Introduction
Heart development is a complicated spatio-temporal
process of organ formation. The eventual anatomic formation of the
heart crescent, linear heart tube, looped heart tube, and
multi-chambered heart during the process of heart development
depends on the coordination of regulatory mechanisms at the
molecular level. Precise expression of heart genes is critical in
specific events of cardiogenesis, and thus dysregulated gene
expression can lead to a variety of heart defects (1). Although many studies have been
conducted to investigate the genetic factors of heart development,
the understanding of epigenetic mechanisms is currently
limited.
microRNAs (miRNAs), as one of the epigenetic
factors, have become acknowledged as new indirect regulators in
heart development. miRNAs are endogenous ~22 nucleotide RNA species
that target the mRNAs of protein-coding genes to direct repression
activities at the post-translational level (2). Based on predictions of the target
genes by bioinformatic analysis, it is estimated that miRNAs
regulate at least 20% of human genes (3).
A number of studies have identified specific miRNAs
in animal models that play distinct roles during heart development
(4–7). Multiple miRNAs have been reported to
play a vital role by regulating heart gene expression in heart
development. For example, miR-1 and miR-133a, co-transcribed in
heart cells, can occupy the Hand2 3′-UTR concurrently, regulating
the expression of Hand2, an essential gene for heart development
(8). In mouse models, miR-27b
exhibits obvious myocardial expression during ventricular chamber
formation by targeting the MEF2c gene. In zebrafish embryos,
miR-218 is involved in the onset of heart malformation as a crucial
mediator of Tbx5, a key gene mediating vertebrate heart development
(9).
Notably, miRNAs have distinct expression patterns at
different stages of development (10). miRNAs in zebrafish and rodent
organs are reported to be expressed in a stage-specific manner
(11,12). There is evidence for a
stage-specific role of miRNAs in heart development: mice with
heart-specific deficiency of Dicer, a key miRNA-processing enzyme,
have different abnormal heart phenotypes at different heart
developmental stages (13,14).
Therefore, stage-specific miRNA expression patterns
are important for better predicting the roles for miRNAs in heart
development. In this study, we aimed to establish the
stage-dependent expression patterns of miRNAs during human fetal
heart development to provide valuable information for further
investigations of congenital heart defects.
Materials and methods
Sample collection
Heart tissue from different weeks of gestation was
obtained from aborted fetuses. The ages of the embryos and fetuses
were carefully calculated after conception based on the last
menstrual period, adjusting for ultrasound measurements of fetal
biparietal diameter or crown rump length. Tissue at 5, 7 and 9
weeks of gestation (5W, 7W and 9W) was obtained from whole embryo
hearts with the help of a dissecting microscope (Leica DFC290;
Danaher Corp., Washington, DC, USA) and sets of 4 of the 5W samples
were pooled prior to processing because the amount of embryonic
heart tissue at this early time-point was minimal. The other
time-points were processed as independent replicates. No obvious
anatomical abnormalities were observed. Fetal heart tissue at 23
weeks of gestational age (23W) was isolated at the conjunction site
of the outflow tract, and ventricles and heart anatomy was
confirmed normal by abdominal fetal echocardiography. To account
for biological variability, a single pool of 4 samples at 5W, and
4, 4 and 2 independent biological replicates of myocardium tissue
at 7W, 9W and 23W were processed for microarray analysis. An
additional pool of 4 samples, and 3, 3 and 3 replicates were used
for qRT-PCR validation analysis. The study was approved by the
Ethics Committee of the Obstetrics and Gynecology Hospital of Fudan
University. All of the donors provided informed consent.
RNA extraction and quality control
Fetal myocardium tissue was incubated in RNA later
solution (Qiagen, Valencia, CA, USA) at room temperature for 12 h,
and then stored at −80°C. RNA was extracted with TRIzol reagent
(Invitrogen Life Technologies, Carlsbad, CA, USA) from 100 to 200
mg of frozen tissue. An RNA quality control assessment was strictly
performed prior to microarray and RT-PCR experiments as follows:
RNA purity of A260/280≥1.90 was confirmed using a spectrophotometer
(NanoDrop 1000; Thermo Scientific, Wilmington, MA, USA). The
integrity of total RNA was verified by agarose gel electrophoresis:
rRNA 28S/18S band brightness ≥2:1 or 1:1. The yield of total RNA
for microarray experiments was verified to be ≥5 μg for each sample
as measured by spectrophotometry (Thermo Scientific NanoDrop 1000).
A total of 2 pools of 5W and 7, 7 and 5 individual samples from 7W,
9W and 23W passed this screen and were used in subsequent
assays.
Microarray analysis
Microarray analysis was performed by CapitalBio
Corp. (Beijing, China). After a brief tailing reaction with polyA,
RNA samples were labeled by FlashTag ligation biotin mix. Labeled
RNAs were hybridized overnight to Affymetrix GeneChip®
miRNA 2.0 Arrays containing probes for 15,644 mature miRNAs derived
from the Sanger miRBase V15 (from 131 organisms). Each array
included probes for 1,105 human mature miRNAs. After hybridization,
arrays were washed and stained according to standard Affymetrix
protocol and then scanned on an Affymetrix GeneChip®
Scanner 3000. Microarray data were preprocessed by extraction of
the intensities for each individual miRNA followed by detection
calls based on the Wilcoxon rank-sum test, background subtraction
based on GC content of the anti-genomic probes, transformation of
values through the addition of a small constant (value 16),
quantile normalization and finally median summarization of all
probe sets for each miRNA. The detection and background adjustments
were conducted using the Affymetrix miRNA QC Tool, and the
remaining workflow was performed under R programming environment
(https://www.r-project.org). Reported intensity data were log2
transformed, and P-values were calculated by the two-sided
Student’s t-test. P≥0.06 was considered to represent a higher than
background probe signal, indicating the expression of the miRNA in
fetal heart tissue.
Identification of differentially
expressed miRNAs
miRNA expression levels in fetal heart tissue were
compared for each of the six pairs of gestational ages (5W vs. 7W,
5W vs. 9W, 5W vs. 23W, 7W vs. 9W, 7W vs. 23W and 5W vs. 23W).
Differentially expressed miRNAs were detected by limma, an R
package based on linear regression. P-values were adjusted by the
false discovery rate, and changes in miRNAs with P<0.05 in any
one of the comparisons was considered to indicate statistical
significance. Expression profiles of 288 dynamically regulated
miRNAs were determined by applied hierarchical clustering, and the
miRNAs were grouped into 5 clusters with distinct patterns of
expression during fetal heart morphogenesis. The above processes
were accomplished using a self-designed R script.
Identification of miRNA families and
miRNA genomics clustering
Enrichment analysis was performed using the Fisher’s
exact test to compare the identified miRNA clusters to the miRNA
family dataset in miRFam (http://admis.fudan.edu.cn/projects/miRFam.htm), which
classifies 748 human miRNAs into 438 families. The four testing
numbers were: total number of miRNAs annotated with a miRNA family;
number of miRNAs in one of the miRNA clusters; number of miRNAs in
a specific miRNA family; and number of miRNAs in the miRNA cluster
also annotated within the specific miRNA family. Significance was
set at P=0.01. Since miRNAs located in close proximity to each
other are highly co-expressed (15), we examined the genomic position
for miRNAs. For each cluster, miRNAs located within 10 kb were
treated as a single miRNA genomic cluster.
Target gene prediction and Gene Ontology
(GO) enrichment
We predicted the target genes of the 288 miRNAs that
were differentially expressed across four gestational ages with
three online prediction tools: Target Scan (http://www.targetscan.org/), miRNAMap2 (http://www.targetscan.org/) and miRDB (http://mirnamap.mbc.nctu.edu.tw/). For each
miRNA, target genes found in any one of the three online databases
were considered for further analysis. GO enrichment analysis was
performed for the target genes for each miRNA. We focused on
biological process (BP), molecular function (MF) and cellular
component (CC) branch GO terms that have 30–300 annotated genes.
Significant P-values were obtained by Fisher’s exact test in R,
adjusted by the false discovery rate using a cut-off value of
0.001.
Quantitative reverse
transcription-polymerase chain reaction (qRT-PCR)
To quantify the differential expression of gestation
age-specific miRNAs, poly-A tails were added to total RNA samples
from different gestation ages using E. coli polyA polymerase
(NEB), as described previously (16). Then, ~2 μg of the tailed total RNA
was reverse transcribed with ImProm-II (Promega, Madison, WI, USA).
SYBR-Green (Takara, Shiga, Japan) qRT-PCR was performed using the
Applied Biosystems 7900 real-time PCR system to assess miRNA
expression with a specific forward primer and a universal reverse
primer complementary to the anchor primer. The normalizer gene in
this analysis was 18S rRNA. The primers used are shown in Table I.
 | Table ImiRNA primer sequences used for
qRT-PCR. |
Table I
miRNA primer sequences used for
qRT-PCR.
| miRNA | Primer
sequences |
|---|
| miRNA-20b |
GGTAGCAAAGTGCTCATAGTGCAGGTAG |
| miRNA-504 |
CTATCAGACCCTGGTCTGCACTCTATC |
| miRNA-302d |
AGTGTTAAGTGCTTCCATGTTTGAGTGT |
| let-7a |
TAGTTTGAGGTAGTAGGTTGTATAGTT |
| let-7b |
TGGTTTGAGGTAGTAGGTTGTGTGGTT |
| let-7c |
TGGTTTGAGGTAGTAGGTTGTATGGTT |
| let-7d |
TAGTTAGAGGTAGTAGGTTGCATAGTT |
| 18SpolyAF |
AGTCGTAACAAGGTTTCCGTAGGTG |
| Universal reverse
primer |
| miR-Hi-RE |
CCAGTCTCAGGGTCCGAGGTATTC |
Statistical analysis
Normality of the data distribution was verified by
the Kolmogorov-Smirnov test. Differences in the expression level of
selected miRNAs in the fetal heart tissue from four gestational
ages were validated using the t-test. Relative expression levels
are expressed as the means ± standard deviation (SD). Statistical
significance was set at the 95% level (P<0.05).
Results
Differential miRNA expression profiling
during fetal heart development
To identify miRNAs differentially expressed in the
fetal heart during development, we performed expression profiling
using Affymetrix Genechip® Arrays (Affymetrix Inc.,
Santa Clara, CA, USA) with 5, 7, 9 and 23 week-old fetal heart
tissue. A total of 703 miRNAs were found to be expressed in
developing fetal heart tissue. The 20 most highly expressed miRNAs
over the four distinct gestational ages are listed in Table II. The expression of most miRNAs
was not significantly altered throughout the fetal heart
morphogenesis period; however, marked changes from 5 to 23 weeks of
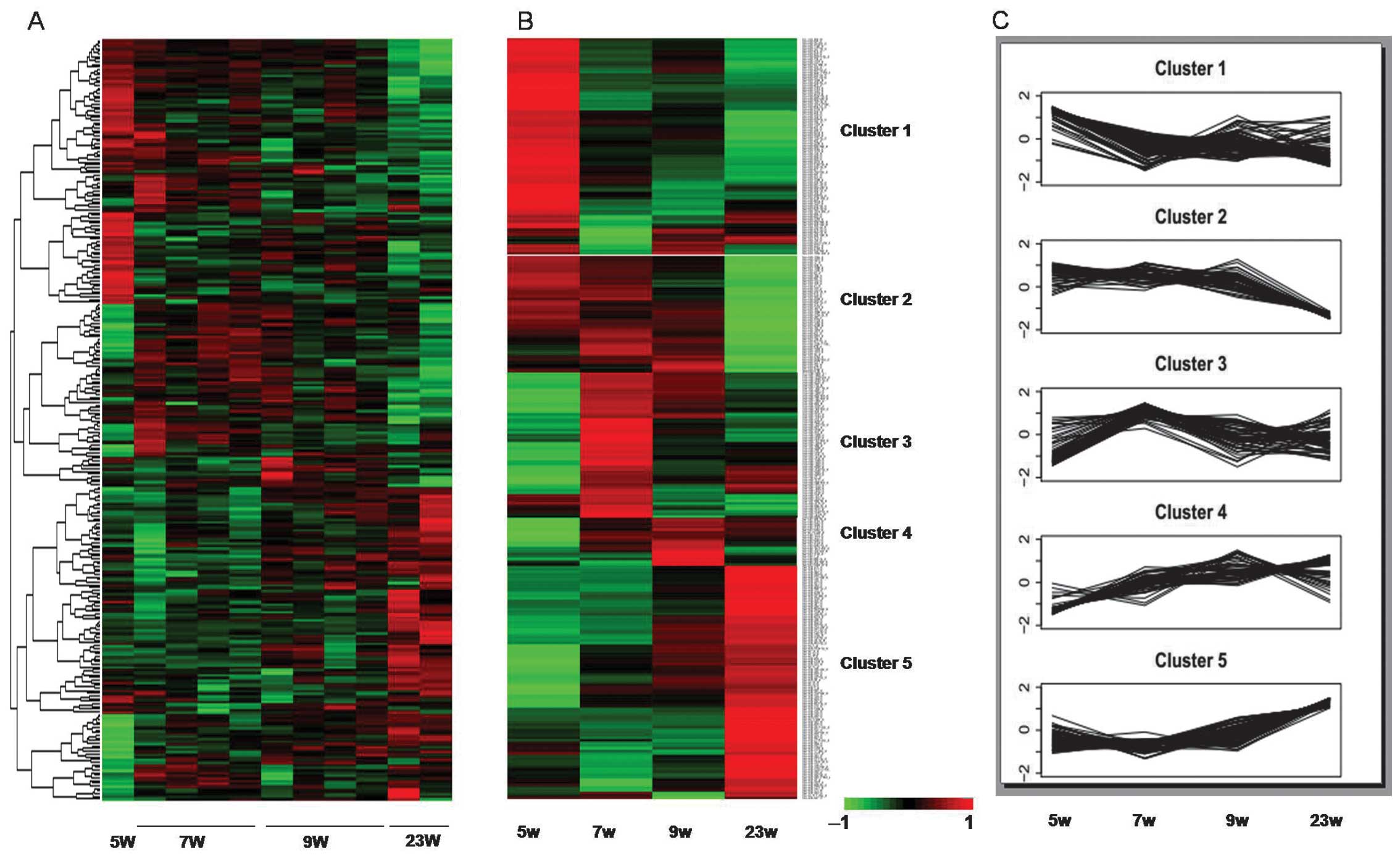
gestation age were observed in a subset of 288 miRNAs (Fig. 1A).
 | Table IIThe top 20 miRNAs ranked by
expression value in four distinct gestational ages. |
Table II
The top 20 miRNAs ranked by
expression value in four distinct gestational ages.
| Gestation age | miRNA name | Signal value |
|---|
| 5W | hsa-miR-103 | 15006.54 |
| hsa-miR-26a | 13228.73 |
| hsa-miR-145 | 12726.34 |
| hsa-miR-17 | 12547.82 |
| hsa-miR-106a | 11815.90 |
| hsa-miR-24 | 11472.96 |
| hsa-miR-107 | 11252.73 |
| hsa-miR-23b | 10569.70 |
| hsa-miR-143 | 10455.16 |
| hsa-miR-92a | 10273.35 |
| hsa-miR-16 | 9893.96 |
| hsa-miR-20a | 9558.97 |
| hsa-miR-125b | 9121.01 |
| hsa-let-7e | 9103.76 |
| hsa-miR-23a | 8884.30 |
| hsa-miR-126 | 8827.87 |
| hsa-miR-99b | 8244.29 |
| hsa-miR-93 | 7993.56 |
| hsa-miR-181a | 7595.30 |
|
hsa-miR-125a-5p | 7258.14 |
| 7W | hsa-miR-26a | 14296.97 |
| hsa-miR-103 | 13448.74 |
| hsa-miR-145 | 11919.23 |
| hsa-miR-143 | 11494.30 |
| hsa-miR-107 | 11381.93 |
| hsa-miR-24 | 10984.56 |
| hsa-let-7e | 10704.38 |
| hsa-miR-17 | 10662.18 |
| hsa-miR-23b | 10160.69 |
| hsa-miR-125b | 10079.82 |
| hsa-miR-106a | 9949.90 |
| hsa-let-7a | 9583.51 |
| hsa-miR-16 | 9484.89 |
| hsa-miR-126 | 8883.17 |
| hsa-let-7c | 8649.26 |
| hsa-miR-23a | 8523.80 |
| hsa-miR-20a | 8444.86 |
| hsa-miR-92a | 7580.87 |
| hsa-miR-1826 | 7473.76 |
| hsa-miR-3196 | 6661.93 |
| 9W | hsa-miR-26a | 14567.85 |
| hsa-miR-145 | 12769.74 |
| hsa-miR-24 | 12584.92 |
| hsa-miR-143 | 11991.22 |
| hsa-miR-23b | 11601.83 |
| hsa-let-7e | 11396.80 |
| hsa-miR-103 | 11348.86 |
| hsa-let-7a | 10556.03 |
| hsa-miR-17 | 9974.02 |
| hsa-miR-16 | 9871.69 |
| hsa-miR-23a | 9664.65 |
| hsa-let-7c | 9632.56 |
| hsa-miR-106a | 9414.99 |
| hsa-miR-107 | 9036.46 |
| hsa-miR-125b | 8924.02 |
| hsa-miR-126 | 8227.37 |
| hsa-let-7b | 8210.81 |
| hsa-let-7d | 8140.66 |
| hsa-miR-20a | 7818.96 |
| hsa-miR-92a | 7655.32 |
| 23W | hsa-miR-26a | 14905.89 |
| hsa-let-7a | 12766.60 |
| hsa-let-7b | 12451.81 |
| hsa-let-7c | 12401.25 |
| hsa-miR-23b | 12350.40 |
| hsa-miR-24 | 11917.27 |
| hsa-miR-145 | 11532.50 |
| hsa-miR-143 | 10717.41 |
| hsa-miR-16 | 10305.78 |
| hsa-let-7d | 10037.60 |
| hsa-miR-125b | 9849.52 |
| hsa-let-7e | 9538.46 |
| hsa-miR-103 | 9532.92 |
| hsa-miR-23a | 9348.89 |
| hsa-miR-126 | 8936.63 |
| hsa-miR-107 | 8528.02 |
| hsa-miR-1826 | 6534.73 |
| hsa-miR-17 | 6519.36 |
| hsa-miR-1975 | 6153.68 |
| hsa-miR-106a | 6004.38 |
Hierarchical clustering analysis was performed to
compare expression profiles of all miRNAs markedly regulated over
the four time periods. Five distinguishable clusters were
identifiable (Fig. 1B and C).
Cluster 1 included 82 miRNAs that were highly expressed at 5 weeks
of gestation, and then decreased with a fluctuating,
uncharacteristic trend in the following three time-points. The 44
miRNAs in Cluster 2 exhibited a high expression across the first
three time-points, followed by a low expression at 23 weeks of
gestation. miRNAs in Clusters 3 and 4 contained 55 and 18 miRNAs
with a high expression level at 7W and 9W, respectively. The 89
miRNAs in Cluster 5 increased in expression, with the highest level
at 23 weeks of gestational age. The miRNAs in the 5 different
clusters are shown in Table
III.
 | Table IIImiRNAs in each cluster. |
Table III
miRNAs in each cluster.
| Cluster no. | miRNA name |
|---|
| Cluster 1 | hsa-miR-509-3-5p,
hsa-miR-769-3p, hsa-miR-1226, hsa-miR-18a*,
hsa-miR-93*, hsa-miR-149 hsa-miR-1307, hsa-miR-935,
hsa-miR-181a-2*, hsa-miR-346, hsa-miR-514b-5p,
hsa-miR-129-3p, hsa-miR-1180, hsa-miR-532-3p,
hsa-miR-99b*, hsa-miR-509-3p, hsa-miR-425, hsa-miR-302d,
hsa-miR-20b*, hsa-miR-424*, hsa-miR-874,
hsa-miR-92a-1*, hsa-miR-339-5p, hsa-miR-127-3p,
hsa-miR-501-5p, hsa-miR-431, hsa-miR-134, hsa-miR-2276,
hsa-miR-500*, hsa-miR-99a*, hsa-miR-1270,
hsa-miR-200c, hsa-miR-654-5p, hsa-miR-551a, hsa-miR-532-5p,
hsa-miR-433, hsa-miR-99b, hsa-miR-500, hsa-miR-504, hsa-miR-1251,
hsa-miR-3143, hsa-miR-1265, hsa-miR-342-3p, hsa-miR-409-5p,
hsa-miR-103-as, hsa-miR-652, hsa-miR-421, hsa-miR-25*,
hsa-miR-589*, hsa-miR-3139, hsa-miR-520c-5p,
hsa-miR-330-3p, hsa-miR-766, hsa-miR-891a, hsa-miR-18b,
hsa-miR-508-5p, hsa-miR-501-3p, hsa-miR-301b, hsa-miR-877,
hsa-miR-1201, hsa-miR-432, hsa-miR-1301, hsa-miR-181a*,
hsa-miR-484, hsa-miR-628-3p, hsa-miR-324-5p,
hsa-miR-518f*, hsa-miR-744, hsa-miR-758, hsa-miR-1296,
hsa-miR-941, hsa-miR-20b, hsa-miR-193b*, hsa-miR-485-5p,
hsa-miR-574-3p, hsa-miR-216a, hsa-miR-340*,
hsa-miR-30c-2*, hsa-miR-154, hsa-miR-3201, hsa-miR-379,
hsa-miR-3200 |
| Cluster 2 | hsa-miR-1910,
hsa-miR-18a, hsa-miR-31, hsa-miR-708, hsa-miR-183, hsa-miR-182,
hsa-miR-130b, hsa-miR-370, hsa-miR-1275, hsa-miR-3178, hsa-miR-887,
hsa-miR-409-3p, hsa-miR-106b*, hsa-miR-106a,
hsa-miR-383, hsa-miR-93, hsa-miR-181b, hsa-miR-20a,
hsa-miR-125b-1*, hsa-miR-638, hsa-miR-1915, hsa-miR-17,
hsa-miR-2861, hsa-miR-4298, hsa-miR-106b, hsa-miR-1285,
hsa-miR-205, hsa-miR-631, hsa-miR-671-5p, hsa-miR-100,
hsa-miR-2277, hsa-miR-1469, hsa-miR-510, hsa-miR-374a,
hsa-miR-4304, hsa-miR-103, hsa-miR-19a, hsa-miR-34c-3p, hsa-miR-32,
hsa-miR-376b, hsa-miR-675, hsa-miR-125a-3p,
hsa-miR-200b*, hsa-miR-155 |
| Cluster 3 | hsa-miR-4299,
hsa-miR-4286, hsa-miR-92b*, hsa-miR-1908, hsa-miR-1909,
hsa-miR-1184, hsa-miR-1228*, hsa-miR-663, hsa-miR-572,
hsa-miR-1274b, hsa-miR-3172, hsa-miR-3141, hsa-miR-720,
hsa-miR-1268, hsa-miR-4281, hsa-miR-149*,
hsa-miR-1225-5p, hsa-miR-3180-3p, hsa-miR-762, hsa-miR-3196,
hsa-miR-1207-5p, hsa-miR-3126-5p, hsa-miR-1260b, hsa-miR-1973,
hsa-miR-1280, hsa-miR-4284, hsa-miR-3197, hsa-miR-4269,
hsa-miR-1308, hsa-miR-4324, hsa-miR-21, hsa-miR-921, hsa-miR-3162,
hsa-miR-1246, hsa-miR-1972, hsa-miR-939, hsa-miR-218, hsa-miR-489,
hsa-miR-374b, hsa-miR-150*, hsa-miR-513b, hsa-miR-4257,
hsa-miR-488*, hsa-miR-886-5p, hsa-miR-1274a,
hsa-miR-886-3p, hsa-miR-513a-5p, hsa-miR-3175, hsa-miR-1912,
hsa-miR-107, hsa-miR-3195, hsa-miR-663b, hsa-miR-1260,
hsa-miR-187*, hsa-miR-4310 |
| Cluster 4 | hsa-miR-1272,
hsa-let-7d*, hsa-miR-3152, hsa-miR-548*,
hsa-miR-1273, hsa-miR-499-5p, hsa-miR-3124, hsa-miR-320e,
hsa-miR-1183, hsa-miR-548u, hsa-miR-885-3p, hsa-miR-548c-3p,
hsa-miR-3128, hsa-miR-548a-5p, hsa-miR-363*, hsa-miR-7,
hsa-miR-16-2*, hsa-miR-155*, |
| Cluster 5 | hsa-miR-215,
hsa-miR-1, hsa-miR-26b, hsa-miR-297, hsa-miR-195*,
hsa-let-7i, hsa-let-7a, hsa-miR-204, hsa-miR-10a*,
hsa-let-7g, hsa-let-7f, hsa-miR-3154, hsa-let-7d, hsa-let-7b,
hsa-miR-224, hsa-miR-139-5p, hsa-miR-98, hsa-miR-193a-5p
hsa-miR-424, hsa-miR-30e, hsa-miR-422a, hsa-let-7c, hsa-miR-483-3p,
hsa-miR-605 hsa-miR-452, hsa-miR-224*, hsa-miR-647,
hsa-miR-150, hsa-miR-10a, hsa-miR-195, hsa-miR-497,
hsa-miR-22*, hsa-miR-10b, hsa-miR-146a, hsa-miR-483-5p,
hsa-miR-486-3p, hsa-miR-376c, hsa-miR-664*,
hsa-miR-193a-3p, hsa-miR-371-5p, hsa-miR-22, hsa-miR-28-3p,
hsa-miR-486-5p, hsa-miR-1827, hsa-miR-139-3p, hsa-miR-378c,
hsa-miR-371-3p, hsa-miR-372, hsa-miR-373, hsa-miR-933, hsa-miR-29a,
hsa-miR-3148, hsa-miR-381, hsa-miR-30d, hsa-miR-411,
hsa-miR-338-5p, hsa-let-7i*, hsa-miR-132*,
hsa-miR-30b, hsa-miR-15a, hsa-miR-30a, hsa-miR-99a, hsa-miR-584,
hsa-miR-125b-2*, hsa-miR-337-5p, hsa-miR-1277,
hsa-miR-4306, hsa-miR-3169, hsa-miR-24-1*, hsa-miR-363,
hsa-miR-10b*, hsa-miR-192, hsa-miR-152, hsa-miR-760,
hsa-miR-455-5p, hsa-miR-542-5p, hsa-miR-223,
hsa-miR-20a*, hsa-miR-27b, hsa-miR-595, hsa-miR-451,
hsa-miR-17*, hsa-miR-29b-2*, hsa-miR-299-5p,
hsa-miR-1271, hsa-miR-2115*, hsa-miR-185,
hsa-let-7f-1*, hsa-miR-625 |
To assess the patterns of expression of miRNAs that
have previously been reported to be associated with heart
development, we identified relevant published studies by searching
‘heart’ and ‘miRNA’ on PubMed. Thirty-four of the miRNAs were
associated with heart development (Table IV).
 | Table IVThe expression values of 34 miRNAs
reported to be associated with heart in our microarray data. |
Table IV
The expression values of 34 miRNAs
reported to be associated with heart in our microarray data.
| Expression value in
our microarray data
Weeks of gestation | |
|---|
|
| |
|---|
| miRNA name | 5W | 7W | 9W | 23W |
Authors/(Refs.) |
|---|
| miRNA-497 | 106.46 | 74.52 | 79.06 | 243.94 | Porrello (53) |
| miRNA-195 | 472.86 | 469.39 | 482.04 | 1791.19 | Porrello (53) |
| miRNA-15a | 706.54 | 642.36 | 729.14 | 1087.14 | Porrello (53) |
| miRNA-15b | 3752.69 | 3619.41 | 3321.15 | 3095.05 | Porrello (53) |
| miRNA-155 | 609.10 | 588.46 | 719.53 | 451.60 | Porrello (53) |
| miRNA-17 | 12547.82 | 10662.18 | 9974.02 | 6519.36 | Porrello (53) |
| miRNA-93 | 7993.56 | 6524.40 | 5446.11 | 3577.60 | Porrello (53) |
| miRNA-208b | 261.65 | 208.86 | 243.61 | 166.51 | Porrello (53) |
| miRNA-25 | 1699.03 | 1537.65 | 1264.74 | 1373.08 | Ventura et
al (54) |
| miRNA-363 | 468.42 | 427.85 | 411.53 | 717.82 | Ventura et
al (54) |
| let-7c | 5919.04 | 8649.26 | 9632.56 | 12401.25 | Vacchi-Suzzi et
al (55) |
| miRNA-125b | 9121.01 | 10079.82 | 8924.02 | 9849.52 | Vacchi-Suzzi et
al (55) |
| miRNA-744 | 720.20 | 514.15 | 483.37 | 274.01 | Vacchi-Suzzi et
al (55) |
| miRNA-328 | 42.56 | 36.72 | 42.25 | 41.47 | Vacchi-Suzzi et
al (55) |
| miRNA-199a-3p | 3252.41 | 5217.02 | 5019.48 | 5159.05 | Vacchi-Suzzi et
al (55) |
| miRNA-99b | 8244.29 | 5172.97 | 4812.78 | 3657.86 | Vacchi-Suzzi et
al (55) |
| miRNA-30e | 322.33 | 344.42 | 501.56 | 712.99 | Vacchi-Suzzi et
al (55) |
|
miRNA-30e* | 112.36 | 146.29 | 161.25 | 180.60 | Vacchi-Suzzi et
al (55) |
| miRNA-21 | 217.27 | 393.67 | 321.76 | 337.54 | Huang et al
(56) |
| miRNA-22 | 2223.63 | 1629.95 | 2363.51 | 3026.23 | Tu et al
(57) |
| miRNA-126 | 8827.87 | 8883.17 | 8227.37 | 8936.63 | Stankunas et
al (58) |
| miRNA-452 | 36.70 | 53.57 | 69.68 | 118.69 | Sheehy et al
(59) |
| miRNA-378 | 2174.08 | 2255.47 | 3417.05 | 4984.94 | Nagalingam et
al (60) |
| miRNA-138 | 51.16 | 43.98 | 53.24 | 72.48 | Morton et al
(6) |
| miRNA-34a | 213.74 | 164.51 | 170.12 | 191.67 | Boon et al
(61) |
| miRNA-181c | 154.76 | 151.84 | 165.73 | 167.21 | Li et al
(62) |
| miRNA-204 | 48.61 | 87.33 | 86.48 | 182.21 | Xiao et al
(63) |
| miRNA-133b | 1778.10 | 1685.77 | 1891.50 | 2353.18 | Townley-Tilson
et al (64) |
| miRNA-133a | 3793.49 | 3548.94 | 3533.02 | 4250.38 | Townley-Tilson
et al (64) |
| miRNA-206 | 149.01 | 178.31 | 207.56 | 171.54 | Townley-Tilson
et al (64) |
| miRNA-1 | 665.07 | 1591.58 | 1655.17 | 1996.21 | Townley-Tilson
et al (64) |
| miRNA-143 | 10455.16 | 11494.30 | 11991.22 | 10717.41 | Deacon et al
(4) |
| miRNA-218 | 71.71 | 129.09 | 90.52 | 86.99 | Chiavacci et
al (9) |
| miRNA-208a | 36.03 | 32.93 | 37.72 | 44.18 | Oliveira-Carvalho
et al (65) |
miRNA families and genomic clusters in 5
differentially expressed clusters
Co-expression of miRNAs is associated with sequence
similarity and genomic co-localization (17). To determine whether patterns of
expression correlate with genomic co-localization, we examined
whether the miRNAs within expression clusters were localized within
common miRNA families or genomic clusters. Five miRNA families with
multiple differentially expressed miRNAs were identified (Table V), while many common genomic
clusters were also observed (Table
VI). These results support the possibility of the co-regulation
of clustered miRNAs within miRNA families and genomic clusters.
 | Table VmiRNA families within expression
clusters. |
Table V
miRNA families within expression
clusters.
| Cluster ID | miRNA family | Members of miRNA
family | P-value |
|---|
| Cluster 2 | mir-17 | hsa-miR-20a,
hsa-miR-18a, hsa-miR-93, hsa-miR-106a, hsa-miR-106b,
hsa-miR-17 | 6.59E-08 |
| Cluster 3 | mir-1274 | hsa-miR-1274a,
hsa-miR-1274b | 9.06E-04 |
| mir-663 | hsa-miR-663,
hsa-miR-663b | 9.06E-04 |
| Cluster 4 | let-7 | hsa-miR-98,
hsa-let-7g, hsa-let-7b, hsa-let-7d, hsa-let-7c, hsa-let-7i | 1.89E-07 |
| Cluster 5 | mir-30 | hsa-miR-30a,
hsa-miR-30b, hsa-miR-30d, hsa-miR-30e | 4.45E-05 |
 | Table VImiRNA genomic-clusters in each
expression cluster. |
Table VI
miRNA genomic-clusters in each
expression cluster.
| Cluster ID | Chromosome | Position at 5′ | Position at 3′ | Members of
genomic-clusters |
|---|
| Cluster 1 | chr14 | 101350820 | 101350913 | hsa-miR-432,
hsa-miR-433, hsa-miR-127-3p, hsa-miR-431 |
| chr14 | 101492357 | 101492444 | hsa-miR-758,
hsa-miR-379 |
| chr14 | 101526092 | 101526175 | hsa-miR-485-5p,
hsa-miR-134, hsa-miR-154 |
| chr19 | 52195865 | 52195934 |
hsa-miR-99b*, hsa-miR-99b |
| chr19 | 54210707 | 54210793 |
hsa-miR-518f*,
hsa-miR-520c-5p |
| chr7 | 99691391 | 99691470 |
hsa-miR-93*,
hsa-miR-25* |
| chrX | 49774330 | 49774413 | hsa-miR-501-3p,
hsa-miR-532-5p, hsa-miR-500*, hsa-miR-532-3p,
hsa-miR-501-5p, hsa-miR-500 |
| chrX | 133304071 | 133304141 |
hsa-miR-20b*, hsa-miR-20b,
hsa-miR-18b |
| chrX | 146341170 | 146341244 | hsa-miR-514b-5p,
hsa-miR-509-3-5p |
| Cluster 2 | chr13 | 92003319 | 92003389 | hsa-miR-17,
hsa-miR-20a, hsa-miR-18a, hsa-miR-19a |
| chr7 | 99691616 | 99691697 | hsa-miR-93,
hsa-miR-106b*, hsa-miR-106b |
| chr7 | 129414745 | 129414854 | hsa-miR-182,
hsa-miR-183 |
| Cluster 3 | chr5 | 135416177 | 135416297 | hsa-miR-886-5p,
hsa-miR-886-3p |
| Cluster 4 | chr17 | 46657200 | 46657309 |
hsa-miR-10a*, hsa-miR-10a |
| chr9 | 96941116 | 96941202 | hsa-let-7d,
hsa-let-7d* |
| Cluster 5 | chr11 | 72326107 | 72326174 | hsa-miR-139-5p,
hsa-miR-139-3p |
| chr13 | 92003319 | 92003389 |
hsa-miR-20a*, hsa-miR-17* |
| chr14 | 101490131 | 101490193 | hsa-miR-411,
hsa-miR-299-5p |
| chr14 | 101512257 | 101512331 | hsa-miR-381,
hsa-miR-376c |
| chr17 | 1617197 | 1617281 | hsa-miR-22,
hsa-miR-22* |
| chr17 | 6921230 | 6921341 | hsa-miR-497,
hsa-miR-195 |
| chr19 | 54291959 | 54292027 | hsa-miR-372,
hsa-miR-371-3p, hsa-miR-373, hsa-miR-371-5p |
| chr2 | 177015031 | 177015140 | hsa-miR-10b,
hsa-miR-10b* |
| chr8 | 41517959 | 41518026 | hsa-miR-486-3p,
hsa-miR-486-5p |
| chr8 | 135817119 | 135817188 | hsa-miR-30d,
hsa-miR-30b |
| chrX | 151128100 | 151128184 |
hsa-miR-224*, hsa-miR-452 |
Verification of miRNA expression patterns
by qRT-PCR
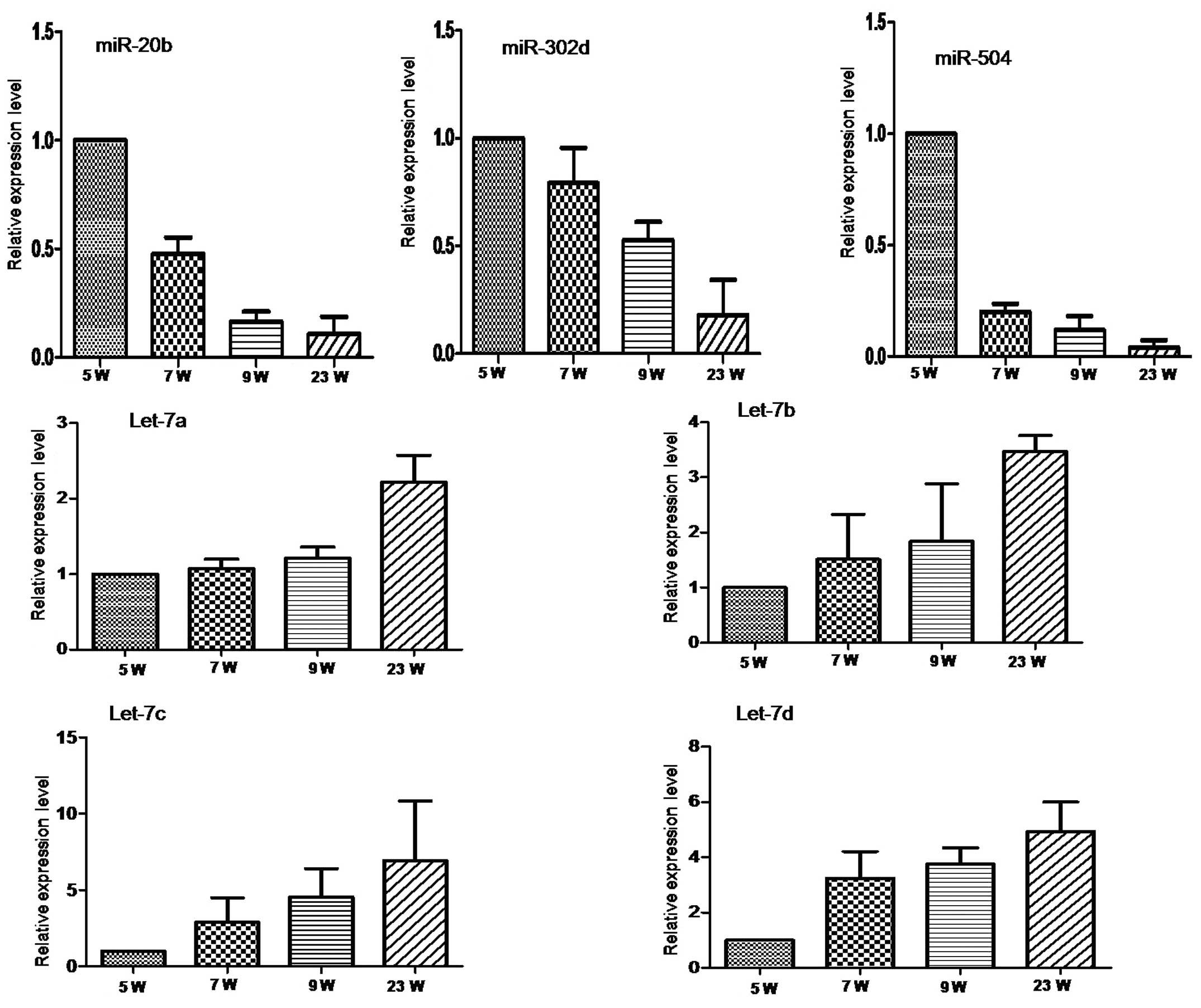
To validate the microarray results, seven miRNAs
predicted to be involved in heart development were selected for
qRT-PCR based on their representation in two distinctive clusters
and in a well-characterized miRNA family for the let-7 miRNAs. This
validation was analyzed in 1 pool from 5W and 3, 3 and 3 individual
samples from 7W, 9W and 23W that we collected separately.
miRNA-20b, miR-504 and miR-302d from Cluster 1 were expressed with
a decreasing trend with gestational age (Fig. 2A). Conversely, the let-7 family
miRNAs, let-7a, let-7b, let-7c and let-7d from Cluster 5 were
expressed with a gradually increasing trend with gestational age
(Fig. 2B). These trends are in
agreement with the microarray results.
Function associations of miRNAs from 5
different expression clusters
To understand how differentially regulated miRNAs
may contribute to fetal cardiogenesis and heart development, we
analyzed the predicted functions of the miRNAs by enriching for
predicted GO functions of target genes using online databases.
We focused on miRNAs with predicted roles in heart
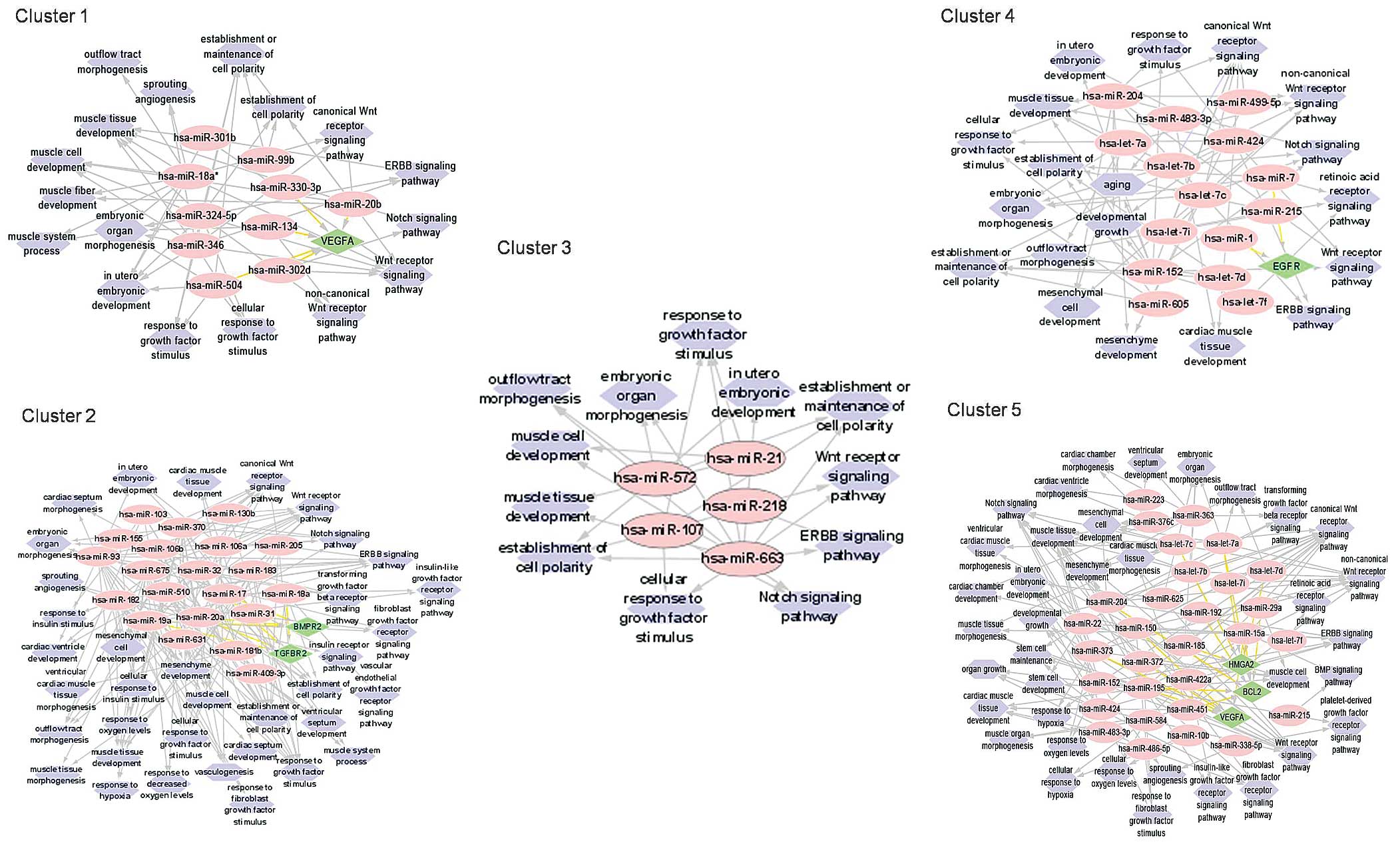
formation and development to obtain a complete network diagram
(data not shown). The miRNAs within several clusters were predicted
to target common genes (Fig. 3).
This included the gene encoding vascular endothelial growth factor
α (VEGFA) in Cluster 1, the bone morphogenetic protein receptor 2
(BMPR2) and transforming growth factor β receptor 2 (TGFBR2) genes
in Cluster 2, and epidermal growth factor receptor (EGFR) in
Cluster 4. The miRNAs in Cluster 5 were predicted to target the
high mobility group (HMGA2), Bcl-2 and VEGFA genes. These genes
have associated roles in heart muscle tissue development,
angiogenesis, outflow tract development, ventricle septum, heart
chamber and ventricle morphogenesis. Common cellular events vital
to cardiogenesis, such as the establishment and maintenance of cell
polarity, cell response to growth factor, cell response to hypoxia,
mesenchymal cell development and stem cell maintenance were also
suggested by the GO annotation analysis. Furthermore, most targeted
mRNAs were associated with cardiogenesis-related molecular
signaling pathways, such as the Wnt, Notch, ERBB, PDGF, FGFR and
retinoic acid receptor (RXR) signaling pathways (Fig. 3).
Discussion
We have characterized miRNA expression in the
developing fetal heart over a period of 5–23 weeks of gestation. We
identified 288 differentially expressed miRNAs, which clustered
into 5 different expression patterns. Evidence was presented for
the co-regulation of multiple miRNAs based on their categorization
within miRNA families or localization within the genome. Based on
GO, these miRNAs were predicted to target several common heart
genes and to be associated with molecular events and signaling
pathways during heart development.
To the best of our knowledge, this is the first
study addressing miRNA expression profiling in human fetal heart
tissue as early as 5 weeks of gestation. At 5 weeks of gestation
the torsion and looping processing of the human heart is completed,
the aortic sac is divided into two conducts, and the left ventricle
begins to acquire its outflow tract (18). Therefore, miRNAs that are highly
expressed at this relatively early time-point (Clusters 1 and 2)
may be essential for the anatomical orchestration of heart
structures. The other selected developmental time-points cover a
period of maturation that occurs after the formation of the major
heart anatomical components. At a later stage in gestation, after
the establishment of heart anatomy, the fetal heart continues to
develop. For example, cardiomyocytes proliferate and enlarge to
keep the heart growing (19), and
atrioventricular and semilunar valves remodel into thin fibrous
leaflets capable of enduring constantly changing haemodynamic
forces (20). We suggest that
miRNAs of Clusters 3, 4 and 5, which increase in expression and
reach a peak later in gestation, may be associated with late heart
development. Consistent with the age-dependent expression of miRNAs
revealed in our study, the differential expression of miRNAs was
observed in the hearts of young adult and old mice (21). However, we have provided
additional information regarding the role of miRNAs in early heart
development by assessing time-dependent expression alterations in
the embryo. These collective findings suggest that different miRNAs
may be required during key stages of fetal heart morphogenesis and
development that extend beyond anatomical formation.
Within the 5 clusters, we have identified several
miRNAs that were members of the same miRNA family or shared a
chromosomal proximity. Several members of the let-7 family of
conserved miRNAs were identified in Cluster 4. The let-7 family has
diverse biological activities. We confirmed the high expression of
let-7a, let-7b, let-7c and let-7d in human fetal heart tissue by
qRT-PCR. We also confirmed the expression patterns of miRNA-20b,
miRNA-504 and miRNA-302d. The functional analysis of these miRNAs
may help to specify their roles in fetal heart development.
Results of our study suggest a role for miRNAs in
the morphogenesis of the heart chamber, ventricle septum and
outflow tract, as supported by previous studies. miRNA-143 is known
to be essential for chamber formation and function through the
active adjustment of myocardial cell morphology in zebrafish lines
(4). Furthermore, miRNA-1 can
indirectly control the balance between muscle differentiation and
proliferation during cardiogenesis (22). Deletion of miRNA-133 in mice
results in late embryonic or neonatal lethality due to ventricle
septum defects, accompanied by abnormalities in cardiomyocyte
proliferation, apoptosis and the aberrant expression of smooth
muscle genes in the heart (5). In
the present study, we have identified new miRNAs that are likely to
be involved in the heart chamber, septum and outflow tract
development through regulating the biological behavior of the
muscle system.
Cell polarity is a feature in early organ patterning
of the embryo (23,24). It regulates the polarization of
cells in a variety of contexts, allowing cells to change shape and
position and to sense their orientation within a mass of tissue
(25). The disruption of cell
polarity is a known mechanism of heart defect (26). Several miRNAs identified in
Clusters 1, 2, 3 and 4 may function in the establishment and
maintenance of cell polarity. In addition, some miRNAs in Clusters
2 and 5 are regulated in response to hypoxia or oxygen levels.
Thus, when the fetal heart is formed, it undergoes a stage of rapid
growth and maturation where oxygen tension plays a vital role, and
physiological normal hypoxia (lower oxygen tension in the fetus as
compared with the adult) may be helpful in heart development
(27).
The present study has identified genes previously
involved in heart development as predicted targets of
differentially expressed miRNAs. For example, VEGF-A, BMPR2,
TGFBR2, EGFR, HMGA2, Bcl-2 were identified as target genes for
multiple miRNAs within specific clusters. VEGF is involved in
coronary vasculature, septation and outflow tract formation and
influences cardiomyocyte survival (28). VEGF expression, at either the mRNA
or protein level, has been observed in rat hearts from the first
embryonic day of myocardial vascular tube formation through the
entire pregnancy (29).
Therefore, miRNAs from Clusters 1 and 5 may regulate heart
development by targeting VEGF. In animal models, inactivation of
BMPR2, TGFBR2 and EGFR causes different subtypes of heart defects
(30–32). Furthermore, HMGA2, a member of the
HMGA sub-family of HMG proteins has a critical function for normal
heart development (33).
Accumulating evidence has revealed focal apoptosis in multiple
cells of developing heart, contributing to normal development of
embryonic outflow tract, heart valves, heart vascular system, and
the conducting system (34).
Bcl-2 is a common mediator of apoptosis that resides within the
mitochondria and regulates cytochrome c release and caspase
activation in the intrinsic apoptotic pathway (35). Several miRNAs, including let-7a,
miRNA-204, miRNA-15a and miRNA-195 regulate the expression of Bcl-2
to influence apoptosis in certain diseases (36–39). Thus, the expression of these
miRNAs in the fetal heart may have a similar function in
influencing apoptosis.
Heart formation and development is known to be a
complex process including numerous signaling pathways and their
interactions. Through network and GO analysis, the differentially
regulated miRNAs are shown to have a putative role in the
regulation of heart development-associated signaling pathways, such
as the canonical and non-canonical Wnt signaling pathway (40), ERBB (41), Notch (28,42), TGF-β (43), retinoic acid receptor (44), BMP (43,45), PDGF (46), FGF (47) and insulin-like growth factor
receptor signaling pathways (48). Previous studies have reported a
connection between miRNAs and many of these signaling pathways. For
instance, miRNA-499 induces rat bone marrow-derived mesenchymal
stem cell differentiation in cardiomyocyte-like cells through the
Wnt/β-catenin signaling pathway (49). Extensive cross talk between miRNAs
and the Notch signaling pathway determines stem cell fates
(50). In addition, several
miRNAs are shown to regulate molecular members of the ERBB
signaling pathway in various types of cancer (51). The Wnt, ERBB and TGF-β signaling
pathways have also been predicted to be regulated by miR-335 in
gastric cancer (52). Our results
provide insight into additional miRNAs that may regulate heart
development by these predicted signaling pathways.
In summary, we have identified a set of miRNAs that
are expressed in a time-specific manner during the fetal period in
the human developing heart. Using clustering and GO analyses, we
have predicted the functions of differentially expressed miRNAs.
These data elucidate the potential role of a network of miRNAs in
anatomical and post-anatomical heart development and may provide
insight into potential treatments of heart defects.
Acknowledgements
This study was supported by the National Basic
Research Program of China (973 program) (2010CB529500) and the
National Science Fund of China (81270712, 81300506, 81200449 and
81200448). We gratefully acknowledge the assistance provided by the
National Academic-Specific Program of Health Care, the Public
Health Care Program of Shanghai (12GWZX0301) from Shanghai
Municipal Health Bureau, and the training program of the Shanghai
Academic Leading Talent by the Shanghai Committee of Science and
Technology and Shanghai Bureau of Human Resources.
Abbreviations:
|
miRNAs
|
microRNAs
|
|
MEF2c
|
myocyte enhancer factor 2c
|
|
Tbx5
|
T-box 5
|
|
VEGFA
|
vascular endothelial growth factor
α
|
|
BMPR2
|
bone morphogenetic protein receptor
2
|
|
TGFBR2
|
transforming growth factor β receptor
2
|
|
EGFR
|
epidermal growth factor receptor
|
|
HMGA2
|
high mobility group A2
|
|
Bcl-2
|
B cell lymphoma/lewkmia-2
|
|
PDGF
|
platelet-derived growth factor
|
|
GO
|
Gene Ontology
|
|
RT-PCR
|
reverse transcription-polymerase chain
reaction
|
References
|
1
|
Huang JB, Liu YL, Sun PW, Lv XD, Du M and
Fan XM: Molecular mechanisms of congenital heart disease.
Cardiovasc Pathol. 19:e183–e193. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bartel DP: microRNAs: target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Xie X, Lu J, Kulbokas E, et al: Systematic
discovery of regulatory motifs in human promoters and 3′ UTRs by
comparison of several mammals. Nature. 434:338–345. 2005.
|
|
4
|
Deacon DC, Nevis KR, Cashman TJ, et al:
The miR-143-adducin3 pathway is essential for cardiac chamber
morphogenesis. Development. 137:1887–1896. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liu N, Bezprozvannaya S, Williams AH, et
al: microRNA-133a regulates cardiomyocyte proliferation and
suppresses smooth muscle gene expression in the heart. Genes Dev.
22:3242–3254. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Morton SU, Scherz PJ, Cordes KR, Ivey KN,
Stainier DY and Srivastava D: microRNA-138 modulates heart
patterning during embryonic development. Proc Natl Acad Sci USA.
105:17830–17835. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lagendijk AK, Goumans MJ, Burkhard SB and
Bakkers J: MicroRNA-23 restricts heart valve formation by
inhibiting Has2 and extracellular hyaluronic acid production. Circ
Res. 109:649–657. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Vo NK, Dalton RP, Liu N, Olson EN and
Goodman RH: Affinity purification of microRNA-133a with the heart
transcription factor, Hand2. Proc Natl Acad Sci USA.
107:19231–19236. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chiavacci E, Dolfi L, Verduci L, et al:
MicroRNA 218 mediates the effects of Tbx5a over-expression on
zebrafish heart development. PLoS One. 7:e505362012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bartel DP: microRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wienholds E, Kloosterman WP, Miska E, et
al: MicroRNA expression in zebrafish embryonic development.
Science. 309:310–311. 2005. View Article : Google Scholar
|
|
12
|
Mukhopadhyay P, Brock G, Pihur V, Webb C,
Pisano MM and Greene RM: Developmental microRNA expression
profiling of murine embryonic orofacial tissue. Birth Defects Res A
Clin Mol Teratol. 88:511–534. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen JF, Murchison EP, Tang R, et al:
Targeted deletion of Dicer in the heart leads to dilated
cardiomyopathy and heart failure. Proc Natl Acad Sci USA.
105:2111–2116. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Saxena A and Tabin CJ: miRNA-processing
enzyme Dicer is necessary for heart outflow tract alignment and
chamber septation. Proc Natl Acad Sci USA. 107:87–91. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Marco A, Ninova M, Ronshaugen M and
Griffiths-Jones S: Clusters of microRNAs emerge by new hairpins in
existing transcripts. Nucleic Acids Res. 41:7745–7752. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang J, Du YY, Lin YF, et al: The cell
growth suppressor, mir-126, targets IRS-1. Biochem Biophys Res
Commun. 377:136–140. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Stäehler CF, Keller A, Leidinger P, et al:
Whole miRNome-wide differential co-expression of microRNAs.
Genomics Proteomics Bioinformatics. 10:285–294. 2012.PubMed/NCBI
|
|
18
|
Marcela SG, Cristina RM, Angel PG, et al:
Chronological and morphological study of heart development in the
rat. Anat Rec (Hoboken). 295:1267–1290. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jonker SS, Zhang L, Louey S, Giraud GD,
Thornburg KL and Faber JJ: Myocyte enlargement, differentiation,
and proliferation kinetics in the fetal sheep heart. J Appl
Physiol. 102:1130–1142. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Butcher JT and Markwald RR:
Valvulogenesis: the moving target. Philos Trans R Soc Lond B Biol
Sci. 362:1489–1503. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang X, Azhar G and Wei JY: The
expression of microRNA and microRNA clusters in the aging heart.
PLoS One. 7:e346882012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhao Y, Samal E and Srivastava D: Serum
response factor regulates a muscle-specific microRNA that targets
Hand2 during cardiogenesis. Nature. 436:214–220. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sinha T, Wang B, Evans S, Wynshaw-Boris A
and Wang J: Disheveled mediated planar cell polarity signaling is
required in the second heart field lineage for outflow tract
morphogenesis. Dev Biol. 370:135–144. 2012. View Article : Google Scholar
|
|
24
|
Zagris N, Gilipathi K, Soulintzi N and
Konstantopoulos K: Decorin developmental expression and function in
the early avian embryo. Int J Dev Biol. 55:633–639. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Henderson DJ and Chaudhry B: Getting to
the heart of planar cell polarity signaling. Birth Defects Res A
Clin Mol Teratol. 91:460–467. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rhee DY, Zhao XQ, Francis RJ, Huang GY,
Mably JD and Lo CW: Connexin 43 regulates epicardial cell polarity
and migration in coronary vascular development. Development.
136:3185–3193. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Patterson AJ and Zhang L: Hypoxia and
fetal heart development. Curr Mol Med. 10:653–666. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
van den Akker NM, Caolo V and Molin DG:
Cellular decisions in heart outflow tract and coronary development:
an act by VEGF and NOTCH. Differentiation. 84:62–78.
2012.PubMed/NCBI
|
|
29
|
Tomanek RJ, Ratajska A, Kitten GT, Yue X
and Sandra A: Vascular endothelial growth factor expression
coincides with coronary vasculogenesis and angiogenesis. Dev Dyn.
215:54–61. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Beppu H, Malhotra R, Beppu Y, Lepore JJ,
Parmacek MS and Bloch KD: BMP type II receptor regulates
positioning of outflow tract and remodeling of atrioventricular
cushion during cardiogenesis. Dev Biol. 331:167–175. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Langlois D, Hneino M, Bouazza L, et al:
Conditional inactivation of TGF-β type II receptor in smooth muscle
cells and epicardium causes lethal aortic and heart defects.
Transgenic Res. 19:1069–1082. 2010.
|
|
32
|
Goishi K, Lee P, Davidson AJ, Nishi E, Zon
LI and Klagsbrun M: Inhibition of zebrafish epidermal growth factor
receptor activity results in cardiovascular defects. Mech Dev.
120:811–822. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Monzen K, Ito Y, Naito AT, et al: A
crucial role of a high mobility group protein HMGA2 in
cardiogenesis. Nat Cell Biol. 10:567–574. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Fisher SA, Langille BL and Srivastava D:
Apoptosis during cardiovascular development. Circ Res. 87:856–864.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Vogler M: BCL2A1: the underdog in the BCL2
family. Cell Death Differ. 19:67–74. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Vasilatou D, Papageorgiou SG, Kontsioti F,
et al: Expression analysis of mir-17–5p, mir-20a and let-7a
microRNAs and their target proteins in CD34+ bone marrow
cells of patients with myelodysplastic syndromes. Leuk Res.
37:251–258. 2013.
|
|
37
|
Ryan J, Tivnan A, Fay J, et al:
MicroRNA-204 increases sensitivity of neuroblastoma cells to
cisplatin and is associated with a favourable clinical outcome. Br
J Cancer. 107:967–976. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Diniz MG, Gomes CC, de Castro WH, et al:
miR-15a/16–1 influences BCL2 expression in keratocystic odontogenic
tumors. Cell Oncol (Dordr). 35:285–291. 2012.
|
|
39
|
Chen YQ, Wang XX, Yao XM, et al:
MicroRNA-195 promotes apoptosis in mouse podocytes via enhanced
caspase activity driven by BCL2 insufficiency. Am J Nephrol.
34:549–559. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Cohen ED, Tian Y and Morrisey EE: Wnt
signaling: an essential regulator of cardiovascular
differentiation, morphogenesis and progenitor self-renewal.
Development. 135:789–798. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Odiete O, Hill MF and Sawyer DB:
Neuregulin in cardiovascular development and disease. Circ Res.
111:1376–1385. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Jain R, Rentschler S and Epstein JA: Notch
and heart outflow tract development. Ann NY Acad Sci. 1188:184–190.
2010.PubMed/NCBI
|
|
43
|
Kruithof BP, Duim SN, Moerkamp AT and
Goumans MJ: TGFβ and BMP signaling in heart cushion formation:
lessons from mice and chicken. Differentiation. 84:89–102.
2012.
|
|
44
|
Li P, Pashmforoush M and Sucov HM:
Retinoic acid regulates differentiation of the secondary heart
field and TGFbeta-mediated outflow tract septation. Dev Cell.
18:480–485. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhang J, Chang JY, Huang Y, et al: The
FGF-BMP signaling axis regulates outflow tract valve primordium
formation by promoting cushion neural crest cell differentiation.
Circ Res. 107:1209–1219. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Van den Akker NM, Winkel LC, Nisancioglu
MH, et al: PDGF-B signaling is important for murine heart
development: its role in developing atrioventricular valves,
coronaries, and heart innervation. Dev Dyn. 237:494–503.
2008.PubMed/NCBI
|
|
47
|
Lavine KJ, Yu K, White AC, et al:
Endocardial and epicardial derived FGF signals regulate myocardial
proliferation and differentiation in vivo. Dev Cell. 8:85–95. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Knezevic I, Patel A, Sundaresan NR, et al:
A novel cardiomyocyte-enriched microRNA, miR-378, targets
insulin-like growth factor 1 receptor: implications in postnatal
heart remodeling and cell survival. J Biol Chem. 287:12913–12926.
2012. View Article : Google Scholar
|
|
49
|
Zhang LL, Liu JJ, Liu F, et al: MiR-499
induces heart differentiation of rat mesenchymal stem cells through
wnt/beta-catenin signaling pathway. Biochem Biophys Res Commun.
420:875–881. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Ichimura A, Ruike Y, Terasawa K and
Tsujimoto G: miRNAs and regulation of cell signaling. FEBS J.
278:1610–1618. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Barker A, Giles KM, Epis MR, Zhang PM,
Kalinowski F and Leedman PJ: Regulation of ErbB receptor signalling
in cancer cells by microRNA. Curr Opin Pharmacol. 10:655–661. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Yan Z, Xiong Y, Xu W, et al:
Identification of hsa-miR-335 as a prognostic signature in gastric
cancer. PLoS One. 7:e400372012. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Porrello ER: microRNAs in heart
development and regeneration. Clin Sci (Lond). 125:151–166. 2013.
View Article : Google Scholar
|
|
54
|
Ventura A, Young AG, Winslow MM, et al:
Targeted deletion reveals essential and overlapping functions of
the miR-17 through 92 family of miRNA clusters. Cell. 132:875–886.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Vacchi-Suzzi C, Hahne F, Scheubel P, et
al: Heart structure-specific transcriptomic atlas reveals conserved
microRNA-mRNA interactions. PloS One. 8:e524422013. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Huang ZP, Chen JF, Regan JN, et al: Loss
of microRNAs in neural crest leads to cardiovascular syndromes
resembling human congenital heart defects. Arterioscler Thromb Vasc
Biol. 30:2575–2586. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Tu Y, Wan L, Bu L, et al: MicroRNA-22
downregulation by atorvastatin in a mouse model of heart
hypertrophy: a new mechanism for antihypertrophic intervention.
Cell Physiol Biochem. 31:997–1008. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Stankunas K, Ma GK, Kuhnert FJ, Kuo CJ and
Chang CP: VEGF signaling has distinct spatiotemporal roles during
heart valve development. Dev Biol. 347:325–336. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Sheehy NT, Cordes KR, White MP, Ivey KN
and Srivastava D: The neural crest-enriched microRNA miR-452
regulates epithelial-mesenchymal signaling in the first pharyngeal
arch. Development. 137:4307–4316. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Nagalingam RS, Sundaresan NR, Gupta MP,
Geenen DL, Solaro RJ and Gupta M: A heart-enriched microRNA,
miR-378, blocks heart hypertrophy by targeting Ras signaling. J
Biol Chem. 288:11216–11232. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Boon RA, Iekushi K, Lechner S, et al:
MicroRNA-34a regulates heart ageing and function. Nature.
495:107–110. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Li J, Cao Y, Ma X-j, et al: Roles of
miR-1–1 and miR-181c in ventricular septal defects. Int J Cardiol.
168:1441–1446. 2013.
|
|
63
|
Xiao J, Liang D, Zhang H, et al:
MicroRNA-204 is required for differentiation of human-derived
cardiomyocyte progenitor cells. J Mol Cell Cardiol. 53:751–759.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Townley-Tilson W, Callis TE and Wang D:
MicroRNAs 1, 133, and 206: critical factors of skeletal and heart
muscle development, function, and disease. Int J Biochem Cell Biol.
42:1252–1255. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Oliveira-Carvalho V, Carvalho VO and
Bocchi EA: The emerging role of miR-208a in the heart. DNA Cell
Biol. 32:8–12. 2013. View Article : Google Scholar : PubMed/NCBI
|