Introduction
Prion diseases or transmissible spongiform
encephalopathies (TSEs) are a group of fatal and infectious
neurodegenerative diseases that can affect humans and animals,
including Creutzfeldt-Jakob disease (CJD),
Gerstmann-Sträussler-Scheinker syndrome (GSS), fatal familial
insomnia (FFI), Kuru in humans, bovine spongiform encephalopathy
(BSE) in cattle, scrapie in sheep and goats, transmissible mink
encephalopathy (TME) in mink and chronic wasting disease (CWD) in
elk and deer (1). The pathogen
for these diseases is believed to be prion, whose infectious
isoform termed PrPSc is derived from the cellular
isoform termed PrPC (2,3).
The conversion from PrPC to PrPSc is the
central event in prion diseases which may occur spontaneously and
can be acquired or induced by autosomal dominant mutations of the
PRNP gene (4).
Nitric oxide (NO) and other reactive nitrogen
species (RNS) are biologically active small molecules involved in
the pathogeneses of a series of neurodegenerative diseases
(5). The transfer of a NO group
to cysteine sulfhydryls on proteins is known as
S-nitrosylation. These S-nitrosylated proteins are
thus referred to as SNO proteins (6–8).
Similar to other post-translational modifications,
S-nitrosylation can activate or inhibit the activity of the
target protein, alter protein interactions with other molecules and
affect protein aggregation or localization under physiological
conditions, thus influencing or balancing cell signal transduction
pathways and cellular functions in a number of biological processes
(9). Under pathological
conditions, aberrant S-nitrosylation can occur in response
to nitrosative stress and may stimulate cell destructive processes,
contributing to neurodegeneration through the disruption of a
number of pathways (5).
Furthermore, the temporal and spatial dynamics of SNO proteins may
critically affect their modulatory role in response to nitrosative
stress. For instance, in the brains of patients with Alzheimer’s
disease (AD), S-nitrosylated dynamin-related protein
(SNO-Drp1) is related to the formation of β-amyloid and subsequent
mitochondrial fission activation (10,11). S-nitrosylated Parkin
(SNO-Parkin) and S-nitrosylated peroxiredoxin 2 (SNO-Prx2)
are associated with the pathogenesis of sporadic Parkinson’s
disease (PD), in which SNO-Parkin causes proteasomal dysfunction
(12), and SNO-Prx2 promotes
oxidative stress-induced neuronal cell death (13). In addition, a number of biological
proteins involved in cellular apoptosis [glyceraldehyde-3-phosphate
dehydrogenase (GAPDH)-Siah, X-linked inhibitor of apoptosis (XIAP)
and caspase-3], phosphorylation [phosphatase with sequence homology
to tensin (PTEN)], neuroinflammation [cyclooxygenase-2 (COX-2)] and
autophagy [c-Jun N-terminal kinase (JNK)1 and IκB kinase (IKKβ)]
are also S-nitrosylated (14). In addition to AD and PD (10,15–17), we have also reported that the
expression levels of some SNO proteins, such as
S-nitrosylated protein disulphide isomerase (SNO-PDI)
(18) and 14-3-3 (19), are abnormally increased in the
brains of scrapie-infected rodents at the terminal stage,
highlighting the significance of SNO proteins in prion
diseases.
Greatly improved proteomics technologies coupled
with bioinformatics provide the potential for hundreds of proteins
to be discovered and verified experimentally (20–23), which makes it possible to analyze
the S-nitrosoproteome for prion diseases. Since the
S-nitrosylated isoform may usually occupy only a small
fraction of the relevant protein at a given time, methods to
effectively enrich SNO proteins are in great need. Although studies
have dealt with SNO protein isolation and enrichment through
multiple methods (22,24–26), direct usage in mass spectrometry
(MS)-based proteomics remains quite problematic. In this study, we
describe an optimized protocol for the isolation and enrichment of
SNO proteins from brain tissue, based on a commercial SNO protein
detection assay kit. Several essential parameters were carefully
evaluated and optimized. We also provide evidence that SNO proteins
isolated using such a protocol from the brain tissues of humans
with various prion diseases can be used in further assays, such as
Western blot analysis and iTRAQ-based proteomics.
Materials and methods
Antibodies
The following antibodies were used in this study,
including anti-actin monoclonal antibody (mAb) (Thermo Fisher
Scientific, Rockford, IL, USA), anti-14-3-3 polyclonal antibody
(pAb), anti-glycogen synthase kinase (GSK)-3β pAb, anti-heat shock
protein 27 (Hsp27) mAb and anti-sirtuin 1 (Sirt1) pAb (all from
Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA), anti-GAPDH
and horseradish peroxidase (HRP)-conjugated anti-rabbit or
anti-mouse immunoglobulin G (Thermo Fisher Scientific).
Preparation of brain tissue samples
The stored frozen brain tissue from normal healthy
hamsters (n=3) and hamsters inoculated intracerebrally with
hamster-adapted scrapie agent 263K (n=3), as well as the
post-mortem cortex and cerebellum of patients with sporadic CJD
(sCJD, n=1), FFI (n=3), G114V genetic CJD (G114V gCJD, n=1) and a
healthy subject (n=1) were enrolled in this study. Brain
homogenates (10%, w/v) were prepared based on a previously
described protocol (27).
Briefly, brain tissues were homogenized in lysis buffer (100 mM
NaCl, 10 mM EDTA, 0.5% Nonidet P-40, 0.5% sodium deoxycholate, 10
mM Tris, pH 7.5) containing 1% protease inhibitor cocktail (Abcam,
Cambridge, MA, USA). The tissue debris was removed by low-speed
centrifugation at 2,000 × g for 10 min and the supernatants were
collected for further analysis.
Use of the biotin-switch technique (BST)
for the analysis of the S-nitrosylation of proteins
SNO proteins in the brain tissue were determined
using a commercial SNO protein detection assay kit (Cayman
Chemical, Inc., Ann Arbor, MI, USA) following the manufacturer’s
instructions. Briefly, all tested samples were normalized to 100 mg
(approximately 1 ml 10% brain homogenate/sample) total brain
proteins and transferred to 1.5-ml Eppendorf tubes. The brain
samples were homogenized in 0.5 ml buffer A containing blocking
reagent (thiol-blocking reagent at 2 mg/ml). Following incubation
at 4°C for 30 min in the dark, the samples were clarified by
centrifugation at 4°C for 10 min. The supernatants were transferred
to 2-ml tubes and mixed with pre-cooling acetone (1:4, v/v), and
total proteins were precipitated at −20°C for 2 h. Following
centrifugation at 4°C for 15 min, the pellet was resuspended in 0.5
ml buffer B containing reducing and labeling reagents, and further
incubated at room temperature for 2 h on a rocker with gentle
agitation. Acetone precipitation was conducted again as described
above. The protein pellet was thoroughly dissolved in dissolving
buffer (8 M urea, 30 mM HEPES, 10 mM DTT, 2 mM EDTA and 1 mM PMSF,
pH 8.2) at the same volume as the brain homogenates initially used.
The products were stored for further analysis.
Binding to streptavidin beads
To pull-down SNO proteins, streptavidin-conjugated
magnetic beads (2.8 μm, Dynal magnetic beads; Invitrogen Life
Technologies Corporation, Carlsbad, CA, USA) were used according to
the manufacturer’s instructions. Briefly, the beads were washed 3
times with phosphate-buffered saline (PBS, pH 7.4) containing 0.01%
(v/v) Tween-20 and added to the samples mentioned above. The
mixture was rotated at room temperature for 1 h. Bound beads were
washed 5 times in PBS containing 0.1% bovine serum albumin (BSA)
(w/v) and then resuspended in the same volume of PBS as the initial
volume of beads used.
Optimization of the working conditions
for elution
Four different elution buffers with different
elution conditions were tested for releasing SNO proteins from
streptavidin beads: i) incubation in non-ionic water at 75°C for 5
min as previously described by Holmberg et al (28); ii) incubation in 10 mM EDTA pH 8.2
with 95% formamide at 90°C for 2 min as described in the
instructions provided with the Dynabeads® M-280 (Life
Technologies); iii) incubation in HEPES elution buffer (20 mM
HEPES-NaOH, 100 mM NaCl, 1 mM EDTA, 100 mM 2-ME, pH 7.7) at room
temperature for 20 min as previously described by Xu et al
(29); iv) incubation with
various concentrations of sodium dodecyl sulfate (SDS; 0.1, 0.2,
0.5 and 1% SDS) at 100°C for 5 min. SNO proteins eluted with 1.0 ml
of various elution buffers were precipitated with acetone at −20°C
for 2 h. Following centrifugation at 10,000 × g at 4°C for 15 min,
the pellets were resuspended with dissolving buffer at the same
volume as the brain homogenates initially used.
Optimization of the working conditions
for incubation
Three different bead/homogenate ratios (v/v) were
tested, including 1:6, 1:3 and 1:1. In addition, 3 different
incubation times at 2 incubation temperatures were evaluated: 4°C
for 30, 60 or 120 min; and 25°C for 30, 60 or 120 min. All other
experimental conditions were maintained invariable.
Determination of protein
concentration
The protein concentration of the redissolved
products was determined in triplicate using the bicinchoninic acid
(BCA) protein assay kit (Thermo Fisher Scientific) according to the
manufacturer’s instructions.
Western blot analysis
SNO protein samples were mixed with 5X loading
buffer and boiled for 8 min. Proteins were separated in 15%
SDS-polyacrylamide gel electrophoresis (PAGE) and transferred onto
nitrocellulose (NC) membranes (Whatman, Pittsburgh, PA, USA) by the
semi-dry method in transfer buffer and immunoblotted with
anti-biotin antibody (Cayman Chemical, Inc.). Individual SNO
proteins were measured by specific antibodies, including
anti-14-3-3 used at 1:1,000, anti-GSK-3β used at 1:1,500,
anti-Hsp27 used at 1:2,000, anti-Sirt1 used at 1:800, anti-β-actin
used at 1:5,000 and anti-GAPDH antibodies used at 1:2,000. Reactive
signals were visualized using an enhanced chemiluminescence (ECL)
kit (Amersham-Pharmacia Biotech, Piscataway, NJ, USA).
Dot blot for the detection of SNO
proteins
One microliter of the isolated SNO proteins was
dotted onto NC strips, which were allowed to dry at room
temperature. The strips were blocked with 2% BSA (w/v) in
Tris-buffered saline containing 0.5% Tween-20 (TBST, w/v) and
incubated overnight at 4°C to block the residual binding sites on
the paper. Subsequently, the strips were further incubated for 1 h
at 37°C with anti-biotin antibody diluted 1:1,000 in 2% BSA. The
strips were washed 3 times with TBST and reactive signals were
visualized using an ECL kit.
iTRAQ labeling and MS analysis
iTRAQ labeling for human brain specimens was carried
out using the iTRAQ® Reagent-8Plex kit (AB Sciex,
Framingham, MA, USA) according to the manufacturer’s instructions.
The purified SNO proteins of the cerebellum and cortex from the
normal control were labeled with iTRAQ labeling reagent 113 and
117, while those of the cerebellum and cortex from the patients
with sCJD, FFI and gCJD were labeled with iTRAQ labeling reagents
114 and 118, 115 and 119, 116 and 121. The amounts of the purified
SNO proteins were 100 μg/label. The labeled products were digested
with trypsin at a ratio of 1:20 (w/w, trypsin/protein) at 37°C for
36 h. The digested peptides were subsequently dried and
reconstituted with 0.5 M triethylammonium bicarbonate (TEAB; Sigma,
St. Louis, MO, USA) and 0.1% SDS. The dried peptides were then
labeled with respective isobaric tags, and incubated at room
temperature for 1 h before being combined.
To remove all interfering substances, such as
dissolving buffer, ethanol, acetonitrile, SDS, excess iTRAQ
reagents, strong cation exchange chromatography (SCX) was carried
out for the combined iTRAQ-labeled peptides using the cation
exchange system provided in the iTRAQ method development kit (AB
Sciex). The eluted fraction was desalted using Sep-Pak C18
cartridges, dried and then reconstituted with 0.1% formic acid (FA)
for nano-flow liquid chromatography-tandem mass spectrometry
(LC-MS/MS) analysis coupled with the ultimate LC system using
Q-Exactive Mass Spectrometer (Thermo Fisher Scientific). Peptide
separations were performed in a 100×75 mm column (BEH130 C18) using
mobile phase A (0.1% FA in LC-MS grade water) and mobile phase B
(0.1% FA in LC-MS grade ACN). The flow rate was set at 400 nl/min.
The LC fluent was directed to the electrospray ionization (ESI)
source for quadrupole mass spectrometry (Q-MS) analysis, using
precursor ions that were selected across the mass range of
350–2,000 m/z with 250 msec accumulation time/spectrum. A maximum
of 20 precursors per cycle from each MS spectra was selected for
MS/MS analyses with 100 msec minimum accumulation time for each
precursor and dynamic exclusion for 15 sec.
The software used for data interpretation was
proteome discoverer version 1.3 (Thermo Fisher Scientific) and
Mascot version 2.3.0. The database searched was the peptide
sequence library in the Swissport database restricted to the human
protein sequence data set.
Ethics statement
Usage of the stored human and animal brain specimens
in this study was approved by the Ethics Committee of the National
Institute for Viral Disease Prevention and Control, China CDC under
protocol 2009ZX10004-101. All Chinese golden hamsters were
maintained under clean grade. Housing and experimental protocols
were in accordance with the Chinese Regulations for the
Administration of Affairs Concerning Experimental Animals.
Results
Optimization of the eluting conditions
for SNO proteins from the beads
Several methods have been described for eluting
biotin-labeled biological macromolecules from streptavidin beads,
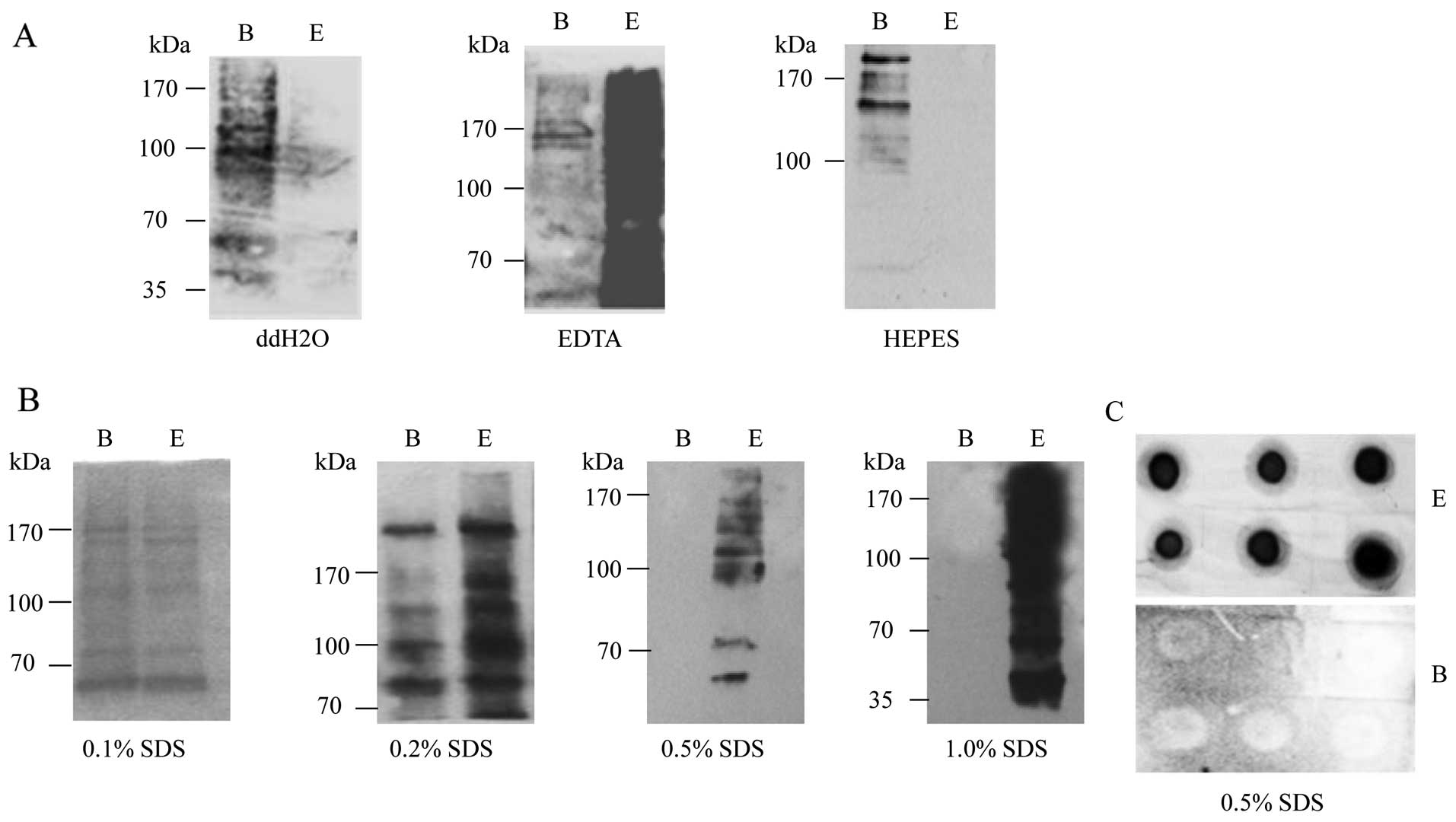
such as ddH2O, EDTA, HEPES or SDS. To evaluate the
eluting abilities of the different methods, 30 μl of brain
homogenate of normal healthy hamsters (roughly 30 μg total
proteins) were subjected to BST for biotin-labeling and bound with
streptavidin-conjugated beads. The biotin-labeled SNO proteins
bound on the beads were separately eluted with various eluting
conditions and the biotin signals in the eluting fractions and
beads were examined by western blot analysis for biotin. After
being eluted with ddH2O, only a small portion of
biotin-positive signals was detected in the fraction of elution
(Fig. 1A, left panel). More
biotin signals were observed in the elution fraction following
treatment with 10 mM EDTA with 95% formamide (pH 8.2), but there
were still biotin signals left in the beads (Fig. 1A, middle panel). In the
preparation of HEPES, almost all biotin signals were observed in
the fraction of beads (Fig. 1A,
right panel). These results indicate that it is not possible to
completely elute the bound biotin-SNO protein from streptavidin
beads with these 3 buffers.
Subsequently, the eluting capacity of the SDS buffer
was evaluated based on the same experimental conditions. Following
treatment with various concentrations of SDS buffer, the SNO
proteins in the elution fraction and beads were evaluated by
biotin-specific western blot analysis. The results revealed that
the biotin signals were distributed almost equally in the fractions
of elution and beads in the reaction of 0.1% SDS, but increased
significantly in the fraction of elution in the reaction of 0.2%
SDS. In the reactions of 0.5 and 1.0% SDS, all biotin signals were
observed in the fraction of elution (Fig. 1B). Considering that a high
concentration of SDS may affect the subsequent MS analysis
(30) and other assays, we used
0.5% SDS as the elution buffer to break biotin-streptavidin
interaction for further experiments.
To simplify the detection of SNO proteins, we used
the biotin-specific dot blot technique instead of western blot
analysis. In the reaction of 0.5% SDS, biotin signals were merely
observed in the eluting products, but not in the beads (Fig. 1C); these results were consistent
with those of western blot analysis.
Optimization of the incubation ratio of
streptavidin beads and biotin-labeled brain homogenate
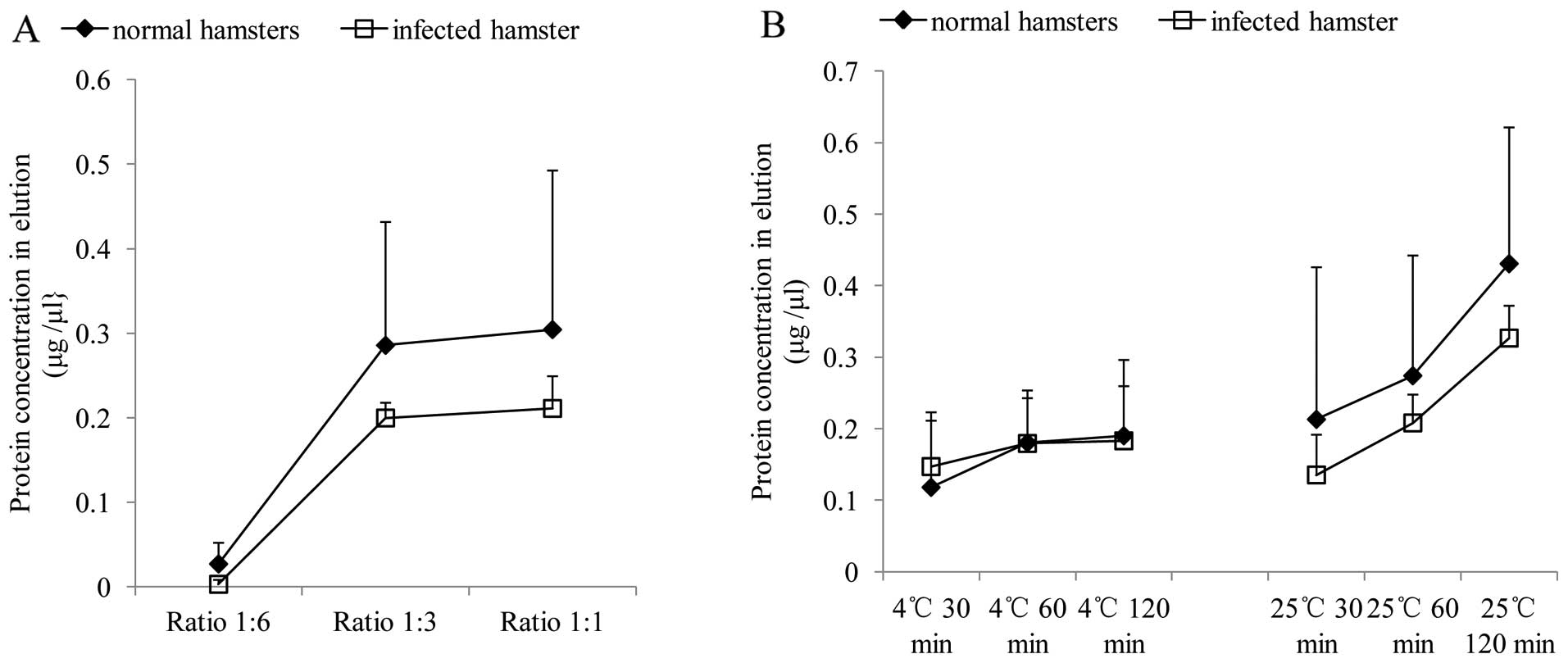
To optimize the volume ratio of the streptavidin
beads to the brain homogenate (10% w/v), the protein capturing
abilities of the different working volume ratios (bead/homogenate,
1:6, 1:3 and 1:1) were examined. Different amounts of bead were
mixed with 30 μl brain homogenates prepared from normal or 263K
scrapie-infected hamsters after being labeled with biotin by BST.
After being eluted with 0.5% SDS buffer, the protein concentrations
in the eluted products were measured. As shown in Fig. 2A, only very small amounts of
protein were detected in the eluted products at a ratio of 1:6,
whereas a markedly larger amount of protein was eluted in the
reactions at a ratio of 1:3 and 1:1. The protein-capturing
abilities were approximately the same between the reactions of
ratios 1:3 and 1:1 in either the normal or infected brain
homogenates. Based on higher cost and effective ratio, we selected
the ratio of 1:3 (bead/homogenate) as the optimal working volume
ratio.
Optimization of the incubation
temperature and incubation time of streptavidin beads and
biotin-labeled brain homogenate
To optimize the incubation temperature and
incubation time for the streptavidin beads with biotin-labeled
brain homogenate, 10 μl of beads were mixed with 30 μl normal or
263K-infected hamster brain homogenates at the volume ratio of 1:3
(bead/homogenate). Following incubation at 2 different temperatures
(4 and 25°C) for 3 different periods of time (30, 60 and 120 min),
the biotin-labeled proteins were eluted from the beads with 0.5%
SDS buffer and the protein amounts were evaluated using the BCA
method. The results revealed similar alternative curves of the
eluted protein contents in both the normal and infected
homogenates, along with the changes in incubation temperature and
duration (Fig. 2B). It was
apparent that incubation at 25°C yielded higher protein
concentrations in the eluted products than at 4°C. The increase in
the incubation time, particularly with the incubation temperature
of 25°C, yielded more proteins in the elution products. Based on
these data, incubation at 25°C for 120 min was regarded as the
optimal working condition.
Optimization of the number of incubations
for recovery of the whole SNO proteins in brain homogenates
To optimize the number of incubations in order to
recover all SNO proteins from the brain homogenates, 15 μl of
BST-treated brain homogenates of normal hamsters (roughly 15 μg
total proteins) were incubated with 5 μl of streptavidin beads at
the optimized working conditions. Following elution and the
discarding of used beads, the brain homogenates were continually
incubated with newly input beads. This process was repeated for at
least 15 rounds and 3 adjacent elution products were pooled for
further assays. Measurement of the protein concentration in the
pooled elution products revealed that the protein contents
gradually decreased along with the number of incubations, in which
almost no protein was detected in the last pooled elution sample
(incubation number 13–15) (Fig.
3A). The biotin-labeled proteins in various pooled samples were
assayed with biotin-specific dot blots. In line with the results of
the protein measurement, biotin-specific signals were observed in
the first 4 pooled elution samples from 3 individual brain
homogenates, but not in the last one (Fig. 3B). This indicates that at least 11
rounds of continual incubations at this experimental condition are
required in order to recover all SNO proteins from the rear brain
extracts of the hamsters.
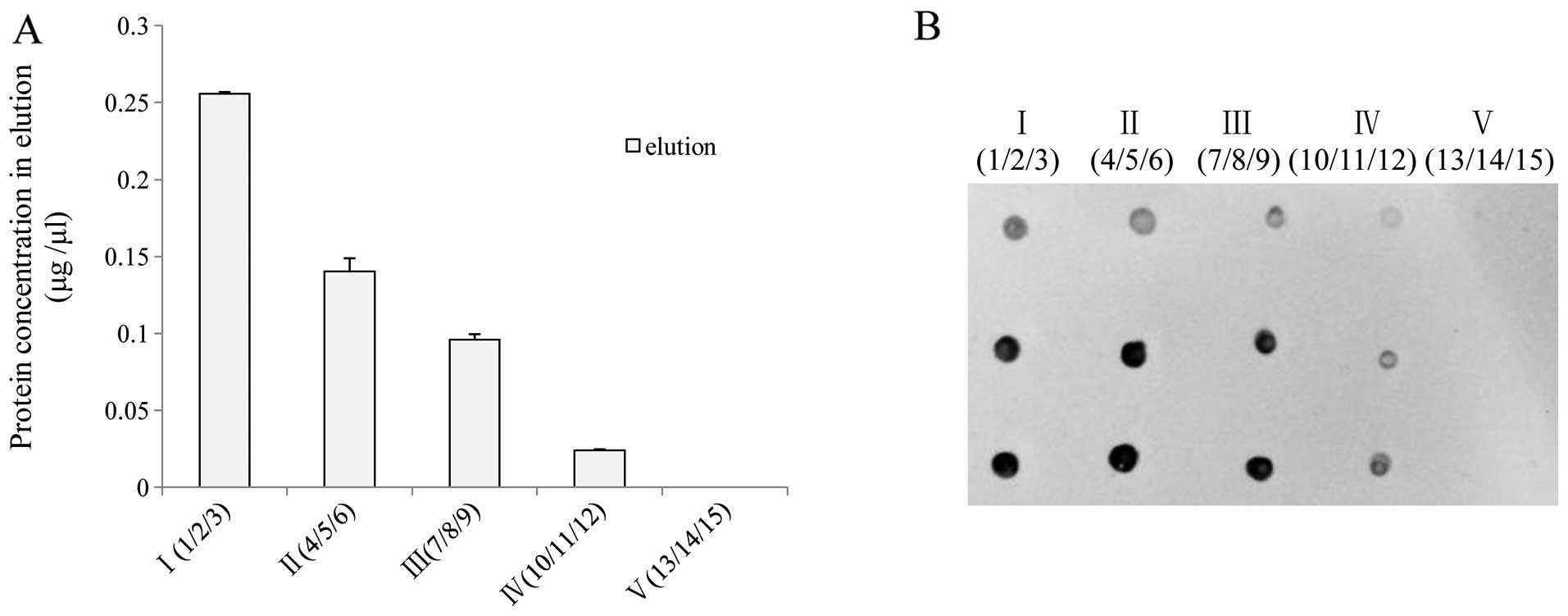 | Figure 3Recovery of whole
S-nitrosylated (SNO) proteins from rodent brain homogenates
by successive incubation with streptavidin beads. Fifteen
microliters of biotin switch technique (BST)-treated brain
homogenates from normal healthy hamsters were incubated with 5 μl
of streptavidin beads. Following elution and discarding of used
beads, the brain homogenates were continually incubated with newly
input beads. This process was repeated for at least 15 rounds and 3
adjacent elution products were pooled for further assays. (A)
Measurement of protein concentrations. (B) Biotin-specific dot
blots; 0.5 μl (upper panel) and 1 μl (middle and bottom panels) of
eluted products were separately dotted on nitrocellulose (NC)
membrane. I (1/2/3), the eluting products pooled from the 1st, 2nd
and 3rd round of incubation; II (4/5/6), the eluting products
pooled from the 4th, 5th and 6th round of incubation; III (7/8/9),
the eluting products pooled from the 7th, 8th and 9th round of
incubation; IV (10/11/12), the eluting products pooled from the
10th, 11th and 12th round of incubation; V (13/14/15), the eluting
products pooled from the 13th, 14th and 15th round of
incubation. |
Furthermore, the number of incubations for the
recovery of the whole SNO proteins from human brain homogenates
were assessed with the same protocol, including the brain
homogenates prepared from the cortex and cerebellum of the healthy
subject, and patients with sCJD, FFI and G114V gCJD. In agreement
with the results obtained with the hamster brain homogenates,
biotin-specific western blot analysis confirmed that 0.5% SDS was
able to elute almost all biotin-labeled proteins from the bound
beads in human brain extracts (Fig.
4A). A total of 15 rounds of successive incubations of the
individual BST-treated brain homogenates with beads were conducted
under optimized working conditions. In agreement with the
observations of the hamster brain homogenates, almost no protein
was detectable in the elution sample pooled from 13 to 15 rounds in
all tested extracts (Fig. 4B).
Dot blots identified similar patterns, according to which the
intensities of the biotin signals gradually decreased in the first
4 pooled samples and diminished in the last of all tested samples
(Fig. 4C). This suggests that the
optimized working conditions based on brain homogenates of hamsters
are suitable for the purification of SNO proteins from human brain
extracts, from either healthy subjects or patients with various
prion diseases.
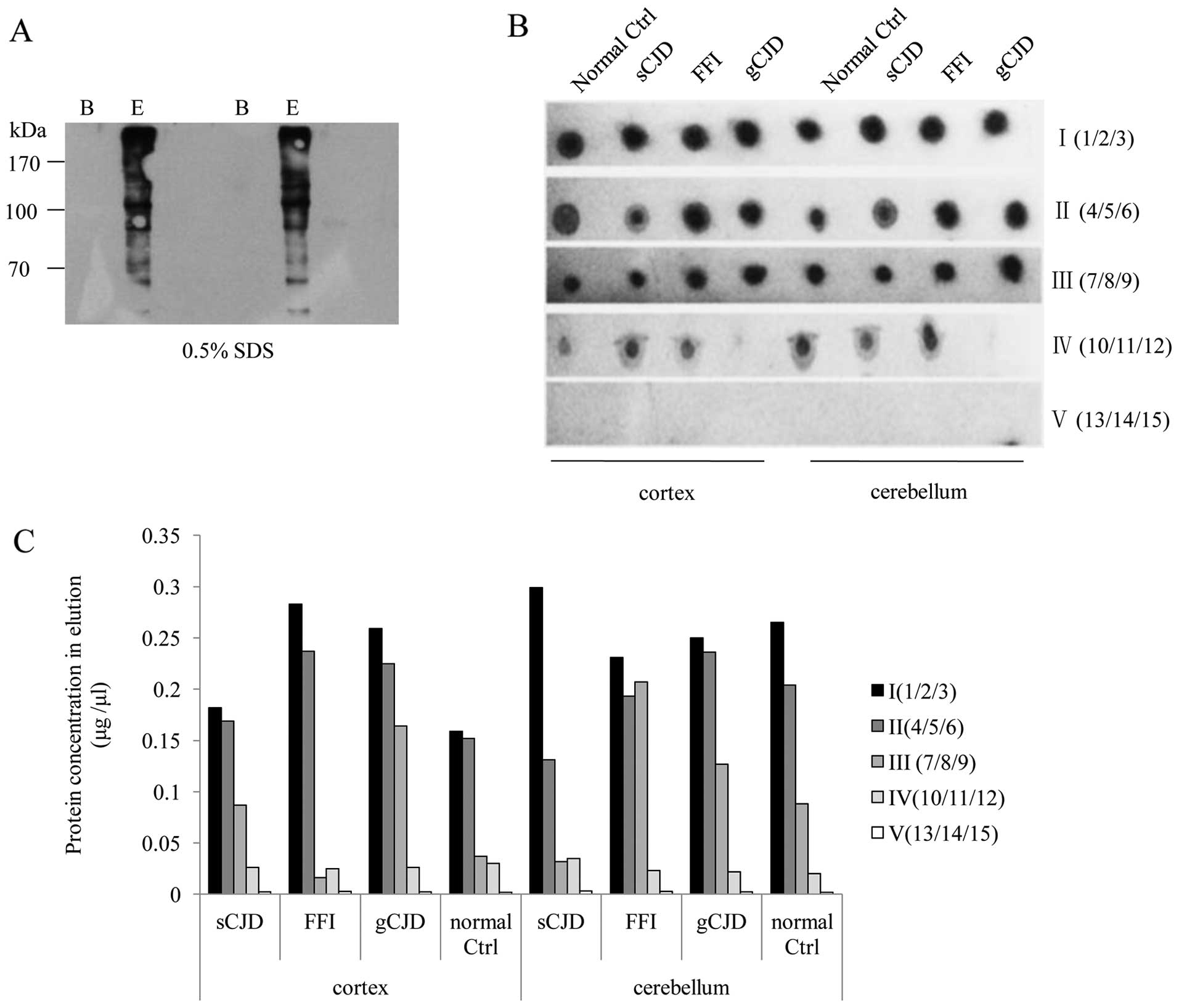 | Figure 4Recovery of S-nitrosylated
(SNO) proteins from human brain homogenates through successive
incubations with streptavidin beads. (A) Western blots of SNO
proteins eluted by 0.5% sodium dodecyl sulfate (SDS). B,
streptavidin beads following elution; E, eluting products. (B) Dot
blots of SNO proteins in the eluting products after being
successively incubated with streptavidin beads. (C) Measurement of
protein concentrations in the eluting products after being
successively incubated with streptavidin beads. Normal ctrl;
healthy control subject; sCJD, sporadic Creutzfeldt-Jakob disease;
FFI, fatal familial insomnia; gCJD, genetic Creutzfeldt-Jakob
disease; I (1/2/3), the eluting products pooled from the 1st, 2nd
and 3rd round of incubation; II (4/5/6), the eluting products
pooled from the 4th, 5th and 6th round of incubation; III (7/8/9),
the eluting products pooled from the 7th, 8th and 9th round of
incubation; IV (10/11/12), the eluting products pooled from the
10th, 11th and 12th round of incubation; V (13/14/15), the eluting
products pooled from the 13th, 14th and 15th round of
incubation. |
Evaluation of the usage of the purified
SNO proteins from human brain homogenates
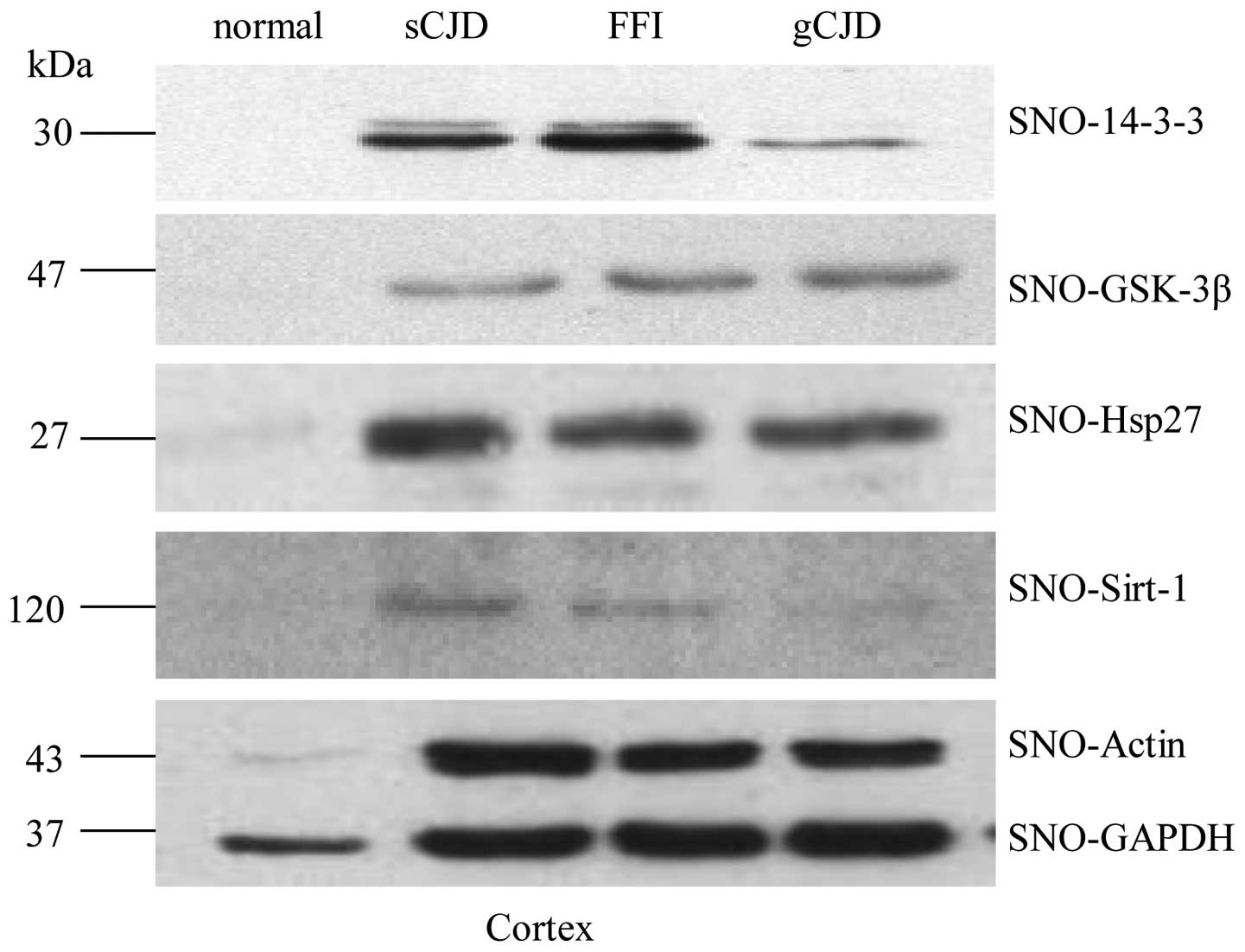
To address whether the isolated SNO proteins with
the newly optimized working conditions can be used for further
experiments, the immunoreactivities of the purified SNO proteins
from human cortex regions were subjected to several specific
western blot analyses, including for 14-3-3, GSK-3β, Hsp27, Sirt-1,
actin and GAPDH. Specific bands were observed at the expected
positions in all tested blots (Fig.
5), strongly indicating that the SNO proteins extracted under
the newly optimized working conditions possess reliable
immunoreactivities. The signal intensities of all tested proteins
in the samples of prion diseases were significantly stronger than
those of the normal control.
Subsequently, the isolated SNO proteins from the
human cortex and cerebellum under the newly optimized working
conditions were subjected to iTRAQ-based quantitative proteomics
analysis. A total of 69,896 spectra was matched from a total of
448,298 spectra. After searching with Mascot software, a total of
1,509 proteins was identified from 9,265 unique peptides with high
confidence (FDR <1%). Fig. 6
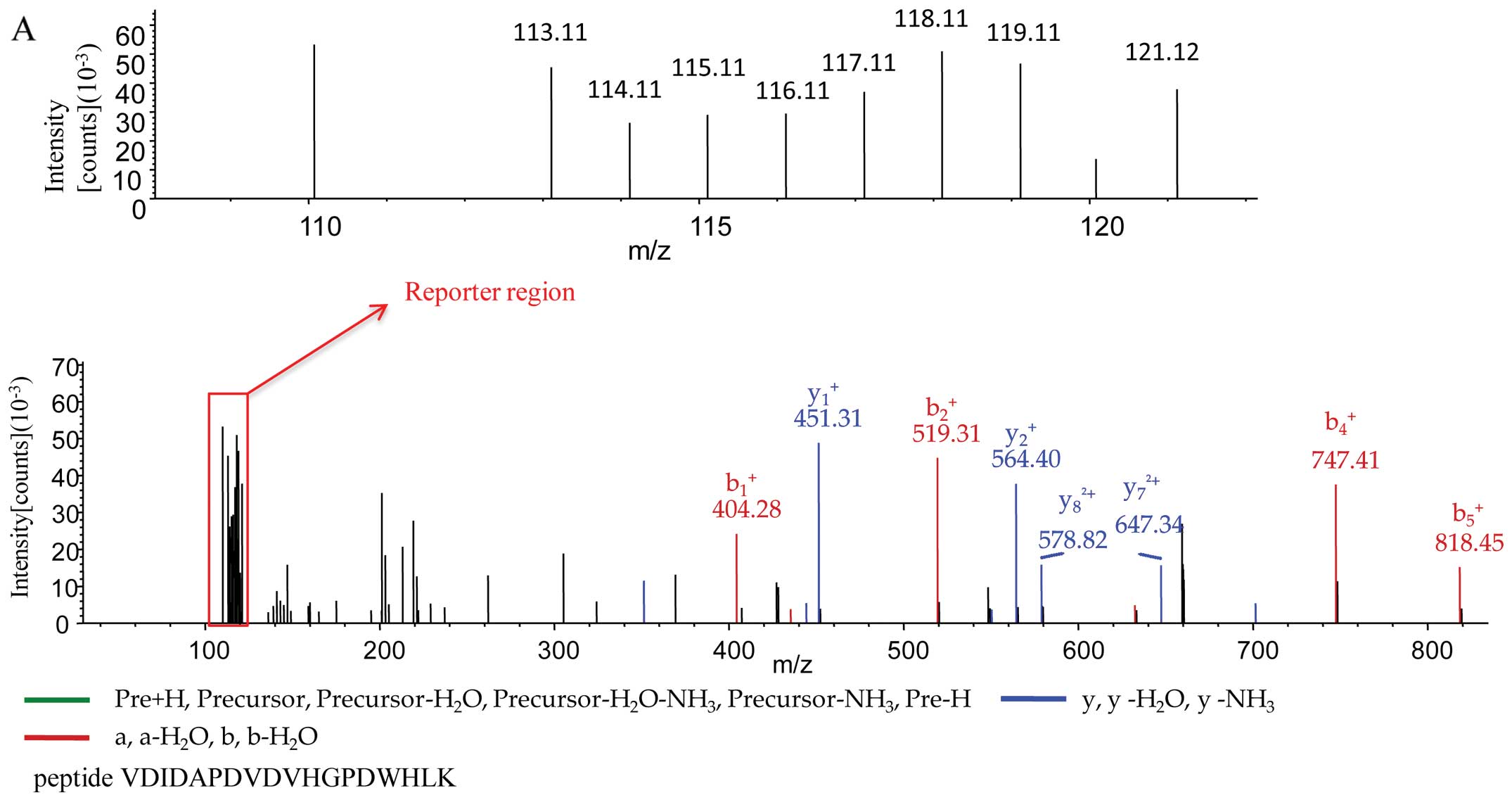
illustrates the MS/MS spectra of the peptides of 3 different
proteins, representing neuroblast differentiation-associated
protein AHNAK isoform 1 (peptide sequence, VDIDAPDVDVHGPDWHLK) with
a molecular weight of 628.7 kDa (Fig.
6A), heat shock protein gp96 precursor (peptide sequence,
GVVDSDDLPLNVSR) with a molecular weight of 90.1 kDa (Fig. 6B) and S100-B (peptide sequence,
AMVALIDVFHQYSGR) with a molecular weight of 10.7 kDa (Fig. 6C). The reporter region of each
MS/MS spectrum illustrated the signals of the peptides labeled with
the different iTRAQ tags in various brain extracts at the expected
positions (m/z), including 113 and 117 for the normal control, 114
and 118 for sCJD, 115 and 119 for FFI, and 116 and 121 for G114V
gCJD. This indicates that the SNO proteins purified with the
present modified method are suitable for proteomics analysis.
Discussion
As a regular process of post-translational
modification, the S-nitrosylation of proteins occurs
biologically and pathologically in different tissues. Usually, SNO
proteins are in low abundance in the context of whole proteins;
thereby, the isolation and enrichment of SNO proteins from tissue
extracts are required prior to teh further evaluation of SNO
proteins. In this study, we describe a modified protocol for the
isolation and enrichment of SNO proteins from brain tissues based
on a commercial SNO protein detection assay kit. Using brain
homogenates, we optimized the elution reagents and the working
conditions for incubation. The incubation ratio, incubation time
and temperature of biotin-labeled brain homogenates with
streptavidin beads were also optimized in order to obtain the
opitmal efficacy/cost ratio. The workflow of such an efficient
protocol was set up for SNO protein isolation and enrichment from
brain extracts (Fig. 7). Briefly,
aliquots of 10% brain homogenates were subjected into a
biotin-switch technique kit. After being labeled with biotin, the
specimens were mixed with streptavidin beads at a ratio of 3:1
(v/v) and incubated at 25°C for 120 min. The SNO proteins are
finally eluted from beads with buffer containing 0.5% SDS.
We also observed that SNO proteins in brain extracts
are not able to be thoroughly isolated by one round of
purification. Instead, the signals of SNO proteins in the elution
products are totally undetectable after 10 to 12 rounds of
purification both in rodent and human brain homogenates. Both
biotin-specific dot blots and protein assays revealed large amounts
of SNO proteins in the elution products of the first 6 rounds of
purification. Although the distributions of SNO proteins in various
eluting fractions may differ based on the molecular weights,
isoelectric points or other biochemical characteristics, the
present data indicate that the complete recovery of SNO proteins in
10% brain homogenates requires at least 6 rounds of purification
under our experimental conditions.
Our data also highlighted that the combined usage of
protein-content determination and biotin-specific dot blot may
precisely reflect the alterations in SNO proteins in eluting
products. Although these 2 assays do not directly represent the
contents of SNO proteins and the possibility of the presence of
non-SNO proteins and unlabeled free biotin in eluting products
cannot be completely excluded, a combination of those 2 assays in
detecting the protein contents and biotin signals in eluting
products strongly suggests the reliability and feasibility of the
combined use of these simple assays in monitoring the recovery of
SNO proteins.
Compared with the protocols described in previous
studies (28,29) or suggested by the manufacturer of
Dynabeads® M-280 (Life Technologies), the denaturing
conditions with 0.5% SDS were effective on the release of
biotin-labeled SNO proteins from streptavidin beads, which allowed
us to obtain almost all biotin-labeled proteins from bound beads
with one elution time. Further biotin-specific western blot
analysis of the elution products verified a number of positive
protein bands with various molecular weights. This suggests that
protein free thiols covalently labeled with maleimide-biotin are
not damaged under 0.5% SDS buffer. In fact, such a concentration of
SDS (0.5%) is widely used for tissue and cell lysis, nucleotide
acid and protein extraction, which usually does not affect further
protein assays. Good immunoreactivities of the final elution
products from human brain homogenates in western blot analysis with
several specific antibodies confirm that the purified SNO proteins
from brain tissues obtained by the optimized method in this study
are suitable for further experiments.
iTRAQ-based proteomics consists of a series of
complex processes, such as iTRAQ labeling, trypsin digestion,
multiple peptide purifications and peptide identification. The data
of iTRAQ-based proteomics demonstrate that the SNO proteins
purified from various human brain homogenates with our optimized
protocol can directly undergo the aforementioned processes for
proteomics analysis. It should be noted that the identified
peptides with high confidence by the iTRAQ-based proteomics cover
large amounts of protein with a wide range of molecular weights.
This highlights a broad applicability on the
S-nitrosoproteome for a wide array of biological samples,
including those derived from clinical materials.
Acknowledgements
This study was supported by grants from the Chinese
National Natural Science Foundation Grants (81301429), the China
Mega-Project for Infectious Disease (2011ZX10004-101 and
2012ZX10004215), the Young Scholar Scientific Research Foundation
of China CDC (2012A102) and the SKLID Development Grant
(2012SKLID102, 2011SKLID104 and 2011SKLID211).
References
|
1
|
Masters CL and Richardson EP Jr: Subacute
spongiform encephalopathy (Creutzfeldt-Jakob disease). The nature
and progression of spongiform change. Brain. 101:333–344. 1978.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Prusiner SB: Prions causing degenerative
neurological diseases. Annu Rev Med. 38:381–398. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Prusiner SB: Shattuck lecture -
neurodegenerative diseases and prions. N Engl J Med. 344:1516–1526.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Prusiner SB and Scott MR: Genetics of
prions. Annu Rev Genet. 31:139–175. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Nakamura T and Lipton SA: S-nitrosylation
and uncompetitive/fast off-rate (UFO) drug therapy in
neurodegenerative disorders of protein misfolding. Cell Death
Differ. 14:1305–1314. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nakamura T and Lipton SA: Emerging roles
of S-nitrosylation in protein misfolding and neurodegenerative
diseases. Antioxid Redox Signal. 10:87–101. 2008. View Article : Google Scholar
|
|
7
|
Lipton SA, Choi YB, Pan ZH, et al: A
redox-based mechanism for the neuroprotective and neurodestructive
effects of nitric oxide and related nitroso-compounds. Nature.
364:626–632. 1993. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lei SZ, Pan ZH, Aggarwal SK, et al: Effect
of nitric oxide production on the redox modulatory site of the NMDA
receptor-channel complex. Neuron. 8:1087–1099. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Choi YB, Tenneti L, Le DA, et al:
Molecular basis of NMDA receptor-coupled ion channel modulation by
S-nitrosylation. Nat Neurosci. 3:15–21. 2000. View Article : Google Scholar
|
|
10
|
Cho DH, Nakamura T, Fang J, et al:
S-nitrosylation of Drp1 mediates beta-amyloid-related mitochondrial
fission and neuronal injury. Science. 324:102–105. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Nakamura T, Cieplak P, Cho DH, Godzik A
and Lipton SA: S-nitrosylation of Drp1 links excessive
mitochondrial fission to neuronal injury in neurodegeneration.
Mitochondrion. 10:573–578. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yao D, Gu Z, Nakamura T, et al:
Nitrosative stress linked to sporadic Parkinson’s disease:
S-nitrosylation of parkin regulates its E3 ubiquitin ligase
activity. Proc Natl Acad Sci USA. 101:10810–10814. 2004. View Article : Google Scholar
|
|
13
|
Fang J, Nakamura T, Cho DH, Gu Z and
Lipton SA: S-nitrosylation of peroxiredoxin 2 promotes oxidative
stress-induced neuronal cell death in Parkinson’s disease. Proc
Natl Acad Sci USA. 104:18742–18747. 2007. View Article : Google Scholar
|
|
14
|
Nakamura T, Tu S, Akhtar MW, Sunico CR,
Okamoto S and Lipton SA: Aberrant protein S-nitrosylation in
neurodegenerative diseases. Neuron. 78:596–614. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang P, Yu PC, Tsang AH, et al:
S-nitrosylation of cyclin-dependent kinase 5 (cdk5) regulates its
kinase activity and dendrite growth during neuronal development. J
Neurosci. 30:14366–14370. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Qu J, Nakamura T, Cao G, Holland EA,
McKercher SR and Lipton SA: S-nitrosylation activates Cdk5 and
contributes to synaptic spine loss induced by beta-amyloid peptide.
Proc Natl Acad Sci USA. 108:14330–14335. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Uehara T, Nakamura T, Yao D, et al:
S-nitrosylated protein-disulphide isomerase links protein
misfolding to neurodegeneration. Nature. 441:513–517. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang SB, Shi Q, Xu Y, et al: Protein
disulfide isomerase regulates endoplasmic reticulum stress and the
apoptotic process during prion infection and PrP mutant-induced
cytotoxicity. PLoS One. 7:e382212012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shi Q, Song QQ, Sun P, et al: Infection of
prions and treatment of PrP106–126 alter the endogenous status of
protein 14-3-3 and trigger the mitochondrial apoptosis possibly via
activating Bax pathway. Mol Neurobiol. 49:840–851. 2014. View Article : Google Scholar
|
|
20
|
Forrester MT, Thompson JW, Foster MW,
Nogueira L, Moseley MA and Stamler JS: Proteomic analysis of
S-nitrosylation and denitrosylation by resin-assisted capture. Nat
Biotechnol. 27:557–559. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Garbán HJ, Márquez-Garbán DC, Pietras RJ
and Ignarro LJ: Rapid nitric oxide-mediated S-nitrosylation of
estrogen receptor: regulation of estrogen-dependent gene
transcription. Proc Natl Acad Sci USA. 102:2632–2636. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hao G, Derakhshan B, Shi L, Campagne F and
Gross SS: SNOSID, a proteomic method for identification of cysteine
S-nitrosylation sites in complex protein mixtures. Proc Natl Acad
Sci USA. 103:1012–1017. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Seth D and Stamler JS: The SNO-proteome:
causation and classifications. Curr Opin Chem Biol. 15:129–136.
2011. View Article : Google Scholar :
|
|
24
|
Camerini S, Polci ML, Restuccia U, Usuelli
V, Malgaroli A and Bachi A: A novel approach to identify proteins
modified by nitric oxide: the HIS-TAG switch method. J Proteome
Res. 6:3224–3231. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Greco TM, Hodara R, Parastatidis I, et al:
Identification of S-nitrosylation motifs by site-specific mapping
of the S-nitrosocysteine proteome in human vascular smooth muscle
cells. Proc Natl Acad Sci USA. 103:7420–7425. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tello D, Tarin C, Ahicart P, Breton-Romero
R, Lamas S and Martinez-Ruiz A: A ‘fluorescence switch’ technique
increases the sensitivity of proteomic detection and identification
of S-nitrosylated proteins. Proteomics. 9:5359–5370. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang J, Chen L, Zhang BY, et al:
Comparison study on clinical and neuropathological characteristics
of hamsters inoculated with scrapie strain 263K in different
challenging pathways. Biomed Environ Sci. 17:65–78. 2004.PubMed/NCBI
|
|
28
|
Holmberg A, Blomstergren A, Nord O, Lukacs
M, Lundeberg J and Uhlén M: The biotin-streptavidin interaction can
be reversibly broken using water at elevated temperatures.
Electrophoresis. 26:501–510. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xu L, Han C, Lim K and Wu T: Activation of
cytosolic phospholipase A2alpha through nitric oxide-induced
S-nitrosylation. Involvement of inducible nitric-oxide synthase and
cyclooxygenase-2. J Biol Chem. 283:3077–3087. 2008. View Article : Google Scholar
|
|
30
|
Hustoft HK, Reubsaet L, Greibrokk T,
Lundanes E and Malerod H: Critical assessment of accelerating
trypsination methods. J Pharm Biomed Anal. 56:1069–1078. 2011.
View Article : Google Scholar : PubMed/NCBI
|