abundant in the milk than in
the blood (Fig. 9). The results
suggest that miRNAs in the milk from milk stasis patients may be
more sensitive biomarkers.
Discussion
Previous studies have focused on gaining a better
understanding of breast carcinogenesis; however, the underlying
molecular mechanisms remain to be determined (31,32). It is well recognized that the
tumor microenvironment plays critical roles in breast cancer
development (33,34). Milk acts as an important
microenvironment of breast cancer, however, its role in breast
carcinogenesis is largely unknown. Milk can remain in the breast
over a long period of time after feeding (milk stasis) and the milk
remaining in the breast can influence the microenvironment of the
breast. However, the exact role of milk stasis in breast
carcinogenesis is unknown. To the best of our knowledge, this is
the first study on the abundance of miRNAs in milk remaining in
breast after feeding. Furthermore, we found that miRNA profiling
was different between milk stasis only patients and patients with
milk stasis plus breast neoplasm. The different miRNAs profiles
suggest that miRNAs in milk remaining in breast may contribute to
breast carcinogenesis. We also provided evidence that miRNAs in
milk remaining in breast were more easily detected compared to
miRNAs in the blood.
Milk consists mainly of water, proteins, lactose,
fat, and minerals with wide-ranging chemical, physical and
functional activities. Evidence has shown that milk also contains
miRNAs (35). Weber et al
have reported that miR-509-5p, miR-515-3p, and miR-335 were the
most abundant miRNAs in the majority of the body fluid samples
including milk, while miR-193b was unique in breast milk (36). Zhou et al reported that
miR-148A-3P, miR-30B-5P, let-7f-5p, miR-146B-5P, miR-29A-3P,
let-7a-5p, miR-141-3P, miR-182-5P, miR-200A-3P and miR-378-3P were
the top 10 miRNAs identified in the milk (26). However, Munch et al showed
that miR-148a, let-7a, mir-200c, miR-146b-5p, let-7f, miR-30d,
miR-103, let-7b, let-7g and hsa-mir-21 were the most abundant
miRNAs present in the milk (24).
Consistent with previous studies, we found that lavage from milk
stasis patients contained a variety of miRNAs. As the patients
included in this study stopped feeding at least three years
previously, the miRNAs in the milk were stabilized and affected
breast microenviroment over a long period of time, which may cause
breast cancer. Profiling therefore is a good biomarker for breast
tumors especially the ductal carcinoma in situ.
Kosaka et al systematically analyzed the
expression of miRNAs in milk during the first six months and 6–11
months of lactation, and found that the majority of immune-related
miRNAs were downregulated during lactation. In this study, we found
that the expression of immune-related miRNAs between milk stasis
only and milk stasis plus neoplasm groups was different, suggesting
that the biological feature of milk stasis is different from
lactation. Milk of lactation provides nutrition and immunity to
infants and supports their health. By contrast, milk from milk
stasis potentially acts as a risk factor for breast cancer because
our profile data showed that many oncogenic miRNAs were highly
expressed in the two groups studied and the immune-related miRNAs
were almost downregulated.
The let-7 family miRNAs are the most recognized
miRNAs in milk from healthy patients (17–19). Consistent with previous reports,
we found that let-7 family miRNAs (let-7g-5p, let-7e-5p, let-7a-5p,
let-7b-5p, -let-7c, let-7f-5p, let-7i-5p) were abundant in the
patients with milk stasis only, although their levels were
significantly reduced in patients with milk stasis plus breast
tumor. Let-7 family miRNAs are known to play a tumor suppressor
function (17–19). Furthermore, a number of other
tumor suppressor miRNAs such as miR-29a, miR-146 and miR-223 were
downregulated, while oncogenic miRNAs such as miR-451, miR-486,
miR-107, miR-92 and miR-10 were upregulated in the milk of milk
stasis plus neoplasm patients. These results suggest that the
expression switch of oncogenic and tumor suppressor miRNAs in the
milk of milk stasis patients contributes to breast carcinogenesis.
However, the specific function of individual miRNA identified in
milk stasis in breast carcinogenesis remains to be addressed.
Another significant finding of this study is that
miRNAs in breast lavage of milk stasis patients are more easily
detected than miRNAs in the blood. Thus miRNAs in breast lavage of
milk stasis may be better biomarkers to predict the risk of breast
cancer.
In summary, although the sample size included in the
current study was limited, we provided several novel and noteworthy
findings. To the best of our knowledge, this is the first study on
miRNA prolifing of breast lavage of milk stasis patients. Our data
suggest that miRNAs in milk from milk stasis patients contribute to
breast carcinogenesis and they are more sensitive biomarkers of
breast cancer than miRNAs in the blood.
References
|
1
|
Ballard O and Morrow AL: Human milk
composition: nutrients and bioactive factors. Pediatr Clin North
Am. 60:49–74. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Smetherman DH: Screening, imaging, and
image-guided biopsy techniques for breast cancer. Surg Clin North
Am. 93:309–327. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Langlands FE, Horgan K, Dodwell DD and
Smith L: Breast cancer subtypes: response to radiotherapy and
potential radiosensitisation. Br J Radiol. 86:201206012013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Benson JR and Jatoi I: The global breast
cancer burden. Future Oncol. 8:697–702. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Reeder JG and Vogel VG: Breast cancer
prevention. Cancer Treat Res. 141:149–164. 2008. View Article : Google Scholar
|
|
6
|
Collaborative Group on Hormonal Factors in
Breast Cancer. Breast cancer and breastfeeding: collaborative
reanalysis of individual data from 47 epidemiological studies in 30
countries, including 50302 women with breast cancer and 96973 women
without the disease. Lancet. 360:187–195. 2002. View Article : Google Scholar
|
|
7
|
Russo J and Russo IH: Susceptibility of
the mammary gland to carcinogenesis. II Pregnancy interruption as a
risk factor in tumor incidence. Am J Pathol. 100:497–512.
1980.PubMed/NCBI
|
|
8
|
Yang L and Jacobsen KH: A systematic
review of the association between breastfeeding and breast cancer.
J Womens Health (Larchmt). 17:1635–1645. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Carmichael AR: Obesity as a risk factor
for development and poor prognosis of breast cancer. BJOG.
113:1160–1166. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Warmuth MA, Sutton LM and Winer EP: A
review of hereditary breast cancer: from screening to risk factor
modification. Am J Med. 102:407–415. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Schwab M, Claas A and Savelyeva L: BRCA2:
a genetic risk factor for breast cancer. Cancer Lett. 175:1–8.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Narod SA: BRCA mutations in the management
of breast cancer: the state of the art. Nat Rev Clin Oncol.
7:702–707. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Castañeda CA, Agullo-Ortuño MT, Fresno
Vara JA, Cortes-Funes H, Gomez HL and Ciruelos E: Implication of
miRNA in the diagnosis and treatment of breast cancer. Expert Rev
Anticancer Ther. 11:1265–1275. 2011.
|
|
16
|
Harquail J, Benzina S and Robichaud GA:
MicroRNAs and breast cancer malignancy: an overview of
miRNA-regulated cancer processes leading to metastasis. Cancer
Biomark. 11:269–280. 2012.PubMed/NCBI
|
|
17
|
Hu X, Guo J, Zheng L, et al: The
heterochronic microRNA let-7 inhibits cell motility by regulating
the genes in the actin cytoskeleton pathway in breast cancer. Mol
Cancer Res. 11:240–250. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Qian P, Zuo Z, Wu Z, et al: Pivotal role
of reduced let-7g expression in breast cancer invasion and
metastasis. Cancer Res. 71:6463–6474. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yu F, Yao H, Zhu P, et al: let-7 regulates
self renewal and tumorigenicity of breast cancer cells. Cell.
131:1109–1123. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Castilla MÁ, Díaz-Martín J, Sarrió D, et
al: MicroRNA-200 family modulation in distinct breast cancer
phenotypes. PLoS One. 7:e477092012.PubMed/NCBI
|
|
21
|
Ng EK, Li R, Shin VY, et al: Circulating
microRNAs as specific biomarkers for breast cancer detection. PLoS
One. 8:e531412013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kosaka N, Izumi H, Sekine K and Ochiya T:
microRNA as a new immune-regulatory agent in breast milk. Silence.
1:72010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sun Q, Chen X, Yu J, Zen K, Zhang CY and
Li L: Immune modulatory function of abundant immune-related
microRNAs in microvesicles from bovine colostrum. Protein Cell.
4:197–210. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Munch EM, Harris RA, Mohammad M, et al:
Transcriptome profiling of microRNA by Next-Gen deep sequencing
reveals known and novel miRNA species in the lipid fraction of
human breast milk. PLoS One. 8:e505642013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Song XM, Jiang JF and Jiang YQ: Progress
on miRNA in mammal breast milk. Yi Chuan. 34:1233–1241. 2012.(In
Chinese).
|
|
26
|
Zhou Q, Li M, Wang X, et al:
Immune-related microRNAs are abundant in breast milk exosomes. Int
J Biol Sci. 8:118–123. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Izumi H, Kosaka N, Shimizu T, Sekine K,
Ochiya T and Takase M: Bovine milk contains microRNA and messenger
RNA that are stable under degradative conditions. J Dairy Sci.
95:4831–4841. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Abou-Dakn M, Richardt A, Schaefer-Graf U
and Wöckel A: Inflammatory breast diseases during lactation: milk
stasis, puerperal mastitis, abscesses of the breast, and malignant
tumors-current and evidence-based strategies for diagnosis and
therapy. Breast Care (Basel). 5:33–37. 2010. View Article : Google Scholar
|
|
29
|
Green KA, Nielsen BS, Castellino FJ, Rømer
J and Lund LR: Lack of plasminogen leads to milk stasis and
premature mammary gland involution during lactation. Dev Biol.
299:164–175. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Noble MS and Hurley WL: Effects of
secretion removal on bovine mammary gland function following an
extended milk stasis. J Dairy Sci. 82:1723–1730. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Rakha EA and Chan S: Metastatic
triple-negative breast cancer. Clin Oncol (R Coll Radiol).
23:587–600. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kumar DH and Kutty MK: Review of stem cell
deregulation and breast cancer: an emerging hypothesis. Indian J
Pathol Microbiol. 55:147–153. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ward C, Langdon SP, Mullen P, et al: New
strategies for targeting the hypoxic tumour microenvironment in
breast cancer. Cancer Treat Rev. 39:171–179. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Artacho-Cordón A, Artacho-Cordón F,
Ríos-Arrabal S, Calvente I and Núñez MI: Tumor microenvironment and
breast cancer progression: a complex scenario. Cancer Biol Ther.
13:14–24. 2012.PubMed/NCBI
|
|
35
|
Walker A: Breast milk as the gold standard
for protective nutrients. J Pediatr. 156(2 Suppl): S3–S7. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Weber JA, Baxter DH, Zhang S, et al: The
microRNA spectrum in 12 body fluids. Clin Chem. 56:1733–1741. 2010.
View Article : Google Scholar : PubMed/NCBI
|