Introduction
PAX6 is a member of the PAX gene family and
encodes a conserved transcription factor with two DNA-binding
domains, a paired domain and a paired-type homeodomain. PAX6 serves
as a regulator in the coordination and pattern formation required
for retinogenesis and the development of other ocular tissues
(1,2). A number of previous studies have
revealed the mechanisms involved in the transcriptional control of
PAX6. For example, PAX6 has been found to bind to the proximal
region of the tartrate acid phosphatase (TRAP) gene promoter and to
suppress nuclear factor of activated T cells c1-induced TRAP gene
expression (3). Recently, the
upregulation of PAX6 has been observed in a number of
ghrelin-expressing endocrine cells and plays an essential role in
the adult maintenance of glucose homeostasis and function of the
endocrine pancreas (4).
However, PAX6 has been found to be uniquely required
for eye development. In the retina, PAX6 is involved in the
regulation of the development of retinal progenitor cells into
neurons and glial cells. As previously demonstrated, mice which
were heterozygous carriers of a loss-of-function allele of PAX6 had
defective eye development, while the homozygotes died after birth
with defects in the eyes and brain (5–7).
PAX6 has been found to initiate the multipotency of retinal
progenitor cells. The inactivation of PAX6 restricts the
multipotent potential of retinal progenitor cells, allowing them to
generate only into amacrine interneurons (8). Furthermore, PAX6 has been shown to
directly control the activation of retinogenic basic
helix-loop-helix (bHLH) factors, influencing the differentiation of
a subset of retinal progenitor cells. Emerging evidence has
indicated that retinoblastoma tumors develop from embryological
retinal photoreceptors (9,10).
However the physiological role of PAX6 in retinal development and
the oncogenesis in retinoblastoma remains largely unknown. The
study by Xu et al demonstrated that retinoblastoma cells
express markers of postmitotic cone precursors, and mouse double
minute 2 (MDM2) and N-Myc are required for the proliferation and
survival of these cells (11).
They further demonstrated MDM2 expression is regulated by the
cone-specific transcription factors, indicating the potential
function of cone-specific signaling circuitry in the oncogenic
effects of RB1 mutations.
Previous studies have indicated that the normal
development of the mammalian eye is dependent on the level of PAX6
and insufficient expression levels of PAX6 lead to pan-ocular
disorders, such as aniridia (12,13). We have previously demonstrated
that the overexpression of PAX6 regulates the growth and apoptosis
of human retinoblastoma cells (14,15). However the limitation of our
previous studies exists in the phenotypes with increased copies
number of PAX6, which may parallel with the phenotypes of a PAX6
haploinsufficiency. Therefore, in the present study, we suppressed
the expression of Pax6 in human retinoblastoma cells and examined
the effects on cell growth and apoptosis. The endogenous PAX6
knockdown was mediated by specific lentiviral PAX6-RNAi and
validated by quantitative reverse transcription-polymerase chain
reaction (RT-qPCR) and western blot analysis. The effects of the
suppression of PAX6 on cell proliferation, cell cycle arrest and
apoptosis were examined by fluorescence-activated cell sorting. The
levels of apoptosis-related and cell cycle-related genes and
proteins were detected by RT-qPCR and western blot analysis.
Materials and methods
Cell lines
Two human retinoblastoma cell lines, SO-Rb50 and
Y79, were used in this study. The SO-Rb50 cell line was established
in the Zhongshan Ophthalmic Center, Sun Yat-Sen University,
Guangzhou, China, as previously described (15,16). The Y79 cell line was obtained from
the American Type Culture Collection (ATCC; Manassas, VA, USA). The
maintenance of these cell lines was carried out as previously
described (17–19). In brief, the cells were cultured
in RPMI-1640 medium (HyClone Co., Logan, UT, USA) supplemented with
10% fetal bovine serum, 100 U/l penicillin, and 100 U/l
streptomycin at 37°C in a humidified atmosphere of 95% air/5%
CO2. The culture medium was replaced every 2 days.
Plasmids
A third generation of the self-inactivating
lentiviral vector containing a cytomegalovirus (CMV)
promoter-driven enhanced green fluorescence protein (eGFP) reporter
was purchased from GeneChem Co., Ltd. (Shanghai, China). The
lentiviral vector system was made from 3 types of plasmids, the
pGCL-GFP vector (5′LTR, 3′LTR and woodchuck hepatitis virus
post-transcriptional regulatory element), the pHelper 1.0 (gag, pol
and rev element) vector and the pHelper 2.0 (VSV-G element)
vectors. The pGCL-GFP vector encoding a sequence targeting the
human PAX6 gene (NCBI Reference Sequence ID: NM_000280.3)
was assembled by GeneChem. PCR and DNA sequencing confirmed the
accurate insertion of the small interfering RNA (siRNA) sequences.
For the preparation of the recombinant lentiviral vectors, the
pHelper 1.0 plasmid (15 μg), the pHelper 2.0 plasmid (10 μg) and
the pGCL-GFP-siRNA or the pGCL-GFP plasmid (negative control, 20
μg) were co-transfected into subconfluent 293T cells in serum-free
medium using Lipofectamine 2000 reagent (Invitrogen Co., Carlsbad,
CA, USA). Untransfected cells were used as blank controls. After 8
h of incubation, the medium was changed to serum-containing medium.
High titers of recombinant lentiviral vectors with PAX6-RNAi were
harvested after the supernatant became concentrated 48 h later.
Before the experiments, 7 human PAX6-specific double-stranded,
siRNA sequences were synthesized and screened (Shanghai Genepharma
Co., Ltd., Shanghai, China). We selected 2 sequences for
co-transfection into the cell lines in our study. The target sites
were CGTCCATCTTTGCTTGGGAAA and TACCAAGCGTGTCATCAATAA.
Target site screening
Before the experiments, 7 human PAX6-specific
double-stranded, siRNA sequences (KD1-KD7) were synthesized and
screened. The details of the target sequences are presented in
Table I. For transfection, the
human retinoblastoma cells were seeded in 48-well plates at a
concentration of 5×105 cells/ml in a volume of 200 μl
for each well. The cells were transfected either with PAX6-RNAi-GFP
(PAX6 inhibition study group) or with GFP lentiviral vectors with
scrambled siRNA (sequence: ‘TTCTCCGAACGTGTCACGT’) in serum-free
medium for 20 h with a total multiplicity of infection (MOI) of 80.
At 4 days after transfection, the reporter GFP gene expression was
examined under a fluorescence microscopy. The transfected human
retinoblastoma cell lines were screened using FACS for the
following experiments.
 | Table ISequences of 7 target sites in
PAX6. |
Table I
Sequences of 7 target sites in
PAX6.
| Name | Sequences of target
sites | Starting site | GC% |
|---|
| KD1 |
CGTCCATCTTTGCTTGGGAAA | 817 | 47.62 |
| KD2 |
CATGGCAAATAACCTGCCTAT | 1541 | 42.86 |
| KD3 |
GCAAGAATACAGGTATGGTTT | 1287 | 38.10 |
| KD4 |
taCCAAGCGTGTCATCAATAA | 883 | 38.10 |
| KD5 |
aaGATTCAGATGAGGCTCAAA | 1117 | 38.10 |
| KD6 |
gaGAGTAGCGACTCCAGAAGT | 761 | 52.38 |
| KD7 |
caCACCTAGTCATATTCCTAT | 1373 | 38.10 |
Cell proliferation
The standard colorimetric cell counting kit-8
(CCK-8; Dojindo Laboratories, Kumamoto, Japan) was used for the
determination of the number of viable cells in the cell
proliferation assays. The transfected retinoblastoma cells were
seeded with a volume of 90 μl cell suspension (5,000 cells/well)
into 96-well plates and 10 μl CCK-8 were added to each well
followed by incubation for 4 h at 37°C. Optical densities were read
using a microplate reader scanning at 450 nm. This procedure was
repeated every 24 h during a 6-day period. Cell survival rates were
measured at 3 time points of the cell growth curve through the log
phase of growth for each cell line.
RT-qPCR
Total RNA was extracted from the cells using TRIzol
reagent [Tiangen Biotech (Beijing) Co., Ltd., Beijing, China; Cat.
no. DP405]. The reverse transcription and the PCR amplification
reactions were performed according to the M-MLV reverse
transcriptase protocol (Tiangen Biotech) (Cat. no. KR104). qPCR
(ABI PRISM 7500, version 1.41) was performed to detect the mRNA
levels of PAX6 and cell cycle-related molecules in both
retinoblastoma cell lines. The details of the primers for qPCR are
presented in Table II and qPCR
was carried out as previously described (20–22). The reaction system was 20 μl,
including 4 μl RNase-free ddH2O, 4 μl cDNA template, 1.7
μl mixed primers, 10 μl SuperReal PreMix and 0.3 μl ROX Reference
Dye l (Tiangen Biotech) (Cat. no. FP204). The PCR running
conditions were as follows: 120 sec at 95°C for the initial
denaturation followed by 45 cycles of 20 sec at 95°C for
denaturation, 25 sec at temperature for annealing, and 30 sec at
95°C for extension. The threshold cycle (Ct value), which is the
cycle number at which the amount of the amplified gene of interest
reaches a fixed threshold, was subsequently determined. Relative
quantification values of the target gene mRNA levels were
normalized to endogenous human β-actin gene levels and calculated
using the 2−ΔΔCt method. Each experiment was performed 3
times.
 | Table IIPrimers used for qPCR. |
Table II
Primers used for qPCR.
| Gene | Gene ID | DNA sequences
(5′→3′) | Tm (°C) |
|---|
| β-actin | NM_001101.3 |
TGGCACCCAGCACAATGAA | |
| |
CTAAGTCATAGTCCGCCTAGAAGCA | 58 |
| PAX6 | NM_000280 |
ATGGGCGGAGTTATGATACCTAC | |
| |
GGAACTTGAACTGGAACTGACA | 58 |
| cdc25 | NM_001789.2 |
CGTGGCTGCCTGCACTCTCA | |
| |
GGCTGTCACAGGTGACTGGGG | 60 |
| CDK2 | NM_001798.3 |
CCAGTACTGCCATCCGAGAG | |
| |
CGGCGAGTCACCATCTCAGC | 60 |
| PCNA | NM_002592.2 |
CTGAGGGCTTCGACACTAC | |
| |
TCACTCCGTCTTTTGCACAG | 55 |
| CDK1 | NM_001786.4 |
AAGCCGGGATCTACCATACC | |
| |
CCTGGAATCCTGCATAAGCAC | 60 |
| p21 | NM_000389.4 |
GGACAGCAGAGGAAGAC | |
| |
GGCGTTTGGAGTGGTAGAA | 55 |
Apoptosis assay
The early apoptosis of the transfected cells was
detected using the PE Annexin V Apoptosis Detection kit I (BD
Pharmingen, San Diego, CA, USA) (Cat. no. 559763). The 2
retinoblastoma cell lines were washed twice with cold PBS and then
re-suspended in 1× binding buffer at a concentration of
1×106 cells/ml. A volume of 100 μl of the solution
(1×105 cells) was transferred to a 5 ml culture tube and
5 μl of PE Annexin V and 5 μl 7-AAD were added. The cells were
gently dispersed and then incubated for 15 min at room temperature
(25°C) in the dark. Finally, 400 μl of 1× binding buffer were added
to each tube. The cells were analyzed using a BD FACSCalibur flow
cytometer (BD Biosciences, San Diego, CA, USA) equipped with a 540
nm exciting laser. The results are shown as percentages of the
count of the early apoptotic cells to the total cell count in the
SO-Rb50 and Y79 retinoblastoma cell lines. Each experiment was
performed 3 times.
Cell cycle assay
A cell suspension corresponding to 1×106
cells was collected and centrifuged. The supernatant was discarded,
1 ml of phosphate-buffered saline (PBS) at room temperature was
added and the cell pellet was re-suspended. The full volume of
re-suspended cells was transferred to 3 ml of absolute ethanol
pre-cooled at −20°C by pipetting the cell suspension slowly into
the ethanol while swirling at top speed. The cells were left in
ethanol at −20°C for 15 min. The cells were centrifuged, the
ethanol was discarded, 4 ml of PBS were added at room temperature
and the cells were re-suspended to allow them to rehydrate for 15
min. Again, the cell suspension was centrifuged and the supernatant
was discarded, 1 ml of DNA staining mixed solution was added and
the cell suspension re-suspended. Following incubation for 30 min
at room temperature, the cells were analyzed by FACS in the
presence of the dye. The cells were then passed through a BD
FACSCalibur flow cytometer equipped with a 488 nm argon laser to
measure the DNA content. The data analysis was performed using Cell
Quest (BD Biosciences) and ModFit LT software (Verity Software
House Inc, Topsham, ME, USA) software. The results are presented as
percentages of the total cell count in different phases of the cell
cycle, namely the G0/G1 phase (diploid cells), S phase (diploid and
tetraploid cells), and G2/M phase (tetraploid cells). Each
experiment was performed 3 times.
Western blot analysis
Approximately 1×107 transfected human
retinoblastoma cells of each of the 2 cell lines was collected into
a 15 ml centrifuge tube and re-suspended in 600 μl cell lysis
buffer [20 mM Tris (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1% Triton
X-100, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM
Na3VO4, 1 μg/ml leupeptin, 1 mM
phenylmethanesulfonyl fluoride (PMSF)]. The cells remained in that
medium on ice for 30 min with re-dispersion every 5 min. The
lysates were then centrifuged at 12,000 rpm for 5 min at 4°C. The
protein concentrations were determined using a NanoPhotometer
(Implen GmbH, München, Germany). The proteins were denatured in 5×
loading buffer for 10 min and were separated by sodium dodecyl
sulfate polyacrylamide gel electropheresis (SDS-PAGE) using 5%
stacking and 12% separating gels. They were then electroplotted
onto polyvinylidene difluoride (PVDF) membranes (0.2 μm,
Immobilon-P; Millipore, Billerica, MA, USA) in the way of a semi
dry process with 20 V for 30 min. After being blocked in a blocking
solution (Beyotime Institute of Biotechnology, Beijing, China; Cat.
no. P0023B) for 30–60 min, the membranes were incubated overnight
at 4°C with primary antibodies, including mouse anti-human β-actin
(diluted at 2,000; Beyotime Institute of Biotechnology; Cat. no.
AA128), mouse anti-human PAX6 (diluted 500; Abcam Co., Hong Kong,
China; Cat. no. Ab78545), rabbit anti-human cyclin-dependent
protein kinase 2 (CDK2) (diluted at 1,000; Cat. no. 2546),
mouse anti-human proliferating cell nuclear antigen (PCNA)
(diluted at 1,000; Cat. no. 2586), mouse anti-human
cyclin-dependent kinase 1 (CDK1) (diluted at 1,000; Cat. no.
9116), rabbit anti-human Bcl-2 (diluted at 1,000; Cat. no.
2870) and rabbit anti-human BAX (diluted at 1,000; Cat. no.
5023) (all from Cell Signaling Technology, Inc., Beverly, MA, USA).
After being rinsed in PBS solution with 0.5%v/v Tween-20 (PBST)
thrice for 10 min, the PVDF membranes were incubated at 37°C for 1
h with the secondary goat anti-mouse IgG(H+L) antibody (diluted at
2,000) (Cat. no. A0216) or goat anti-rabbit IgG(H+L) antibody
(diluted at 2,000) (both from Beyotime Institute of Biotechnology)
(Cat. no. A0208) conjugated with horseradish peroxidase (HRP).
Using PBST, the rinsed PVDF membranes were subjected to enhanced
chemiluminescence (ECL) using an ECL detection kit (Beyotime
Institute of Biotechnology) (Cat. no. P0018) and quantified using
Quantity One software (Bio-Rad Laboratories Inc., Hercules, CA,
USA). Each western blot analysis was performed 3 times. For further
analysis, the means were calculated and the data were
normalized.
Statistical analysis
Statistical analysis was performed using a
commercially available software package, SPSS (version 20.0 for
Windows IBM; SPSS, Inc., Chicago, IL, USA). The distributions of
the parameters were evaluated using the Levene test. The data are
presented as the means ± standard deviation. Statistical analysis
of the differences was carried out using an independent samples
t-test and paired-samples t-test. A P-value of <0.05 was
considered to indicate a statistically significant difference.
Results
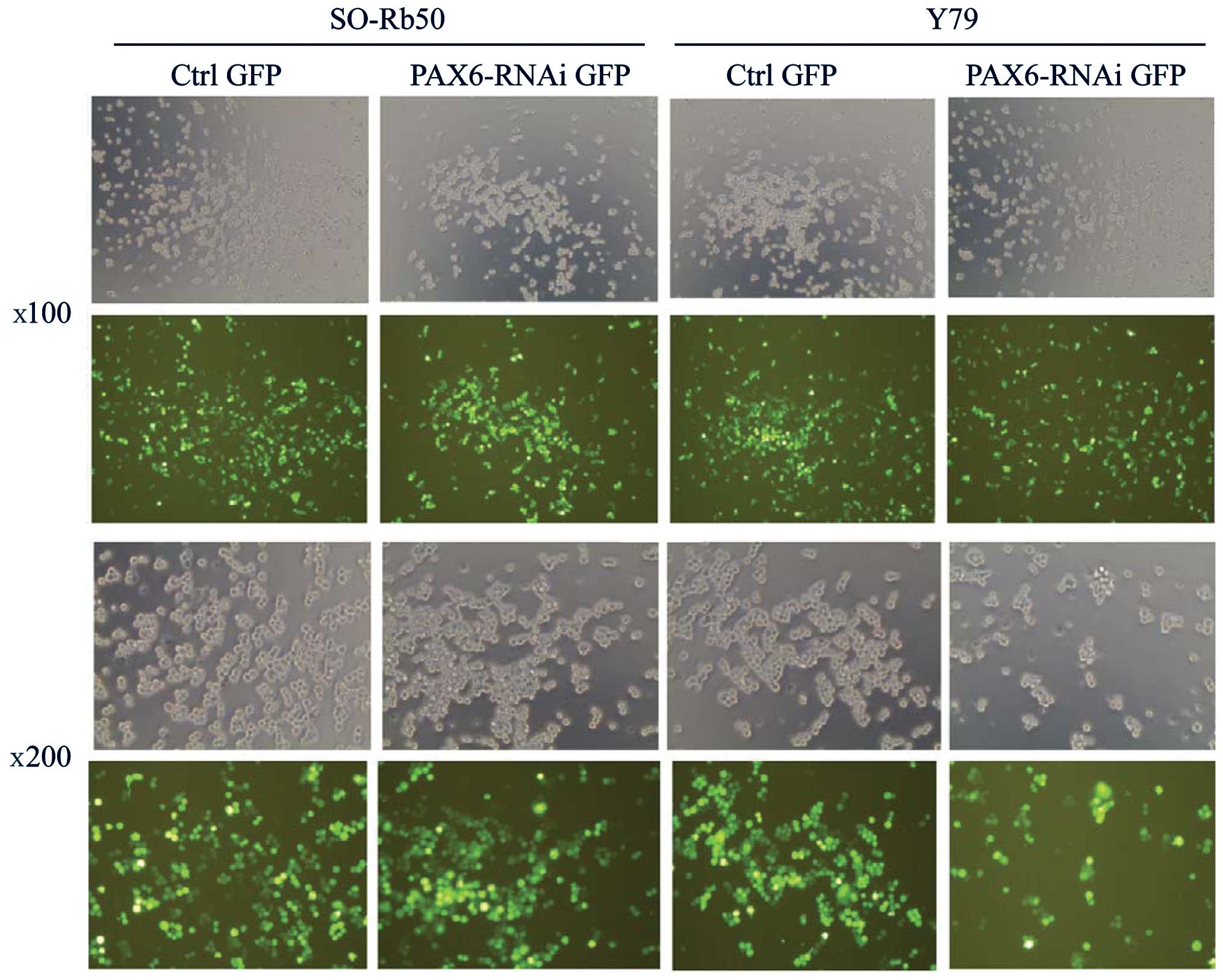
Transfection efficiency of lentiviral
vectors
To investigate the function of PAX6 in both
retinoblastoma cell lines, the gene was silenced by transfecting
the retinoblastoma cells with lentiviral vectors carrying
GFP-PAX6-RNAi sequences (PAX6 inhibition study group). The SO-Rb50
and Y79 cell lines were transfected with the lentiviral vectors at
an MOI of 80. The successfully transfected cells expressed GFP and
were examined under a fluorescence microscope at 4 days after
transfection (Fig. 1). The
efficiency of the infection was approximately 80% at an MOI of 80.
The stably transfected cells were screened by FACS for the next
step of the study.
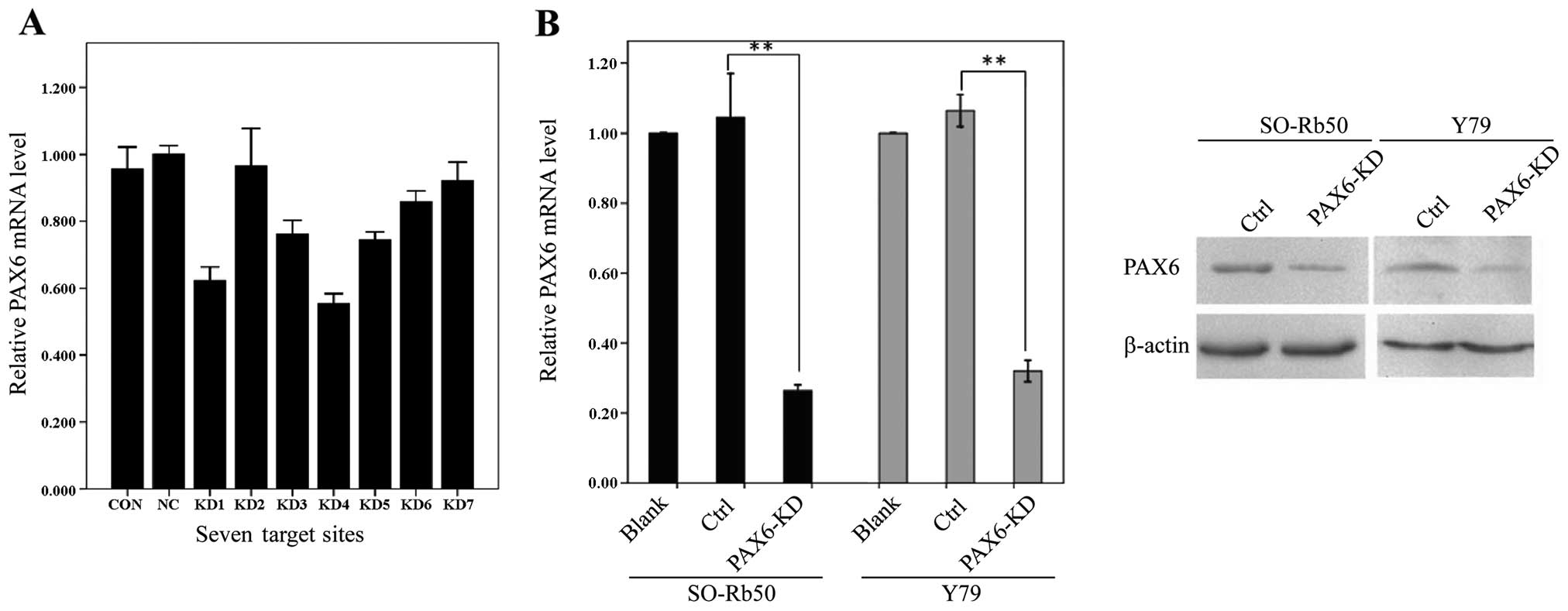
Target site screening
Seven human PAX6-specific small interfering RNA
sequences (KD1-KD7) were primarily screened using RT-qPCR in the
SO-Rb50 cell line. The highest inhibition rate was 45% in KD4,
indicating that the siRNA failed to have sufficient inhibitory
effects (Fig. 2A). To obtain a
higher inhibition rate, 2 combined target sites (KD1 + KD4, KD3 +
KD4 and KD4 + KD5) were used to inhibit PAX6 in the 2 human
retinoblastoma cell lines. The inhibition rate of the KD4 + KD5
group reached 70%, providing sufficeint inhibitory effects for the
following experiments (Fig. 2B,
left panel). The inhibitory effects on endogenous PAX6 expression
were confirmed by western blot analysis. The protein levels of PAX6
in the SO-Rb50 and Y79 retinoblastoma cell lines were significantly
lower in the knockdown groups than in the control groups (Fig. 2B).
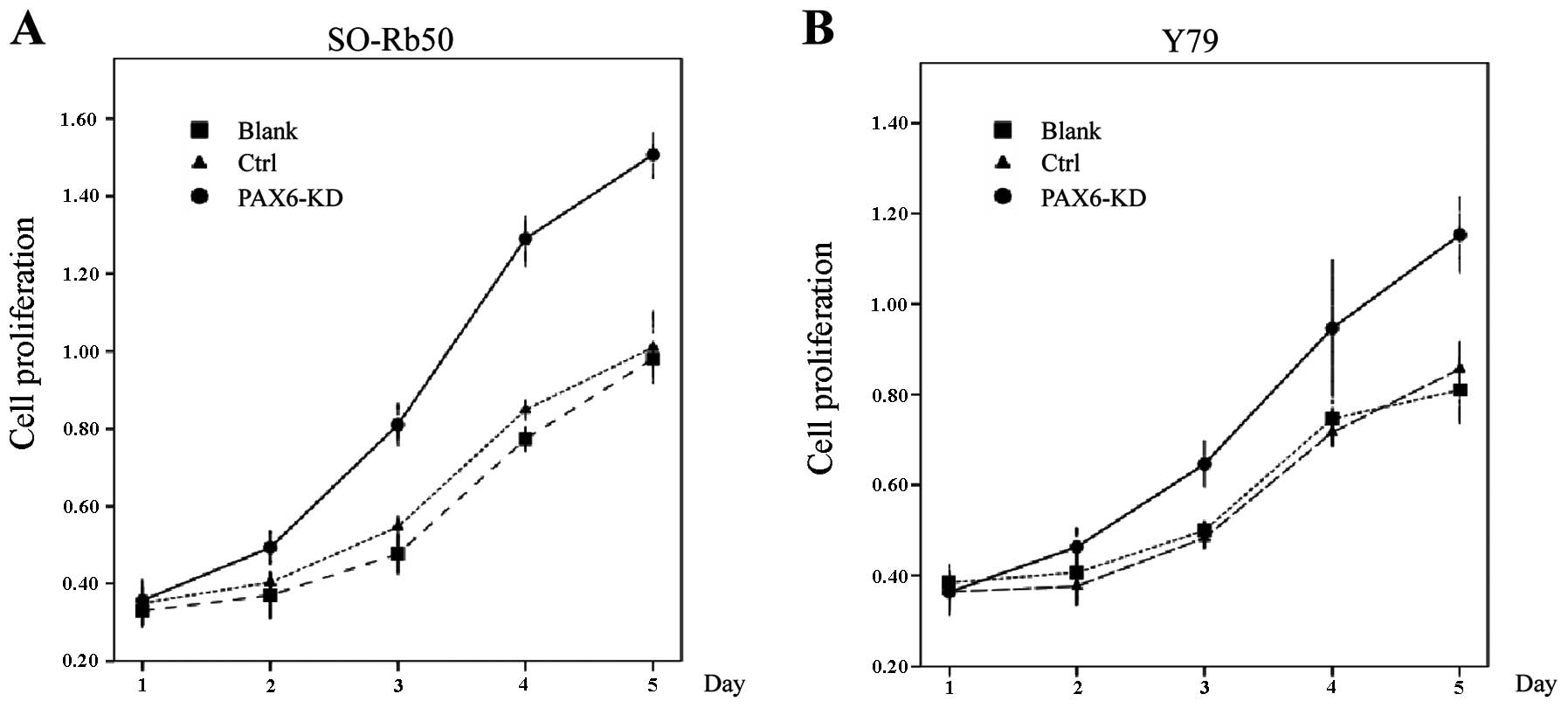
Suppression of PAX6 promotes cell
proliferation
To examined the proliferation of the 2
retinoblastoma cell lines following the inhibition of PAX6, cell
proliferation assay was performed using the standard colorimetric
CCK-8. OD values were measured at 5 time points of the cell growth
curve for each cell line. A significant increase in the cell
survival rates was observed in the knockdown group in these 2 cell
lines. The OD values of the 2 cell lines increased sharply at 1–5
days after cell plating in the knockdown group compared with the
control groups (Fig. 3).
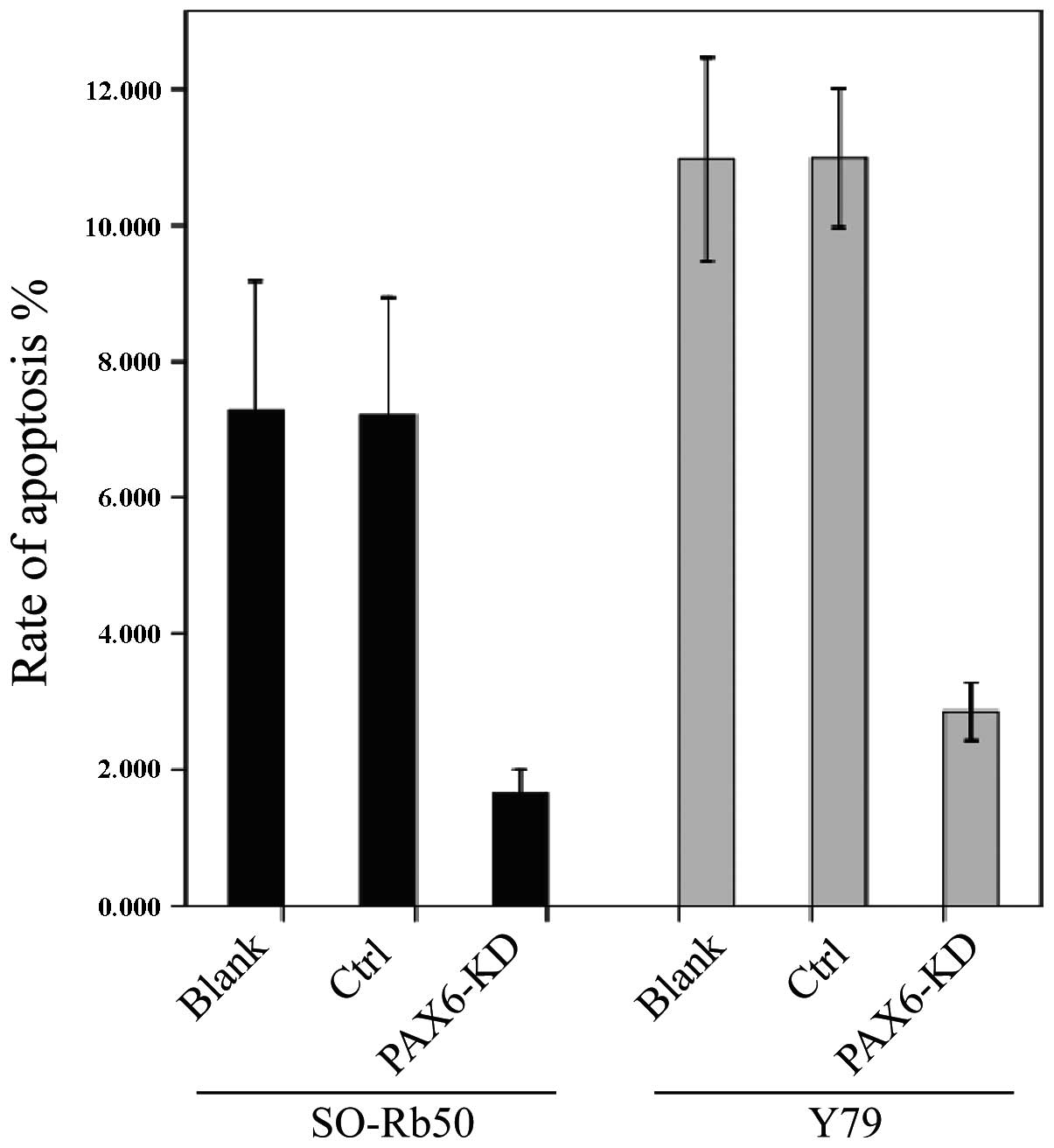
Suppression of PAX6 inhibits
apoptosis
The effects of endogenous PAX6 inhibition on cell
apoptosis were examined using the PE Annexin V Apoptosis Detection
kit I. Flow cytometric analysis revealed a reduced early apoptotic
rate in the PAX6-knockdown groups compared to the control groups.
The percentage of apoptotic cells was significantly lower in the
PAX6-knockdown group than the corresponding negative GFP-control
groups (t=4.036, P>0.05, n=3) and the negative control group
without transfection (t=7.948, P<0.05, n=3) (Fig. 4).
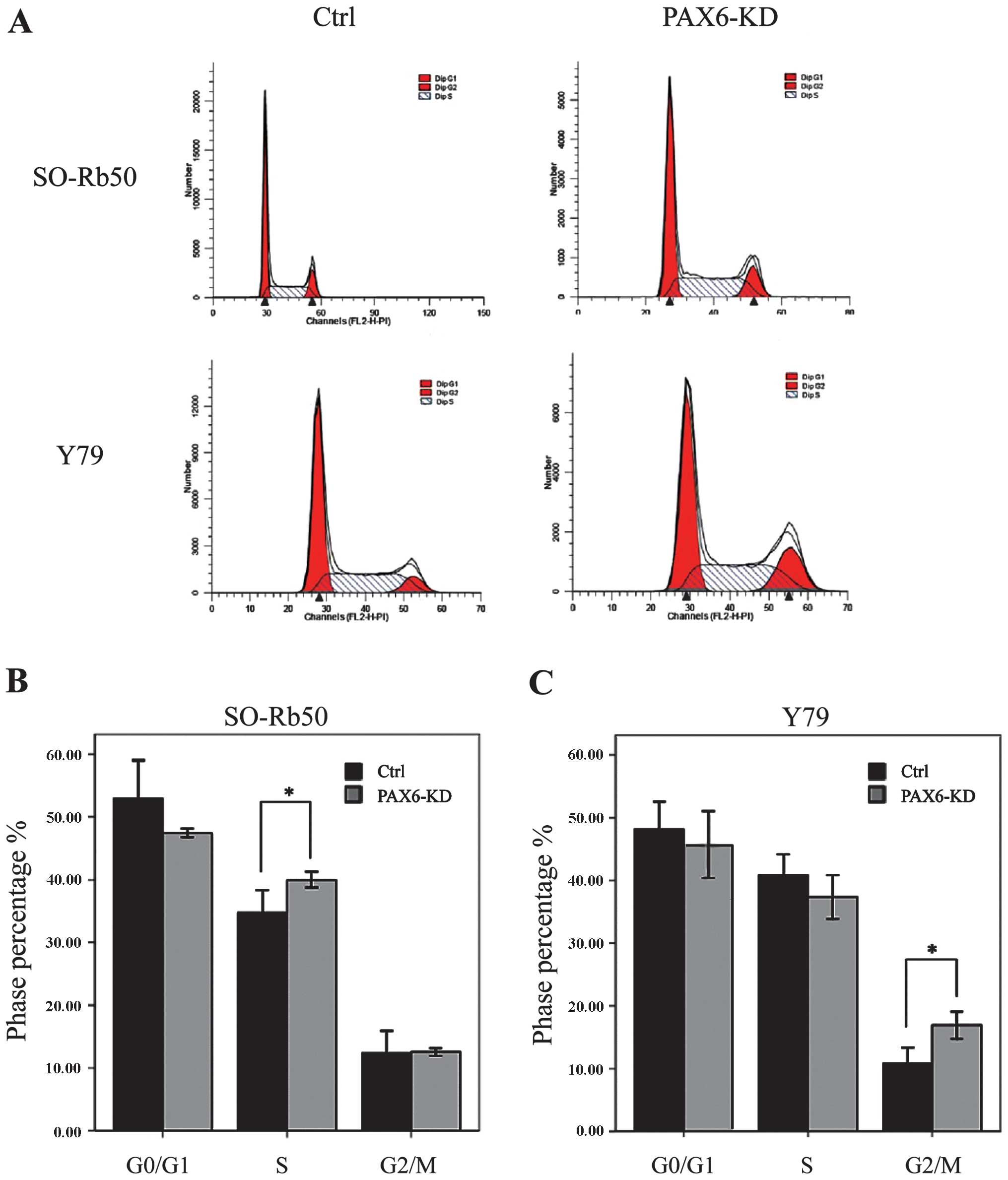
Suppression of PAX6 regulates cell cycle
distribution
We then determined the effects of the inhibition of
endogenous PAX6 on the cell cycle by FACS. For the SO-Rb50 cell
line, the percentage cell count in the S phase was significantly
higher in the PAX6-knockdown group than in the negative GFP-control
group (40.00±1.10 vs. 34.69 ± 3.17%; t=−4.44; P<0.05, n=3). For
the the Y79 cell line, the percentage cell count in the G2/M phase
was significantly higher in the PAX6-knockdown group than in the
negative GFP-control group (16.92±1.89 vs. 10.78±2.23%; t=−8.31,
P<0.05, n=3) (Fig. 5).
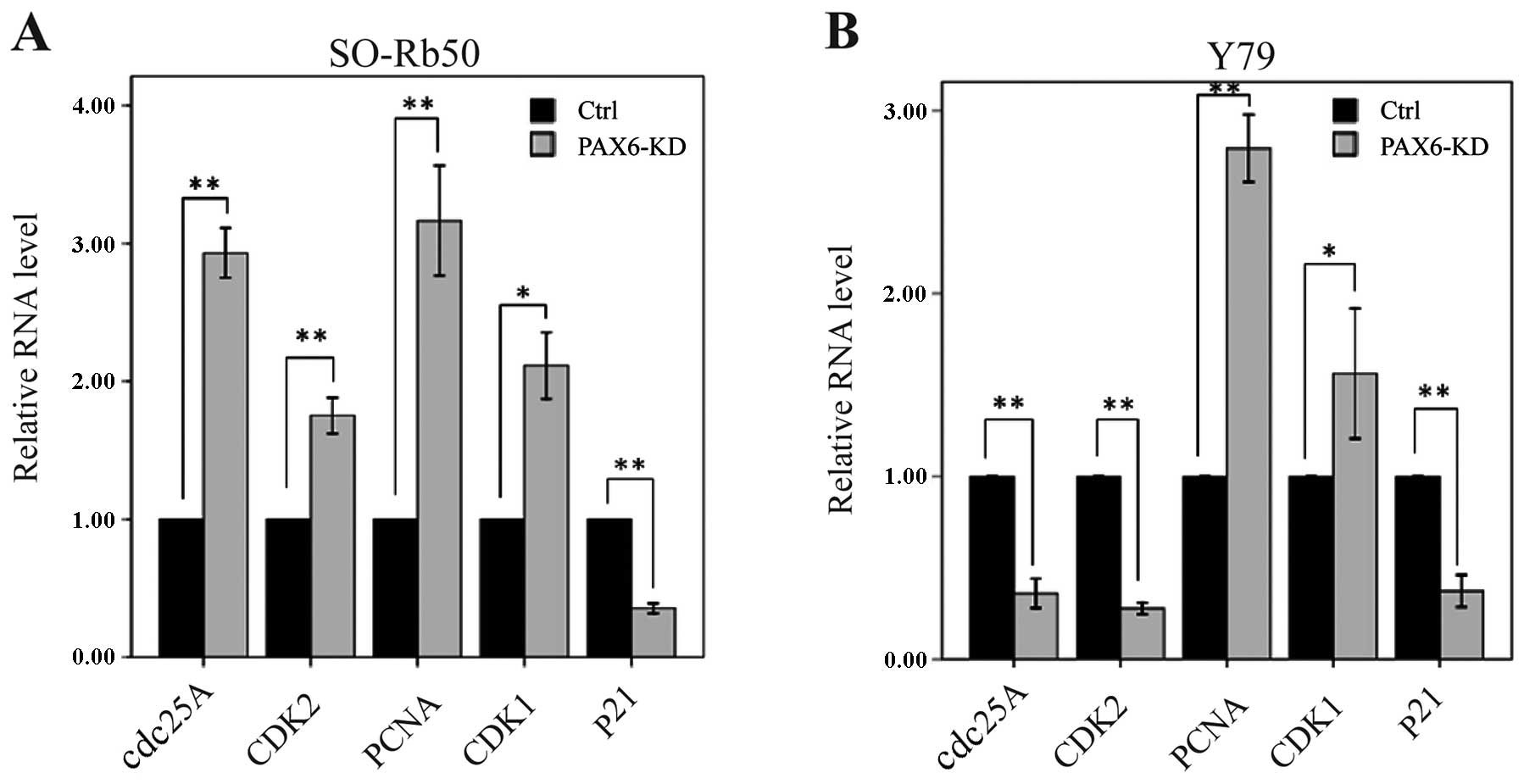
Suppression of PAX6 affects cell
cycle-related gene expression
To determine how the suppression of PAX6 affects
cell cycle distribution, we measured the mRNA levels of cell
cycle-related genes by RT-qPCR. In the SO-Rb50 retinoblastoma cell
line, the mRNA levels of cdc25A, CDK2, PCNA and CDK1
were significantly higher in the PAX6 inhibition study group than
in negative GFP-control group (cdc25A, 2.93±0.16 vs.
1.00±0.00; t=−21.47; P<0.01, n=3; CDK2, 1.75±0.11 vs.
1.00±0.00; t=−11.56; P<0.01, n=3; PCNA, 3.16±0.35 vs.
1.00±0.00; t=−10.86; P<0.01, n=3; CDK1, 2.11±0.21 vs.
1.00±0.00; t=−9.22; P<0.05, n=3) (Fig. 6A). However, the mRNA level of
p21 was significantly lower in the PAX6 inhibition study
group than in negative GFP-control group (0.35±0.03 vs. 1.00±0.00;
t=37.25; P<0.01, n=3) (Fig.
6A). In the Y79 retinoblastoma cell line, the mRNA levels of
cdc25A, CDK2 and p21 were significantly lower in the
PAX6 inhibition study group than in negative GFP-control group
(cdc25A, 0.36±0.07 vs. 1.00±0.00; t=15.91; P<0.01, n=3;
CDK2, 0.28±0.03 vs. 1.00±0.00; t=46.77; P<0.01, n=3;
p21, 0.37±0.08 vs. 1.00±0.00; t=14.37; P<0.01, n=3)
(Fig. 6B). The mRNA levels of
PCNA and CDK1 were significantly higher in the PAX6
inhibition study group than in negative GFP-control group
(PCNA, 2.79±0.16 vs. 1.00±0.00; t=−19.47; P<0.01, n=3;
CDK1, 1.56±0.31 vs. 1.00±0.00; t=−3.15; P<0.05, n=3)
(Fig. 6B).
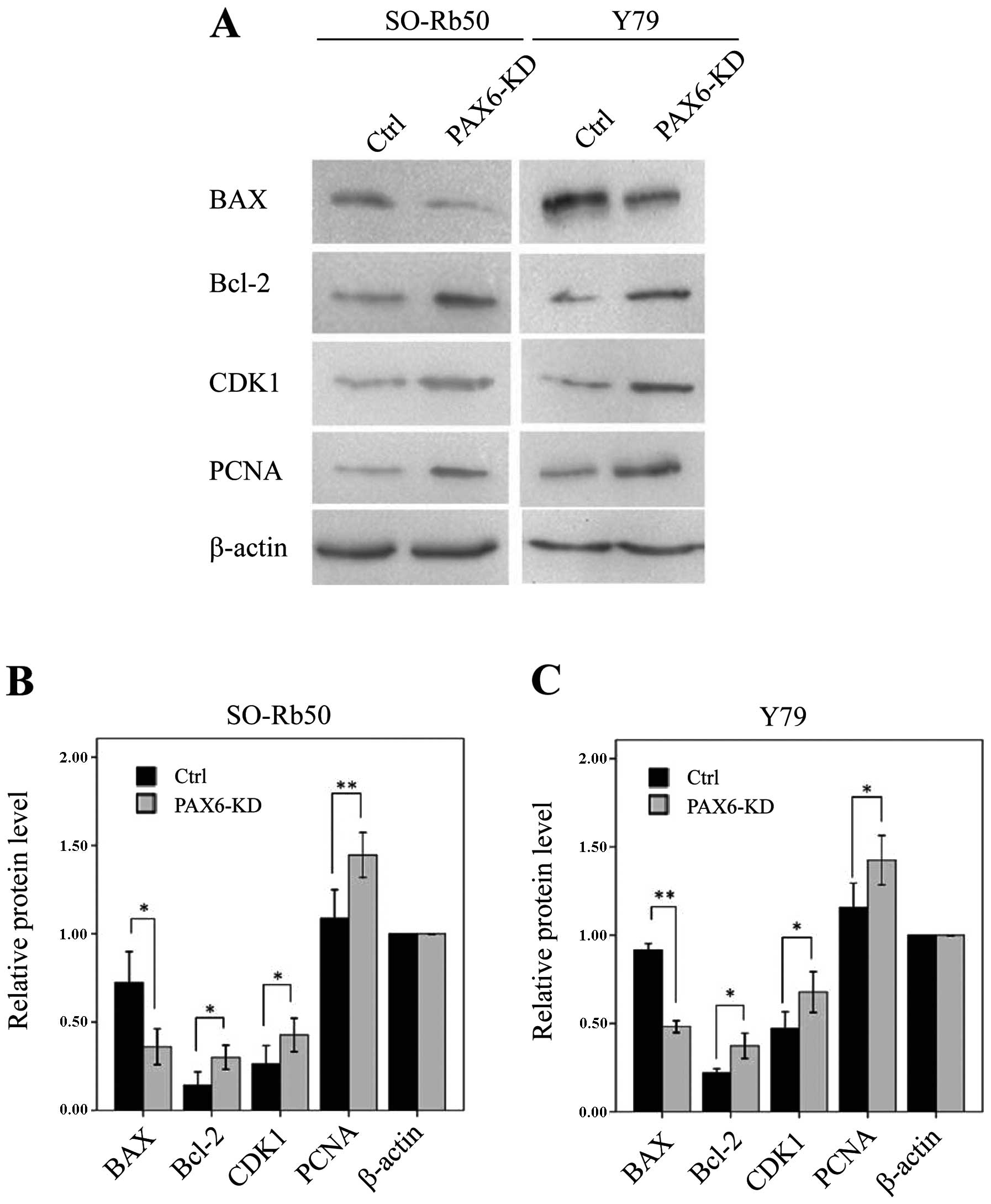
Suppression of PAX6 affects proteins
related to apoptosis and the cell cycle
To determine the molecular mechanisms involved in
the inhibition of PAX6 expression, as well as the signaling
pathways associated with the cell cycle and apoptosis, we measured
the expression of apoptosis-related Bcl-2 and BAX and
the cell cycle regulatory proteins, CDK1 and PCNA, by
western blot analysis. In both retinoblastoma cell lines, the
levels of Bcl-2, CDK1 and PCNA were significantly
upregulated in the PAX6 inhibition study groups than in negative
GFP-control groups (Bcl-2, t=−5.90, P<0.05 and t=−4.86,
P<0.05, resp. n=3; CDK1, t=−5.27, P<0.05; and t=−7.60,
P<0.05, resp. n=3; PCNA, t=−11.49, P<0.01; and
t=−4.32, P=0.05, resp. n=3) (Fig.
7). However, the level of BAX was downregulated in both
retinoblastoma cell lines (t=4.51, P<0.05; and t=67.96,
P<0.01, resp. n=3) (Fig.
7).
Discussion
Using a lentiviral vector-mediated transfection of
human retinoblastoma cells, our study demonstrates that the
inhibition of endogenous PAX6 expression results in decreased
retinoblastoma cell apoptosis in vitro. First, we
transfected 2 retinoblastoma cell lines with a lentiviral pGCL-GFP
vector encoding a PAX6 siRNA sequence. The transfection was
confirmed by measuring the GFP expression using a fluorescence
microscope at 4 days after transfection (Fig. 1). We then confirmed the silencing
effect on PAX6 by RT-qPCR. The results revealed that the mRNA
levels of endogenous PAX6 were significantly lower in the
PAX6-knockdown groups than in the control groups. Furthermore, we
observed a phenotype with a reduced early apoptotic rate in the
PAX6-knockdown groups by flow cytometric analysis. We also found
that the percentage of cells in the S phase or the G2/M phase of
the cell cycle was significantly higher in the PAX6-knockdown
groups than in the control groups (Fig. 6). These changes in the cell cycle
were paralleled with the changes in the mRNA levels of the cell
cycle-associated genes, such as cdc25A, CDK2,
PCNA, CDK1 and p21. The upregulation of
anti-apoptotic proteins and the downregulation of pro-apoptotic
proteins was also confirmed by western blot analysis.
Apoptosis is an important biological phenomenon and
plays a key role in organic evolution, the maintenance of the
internal environment and the development of tumors (23). Proto-oncogenes and tumor
suppressor genes associated with proliferation are found to
participate in the regulation of apoptosis. These conserved genes
contain the Bcl-2 gene family, the caspase gene family and
p53. The Bcl-2 gene family is divided into two
groups; one group includes inhibiting factors of apoptosis, such as
Bcl-2, Bcl-xL, Bcl-w and mcl-1, while the other group
includes apoptosis-promoting factors, such as BAX, Bcl-xs,
Bak and Bad. The proportion of Bcl-2 and BAX determines
the fate of the cells (24). In
this study, we found that inhibition of PAX6 in human
retinoblastoma cell lines led to a low apoptotic rate which was
supported by the changes in the levels of apoptosis-related
proteins (Bcl-2 and BAX). Consistently, Kashiwagi et
al reported that cotylenin A induced apoptosis and inhibited
cell proliferation by upregulating the mRNA expression of p21 and
PAX6 in the retinoblastoma cell lines, Y79 and WERI-Rb1 (25). Ouyang et al reported that
PAX6 suppressed the proliferation of corneal epithelial cells and
led to apoptosis (26). Zhang
et al reported that the inhibition of PAX6 blocked
neuroectoderm specification from human embryonic stem cells
(27). In the same manner, the
overexpression of PAX6(a) has been shown to contribute to the
differentiation of human embryonic stem cells. Davis et al
found that the upregulation of PAX6 inhibited cell proliferation
(28). The lack PAX6 in knockout
models has been shown to increase cell proliferation in the
cerebral cortex by shortening the cell cycle of progenitor cells at
the onset of corticogenesis (29–31). Berger et al demonstrated
that the activation of PAX6 inhibited proliferation and induced the
apoptosis of cortical progenitors in the developing cortex
(32).
However, the function of PAX6 in proliferation and
apoptosis remains controversial. Certain studies have shown that a
high expression of PAX6 is found in proliferating lens epithelial
cells and that PAX6 expression is essential for maintaining the
multipotency of retinal precursor cells (8,33–37). For instance, Maulbecker and Gruss
found that PAX6 induced tumor formation in mice (9). Yamaoka et al demonstrated
that the overexpression of PAX6 increased the proliferation of
ductal epithelial cells in the mouse pancreas and led to the
development of cystic pancreatic adenomas (38). In our previous study, we reported
that the lentiviral vector-mediated overexpression of PAX6 in human
retinoblastoma cells was associated with an increased cell
proliferation parallel to a reduced caspase-3-dependent apoptotic
rate, and with a change in the p53-regulated cell cycle (14). All these controversial data
suggest that either an abnormally low level or high level of PAX6
is associated with a reduced rate of apoptosis and an increased
rate of proliferation of (retinoblastoma) tumor cells. Furthermore,
another study suggested that the quantitative and spatiotemporal
expression of PAX6 influences its effect and functions (39). Accordingly, Zhang et al
demonstrated that inducible PAX6(+5a) expression showed a biphasic
and dose-dependent regulation of δ-catenin (a neural specific
member of the armadillo/β-catenin superfamily) expression and cell
fates. A moderate upregulation of Pax6(+5a) promoted δ-catenin
expression and induced neurite-like cellular protrusions, but
increasing the expression of PAX6(+5a) reversed these processes
(40).
Apart from the decreased tumor cell apoptosis after
the inhibition of endogenous PAX6 in the human retinoblastoma
cells, we found that the inhibition of endogenous PAX6 led to an
induction into the cell cycle S phase of the SO-Rb50 cell line and
an induction into the cell cycle G2/M phase of the Y79
retinoblastoma cell line. Previous studies have suggested that PAX6
regulates cell proliferation and apoptosis by controlling the cell
cycle in a cell type-specific manner. Duparc et al showed
that PAX6 null retinal spheres over-proliferated and displayed
reduced expression levels of several negative regulators of the
cell cycle, such as p16, p27 and p21 (41). They also found that the gain of
function of PAX6 suppressed cellular proliferation and secondary
sphere formation. Accodingly, our present results indicated that
the level of p21 was significantly lower in the PAX6-knockdown
groups than in the control groups (Fig. 7). Similarly, other studies have
shown that the suppression of PAX6 is associated with a lengthening
of the cell cycle S phase (29,30,42). Shaham et al reported that
PAX6 negatively regulates SOX2 [(sex determining region Y)-box 2]
in the embryonic lens and suggested that PAX6 is a crucial factor
in cell cycle exit (43). Ouyang
et al reported that PAX6 overexpression retards the cell
cycle in corneal epithelial cells (26). Hsieh et al demonstrated
that maintaining a relatively low level of PAX6 is necessary for
the S phase re-entry (5). All
these data are in accordance with those of our study and support
the hypothesis that PAX6 affects the proliferation and apoptosis of
cells by regulating the cell cycle.
To determine the possible mechanisms of how PAX6
regulates cell cycle progression, we examined the mRNA and protein
levels of several cell cycle regulatory genes, such as cdc25A,
CDK2, PCNA CDK1 and p21. In the PAX6-knockdown cells,
CDK2 expression was increased in the SO-Rb50 retinoblastoma cells,
suggesting an induction into the S phase. This finding was
supported by an increased level of cdc25A, an upstream factor which
promotes the expression of CDK2 and shortens the cell cycle. The
mRNA level of PCNA was elevated in the PAX6-knockdown groups. PCNA
is a DNA polymerase accessory factor which is involved in DNA
replication and repair during the S phase of the cell cycle. In a
parallel manner, the mRNA level of p21 was decreased in the study
groups. p21 may block the ability of PCNA to bind with Gadd45
(44,45). At the G2/M checkpoint, CDK1
controlling the entry into the M phase is pivotal in regulating
this transition. As an inhibitor of CDK1, p21 may be turned off by
the activation of the CDK1/cyclin B complex (46–48). In our study, the level of CDK1 was
upregulated and correspondingly, the level of p21 was downregulated
in the study groups. These findings may explain the induction of
tumor cell G2/M phase in the Y79 cells. Our results suggested that
PAX6 was associated with the cell cycle through
cdc25A/CDK2-dependent pathways and p53/p21/CDK1-dependent pathway.
This was supported by parallel findings on the concentration of S
phase- and G2/M phase-related proteins (PCNA and CDK) and
apoptosis-related proteins (Bcl-2 and BAX).
In conclusion, our data demonstrate that the
lentiviral-vector-mediated inhibition of endogenous PAX6 expression
in human retinoblastoma cells is associated with an increased
proliferation and a decreased apoptosis of human retinoblastoma
cells in vitro, paralleled by corresponding changes in cell
cycle-related and apoptosis-related molecules.
Acknowledgements
Firstly, we sincerely thank Professor Jost B. Jonas
(Department of Ophthalmology, Medical Faculty Mannheim,
Ruprecht-Karls-University of Heidelberg, Mannheim, Germany) for
assisting in the revision process of this manuscript. Secondly, we
sincerely thank Professor An Jing (Department of Microbiology,
School of Basic Medical Sciences, Capital Medical University,
Beijing, China), Professor Qingjun Lu (Beijing School of
Ophthalmology, Beijing Institute of Ophthalmology, Beijing Tongren
Hospital, Capital Medical University) and Dr Liang Li (Beijing
Institute of Ophthalmology, Beijing Tongren Hospital, Capital
Medical University, Beijing, China) for providing technical
assistance. This study was supported by the National Natural
Science Foundation of China (grant no. 81172393, 2012).
References
|
1
|
Jordan T, Hanson I, Zaletayev D, et al:
The human PAX6 gene is mutated in two patients with aniridia. Nat
Genet. 1:328–332. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Quiring R, Walldorf U, Kloter U and
Gehring WJ: Homology of the eyeless gene of Drosophila to
the small eye gene in mice and aniridia in humans. Science.
265:785–789. 1994.PubMed/NCBI
|
|
3
|
Kogawa M, Hisatake K, Atkins GJ, et al:
The paired-box homeodomain transcription factor Pax6 binds to the
upstream region of the TRAP gene promoter and suppresses receptor
activator of NF-kappaB ligand (RANKL)-induced osteoclast
differentiation. J Biol Chem. 288:31299–31312. 2013. View Article : Google Scholar
|
|
4
|
Hart AW, Mella S, Mendrychowski J, van
Heyningen V and Kleinjan DA: The developmental regulator Pax6 is
essential for maintenance of islet cell function in the adult mouse
pancreas. PLoS One. 8:e541732013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hsieh YW and Yang XJ: Dynamic Pax6
expression during the neurogenic cell cycle influences
proliferation and cell fate choices of retinal progenitors. Neural
Dev. 4:322009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Livesey FJ and Cepko CL: Vertebrate neural
cell-fate determination: lessons from the retina. Nat Rev Neurosci.
2:109–118. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shaham O, Menuchin Y, Farhy C and
Ashery-Padan R: Pax6: a multi-level regulator of ocular
development. Prog Retin Eye Res. 31:351–376. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Marquardt T, Ashery-Padan R, Andrejewski
N, Scardigli R, Guillemot F and Gruss P: Pax6 is required for the
multipotent state of retinal progenitor cells. Cell. 105:43–55.
2001. View Article : Google Scholar
|
|
9
|
Maulbecker CC and Gruss P: The oncogenic
potential of Pax genes. EMBO J. 12:2361–2367. 1993.PubMed/NCBI
|
|
10
|
Sachdeva UM and O’Brien JM: Understanding
pRb: toward the necessary development of targeted treatments for
retinoblastoma. J Clin Invest. 122:425–434. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xu XL, Fang Y, Lee TC, et al:
Retinoblastoma has properties of a cone precursor tumor and depends
upon cone-specific MDM2 signaling. Cell. 137:1018–1031. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cvekl A, Yang Y, Chauhan BK and Cveklova
K: Regulation of gene expression by Pax6 in ocular cells: a case of
tissue-preferred expression of crystallins in lens. Int J Dev Biol.
48:829–844. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Schedl A, Ross A, Lee M, et al: Influence
of PAX6 gene dosage on development: overexpression causes severe
eye abnormalities. Cell. 86:71–82. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bai SW, Li B, Zhang H, et al: Pax6
regulates proliferation and apoptosis of human retinoblastoma
cells. Invest Ophthalmol Vis Sci. 52:4560–4570. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li L, Li B, Zhang H, et al: Lentiviral
vector-mediated PAX6 overexpression promotes growth and inhibits
apoptosis of human retinoblastoma cells. Invest Ophthalmol Vis Sci.
52:8393–8400. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xin GH, Zhao XH, Liu D, et al: Effect of
VEGF-targeted antisense gene therapy on retinoblastoma cell line
SO-RB50 in vitro and in vivo. Int J Ophthalmol. 5:440–447.
2012.PubMed/NCBI
|
|
17
|
Sreenivasan S, Thirumalai K and
Krishnakumar S: Expression profile of genes regulated by curcumin
in Y79 retinoblastoma cells. Nutr Cancer. 64:607–616. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhao B, Li B, Bai S, et al: Effects of
matrine on proliferation and apoptosis of cultured retinoblastoma
cells. Graefes Arch Clin Exp Ophthalmol. 250:897–905. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Reid TW, Albert DM, Rabson AS, et al:
Characteristics of an established cell line of retinoblastoma. J
Natl Cancer Inst. 53:347–360. 1974.PubMed/NCBI
|
|
20
|
Marconett CN, Morgenstern TJ, San Roman
AK, Sundar SN, Singhal AK and Firestone GL: BZL101, a phytochemical
extract from the Scutellaria barbata plant, disrupts
proliferation of human breast and prostate cancer cells through
distinct mechanisms dependent on the cancer cell phenotype. Cancer
Biol Ther. 10:397–405. 2010.PubMed/NCBI
|
|
21
|
Shi T, Mazumdar T, Devecchio J, et al:
cDNA microarray gene expression profiling of hedgehog signaling
pathway inhibition in human colon cancer cells. PLoS One. 5:pii:
e13054. 2010.PubMed/NCBI
|
|
22
|
Wang T, Zhao R, Wu Y, et al: Hepatitis B
virus induces G1 phase arrest by regulating cell cycle genes in
HepG2.2.15 cells. Virol J. 8:2312011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Johnstone RW, Ruefli AA and Lowe SW:
Apoptosis: a link between cancer genetics and chemotherapy. Cell.
108:153–164. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fulda S: Tumor resistance to apoptosis.
Int J Cancer. 124:511–515. 2009. View Article : Google Scholar
|
|
25
|
Kashiwagi Y, Kato N, Sassa T, et al:
Cotylenin A inhibits cell proliferation and induces apoptosis and
PAX6 mRNA transcripts in retinoblastoma cell lines. Mol Vis.
16:970–982. 2010.PubMed/NCBI
|
|
26
|
Ouyang J, Shen YC, Yeh LK, et al: Pax6
overexpression suppresses cell proliferation and retards the cell
cycle in corneal epithelial cells. Invest Ophthalmol Vis Sci.
47:2397–2407. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang X, Huang CT, Chen J, et al: Pax6 is
a human neuroectoderm cell fate determinant. Cell Stem Cell.
7:90–100. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Davis N, Yoffe C, Raviv S, et al: Pax6
dosage requirements in iris and ciliary body differentiation. Dev
Biol. 333:132–142. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Estivill-Torrus G, Pearson H, van
Heyningen V, Price DJ and Rashbass P: Pax6 is required to regulate
the cell cycle and the rate of progression from symmetrical to
asymmetrical division in mammalian cortical progenitors.
Development. 129:455–466. 2002.PubMed/NCBI
|
|
30
|
Gotz M, Stoykova A and Gruss P: Pax6
controls radial glia differentiation in the cerebral cortex.
Neuron. 21:1031–1044. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Warren N, Caric D, Pratt T, et al: The
transcription factor, Pax6, is required for cell proliferation and
differentiation in the developing cerebral cortex. Cereb Cortex.
9:627–635. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Berger J, Berger S, Tuoc TC, et al:
Conditional activation of Pax6 in the developing cortex of
transgenic mice causes progenitor apoptosis. Development.
134:1311–1322. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Duncan MK, Haynes JI II, Cvekl A and
Piatigorsky J: Dual roles for Pax-6: a transcriptional repressor of
lens fiber cell-specific beta-crystallin genes. Mol Cell Biol.
18:5579–5586. 1998.PubMed/NCBI
|
|
34
|
Grindley JC, Davidson DR and Hill RE: The
role of Pax-6 in eye and nasal development. Development.
121:1433–1442. 1995.PubMed/NCBI
|
|
35
|
Richardson J, Cvekl A and Wistow G: Pax-6
is essential for lens-specific expression of zeta-crystallin. Proc
Natl Acad Sci USA. 92:4676–4680. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Walther C and Gruss P: Pax-6, a murine
paired box gene, is expressed in the developing CNS. Development.
113:1435–1449. 1991.PubMed/NCBI
|
|
37
|
Zhang W, Cveklova K, Oppermann B, Kantorow
M and Cvekl A: Quantitation of PAX6 and PAX6(5a) transcript levels
in adult human lens, cornea, and monkey retina. Mol Vis. 7:1–5.
2001.
|
|
38
|
Yamaoka T, Yano M, Yamada T, Matsushita T,
Moritani M, Ii S, Yoshimoto K, Hata J and Itakura M: Diabetes and
pancreatic tumours in transgenic mice expressing Pax6.
Diabetologia. 43:332–339. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kleinjan DA, Seawright A, Mella S, et al:
Long-range downstream enhancers are essential for Pax6 expression.
Dev Biol. 299:563–581. 2006. View Article : Google Scholar
|
|
40
|
Zhang J, Lu JP, Suter DM, et al: Isoform-
and dose-sensitive feedback interactions between paired box 6 gene
and delta-catenin in cell differentiation and death. Exp Cell Res.
316:1070–1081. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Duparc RH, Abdouh M, David J, Lepine M,
Tetreault N and Bernier G: Pax6 controls the proliferation rate of
neuroepithelial progenitors from the mouse optic vesicle. Dev Biol.
301:374–387. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Haubst N, Berger J, Radjendirane V, et al:
Molecular dissection of Pax6 function: the specific roles of the
paired domain and homeodomain in brain development. Development.
131:6131–6140. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Shaham O, Smith AN, Robinson ML, Taketo
MM, Lang RA and Ashery-Padan R: Pax6 is essential for lens fiber
cell differentiation. Development. 136:2567–2578. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Cayrol C, Knibiehler M and Ducommun B: p21
binding to PCNA causes G1 and G2 cell cycle arrest in p53-deficient
cells. Oncogene. 16:311–320. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Frouin I, Maga G, Denegri M, et al: Human
proliferating cell nuclear antigen, poly(ADP-ribose) polymerase-1,
and p21waf1/cip1. A dynamic exchange of partners. J Biol Chem.
278:39265–39268. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Bunz F, Dutriaux A, Lengauer C, et al:
Requirement for p53 and p21 to sustain G2 arrest after DNA damage.
Science. 282:1497–1501. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Møller MB: P27 in cell cycle control and
cancer. Leuk Lymphoma. 39:19–27. 2000.
|
|
48
|
Taylor WR and Stark GR: Regulation of the
G2/M transition by p53. Oncogene. 20:1803–1815. 2001. View Article : Google Scholar : PubMed/NCBI
|