Atrial fibrillation (AF), a supraventricular
tachyarrhythmia characterized by chaotic atrial electrical activity
and consequently ineffective atrial contraction, is the most common
type of cardiac arrhythmia observed in clinical settings,
accounting for approximately one-third of hospitalizations for
cardiac rhythm disturbances (1).
The prevalence of AF is approximately 1% in the general population,
and increases strikingly with advancing age, rising from >1% in
subjects under 60 years of age to nearly 10% in individuals aged
over 80 years (2). According to a
report from the Framingham Heart Study, individuals older than 40
years have an approximately 25% chance of developing AF during
their lifetime (3). The number of
individuals with AF in the United States is presently estimated at
2.3 million and is projected to exceed 15.9 million by the year
2050, mainly due to the aging population and improved
cardiovascular survival (4). The
condition is associated with severe complications, such as chronic
heart failure and stroke, significantly contributing to
cardiovascular morbidity and mortality. Compared to sinus rhythm,
AF confers a 2-fold increased risk of death (5) and a 5-fold increased risk of
thromboembolic stroke (6). Of
note, the annual incidence of ischemic stroke resulting from AF
increases considerably with age, ranging from 1.5% of adults in
their fifties to 23.5% in octogenarians (6). Additionally, AF may result in a
degraded quality of life, reduced exercise tolerance, cognitive
dysfunction or dementia, worsened renal function,
tachycardia-induced cardiomyopathy, myocardial infarction and left
ventricular dysfunction or even congestive heart failure (7–16).
Therefore, AF represents an increasing public health challenge with
profound socioeconomic implications. The appraised cost of
treatment for AF is <1% of the health care expenditure (17) and only in the United States, the
direct costs of treating nonvalvular AF are in excess of $6.4
billion per year (18). Despite
the high prevalence and substantial clinical significance, the
molecular basis underlying AF remains largely unclear.
AF often occurs secondary to various cardiovascular
and systemic diseases and surgical procedures, such as coronary
artery disease, congenital heart disease, valvular heart disease,
cardiac surgery, cardiomyopathy, myocarditis, pulmonary heart
disease, left ventricular dysfunction, hypertension,
hyperthyroidism, diabetes mellitus and electrolyte imbalance. Other
potential risk factors responsible for the development of AF
include age, male gender, obesity, high-level physical training,
prehypertension, increased pulse pressure, diastolic dysfunction,
obstructive sleep apnea, hyperuricemia, kidney disease, systemic
inflammation, pericardial fat, tobacco use and alcohol consumption
(19–23). However, in 30–45% of the total
number of patients, AF is diagnosed in the absence of the
above-mentioned associated diseases or predisposing factors, a
condition referred to as idiopathic AF or lone AF, and up to 15% of
these patients with lone AF have a clearly established positive
family history, and are thus defined as familial AF (1). Aggregating epidemiological studies
have demonstrated the pronounced clustering of AF in families and
the markedly increased incidence of AF in the close relatives of AF
patients, indicating a pivotal role of genetic factors in the
pathogenesis of familial AF (24–31). In previous studies, genome-wide
genetic linkage analysis with highly polymorphic microsatellite
markers mapped AF-susceptibility loci on human chromosomes
10q22-24, 11p15.5, 6q14-16, 5p13, 10p11-q21 and 5p15, of which
AF-causing mutations in 2 genes, including potassium voltage-gated
channel, KQT-like subfamily, member 1 (KCNQ1) on chromosome
11p15.5 and nucleoporin 155 kDa (NUP155) on chromosome 5p13,
were identified and functionally deciphered (32–37). The direct analysis of candidate
genes has identified an increasing number of AF-related genes,
including potassium voltage-gated channel, Isk-related family,
member 1 (KCNE1-5), potassium voltage-gated channel,
subfamily H (eag-related), member 2 (KCNH2),
potassium voltage-gated channel, shaker-related subfamily, member 5
(KCNA5), potassium voltage-gated channel,
Shal-related subfamily, member 3 (KCND3), potassium
inwardly-rectifying channel, subfamily J (KCNJ2), KCNJ8, gap
junction protein, alpha 1, 43 kDa (GJA1), gap
junction protein, alpha 5, 40 kDa (GJA5), atrial
natriuretic peptide (ANP), sodium channel, voltage-gated,
type V, alpha subunit (SCN5A) and sodium channel,
voltage-gated, type I, beta subunit (SCN1B-4B) (38–63). Nevertheless, these well
established AF-associated genes only explain a small fraction of
cases of AF, and in an overwhelming majority of patients, the
genetic determinants for AF remain unknown.
Recent studies indicate that abnormal embryonic
development and structural remodeling of the cardiovascular system,
particularly the pulmonary veins and the atria, create a major
anatomic substrate liable to AF (64,65). A growing body of evidence
substantiates the crucial role of several cardiac transcription
factors, including NK2 homeobox (NKX2)-5, GATA binding protein
(GATA)4, GATA5, GATA6 and paired-like homeodomain 2 (PITX2)c in
normal cardiovascular morphogenesis (66–87), and multiple mutations in these
transcription factors have been causally linked to AF (88–101). NKX2-6 is another member of the
NK2-family of transcription factors and its expression profile and
functional roles partially overlap with those of NKX2-5 during
cardiovascular development (102–105), which warrants the screening of
NKX2-6 as a preferred candidate gene for the development of
AF.
A cohort of 150 unrelated patients with lone AF was
enrolled from the Han Chinese population. The available relatives
of the index patients were also recruited. A total of 200
ethnically-matched unrelated healthy individuals were enlisted as
the controls. All participants were evaluated by detailed medical
history, physical examination, an electrocardiogram and
echocardiography. Cardiac catheterization, angiography, a chest
X-ray and cardiac magnetic resonance imaging were performed only if
there was a strong clinical indication. Medical records were also
reviewed in the case of deceased or unavailable relatives. The
diagnosis and classification of AF was made in accordance with the
guidelines for the management of patients with AF (1,88).
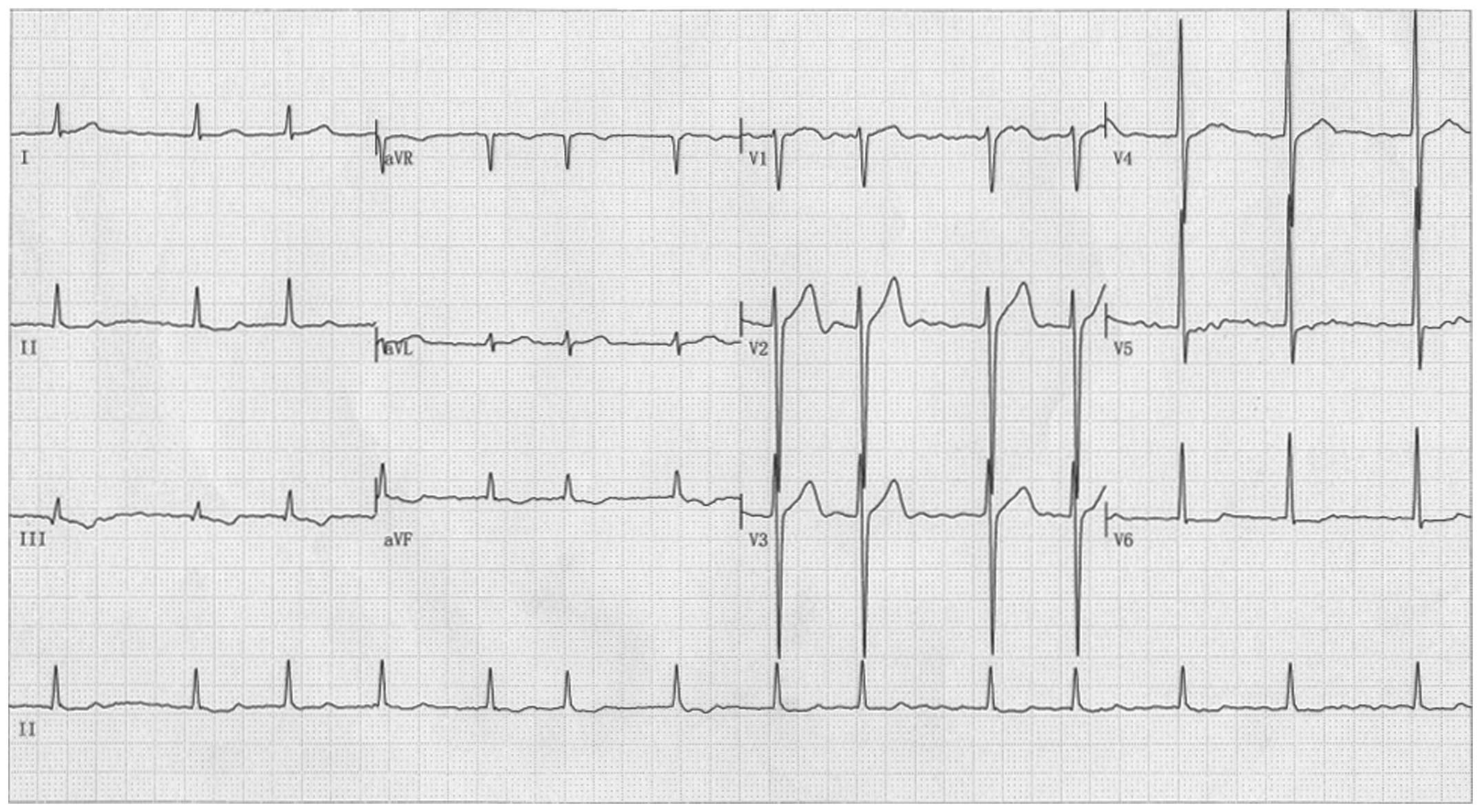
Briefly, AF was diagnosed by a standard 12-lead electrocardiogram
demonstrating no P-waves and irregular R-R intervals regardless of
clinical symptoms. Lone AF was defined as AF occurring in
individuals <60 years of age without other cardiac or systemic
diseases by physical examination, electrocardiogram, transthoracic
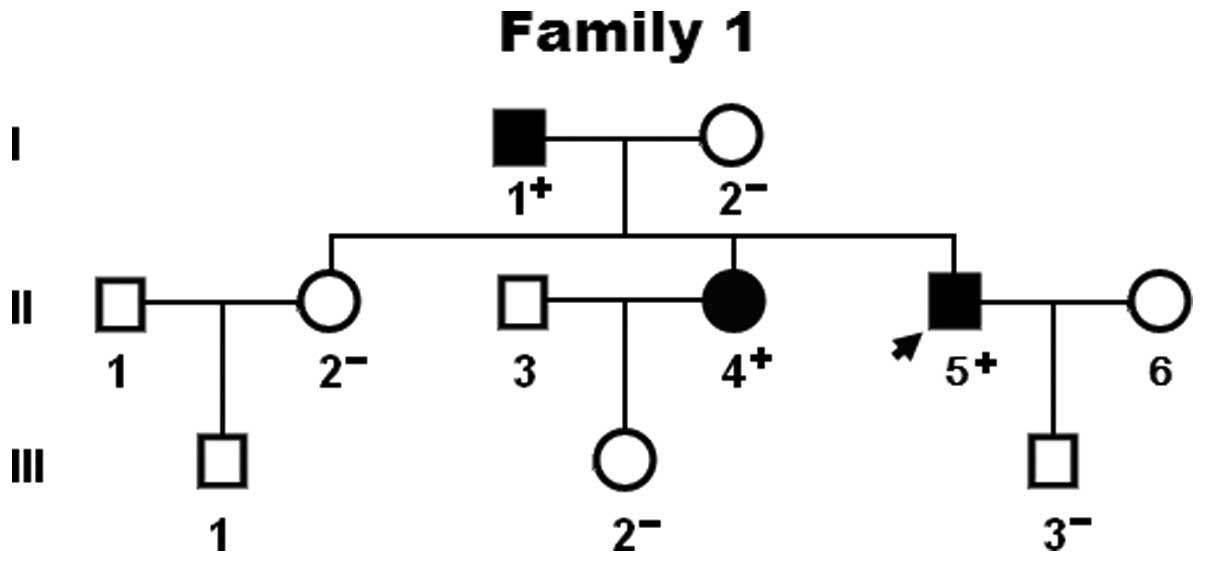
echocardiogram and extensive laboratory tests. Familial AF was
termed when lone AF existed in an additional 2 or more first- or
second-degree relatives. Relatives with AF occurring at any age in
the presence of structural heart disease (hypertensive, ischemic,
myocardial or valvular) were classified as ‘undetermined’ for
having an inherited form of AF. The ‘undetermined’ classification
was also used if documentation of AF on an electrocardiogram
tracing was lacking in relatives with symptoms consistent with AF
(palpitation, dyspnea and light-headedness), or if a screening
electrocardiogram and echocardiogram were not performed,
irrespective of the symptoms. Relatives were classified as
‘unaffected’ if they were asymptomatic and had a normal
electrocardiogram. Paroxysmal AF was defined as AF lasting >30
sec that terminated spontaneously. Persistent AF was defined as AF
lasting >1 week and requiring either pharmacological therapy or
electrical cardioversion for termination. AF that was refractory to
cardioversion or that was allowed to continue was classified as
permanent AF. Peripheral venous blood samples were obtained from
all the participants. The clinical studies were performed with
investigators blinded to the results of the genotypes. This study
conformed to the principles of the Declaration of Helsinki and the
study protocol was approved by the local Institutional Ethics
Committee of Shanghai Chest Hospital, Shanghai Jiao Tong University
(the ethical approval number for cases and controls: KS1101; the
date of the approval: April 12, 2011). Written informed consent was
obtained from all participants prior to enrollment.
Genomic DNA was extracted from the blood lymphocytes
of each participant using the Wizard Genomic DNA Purification kit
(Promega, Madison, WI, USA). The coding exons and exon-intron
boundaries of the NKX2-6 gene were sequenced in the 150
unrelated patients with lone AF. The available relatives of the
index patient harboring an identified NKX2-6 mutation and
the 200 unrelated control individuals were genotyped for
NKX2-6. The referential genomic DNA sequence of
NKX2-6 was derived from GenBank (accession no. NG_030636), a
gene sequence database at the National Center for Biotechnical
Information (NCBI; http://www.ncbi.nlm.nih.gov/). With the help of online
Primer 3 software (http://frodo.wi.mit.edu/), the primer pairs used to
amplify the coding regions and flanking splice junction sites of
NKX2-6 by polymerase chain reaction (PCR) were designed as
described in Table I. PCR was
carried out using HotStar TaqDNA Polymerase (Qiagen, Hilden,
Germany) on a Veriti Thermal Cycler (Applied Biosystems, Foster
City, CA, USA) with standard conditions and concentrations of
reagents. Amplified products were purified with the QIAquick Gel
Extraction kit (Qiagen). Both strands of each amplicon were
sequenced using the BigDye® Terminator v3.1 Cycle
Sequencing kit under an ABI PRISM 3130 XL DNA Analyzer (both from
Applied Biosystems). The sequencing primers were the same as those
mentioned above for the specific region amplifications. DNA
sequences were viewed and analyzed using DNA Sequencing Analysis
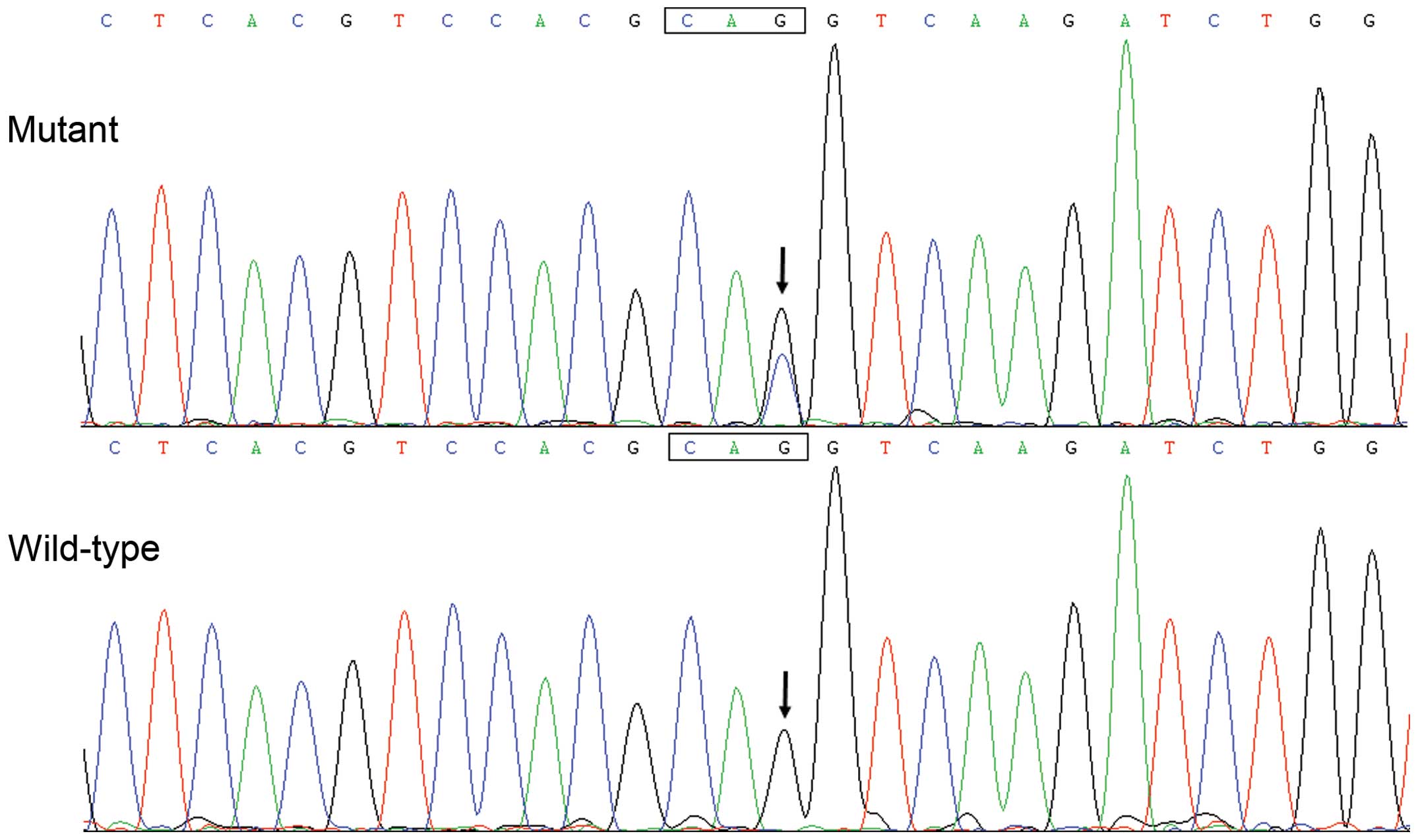
Software v5.1 (Applied Biosystems). The variant was validated by
re-sequencing of an independent PCR-generated amplicon from the
same subject. In addition, for an identified sequence variant, the
single nucleotide polymorphism (SNP; http://www.ncbi.nlm.nih.gov/SNP) and human gene
mutation (HGM; http://www.hgmd.org) databases were
queried to confirm its novelty.
The conservation of the amino acid altered by
missense mutation was appraised by aligning human NKX2-6 to
chimpanzee, monkey, dog, cattle, mouse, rat, fowl, zebrafish and
frog NKX2-6 using the HomoloGene and Show Multiple Alignment links
on the NCBI website (http://www.ncbi.nlm.nih.gov/homologene).
The recombinant expression vector NKX2-5-pEFSA and
the ANF-luciferase (ANF-luc) reporter plasmid, which contains the
2,600 bp 5′-flanking region of the ANF gene, were kindly provided
by Dr. Ichiro Shiojima from Chiba University School of Medicine,
Chiba, Japan. Owing to unknown downstream genes of NKX2-6, NKX2-5
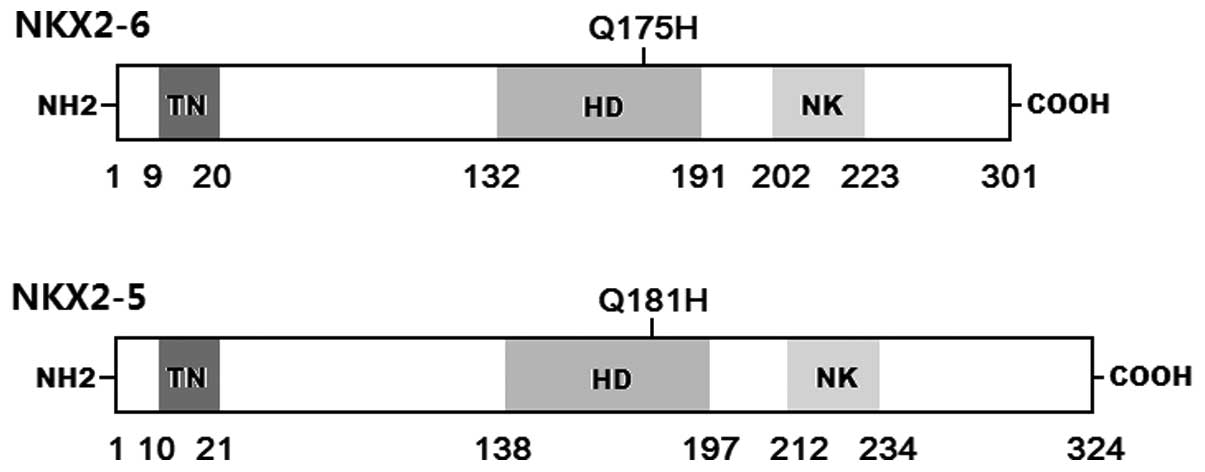
was used as a surrogate in transcriptional analysis to assess the
functional consequences of the Q175H homeodomain substitution
(104). Alignment between the
human NKX2-6 and NKX2-5 proteins illustrated that Q175H-mutant
NKX2-6 was equivalent to Q181H-mutant NKX2-5 (data not shown). The
c.543G>C transition, which was predicted to generate the p.Q181H
mutation, was introduced into wild-type NKX2-5 using a QuickChange
II XL Site-Directed Mutagenesis kit (Stratagene, La Jolla, CA, USA)
with a complementary pair of primers. The mutant was sequenced to
confirm the desired mutation and to exclude any other sequence
variations.
COS-7 cells from our cell bank were cultured in
Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf
serum. The internal control reporter plasmid pGL4.75 (hRluc/CMV,
Promega) was used in transient transfection analyses to evaluate
the transcriptional activity of the NKX2-5 mutant. The COS-7
cells were transfected with 0.4 μg of wild-type or mutant
NKX2-5-pEFSA, 1.0 μg of ANF-luc and 0.04 μg of pGL4.75 using
PolyFect Transfection Reagent (Qiagen). For co-transfection
experiments, 0.2 μg of wild-type NKX2-5-pEFSA, 0.2 μg of mutant
NKX2-5-pEFSA, 1.0 μg of ANF-luc and 0.04 μg of pGL4.75 were used.
Firefly luciferase and Renilla luciferase activities were
measured using the Dual-Glo luciferase assay system (Promega) 48 h
after transfection. The activity of the ANF promoter was
presented as the fold activation of firefly luciferase relative to
Renilla luciferase. Three independent experiments were
performed at minimum for wild-type and mutant NKX2-5.
Data are expressed as the means ± SD. Continuous
variables were tested for normality of distribution and the
Student’s unpaired t-test was used for the comparison of numeric
variables between 2 groups. Comparison of the categorical variables
between 2 groups was performed using Pearson’s χ2 test
or Fisher’s exact test where appropriate. A two-tailed P-value of
<0.05 was considered to indicate a statistical difference.
A total of 150 unrelated patients with lone AF were
clinically evaluated in contrast to 200 control individuals. None
of them had underlying comorbidities or traditional risk factors
for AF. There was no significant difference between the patient and
control groups in baseline characteristics including age, gender,
body mass index, blood pressure, fasting blood glucose, serum lipid
levels, left atrial dimension, left ventricular ejection fraction,
heart rate at rest, as well as lifestyle (data not shown). The
baseline clinical characteristics of the study subjects are
summarized in Table II.
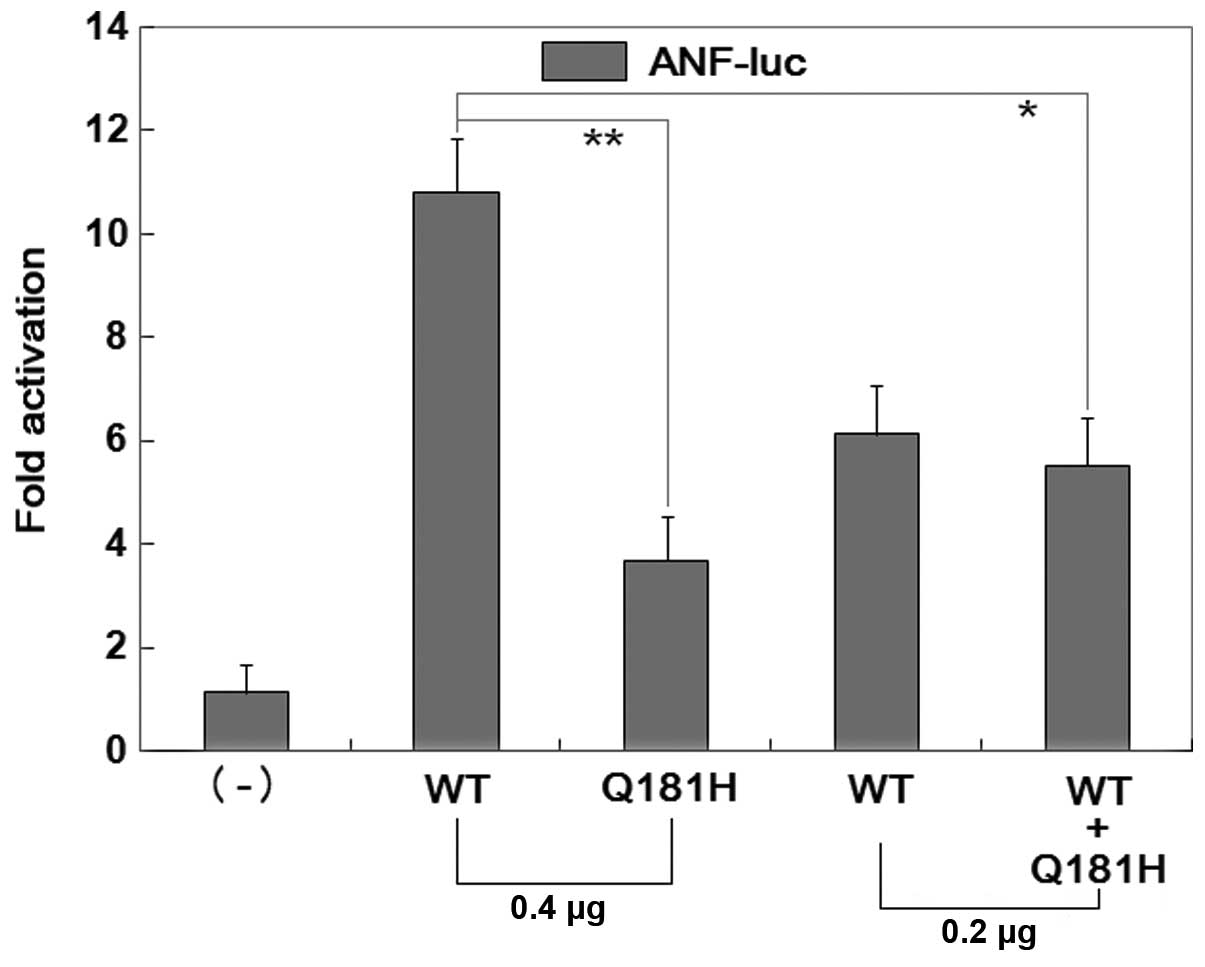
Due to unknown target genes of NKX2-6, Q181H-mutant
NKX2-5 was used as a surrogate to assess the functional sequelae of
Q175H-mutant NKX2-6. As shown in Fig.
6, the same amount (0.4 μg) of wild-type and mutant
NKX2-5 activated the ANF-luciferase (ANF) promoter by
approximately 11- and 4-fold, respectively. When the same amount of
wild-type NKX2-5 (0.2 μg) was co-transfected with mutant NKX2-5
(0.2 μg), the induced activation of the ANF promoter was
approximately 6-fold. These results demonstrate that Q181H-mutant
NKX2-5 is associated with a significantly diminished
transcriptional activity compared with its wild-type counterpart,
suggesting that the homeobox substitution Q175H results in
functional defects of NKX2-5
In order to assess the functional characteristics of
the Q175H homeodomain substitution in NKX2-6, NKX2-5 was selected
as a surrogate based on the following reasons: firstly, no
downstream genes or a binding site recognition sequence for NKX2-6
have been substantiated. Secondly, NKX2-5 and NKX2-6 are 2 members
of the NK2 family of transcription factors, and they share a highly
conserved structural motif, particularly for the homeodomain.
Thirdly, biochemical assays of NKX2-5 have been well characterized.
Finally, there are in vivo studies corroborating that NKX2-5
may compensate for the lack of NKX2-6 during embryogenesis,
therefore suggesting that some DNA binding and transcriptional
regulatory activities are shared (102–104). Consequently, functional analysis
of Q181H-mutant NKX2-5 instead of Q175H NKX2-6 revealed that the
homeodomain substitution significantly diminished transcriptional
activity on a target gene, ANF. These findings suggest that
the haploinsufficiency or dominant-negative effects caused by the
NKX2-6 mutation are potentially alternative molecular
pathological mechanisms responsible for the development of AF.
In conclusion, to the best of our knowledge, the
present study associates mutant NKX2-6 with enhanced susceptibility
to AF for the first time, providing novel insight into the
molecular pathogenesis of AF and suggesting potential strategies
for the antenatal prophylaxis and personalized treatment of AF.
The authors are really thankful to the participants
for their participation in the study. This study was supported in
part by grants from the Key Discipline Development Program of
Health Bureau of Jian-An district, Shanghai, China (JWXK201-201),
the National Natural Science Fund of China (81270161, 81370301 and
81070153) and the Natural Science Fund of Shanghai, China
(13ZR1438400).
|
1
|
Fuster V, Rydén LE, Cannom DS, Crijns HJ,
Curtis AB, Ellenbogen KA, Halperin JL, Kay GN, Le Huezey JY, Lowe
JE, Olsson SB, Prystowsky EN, Tamargo JL, Wann LS, Smith SC Jr,
Priori SG, Estes NA III, Ezekowitz MD, Jackman WM, January CT, Lowe
JE, Page RL, Slotwiner DJ, Stevenson WG, Tracy CM, Jacobs AK,
Anderson JL, Albert N, Buller CE, Creager MA, Ettinger SM, Guyton
RA, Halperin JL, Hochman JS, Kushner FG, Ohman EM, Stevenson WG,
Tarkington LG and Yancy CW: American College of Cardiology
Foundation/American Heart Association Task Force: 2011 ACCF/AHA/HRS
focused updates incorporated into the ACC/AHA/ESC 2006 guidelines
for the management of patients with atrial fibrillation: a report
of the American College of Cardiology Foundation/American Heart
Association Task Force on practice guidelines. Circulation.
123:e269–e367. 2011.
|
|
2
|
Go AS, Hylek EM, Phillips KA, Chang Y,
Henault LE, Selby JV and Singer DE: Prevalence of diagnosed atrial
fibrillation in adults: national implications for rhythm management
and stroke prevention: the AnTicoagulation and Risk Factors in
Atrial Fibrillation (ATRIA) Study. JAMA. 285:2370–2375. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lloyd-Jones DM, Wang TJ, Leip EP, Larson
MG, Levy D, Vasan RS, D’Agostino RB, Massaro JM, Beiser A, Wolf PA
and Benjamin EJ: Lifetime risk for development of atrial
fibrillation: the Framingham Heart Study. Circulation.
110:1042–1046. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Miyasaka Y, Barnes ME, Gersh BJ, Cha SS,
Bailey KR, Abhayaratna WP, Seward JB and Tsang TS: Secular trends
in incidence of atrial fibrillation in Olmsted County, Minnesota,
1980 to 2000, and implications on the projections for future
prevalence. Circulation. 114:119–125. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Benjamin EJ, Wolf PA, D’Agostino RB,
Silbershatz H, Kannel WB and Levy D: Impact of atrial fibrillation
on the risk of death: the Framingham Heart Study. Circulation.
98:946–952. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wolf PA, Abbott RD and Kannel WB: Atrial
fibrillation as an independent risk factor for stroke: the
Framingham Study. Stroke. 22:983–988. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Singh SN, Tang XC, Singh BN, Dorian P,
Reda DJ, Harris CL, Fletcher RD, Sharma SC, Atwood JE, Jacobson AK,
Lewis HD Jr, Lopez B, Raisch DW and Ezekowitz MD: SAFE-T
Investigators: Quality of life and exercise performance in patients
in sinus rhythm versus persistent atrial fibrillation: a Veterans
Affairs Cooperative Studies Program Substudy. J Am Coll Cardiol.
48:721–730. 2006. View Article : Google Scholar
|
|
8
|
Santangeli P, Di Biase L, Bai R, Mohanty
S, Pump A, Cereceda Brantes M, Horton R, Burkhardt JD, Lakkireddy
D, Reddy YM, Casella M, Dello Russo A, Tondo C and Natale A: Atrial
fibrillation and the risk of incident dementia: a meta-analysis.
Heart Rhythm. 9:1761–1768. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chao TF, Tsao HM, Ambrose K, Lin YJ, Lin
WS, Chang SL, Lo LW, Hu YF, Tuan TC, Suenari K, Li CH, Hartono B,
Chang HY, Chung FP, Hanafy DA, Lin WY and Chen SA: Renal
dysfunction and the risk of thromboembolic events in patients with
atrial fibrillation after catheter ablation - the potential role
beyond the CHA2DS2-VASc score. Heart Rhythm. 9:1755–1760. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Calvo N, Bisbal F, Guiu E, Ramos P, Nadal
M, Tolosana JM, Arbelo E, Berruezo A, Sitges M, Brugada J and Mont
L: Impact of atrial fibrillation-induced tachycardiomyopathy in
patients undergoing pulmonary vein isolation. Int J Cardiol.
168:4093–4097. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Naji F, Pagliaruzzi M, Penko M, Kanic V
and Vokac D: Changes in left ventricular filling in patients with
persistent atrial fibrilation. Int J Med Sci. 10:1876–1879. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tayebjee MH, Gilbert K, Macdonald W,
Hogarth AJ, Lewis NT and Tan LB: Atrial fibrillation reduces the
functional REServe of the heart by a fifth: a pilot FRESH-AF study.
Int J Cardiol. 168:4369–4370. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Weijs B, Pisters R, Haest RJ, Kragten JA,
Joosen IA, Versteylen M, Timmermans CC, Pison L, Blaauw Y, Hofstra
L, Nieuwlaat R, Wildberger J and Crijns HJ: Patients originally
diagnosed with idiopathic atrial fibrillation more often suffer
from insidious coronary artery disease compared to healthy sinus
rhythm controls. Heart Rhythm. 9:1923–1929. 2012. View Article : Google Scholar
|
|
14
|
Soliman EZ, Safford MM, Muntner P,
Khodneva Y, Dawood FZ, Zakai NA, Thacker EL, Judd S, Howard VJ,
Howard G, Herrington DM and Cushman M: Atrial fibrillation and the
risk of myocardial infarction. JAMA Intern Med. 174:107–114. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Marijon E, Le Heuzey JY, Connolly S, Yang
S, Pogue J, Brueckmann M, Eikelboom J, Themeles E, Ezekowitz M,
Wallentin L and Yusuf S: RE-LY Investigators: Causes of death and
influencing factors in patients with atrial fibrillation: a
competing-risk analysis from the randomized evaluation of long-term
anticoagulant therapy study. Circulation. 128:2192–2201. 2013.
View Article : Google Scholar
|
|
16
|
Go AS, Mozaffarian D, Roger VL, Benjamin
EJ, Berry JD, Blaha MJ, Dai S, Ford ES, Fox CS, Franco S, Fullerton
HJ, Gillespie C, Hailpern SM, Heit JA, Howard VJ, Huffman MD, Judd
SE, Kissela BM, Kittner SJ, Lackland DT, Lichtman JH, Lisabeth LD,
Mackey RH, Magid DJ, Marcus GM, Marelli A, Matchar DB, McGuire DK,
Mohler ER III, Moy CS, Mussolino ME, Neumar RW, Nichol G, Pandey
DK, Paynter NP, Reeves MJ, Sorlie PD, Stein J, Towfighi A, Turan
TN, Virani SS, Wong ND, Woo D and Turner MB: American Heart
Association Statistics Committee and Stroke Statistics
Subcommittee: Heart disease and stroke statistics - 2014 update: a
report from the American Heart Association. Circulation.
129:e28–e292. 2014. View Article : Google Scholar
|
|
17
|
Ball J, Carrington MJ, McMurray JJ and
Stewart S: Atrial fibrilation: profile and burden of an evolving
epidemic in the 21st century. Int J Cardiol. 67:1807–1824. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Coyne KS, Paramore C, Grandy S, Mercader
M, Reynolds M and Zimetbaum P: Assessing the direct costs of
treating nonvalvular atrial fibrillation in the United States.
Value Health. 9:348–356. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Nattel S: New ideas about atrial
fibrillation 50 years on. Nature. 415:219–226. 2002.PubMed/NCBI
|
|
20
|
Abed HS, Samuel CS, Lau DH, Kelly DJ,
Royce SG, Alasady M, Mahajan R, Kuklik P, Zhang Y, Brooks AG,
Nelson AJ, Worthley SG, Abhayaratna WP, Kalman JM, Wittert GA and
Sanders P: Obesity results in progressive atrial structural and
electrical remodeling: implications for atrial fibrillation. Heart
Rhythm. 10:90–100. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Naruse Y, Tada H, Satoh M, Yanagihara M,
Tsuneoka H, Hirata Y, Ito Y, Kuroki K, Machino T, Yamasaki H,
Igarashi M, Sekiguchi Y, Sato A and Aonuma K: Concomitant
obstructive sleep apnea increases the recurrence of atrial
fibrillation following radiofrequency catheter ablation of atrial
fibrillation: clinical impact of continuous positive airway
pressure therapy. Heart Rhythm. 10:331–337. 2013. View Article : Google Scholar
|
|
22
|
Chao TF, Hung CL, Chen SJ, Wang KL, Chen
TJ, Lin YJ, Chang SL, Lo LW, Hu YF, Tuan TC and Chen SA: The
association between hyperuricemia, left atrial size and new-onset
atrial fibrillation. Int J Cardiol. 168:4027–4032. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Andrade J, Khairy P, Dobrev D and Nattel
S: The clinical profile and pathophysiology of atrial fibrillation:
relationships among clinical features, epidemiology, and
mechanisms. Circ Res. 114:1453–1468. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fox CS, Parise H, D’Agostino RB Sr,
Lloyd-Jones DM, Vasan RS, Wang TJ, Levy D, Wolf P and Benjamin EJ:
Parental atrial fibrillation as a risk factor for atrial
fibrillation in offspring. JAMA. 291:2851–2855. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ellinor PT, Yoerger DM, Ruskin JN and
MacRae CA: Familial aggregation in lone atrial fibrillation. Hum
Genet. 118:179–184. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Arnar DO, Thorvaldsson S, Manolio TA,
Thorgeirsson G, Kristjansson K, Hakonarson H and Stefansson K:
Familial aggregation of atrial fibrillation in Iceland. Eur Heart
J. 27:708–712. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Christophersen IE, Ravn LS,
Budtz-Joergensen E, Skytthe A, Haunsoe S, Svendsen JH and
Christensen K: Familial aggregation of atrial fibrillation: a study
in Danish twins. Circ Arrhythm Electrophysiol. 2:378–383. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yang YQ, Zhang XL, Wang XH, Tan HW, Shi
HF, Fang WY and Liu X: Familial aggregation of lone atrial
fibrillation in Chinese population. Intern Med. 49:2385–2391. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lubitz SA, Yin X, Fontes JD, Magnani JW,
Rienstra M, Pai M, Villalon ML, Vasan RS, Pencina MJ, Levy D,
Larson MG, Ellinor PT and Benjamin EJ: Association between familial
atrial fibrillation and risk of new-onset atrial fibrillation.
JAMA. 304:2263–2269. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zöller B, Ohlsson H, Sundquist J and
Sundquist K: High familial risk of atrial fibrillation/atrial
flutter in multiplex families: a nationwide family study in Sweden.
J Am Heart Assoc. 2:e0033842012.PubMed/NCBI
|
|
31
|
Oyen N, Ranthe MF, Carstensen L, Boyd HA,
Olesen MS, Olesen SP, Wohlfahrt J and Melbye M: Familial
aggregation of lone atrial fibrillation in young persons. J Am Coll
Cardiol. 60:917–921. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Brugada R, Tapscott T, Czernuszewicz GZ,
Marian AJ, Iglesias A, Mont L, Brugada J, Girona J, Domingo A,
Bachinski LL and Roberts R: Identification of a genetic locus for
familial atrial fibrillation. N Engl J Med. 336:905–911. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chen YH, Xu SJ, Bendahhou S, Wang XL, Wang
Y, Xu WY, Jin HW, Sun H, Su XY, Zhuang QN, Yang YQ, Li YB, Liu Y,
Xu HJ, Li XF, Ma N, Mou CP, Chen Z, Barhanin J and Huang W: KCNQ1
gain-of-function mutation in familial atrial fibrillation. Science.
299:251–254. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ellinor PT, Shin JT, Moore RK, Yoerger DM
and MacRae CA: Locus for atrial fibrillation maps to chromosome
6q14-16. Circulation. 107:2880–2883. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Oberti C, Wang L, Li L, Dong J, Rao S, Du
W and Wang Q: Genome-wide linkage scan identifies a novel genetic
locus on chromosome 5p13 for neonatal atrial fibrillation
associated with sudden death and variable cardiomyopathy.
Circulation. 110:3753–3759. 2004. View Article : Google Scholar
|
|
36
|
Volders PG, Zhu Q, Timmermans C, Eurlings
PM, Su X, Arens YH, Li L, Jongbloed RJ, Xia M, Rodriguez LM and
Chen YH: Mapping a novel locus for familial atrial fibrillation on
chromosome 10p11-q21. Heart Rhythm. 4:469–475. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Darbar D, Hardy A, Haines JL and Roden DM:
Prolonged signal-averaged P-wave duration as an intermediate
phenotype for familial atrial fibrillation. J Am Coll Cardiol.
51:1083–1089. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Olesen MS, Bentzen BH, Nielsen JB,
Steffensen AB, David JP, Jabbari J, Jensen HK, Haunsø S, Svendsen
JH and Schmitt N: Mutations in the potassium channel subunit KCNE1
are associated with early-onset familial atrial fibrillation. BMC
Med Genet. 13:242012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yang Y, Xia M, Jin Q, Bendahhou S, Shi J
and Chen Y, Liang B, Lin J, Liu Y, Liu B, Zhou Q, Zhang D, Wang R,
Ma N, Su X, Niu K, Pei Y, Xu W, Chen Z, Wan H, Cui J, Barhanin J
and Chen Y: Identification of a KCNE2 gain-of-function mutation in
patients with familial atrial fibrillation. Am J Hum Genet.
75:899–905. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
40
|
Nielsen JB, Bentzen BH, Olesen MS, David
JP, Olesen SP, Haunsø S, Svendsen JH and Schmitt N:
Gain-of-function mutations in potassium channel subunit KCNE2
associated with early-onset lone atrial fibrillation. Biomark Med.
8:557–570. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lundby A, Ravn LS, Svendsen JH, Hauns S,
Olesen SP and Schmitt N: KCNE3 mutation V17M identified in a
patient with lone atrial fibrillation. Cell Physiol Biochem.
21:47–54. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zeng Z, Tan C, Teng S, Chen J, Su S, Zhou
X, Wang F, Zhang S, Gu D, Makielski JC and Pu J: The single
nucleotide polymorphisms of I(Ks) potassium channel genes and their
association with atrial fibrillation in a Chinese population.
Cardiology. 108:97–103. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ravn LS, Aizawa Y, Pollevick GD,
Hofman-Bang J, Cordeiro JM, Dixen U, Jensen G, Wu Y, Burashnikov E,
Haunso S, Guerchicoff A, Hu D, Svendsen JH, Christiansen M and
Antzelevitch C: Gain of function in IKs secondary to a mutation in
KCNE5 associated with atrial fibrillation. Heart Rhythm. 5:427–435.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Hong K, Bjerregaard P, Gussak I and
Brugada R: Short QT syndrome and atrial fibrillation caused by
mutation in KCNH2. J Cardiovasc Electrophysiol. 16:394–396. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Sinner MF, Pfeufer A, Akyol M, Beckmann
BM, Hinterseer M, Wacker A, Perz S, Sauter W, Illig T, Näbauer M,
Schmitt C, Wichmann HE, Schömig A, Steinbeck G, Meitinger T and
Kääb S: The non-synonymous coding IKr-channel variant KCNH2-K897T
is associated with atrial fibrillation: results from a systematic
candidate gene-based analysis of KCNH2 (HERG). Eur Heart J.
29:907–914. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Olson TM, Alekseev AE, Liu XK, Park S,
Zingman LV, Bienengraeber M, Sattiraju S, Ballew JD, Jahangir A and
Terzic A: Kv1.5 channelopathy due to KCNA5 loss-of-function
mutation causes human atrial fibrillation. Hum Mol Genet.
15:2185–2191. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Yang Y, Li J, Lin X, Yang Y, Hong K, Wang
L, Liu J, Li L, Yan D, Liang D, Xiao J, Jin H, Wu J, Zhang Y and
Chen YH: Novel KCNA5 loss-of-function mutations responsible for
atrial fibrillation. J Hum Genet. 54:277–283. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Christophersen IE, Olesen MS, Liang B,
Andersen MN, Larsen AP, Nielsen JB, Haunsø S, Olesen SP, Tveit A,
Svendsen JH and Schmitt N: Genetic variation in KCNA5: impact on
the atrial-specific potassium current IKur in patients with lone
atrial fibrillation. Eur Heart J. 34:1517–1525. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Olesen MS, Refsgaard L, Holst AG, Larsen
AP, Grubb S, Haunsø S, Svendsen JH, Olesen SP, Schmitt N and Calloe
K: A novel KCND3 gain-of-function mutation associated with
early-onset of persistent lone atrial fibrillation. Cardiovasc Res.
98:488–495. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Xia M, Jin Q, Bendahhou S, He Y, Larroque
MM and Chen Y, Zhou Q, Yang Y, Liu Y, Liu B, Zhu Q, Zhou Y, Lin J,
Liang B, Li L, Dong X, Pan Z, Wang R, Wan H, Qiu W, Xu W, Eurlings
P, Barhanin J and Chen Y: A Kir2.1 gain-of-function mutation
underlies familial atrial fibrillation. Biochem Biophys Res Commun.
332:1012–1019. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Delaney JT, Muhammad R, Blair MA, Kor K,
Fish FA, Roden DM and Darbar D: A KCNJ8 mutation associated with
early repolarization and atrial fibrillation. Europace.
14:1428–1432. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Gollob MH, Jones DL, Krahn AD, Danis L,
Gong XQ, Shao Q, Liu X, Veinot JP, Tang AS, Stewart AF, Tesson F,
Klein GJ, Yee R, Skanes AC, Guiraudon GM, Ebihara L and Bai D:
Somatic mutations in the connexin 40 gene (GJA5) in atrial
fibrillation. N Engl J Med. 354:2677–2688. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Sun Y, Yang YQ, Gong XQ, Wang XH, Li RG,
Tan HW, Liu X, Fang WY and Bai D: Novel germline GJA5/connexin40
mutations associated with lone atrial fibrillation impair gap
junctional intercellular communication. Hum Mutat. 34:603–609.
2013.PubMed/NCBI
|
|
54
|
Shi HF, Yang JF, Wang Q, Li RG, Xu YJ, Qu
XK, Fang WY, Liu X and Yang YQ: Prevalence and spectrum of GJA5
mutations associated with lone atrial fibrillation. Mol Med Rep.
7:767–774. 2013.PubMed/NCBI
|
|
55
|
Thibodeau IL, Xu J, Li Q, Liu G, Lam K,
Veinot JP, Birnie DH, Jones DL, Krahn AD, Lemery R, Nicholson BJ
and Gollob MH: Paradigm of genetic mosaicism and lone atrial
fibrillation: physiological characterization of a connexin
43-deletion mutant identified from atrial tissue. Circulation.
122:236–244. 2010. View Article : Google Scholar
|
|
56
|
Hodgson-Zingman DM, Karst ML, Zingman LV,
Heublein DM, Darbar D, Herron KJ, Ballew JD, de Andrade M, Burnett
JC Jr and Olson TM: Atrial natriuretic peptide frameshift mutation
in familial atrial fibrillation. N Engl J Med. 359:158–165. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Darbar D, Kannankeril PJ, Donahue BS,
Kucera G, Stubblefield T, Haines JL, George AL Jr and Roden DM:
Cardiac sodium channel (SCN5A) variants associated with atrial
fibrillation. Circulation. 117:1927–1935. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Watanabe H, Darbar D, Kaiser DW,
Jiramongkolchai K, Chopra S, Donahue BS, Kannankeril PJ and Roden
DM: Mutations in sodium channel β1- and β2-subunits associated with
atrial fibrillation. Circ Arrhythm Electrophysiol. 2:268–275.
2009.
|
|
59
|
Olesen MS, Holst AG, Svendsen JH, Haunsø S
and Tfelt-Hansen J: SCN1Bb R214Q found in 3 patients: 1 with
Brugada syndrome and 2 with lone atrial fibrillation. Heart Rhythm.
9:770–773. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Wang P, Yang Q, Wu X, Yang Y, Shi L, Wang
C, Wu G, Xia Y, Yang B, Zhang R, Xu C, Cheng X, Li S, Zhao Y, Fu F,
Liao Y, Fang F, Chen Q, Tu X and Wang QK: Functional
dominant-negative mutation of sodium channel subunit gene SCN3B
associated with atrial fibrillation in a Chinese GeneID population.
Biochem Biophys Res Commun. 398:98–104. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Li RG, Wang Q, Xu YJ, Zhang M, Qu XK, Liu
X, Fang WY and Yang YQ: Mutations of the SCN4B-encoded sodium
channel β4 subunit in familial atrial fibrillation. Int J Mol Med.
32:144–150. 2013.
|
|
62
|
Olesen MS, Andreasen L, Jabbari J,
Refsgaard L, Haunsø S, Olesen SP, Nielsen JB, Schmitt N and
Svendsen JH: Very early-onset lone atrial fibrillation patients
have a high prevalence of rare variants in genes previously
associated with atrial fibrillation. Heart Rhythm. 11:246–251.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Weeke P, Parvez B, Blair M, Short L,
Ingram C, Kucera G, Stubblefield T, Roden DM and Darbar D:
Candidate gene approach to identifying rare genetic variants
associated with lone atrial fibrillation. Heart Rhythm. 11:46–52.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Mommersteeg MT, Christoffels VM, Anderson
RH and Moorman AF: Atrial fibrillation: a developmental point of
view. Heart Rhythm. 6:1818–1824. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Mahida S: Transcription factors and atrial
fibrillation. Cardiovasc Res. 101:194–202. 2014. View Article : Google Scholar
|
|
66
|
Schott JJ, Benson DW, Basson CT, Pease W,
Silberbach GM, Moak JP, Maron BJ, Seidman CE and Seidman JG:
Congenital heart disease caused by mutations in the transcription
factor NKX2-5. Science. 281:108–111. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Reamon-Buettner SM and Borlak J: NKX2-5:
an update on this hypermutable homeodomain protein and its role in
human congenital heart disease (CHD). Hum Mutat. 31:1185–1194.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Costa MW, Guo G, Wolstein O, Vale M,
Castro ML, Wang L, Otway R, Riek P, Cochrane N, Furtado M,
Semsarian C, Weintraub RG, Yeoh T, Hayward C, Keogh A, Macdonald P,
Feneley M, Graham RM, Seidman JG, Seidman CE, Rosenthal N, Fatkin D
and Harvey RP: Functional characterization of a novel mutation in
NKX2-5 associated with congenital heart disease and adult-onset
cardiomyopathy. Circ Cardiovasc Genet. 6:238–247. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Garg V, Kathiriya IS, Barnes R,
Schluterman MK, King IN, Butler CA, Rothrock CR, Eapen RS,
Hirayama-Yamada K, Joo K, Matsuoka R, Cohen JC and Srivastava D:
GATA4 mutations cause human congenital heart defects and reveal an
interaction with TBX5. Nature. 424:443–447. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Zhou P, He A and Pu WT: Regulation of
GATA4 transcriptional activity in cardiovascular development and
disease. Curr Top Dev Biol. 100:143–169. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Yang YQ, Wang J, Liu XY, Chen XZ, Zhang W,
Wang XZ, Liu X and Fang WY: Novel GATA4 mutations in patients with
congenital ventricular septal defects. Med Sci Monit.
18:CR344–CR350. 2012.PubMed/NCBI
|
|
72
|
Yang YQ, Wang J, Liu XY, Chen XZ, Zhang W
and Wang XZ: Mutation spectrum of GATA4 associated with congenital
atrial septal defects. Arch Med Sci. 9:976–983. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Yang YQ, Gharibeh L, Li RG, Xin YF, Wang
J, Liu ZM, Qiu XB, Xu YJ, Xu L, Qu XK, Liu X, Fang WY, Huang RT,
Xue S and Nemer G: GATA4 loss-of-function mutations underlie
familial tetralogy of fallot. Hum Mutat. 34:1662–1671. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Li RG, Li L, Qiu XB, Yuan F, Xu L, Li X,
Xu YJ, Jiang WF, Jiang JQ, Liu X, Fang WY, Zhang M, Peng LY, Qu XK
and Yang YQ: GATA4 loss-of-function mutation underlies familial
dilated cardiomyopathy. Biochem Biophys Res Commun. 439:591–596.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Zhao L, Xu JH, Xu WJ, Yu H, Wang Q, Zheng
HZ, Jiang WF, Jiang JF and Yang YQ: A novel GATA4 loss-of-function
mutation responsible for familial dilated cardiomyopathy. Int J Mol
Med. 33:654–660. 2014.PubMed/NCBI
|
|
76
|
Jiang JQ, Li RG, Wang J, Liu XY, Xu YJ,
Fang WY, Chen XZ, Zhang W, Wang XZ and Yang YQ: Prevalence and
spectrum of GATA5 mutations associated with congenital heart
disease. Int J Cardiol. 165:570–573. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Wei D, Bao H, Zhou N, Zheng GF, Liu XY and
Yang YQ: GATA5 loss-of-function mutation responsible for the
congenital ventriculoseptal defect. Pediatr Cardiol. 34:504–511.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Wei D, Bao H, Liu XY, Zhou N, Wang Q, Li
RG, Xu YJ and Yang YQ: GATA5 loss-of-function mutations underlie
tetralogy of fallot. Int J Med Sci. 10:34–42. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Huang RT, Xue S, Xu YJ, Zhou M and Yang
YQ: Somatic GATA5 mutations in sporadic tetralogy of Fallot. Int J
Mol Med. 33:1227–1235. 2014.PubMed/NCBI
|
|
80
|
Shi LM, Tao JW, Qiu XB, Wang J, Yuan F, Xu
L, Liu H, Li RG, Xu YJ, Wang Q, Zheng HZ, Li X, Wang XZ, Zhang M,
Qu XK and Yang YQ: GATA5 loss-of-function mutations associated with
congenital bicuspid aortic valve. Int J Mol Med. 33:1219–1226.
2014.PubMed/NCBI
|
|
81
|
Kodo K, Nishizawa T, Furutani M, Arai S,
Yamamura E, Joo K, Takahashi T, Matsuoka R and Yamagishi H: GATA6
mutations cause human cardiac outflow tract defects by disrupting
semaphorin-plexin signaling. Proc Natl Acad Sci USA.
106:13933–13938. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Zheng GF, Wei D, Zhao H, Zhou N, Yang YQ
and Liu XY: A novel GATA6 mutation associated with congenital
ventricular septal defect. Int J Mol Med. 29:1065–1071.
2012.PubMed/NCBI
|
|
83
|
Wang J, Luo XJ, Xin YF, Liu Y, Liu ZM,
Wang Q, Li RG, Fang WY, Wang XZ and Yang YQ: Novel GATA6 mutations
associated with congenital ventricular septal defect or tetralogy
of fallot. DNA Cell Biol. 31:1610–1617. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Huang RT, Xue S, Xu YJ and Yang YQ:
Somatic mutations in the GATA6 gene underlie sporadic tetralogy of
Fallot. Int J Mol Med. 31:51–58. 2013.PubMed/NCBI
|
|
85
|
Yuan F, Zhao L, Wang J, Zhang W, Li X, Qiu
XB, Li RG, Xu YJ, Xu L, Qu XK, Fang WY and Yang YQ: PITX2c
loss-of-function mutations responsible for congenital atrial septal
defects. Int J Med Sci. 10:1422–1429. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Wang J, Xin YF, Xu WJ, Liu ZM, Qiu XB, Qu
XK, Xu L, Li X and Yang YQ: Prevalence and spectrum of PITX2c
mutations associated with congenital heart disease. DNA Cell Biol.
32:708–16. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Wei D, Gong XH, Qiu G, Wang J and Yang YQ:
Novel PITX2c loss-of-function mutations associated with complex
congenital heart disease. Int J Mol Med. 33:1201–1208.
2014.PubMed/NCBI
|
|
88
|
Huang RT, Xue S, Xu YJ, Zhou M and Yang
YQ: A novel NKX2.5 loss-of-function mutation responsible for
familial atrial fibrillation. Int J Mol Med. 31:1119–1126.
2013.PubMed/NCBI
|
|
89
|
Xie WH, Chang C, Xu YJ, Li RG, Qu XK, Fang
WY, Liu X and Yang YQ: Prevalence and spectrum of Nkx2.5 mutations
associated with idiopathic atrial fibrillation. Clinics (Sao
Paulo). 68:777–784. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Yu H, Xu JH, Song HM, Zhao L, Xu WJ, Wang
J, Li RG, Xu L, Jiang WF, Qiu XB, Jiang JQ, Qu XK, Liu X, Fang WY,
Jiang JF and Yang YQ: Mutational spectrum of the NKX2-5 gene in
patients with lone atrial fibrillation. Int J Med Sci. 11:554–563.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Wang J, Sun YM and Yang YQ: Mutation
spectrum of the GATA4 gene in patients with idiopathic atrial
fibrillation. Mol Biol Rep. 39:8127–8135. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Yang YQ, Wang J, Wang XH, Wang Q, Tan HW,
Zhang M, Shen FF, Jiang JQ, Fang WY and Liu X: Mutational spectrum
of the GATA5 gene associated with familial atrial fibrillation. Int
J Cardiol. 157:305–307. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Wang XH, Huang CX, Wang Q, Li RG, Xu YJ,
Liu X, Fang WY and Yang YQ: A novel GATA5 loss-of-function mutation
underlies lone atrial fibrillation. Int J Mol Med. 31:43–50.
2013.PubMed/NCBI
|
|
94
|
Gu JY, Xu JH, Yu H and Yang YQ: Novel
GATA5 loss-of-function mutations underlie familial atrial
fibrillation. Clinics (Sao Paulo). 67:1393–1399. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Yang YQ, Wang XH, Tan HW, Jiang WF, Fang
WY and Liu X: Prevalence and spectrum of GATA6 mutations associated
with familial atrial fibrillation. Int J Cardiol. 155:494–496.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Yang YQ, Li L, Wang J, Zhang XL, Li RG, Xu
YJ, Tan HW, Wang XH, Jiang JQ, Fang WY and Liu X: GATA6
loss-of-function mutation in atrial fibrillation. Eur J Med Genet.
55:520–526. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Li J, Liu WD, Yang ZL and Yang YQ: Novel
GATA6 loss-of-function mutation responsible for familial atrial
fibrillation. Int J Mol Med. 30:783–790. 2012.PubMed/NCBI
|
|
98
|
Yang YQ, Xu YJ, Li RG, Qu XK, Fang WY and
Liu X: Prevalence and spectrum of PITX2c mutations associated with
familial atrial fibrillation. Int J Cardiol. 168:2873–2876. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Zhou YM, Zheng PX, Yang YQ, Ge ZM and Kang
WQ: A novel PITX2c loss-of-function mutation underlies lone atrial
fibrillation. Int J Mol Med. 32:827–834. 2013.PubMed/NCBI
|
|
100
|
Wang J, Zhang DF, Sun YM and Yang YQ: A
novel PITX2c loss-of-function mutation associated with familial
atrial fibrillation. Eur J Med Genet. 57:25–31. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Qiu XB, Xu YJ, Li RG, Xu L, Liu X, Fang
WY, Yang YQ and Qu XK: PITX2C loss-of-function mutations
responsible for idiopathic atrial fibrillation. Clinics (Sao
Paulo). 69:15–22. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Biben C, Hatzistavrou T and Harvey RP:
Expression of NK-2 class homeobox gene Nkx2-6 in foregut endoderm
and heart. Mech Dev. 73:125–127. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Tanaka M, Schinke M, Liao HS, Yamasaki N
and Izumo S: Nkx2.5 and Nkx2.6, homologs of Drosophila
tinman, are required for development of the pharynx. Mol Cell Biol.
21:4391–4398. 2001. View Article : Google Scholar
|
|
104
|
Heathcote K, Braybrook C, Abushaban L, Guy
M, Khetyar ME, Patton MA, Carter ND, Scambler PJ and Syrris P:
Common arterial trunk associated with a homeodomain mutation of
NKX2.6. Hum Mol Genet. 14:585–593. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Ta-Shma A, El-lahham N, Edvardson S,
Stepensky P, Nir A, Perles Z, Gavri S, Golender J,
Yaakobi-Simhayoff N, Shaag A, Rein AJ and Elpeleg O: Conotruncal
malformations and absent thymus due to a deleterious NKX2-6
mutation. J Med Genet. 51:268–270. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Pradhan L, Genis C, Scone P, Weinberg EO,
Kasahara H and Nam HJ: Crystal structure of the human NKX2.5
homeodomain in complex with DNA target. Biochemistry. 51:6312–6319.
2012. View Article : Google Scholar : PubMed/NCBI
|