Introduction
Human cytomegalovirus (HCMV) is the prototype member
of the betaherpesvirus family (1,2).
HCMV infection is generally asymptomatic in healthy individuals;
however, it can cause severe disease in immunocompromised patients,
such as AIDS and transplant patients (3). Congenital HCMV infection in newborns
may cause multiple malformations and diseases, including jaundice
(associated with hepatitis), mental retardation, blindness and
deafness, involving multiple systems (4). HCMV contains a 240-kb
double-stranded DNA genome which encodes over 165 open reading
frames (ORFs) and several microRNAs (miRNAs) (5–10).
The HCMV genome has three expression phases, i.e., immediate-early
(IE), early (E) and late (L) (2).
To date, fewer than 100 predicted genes and partial
gene segments have been extensively characterized with respect to
their expression patterns, transcript structures and transcription
characteristics (11). Some of
these genes produce multiple gene products due to the alternative
splicing of pre-messenger RNA (pre-mRNA) (12,13). The majority of the alternatively
spliced mRNA species of HCMV originate primarily, but not
exclusively, from the IE gene regions. Some of the alternatively
spliced variants of the IE region of HCMV have been investigated
(14,15). Recently, new evidence of mRNA
splicing was obtained from the RL8A, UL74A, UL124 and UL150A genes
(16). Alternatively, spliced
mRNAs encode a number of gene products with different sizes
(16).
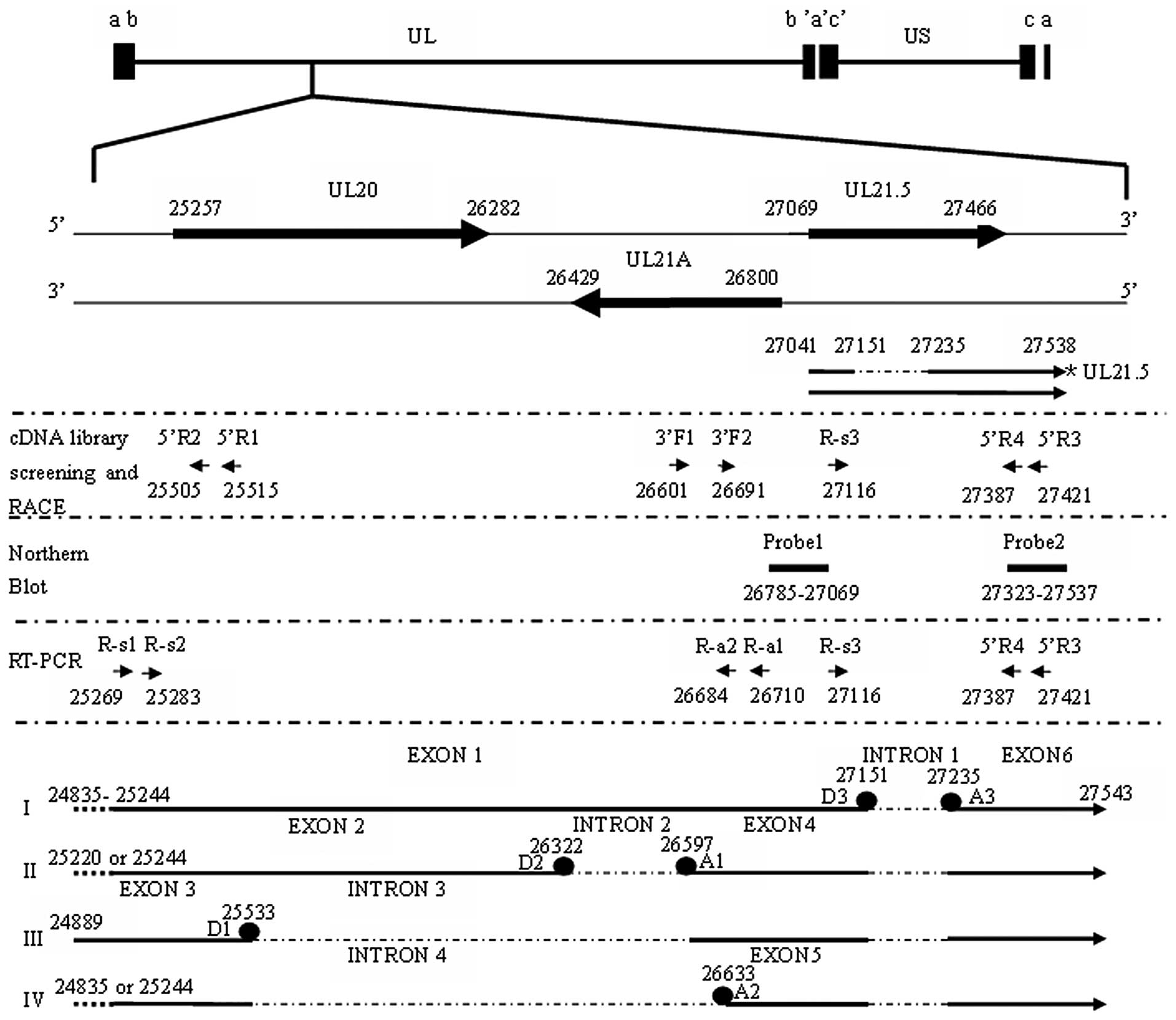
Rawlinson and Barrell (18) found a new spliced transcript and
an unspliced transcript, which were named UL21.5 (9,10,17), from the complementary strand of
the predicted UL21A gene of the HCMV AD169 genome during the late
infection phase. UL21.5 was found to be one of the viral
transcripts packaged within HCMV particles (17). The virion-associated UL21.5 mRNA
is translated into a secreted glycoprotein, which is a viral
chemokine decoy receptor specifically interacting with RANTES
(19).
A spliced transcript that is highly homologous with
the UL21.5 transcript was previously detected from a late HCMV cDNA
library in our laboratory (20).
In the present study, four novel low-abundance spliced transcripts,
termed UL21.5 multiple spliced transcripts (UL21.5 MST), were
identified to be transcribed from the UL21.5 gene region of a
low-passage strain dependent upon different combinations of three
splice donor sites and three splice acceptor sites. Furthermore,
the splice donor and acceptor sites were validated by an in
vitro transcriptional system. The necessary regulatory
sequences of one splice donor site and two splice acceptor sites
were also identified.
Materials and methods
Viruses, cells and specimens
A low-passage HCMV strain H was isolated from a
urine sample of a congenitally HCMV-infected infant at the
Department of Pediatrics at the Affiliated Shengjing Hospital of
China Medical University, Shenyang, China. MRC-5 and HEK293 cells
were obtained from Shanghai Institute of Cell Biology and
Biochemistry (Shanghai, China). The H strain was inoculated into
MRC-5 cells maintained in minimal essential medium (MEM)
supplemented with 2% fetal calf serum (HyClone, Logan UT, USA) and
penicillin-streptomycin in an incubator at 37°C, 5% CO2.
For HCMV IE infection, the cells were harvested at 24 h
post-infection (hpi) in the presence of 100 μl/ml of the protein
synthesis inhibitor, cycloheximide (Sigma, St. Louis, MO, USA). For
E infection, 100 μl/ml of the DNA synthesis inhibitor,
phosphonoacetic acid (Sigma), were added to the medium immediately
following inoculation, and the infected cells were harvested at 48
hpi. For L infection, the cells were harvested at 72 hpi without
any drug treatment.
HEK 293 cells were propagated in Dulbecco’s modified
Eagle’s medium (DMEM) supplemented with 10% fetal calf serum
(HyClone) and penicillin-streptomycin at 37°C in 5%
CO2.
RNA preparation
Total RNA was isolated from HCMV H-infected and
uninfected MRC-5 cells using TRIzol reagent (Invitrogen, Carlsbad,
CA, USA). To remove contaminating DNA, the extracted RNA was
treated with DNA-free reagent (Ambion, Austin, TX, USA). The
quantity and purity of the RNA preparations were estimated by
optical density value detection.
cDNA library screening
A full-length cDNA library of the HCMV H strain was
previously constructed using the pBluescript II SK vector (20). Recombinants of the cDNA library
were transferred into the E. coli JM109 strain (Promega,
Madison, WI, USA). A total of 8,600 cDNA clones were randomly
selected and inoculated into LB medium. A pair of specific primers
for the UL21.5 gene region (R-s3 and 5’R3) (Table I and Fig. 1) were designed and used to screen
UL21.5 gene-specific clones from the cDNA library by graded
polymerase chain reaction (PCR) as described in previous studies
(21,22). The PCR reaction conditions were as
follows: 95°C for 5 min, 30 cycles of 95°C for 45 sec, 56°C for 45
sec and 72°C for 30 sec, followed by a final elongation at 72°C for
10 min. Inserts of the selected clones were sequenced using the
vector primers, M13F and M13R, on an ABI PRISM 3730 DNA analyzer
(Applied Biosystems, Foster City, CA, USA). The screening results
allowed us to obtain cDNA clones containing sequences between the
screening primers from both genome strands.
 | Table IPrimers used in the study. |
Table I
Primers used in the study.
| Experiment | Primer name | Sequence
(5′→3′) | Position
(5′→3′) |
|---|
| cDNA library
screening and RACE | R-s3 |
GGCTTTGGCGGCACCTTCT | 27116–27134 |
| 3′F1 |
GGCAACATCTCTTCATCTCCC | 26601–26621 |
| 3′F2 |
GCGTAGAAAGCCAAGCGG | 26691–26708 |
| 5′R1 |
CCGTTAGGTTAGTCACGCTCCA | 25515–25494 |
| 5′R2 |
AGTCACGCTCCAACTACCCTCA | 25505–25484 |
| 5′R3 |
GTCGGTCTTCTCTTGTGG | 27421–27404 |
| 5′R4 |
CCATCTCCGTCTGTAATCAAAA | 27387–27366 |
| Northern blot
analysis | 21.5-NS1 |
CAGGGCTACCTCCCATCGC | 26785–26803 |
| 21.5-NAS1 |
TGCTGAGGTCTTGTCGGCG | 27069–27051 |
| 21.5-NS2 |
ACCACCGACGGAAACGAAGAT | 27323–27343 |
| 21.5-NAS2 |
GAGCAGAACCTTACAGCTTTTTA | 27537–27515 |
| 99-NS |
CTGGGCTGCGAGTTGCTGGC | 144702–144721 |
| 99-NAS |
GATGGTGGTGATGTTTTGAGGGTT | 145235–145212 |
| RT-PCR | R-s1 |
CGGGCTATGCTGGTGATG | 25269–25286 |
| R-a1 |
CGCCGCTTGGCTTTCTAC | 26710–26693 |
| R-s2 |
GATGCTGGATTACTACTGGA | 25283–25302 |
| R-a2 |
AGCTCGTCGGAGGCTTTT | 26684–26667 |
| R-s3 |
GGCTTTGGCGGCACCTTCT | 27116–27134 |
| 5′R3 |
GTCGGTCTTCTCTTGTGG | 27421–27404 |
| 5′R4 |
CCATCTCCGTCTGTAATCAAAA | 27387–27366 |
| Plasmid
construction | M-S |
acgcGTCGACTGCTGGTGATGCTGGATTACTACT | 25276–25299 |
| M-AS |
aaggaaaaaaGCGGCCGCGTCGGTCTTCTCTTGTGGTT | 27421–27402 |
| M-d-as |
ggaAGATCTTGAGGGTTGGGTTGATGCAACTCTCCGTTAGG | 25530–25508 |
| M-dL-s | ggaAGATCT
CCTATATAACTACCATCTGGCTTCTG | 25540–25565 |
| M-a1-as |
ggaAGATCTGATGTTTTGAGGGTTGCGTCATCGACCGACCTCCGC | 26576–26557 |
| M-a1L-s |
ggaAGATCTATCCGGCAACATCTCTTCATCTCCC | 26597–26621 |
| M-a2-a1 |
ggaAGATCTGATGTTTTGAGGGTTGGAGATGTTGCCGGATCTGCCG | 26612–26591 |
| M-a2L-s1 | ggaAGATCT
CACACGGCGCTGTTCTGGATGTAT | 26633–26656 |
| M-a2-a2 |
ggaAGATCTGATGTTTTGAGGGTTGGGAGACGAAGAGATGTTACCGGATC | 26620–26596 |
| M-a2L-s2 | ggaAGATCT
GCTGTTCTGGATGTATATGATTC | 26641–26663 |
| M-a2-a3 |
ggaAGATCTGGATGGTGGTGATGTTTTGAGGGTTGGAGACGAAGAGATGTTACCGGA | 26620–26598 |
| M-a2L-s3 |
ggaAGATCTATGTATATGATTCTGGAAAAGCCTC | 26651–26675 |
Northern blot analysis
Northern blot analysis was performed using the
digoxigenin (DIG) system (Roche Diagnostics, Indianapolis, IN, USA)
according to the manufacturer’s instructions. Initially, the same
amount of IE, E and L RNA preparations of the HCMV H strain and RNA
from uninfected MRC-5 cells (10 μg RNAs in each lane for detection
by probe 1, and 5 μg RNA in each lane for detection by probe 2 and
UL99 probe) were subjected to denaturing agarose gel [1.5%
(wt/vol)] electrophoresis in the presence of formaldehyde along
with the DIG-labeled RNA molecular weight marker I (Roche
Diagnostics). The probes were synthesized and labeled using a DIG
Northern Starter kit (Roche Diagnostics) and primers 21.5-NS1 and
21.5-NAS1, 21.5-NS2 and 21.5-NAS2, as well as 99-NS and 99-NAS. The
primers were designed according to the sequences of the transcripts
identified by cDNA library screening, the UL21.5 transcript
reported above (Table I and
Fig. 1) and the UL99 gene of the
HCMV Towne strain (GenBank accession no: FJ616285.1). A probe
located from nt 27537 to 27323 complementary to the antisense
strand of the UL21.5 gene locus was synthesized. The separated RNA
was transferred onto a positively charged nylon membrane by
capillary transfer. Subsequently, the nylon membrane was baked at
80°C for 2 h followed by pre-hybridization at 65°C for 30 min using
DIG Easy Hyb buffer and hybridization to the UL21.5 and UL99 probes
at 65°C for 16 h. The hybridized probes were reacted to the
anti-DIG conjugated to alkaline phosphatase and the
chemiluminescence substrate CDP-Star, and were exposed using
ChemiDoc™ XRS+ (Bio-Rad Laboratories, Hercules, CA, USA). To ensure
that equal amounts of RNA were loaded in each experiment, the RNA
preparations were adjusted by comparing them to the quantities of
the 28S and 18S rRNA in the same RNA preparations, which were
estimated by electrophoresis and ethidium bromide staining.
Reverse transcription PCR (RT-PCR)
According to the results of cDNA library screening,
a set of primers, flanking the introns of the spliced transcripts
identified, were designed. RT-PCR was performed using an RNA PCR
kit (AMV) version 3.0 (Takara Biotechnology, Dalian, China).
First-strand cDNA was synthesized with an oligo(dT) primer at 42°C
for 30 min, 95°C for 5 min, 5°C for 5 min using L RNA from the HCMV
H-infected MRC-5 cells. Subsequently, hot-start nested PCR was
performed using two pairs of primers, R-s1 and R-a1, as well as
R-s2 and R-a2 for identification of the spliced transcripts, and
using primers R-s3 and 5′R3, as well as R-s3 and 5′R4 for the
identification of the UL21.5 transcripts (Table I and Fig. 1). All the reactions were carried
out at 94°C for 3 min, 30 cycles of 94°C for 30 sec, 56°C for 30
sec, and 72°C for 2 min, and a final extension at 72°C for 10 min.
A negative control reaction by omitting the reverse transcription
step was carried out concurrently.
Rapid amplification of cDNA ends
(RACE)
To determine the 3′ and 5′ ends of the UL21.5
transcripts, RACE was performed using the 3′-full RACE Core Set
version 2.0 and 5′-full RACE kits (Takara Biotechnology) according
to the manufacturer’s instructions. Using L RNA preparations from
the HCMV H-infected MRC5 cells, first-strand cDNA was synthesized
with M-MLV reverse transcriptase using oligo(dT)-adaptor primer
(3′RACE) and random 9-mer Primer (5′RACE) provided by the
respective kits. In the 5′RACE experiments, reactions without
tobacco acid pyrophosphatase (TAP) and M-MLV, which exclude the
interference caused by the 5′-terminal phosphate of tRNA, rRNA and
incomplete mRNA and exclude the interference caused by DNA
pollution, were performed as two control reactions. Subsequently,
specific sequences of the first-strand cDNA were amplified by PCR
using the specific primers, 3′F1 and 3′F2 for 3′RACE, and the
primers, 5′R1 and 5′R2, as well as 5′R3 and 5′R4, for 5′RACE. The
primers are listed in Table I and
shown in Fig. 1.
Plasmid construction
To construct an expression vector for the UL21.5
gene, the sequence containing all the introns of the HCMV UL21.5
gene region from nt 25276 to 27421 was amplified by PCR using H
strain DNA as a template and a pair of primers, M-S and M-AS, which
introduce a SalI and NotI recognition site into the
ends of the amplicon respectively (Table I). The PCR products were digested
using SalI and NotI, and then cloned into the
SalI and NotI sites of the pCMV-HA vector, resulting
in the plasmid, pCMV-UL21.5 MST (Table II).
 | Table IIResults identified by the in
vitro expression system. |
Table II
Results identified by the in
vitro expression system.
| Plasmids | Mutation
regions | Original
sequences | Mutation
sequences | Detected
transcripts | Undetected
transcriptsa |
|---|
| pCMV-UL21.5
MST | | | | I, II, III, IV | No |
|
pCMV-UL21.5-mut-D1 |
D1(nt25533)/nt25531–39b | GCGGTGAGT | TGAGGGTTG | I, II | III, IV |
|
pCMV-UL21.5-mut-A1 |
A1(nt26597)/nt26577–96 |
GGATTGGGTA
GGGGTTGCAG |
GATCTGATGT
TTTGAGGGTT | I, IV | II, III |
|
pCMV-UL21.5-mut-A2-1 |
A2(nt26633)/nt26613–32 |
TCGTCTCCCT
CACCGACCAG |
GATCTGATGT
TTTGAGGGTT | I, II, III, IV | No |
|
pCMV-UL21.5-mut-A2-2 |
A2(nt26633)/nt26621–40 |
CTCACCGACC
AGCACACGGC |
GATCTGATGT
TTTGAGGGTT | I, II, III, IV | No |
|
pCMV-UL21.5-mut-A2-3 |
A2(nt26633)/nt26621–50 |
CTCACCGACC
AGCACACGGC
GCTGTTCTGG |
GATCTGGATG
GTGGTGATGT
TTTGAGGGTT | I, II, III | IV |
To identify the determinant regulatory region of the
splice sites found in this study, recombinant vectors carrying
mutant sequences adjacent to one splice donor site (D1 at nt 25533)
and two splice acceptor sites (A1 at nt 26597 and A2 at nt 26633)
were generated using specific primers containing mutant
sequences.
To generate an expression vector with a mutant
sequence around the splice donor site D1, a 9-bp sequence from nt
25531 to nt 25539 was mutated from GCGGTGAGT to TGAGGGTTG. First,
one 264-bp SalI-BglII fragment and one 1882-bp
BglII-NotI fragment were amplified using two pairs of
primers, M-S and M-d-as, as well as M-dL-s and M-AS. Following
digestion with SalI and BglII, as well as
BglII and NotI, the fragments were inserted into the
pCMV-HA vector at the SalI and NotI sites, resulting
in pCMV-UL21.5-mut-D1 (Table
II).
To construct an expression vector with a mutant
sequence adjacent to the splice acceptor site A1 at nt 26597, a
20-bp sequence located upstream of the A1 site from nt 26577 to nt
6596 was mutated from GGATTGGGTAGGGGTTGCAG to GATCTGATGTTTTGAGGGTT.
Using two pairs of primers, M-S and M-a1-as, as well as M-a1L-s and
M-AS, one 1,321-bp SalI-BglII fragment and one 825-bp
BglII-NotI fragment were obtained. Following
digestion with SalI and BglII, as well as
BglII and NotI, the fragments were cloned into the
SalI and NotI sites of the pCMV-HA vector, resulting
in pCMV-UL21.5-mut-A1 (Table
II).
To generate an expression vector containing a mutant
sequence around the splice acceptor site A2 at nt 26633, a series
of expression vectors, pCMV-UL21.5-mut-A2-1 containing a 20-bp
mutant sequence located upstream of the A2 site from nt 26613 to nt
26632, pCMV-UL21.5-mut-A2-2 containing a 20-bp mutant sequence
around the A2 site from nt 26621 to nt 26640 and
pCMV-UL21.5-mut-A2-3 containing a mutant sequence from nt 26621 to
nt 26650 were generated using a method similar to the one described
above. In pCMV-UL21.5-mut-A2-3, a 30-bp sequence adjacent to the
splice acceptor site A2 was mutated from CTCACCGACCAGCACACGG
CGCTGTTCTGG to GATCTGGATGGTGGTGATGTTTT GAGGGTT (Table II). The inserts of the plasmids
constructed above were confirmed by sequencing.
Transfection of HEK 293 cells and
detection of transcripts by RT-PCR
The HEK 293 cells were plated onto 6-well plates at
a density of 2×105 cells/well. Twenty-four hours later,
the cells were transfected with 4 μg of each plasmid DNA
(pCMV-UL21.5 MST, pCMV-UL21.5-mut-D1, pCMV-UL21.5-mut-A1,
pCMV-UL21.5-mut-A2-1, pCMV-UL21.5-mut-A2-2 and
pCMV-UL21.5-mut-A2-3) using 10 μl Lipofectamine 2000 (Invitrogen,
Carlsbad, CA, USA) diluted in serum-free medium as recommended by
the manufacturer. The cells were harvested 48 h after transfection,
and the RNA of the transfected cells was prepared as described
above. Subsequently, RT-PCR was performed using the RNA
preparations and one pair of primers, R-s2 and R-a2. The total RNA
isolated from the HEK 293 cells and the pCMV-HA-transfected HEK 293
cells was used as the controls. An RT-PCR reaction without M-MLV in
the reverse transcription step was performed concurrently.
Cloning and sequencing
All the RACE and RT-PCR products were gel-purified
and cloned into the vector, PCR2.1 (Invitrogen). Subsequently, the
inserts of the identified clones were sequenced on the ABI PRISM
3730 DNA analyzer (Applied Biosystems). The nucleotide positions
referred to in the present study are in reference to the sequence
of the HCMV Towne strain (GenBank accession no: FJ616285.1).
Results
UL21.5 spliced transcripts obtained from
the cDNA library
By graded PCR screening, 34 cDNA clones were found
to contain sequences congruent with the UL21.5 gene locus from
8,600 clones of the HCMV cDNA library. The sequencing results
revealed that all 34 cDNA sequences terminated at nt 27543, which
is downstream of a poly(A) signal (ATTAAA) at nt 27513–27518. Among
the identified cDNA sequences, 25 sequences with lengths from 417
nt to 421 nt began at the region from nt 27040 to nt 27044, which
are similar to the spliced UL21.5 transcript reported previously
(18). Five sequences with
lengths from 765 nt to 1386 nt began from nt 26696 to 26082, which
have a similar structure as transcript I (Fig. 1). The sequence with its 5′ end at
nt 24889 comprised three exons and two introns (Fig. 1, transcript III), and was 1473 nt
in length. However, no clone containing an unspliced UL21.5
sequence or a sequence from the antisense strand of the UL21.5 gene
was obtained from the cDNA library.
UL21.5 transcripts identified by northern
blot analysis
To analyze potential transcripts arising from the
UL21.5 gene locus during HCMV infection, northern blot analyses
were performed using RNA preparations from the MRC-5 cells infected
with the HCMV H strain at the IE, E and L phases and RNA from
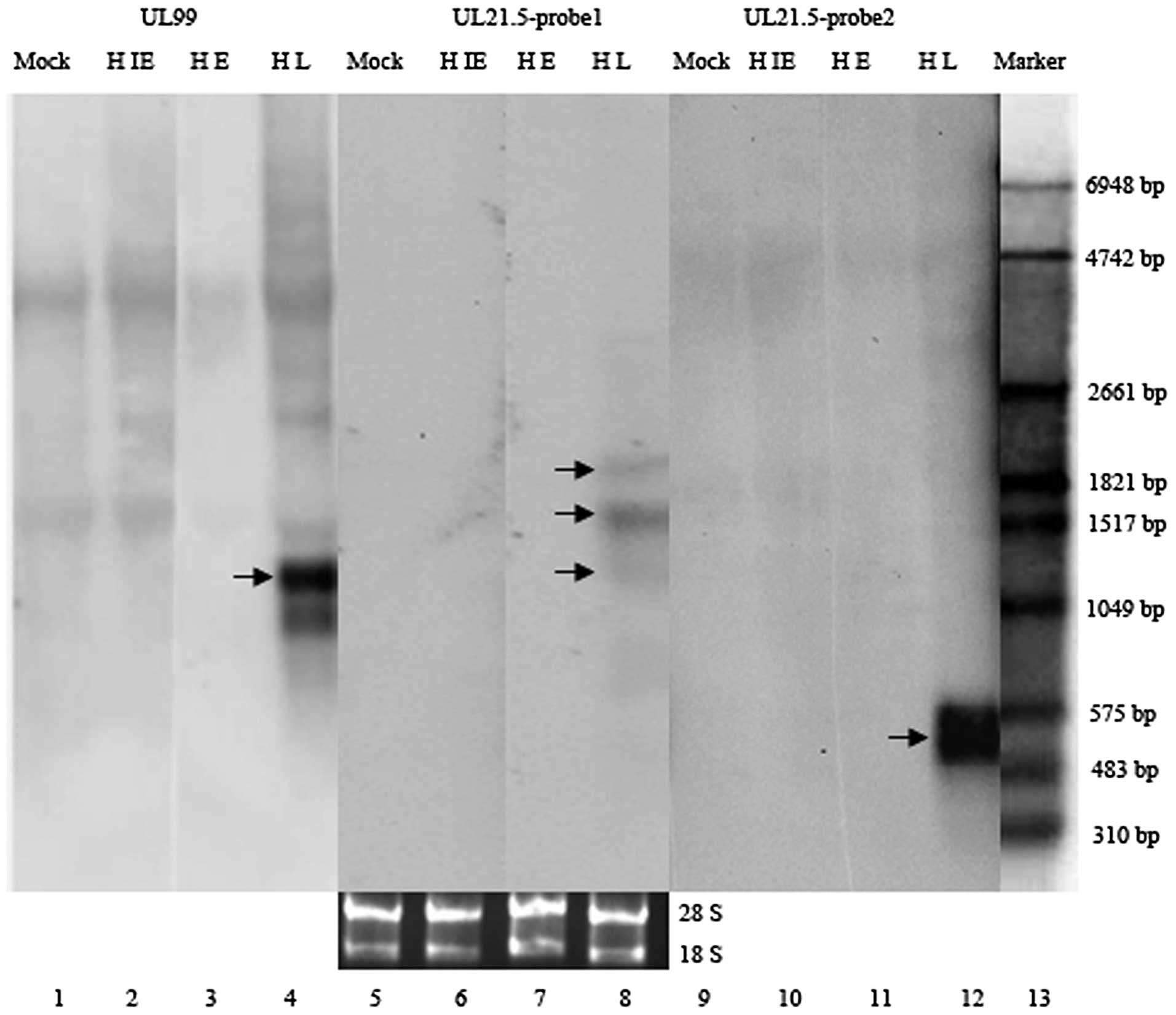
mock-infected cells. Three clusters of low-abundance transcripts
with lengths of ~1250 nt, ~1517 nt and 1900–2100 nt were detected
in the L RNA preparation by the UL21.5-specific probe 1, which is
located at nt 26785–27069 (Fig.
2, lane 8). A cluster of an abundant transcript ~400–600 bp was
detected in the same L RNA by the UL21.5-specific probe 2, which
was located at nt 27323 to 27537 (Fig. 2, lane 12). No specific band to any
of the UL21.5 probes was observed in the IE, E or mock-infected RNA
preparations (Fig. 2, lanes 5–7
and 9–11). A specific band to the UL99 probe, which is a true late
phase gene (23), was detected in
the L RNA preparation but not in the IE and E RNA preparations
(Fig. 2, lanes 1–4). No
transcript was detected by the probe complementary to the antisense
strand of the UL21.5 gene locus (data not shown).
UL21.5 multiple spliced transcripts
confirmed by RT-PCR analyses
To confirm the UL21.5 spliced transcripts obtained
from the cDNA library and identify more spliced transcripts, RT-PCR
was performed using L RNA preparations of the HCMV H strain. The
sequencing results revealed that, apart from the UL21.5 transcripts
(Fig. 3B), four spliced
transcripts (Fig. 1, transcripts
I to IV) were transcribed from the UL21.5 transcription region
(Fig. 3A) mediated by different
combinations of three splice donor sites and three splice acceptor
sites (Fig. 1 and Table III). The UL21.5 gene locus
consisted of six exons and four introns (Fig. 1). The largest intron is intron 4,
which is located between the splice donor site D1 at nt 25533 and
the splice acceptor site A2 at nt 26633, with a length of 1099 nt.
The shortest intron is intron 1 which is located between the splice
donor site D3 at nt 27151 and the splice acceptor site A3 at nt
27235, with a length of 83 nt. Alterable combinations of the six
exons constitute the four spliced transcripts (Fig. 1).
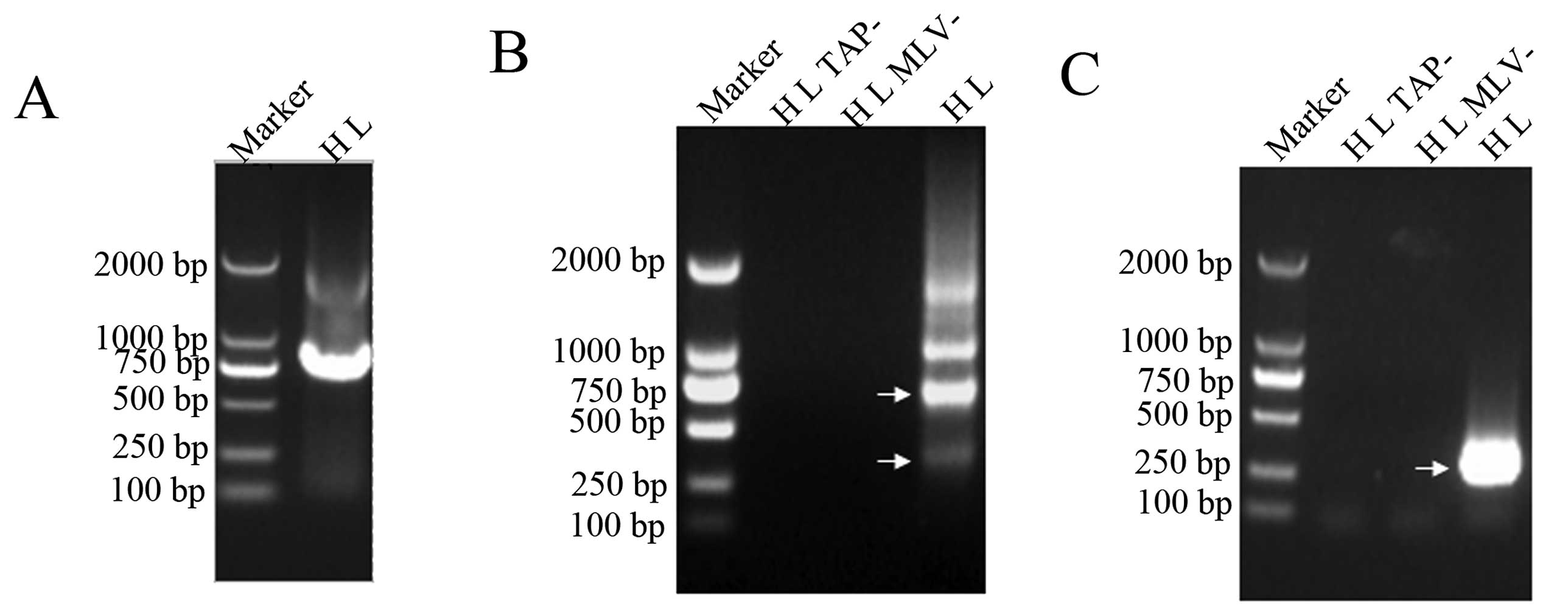
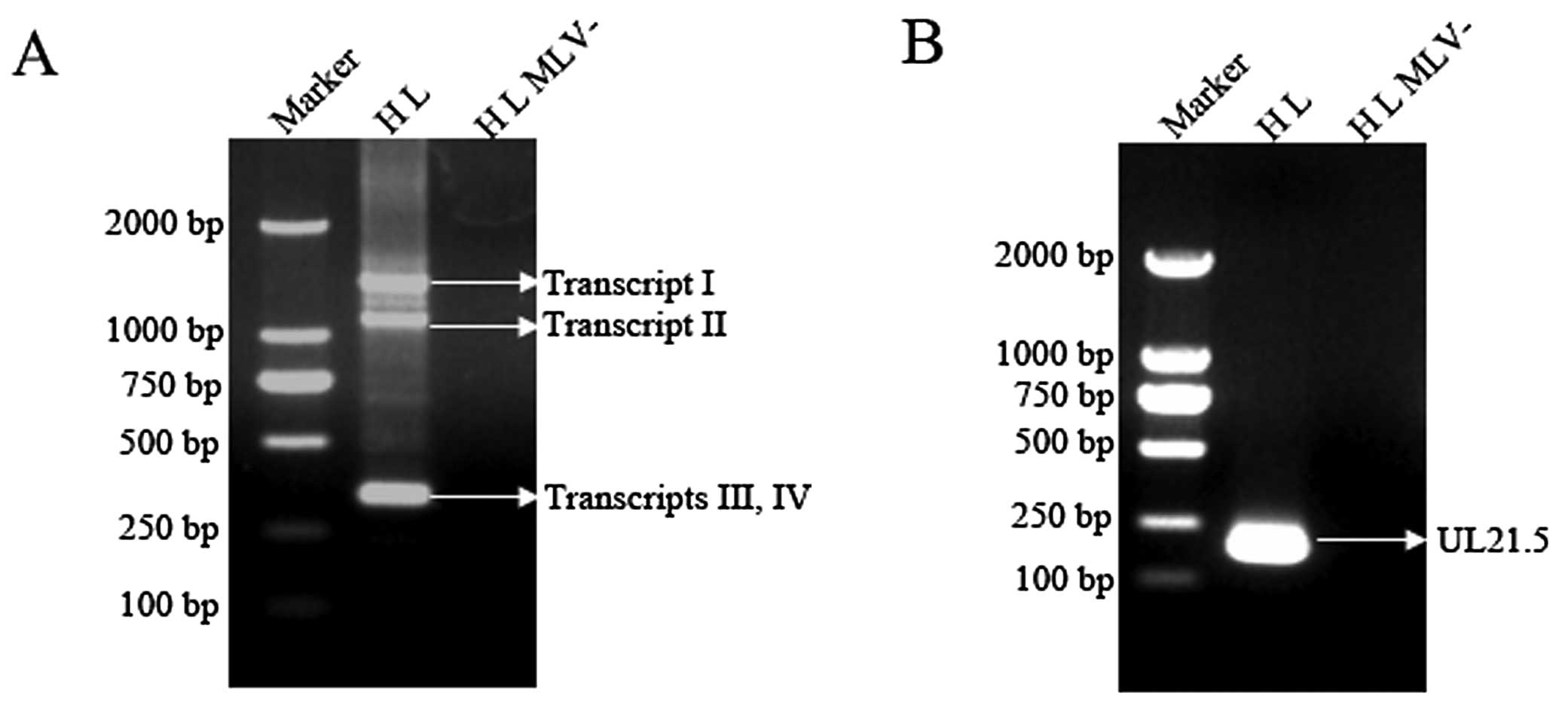 | Figure 3Results of RT-PCR for UL21.5
transcripts. RT-PCR was performed using the late-phase (L) RNA of
the H strain (HL) and two pairs of primers, R-s1 and R-a1, as well
as R-s2 and R-a2, which were located upstream of the splice donor
site D1 at nt 25533 and downstream of the splice acceptor site A2
at nt 26633 (A), and the primers, R-s3 and 5′R3, as well as R-s3
and 5′R4, which span the intron 1 (B). Based on sequencing results,
RT-PCR products of the UL21.5 spliced transcripts are indicated by
arrows with corresponding names. The products of transcripts I, II,
III and IV were amplified by RT-PCR using the primers, R-s1 and
R-a1, as well as R-s2 and R-a2 and were 1402, 1114, 338 and 302 bp
in length, respectively. Those of the UL21.5 transcript were 188 bp
(spliced) and 272 bp (unspliced). To exclude the possibility of DNA
contamination, a reverse reaction without reverse transcriptase
M-MLV (MLV−) was used as a negative control. |
 | Table IIISplicing patterns of UL21.5 MST
mapped in the study. |
Table III
Splicing patterns of UL21.5 MST
mapped in the study.
| Related donor | Related
acceptor | |
|---|
|
|
| |
|---|
| Intron name | Name | Position | Sequence | Name | Position | Sequence | Splicing
pattern |
|---|
| Int 1 | D3 | 27151 | CAG|GTAAAAC | A3 | 27235 |
CGCGGTTATCGTTTTTGCAG|C | GT-AG |
| Int 2 | D2 | 26309 | GCG|GTACGG | A1 | 26597 |
ATGTCGATCTCCATCGGCAG|A | GT-AG |
| Int 3 | D1 | 25533 | GCG|GTGAGT | A1 | 26597 |
ATGTCGATCTCCATCGGCAG|A | GT-AG |
| Int 4 | D1 | 25533 | GCG|GTGAGT | A2 | 26633 |
TCGTCTCCCTCACCGACCAG|C | GT-AG |
3′ and 5′ ends of UL21.5 multiple spliced
transcripts mapped by RACE
To further map the 3′ and 5′ ends of the UL21.5
multiple spliced transcripts (UL21.5MST), RACE experiments were
performed using the L RNA of the HCMV H strain. The sequencing
result of the 3′RACE product (Fig.
4A) identified a 3′ end at nt 27543 downstream of the poly(A)
signal (ATTAAA) at nt 27513–27518, which is consistent with that of
the transcripts identified by cDNA library screening.
In 5′RACE experiments, two specific products were
obtained using 5′R1 and 5′R2 (nested) primers (Table I and Fig. 4B). The sequencing results revealed
four possible 5′ ends located at nt 24835, nt 24895, nt 25220 and
nt 25244. Based on the structures of transcripts identified by
RT-PCR and the overlapping sequences, 16 possible transcript
sequences were obtained by linking the sequences obtained from 5′
and 3′RACE, as well as sequences obtained by RT-PCR (Table IV). Among these, the transcript
lengths with the possible 5′ ends at nt 24835 and nt 24895, and the
possible structures of transcripts III and IV were 1470–1566 nt,
which may correspond to the ~1517 nt transcripts found in northern
blot analysis. By contrast, the lengths of transcripts with the
possible 5′ ends at nt 25220 and nt 25244 and the possible
structures of transcripts III and IV were 1121–1179 nt, which may
correspond to the ~1250 nt transcripts, and those with the possible
5′ ends at nt 25220 and nt 25244, and the possible structures of
transcript II were 1946–1968 nt, which may correspond to the
1900–2100 nt transcripts identified in northern blot analysis. Four
ORFs were predicted in the spliced transcripts identified (Table IV). As the ORF predicted in the
UL21.5 spliced transcript, ORF A (from nt 27069 to nt 27151 and
from nt 27235 to nt 27466), ORF B (from nt 26845 to nt 27093) and
ORF C (from nt 26651 to nt 26929) were exactly the same in
transcripts II, III and IV. And ORF D, E and F were specific for
transcripts II, III and IV, respectively.
 | Table IVAnalyses of the UL21.5 multiple
spliced transcripts based on RACE. |
Table IV
Analyses of the UL21.5 multiple
spliced transcripts based on RACE.
| Transcripts with
possible 5′ ends and structures | Correspond to
transcripts found in northern blot analysis | Positions of the
predicted ORFs of the transcripts |
|---|
|
|---|
| Structure | 5′ ends | Length (nt) |
|---|
| I | nt 24835 | 2629 | --- | |
| nt 24895 | 2569 | --- | |
| nt 25220 | 2242 | --- | |
| nt 25244 | 2217 | --- | |
| II | nt 24835 | 2355 | --- | |
| nt 24895 | 2295 | --- | |
| nt 25220 | 1968 | 1900–2100 nt | A, B, C and D |
| nt 25244 | 1946 | 1900–2100 nt | A, B, C and D |
| III | nt 24835 | 1566 | ~1517 nt | A, B, C and E |
| nt 24895 | 1506 | ~1517 nt | A, B, C and E |
| nt 25220 | 1179 | ~1250 nt | A, B, C and E |
| nt 25244 | 1157 | ~1250 nt | A, B, C and E |
| IV | nt 24835 | 1530 | ~1517 nt | A, B, C and F |
| nt 24895 | 1470 | ~1517 nt | A, B, C and F |
| nt 25220 | 1143 | ~1250 nt | A, B, C and F |
| nt 25244 | 1121 | ~1250nt | A, B, C and F |
In order to confirm the 5′ ends located from nt
27040 to 27044, which were obtained in the cDNA library screening,
two other nested primers, 5′R3 and 5′R4 (Table I and Fig. 1), were used in 5′RACE. One ~300-bp
product was obtained (Fig. 4C).
The sequencing results of this product showed three possible 5′
ends located at nt 27040, 27041 and 27043, respectively; this was
in accordance with the results of cDNA library screening.
Determinant regulatory region for a
splice donor site and two splice acceptor sites
identified by an in vitro expression system
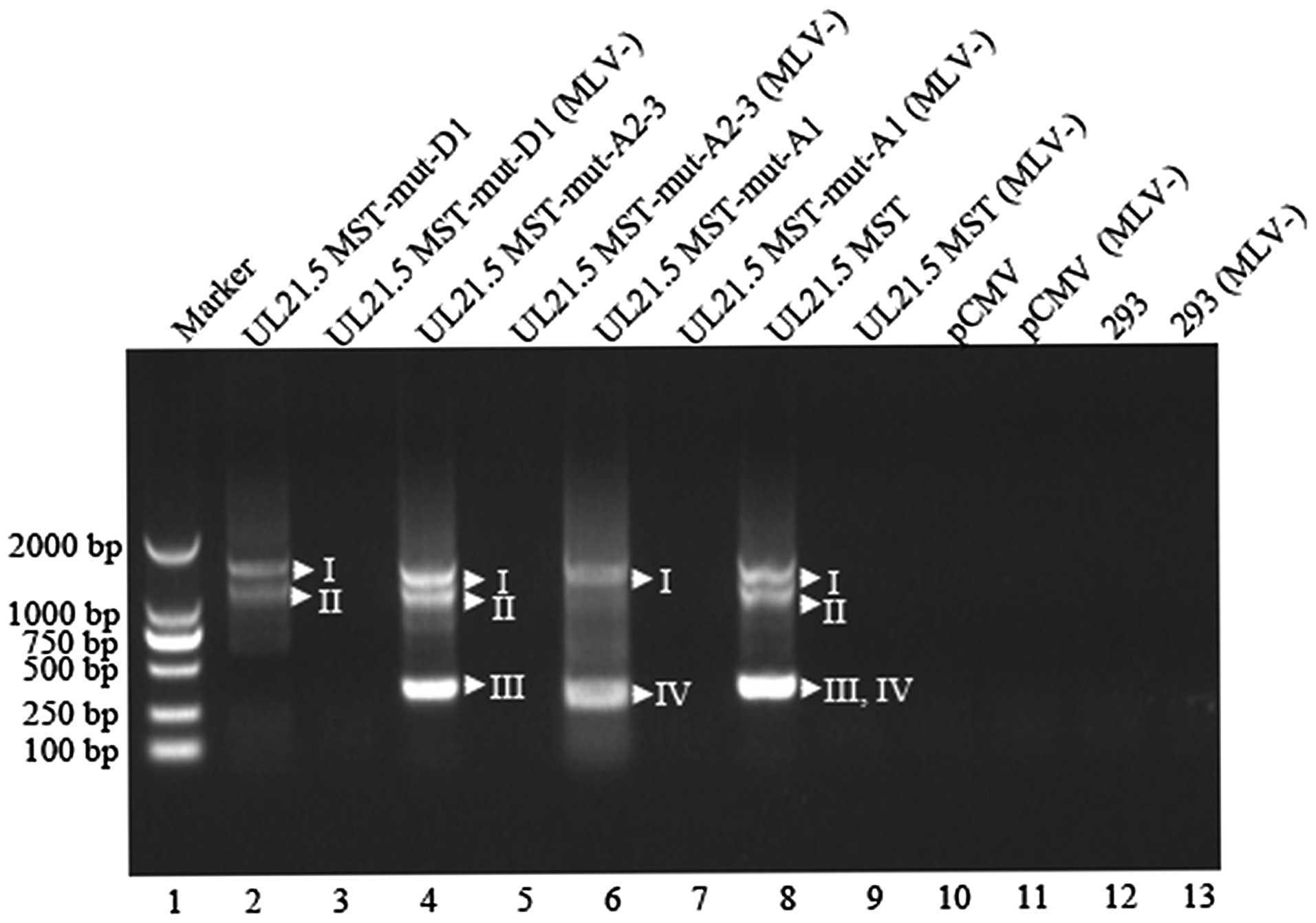
To confirm that the UL21.5 MST was derived from
splice events, the plasmid, pCMV-UL21.5 MST, was constructed. As
shown in lane 8 of Fig. 5, in
accordance with those identified by RT-PCR using the primers, R-s2
and R-a2, four spliced transcripts were confirmed by the sequencing
results of the bands.
To identify the determinant regulatory regions
necessary for splice sites D1 (at nt 25533), A1 (at nt 26597) and
A2 (at nt 26633) (Fig. 1) in the
UL21.5 gene locus, expression vectors containing mutant sequences
around the splice donor site D1 and adjacent to the splice acceptor
sites A1 and A2 were constructed (Table II). RT-PCR analyses were
performed using RNA preparations from the HEK 293 cells transfected
with pCMV-UL21.5-mut-D1, pCMV-UL21.5-mut-A1, pCMV-UL21.5-mut-A2-1,
pCMV-UL21.5-mut-A2-2 and pCMV-UL21.5-mut-A2-3, and one pair of
primers, R-s2 and R-a3. The sequencing results of the RT-PCR
products revealed that transcripts III and IV, which are relevant
to the splice donor site D1, were not obtained from the HEK 293
cells transfected with pCMV-UL21.5-mut-D1 (Fig. 5, lane 2 and Table II) due to a 9-bp sequence
mutation from nt 25531 to nt 25539 around the splice donor site D1.
However, transcripts II and III, which are relevant to the splice
site A1, were not detected in the HEK 293 cells transfected with
pCMV-UL21.5-mut-A1 (Fig. 5, lane
6 and Table II) for a 20-bp
sequence mutation from nt 26577 to nt 26596 just upstream the
splice acceptor site A1. The structures of UL21.5 MST from the HEK
293 cells transfected with pCMV-UL21.5-mut-A2-1 and
pCMV-UL21.5-mut-A2-2 were the same as those from the HEK 293 cells
transfected with pCMV-UL21.5 MST (data not shown). Only transcripts
from the HEK 293 cells transfected with pCMV-UL21.5-mut-A2-3, which
contains a mutant sequence from nt 26621 to nt 26650, showed the
loss of transcript IV (Fig. 5,
lane 4 and Table II), which is
relevant to the splice acceptor site A2, due to a further 10-bp
fragment mutation of GCTGTTCTGG from nt 26641 to 26650, compared to
the pCMV-UL21.5-mut-A2-2.
Discussion
It has been previously believed that HCMV mRNA
splicing is restricted to a relatively small IE region of the HCMV
genome (24), until a group of
spliced L genes were described (18). The UL21.5 transcript, which
consists of a spliced and a non-spliced transcript, was first
reported by Rawlinson and Barrell (18) and later re-termed by Bresnahan and
Shenk (17). The UL21.5
transcript is an important HCMV virion RNA for the virus
replication cycle (25). Together
with the HCMV virion, the UL21.5 transcript is delivered into the
infected cell and is translated to a secreted glycoprotein, which
functions as a viral chemokine decoy receptor specifically
interacting with the RANTES chemokine (17,19). It is also possible that virion-RNA
plays a structural role in the HCMV assembly.
In the present study, transcripts from the HCMV
UL21.5 region were further analyzed using a low passage HCMV
strain. Apart from the higher abundant UL21.5 transcripts reported,
three clusters of low-abundance transcripts were detected by probe
1 in L RNAs in northern blot experiments. Four spliced transcripts
with a 3′ co-terminus at nt 27543 of the UL21.5 gene locus were
identified by cDNA library screening, RACE and RT-PCR. One ORF,
which was the same as that predicted in the UL21.5 spliced
transcript, was predicted in all possible forms of the
low-abundance spliced transcripts II, III and IV. Apart from this
ORF, three transcript-specific ORFs were predicted in the
low-abundance spliced transcripts II, III and IV, respectively
(Table IV).
In general, three classes of transcripts are
expressed from a genome, including high-, intermediate- and
low-abundance transcripts (26).
Although the majority of the high- and intermediate-abundance
transcripts have been identified, it remains a serious challenge to
identify the low-abundance transcripts (27,28). To date, some low-abundance
transcripts have been identified in HCMV. As a low-abundance
spliced transcript from the IE gene region, the IE18 transcript was
previously detected in infected human monocyte-derived macrophages
at the IE phase, but was not detectable during normal infection
(29). The authors concluded that
the IE18 transcript exhibits cell type-specific expression
indicating differential regulation of the major IE gene region in
different permissive cell types (29). By detection in peripheral blood
mononuclear cells of organ transplant recipients with different
viral DNA loads, the natural killer (NK) cell decoy gene, UL18, was
demonstrated to be produced through a low-abundance transcript late
during the infectious cycle at a time coincidental with the
increased risk of NK cell lysis (30). It can be hypothesized that the
existence of the low-abundance UL21.5 spliced transcripts in the
HCMV genome may also play some unknown role during HCMV
infection.
Little was known about splicing situations in HCMV
until deep sequencing was used to bring high resolution to the HCMV
transcriptome (16). Multiple
spliced transcripts have been found in UL37, UL73, UL111A, UL122,
UL123, UL128 and US3 to date (14,15,29,31–39). In the present study, four
previously unrecognized spliced transcripts were identified. Based
on the structures of UL21.5 MST identified in this study and the
findings of Rawlinson and Barrell (18), three splice donor sites and three
splice acceptor sites were proposed in the UL21.5 gene locus.
According to the structures of the four introns indentified in
UL21.5 MST, the splicing pattern is a canonical pattern of
GT-AG.
Three signals are known to direct splicing events,
including the 5′ splice site (5′ss) at the 5′ end of the intron,
the polypyrimidine tract/3′ splice site (PPT-3′ss) at the 3′ end of
the intron and a branch site (BS) upstream of the PPT-3′ss
(40–43). According to previous studies
(40, 41), the 5′ element generally is located
in the position −3 to +6 (the third position upstream the 5′ss and
the sixth position downstream the 5′ss, respectively), and the
essential 3′ element is generally located at nt −3 to −12 upstream
of the 3′ss (44–45). To further validate these canonical
splice sites and find the determinant regulatory region necessary
for splicing events, an in vitro system was used in the
present study. Based on the above principles, the expression
plasmid, pCMV-UL21.5 MST, and plasmids containing mutant 5′ element
or 3′ element of the UL21.5 MST coding sequences were constructed
and used in this system. Similar to those found in HCMV H-infected
MRC5 cells, four spliced transcripts were identified from HEK 293
cells transfected with the plasmid, pCMV-UL21.5 MST. This finding
confirmed that the splicing events occurred during transcription.
However, when mutation occurred adjacent to the splice donor site
D1 at nt 25533 (in plasmid pCMV-UL21.5-mut-D1), D1-related
transcripts (transcripts III and IV) of UL21.5 MST were not
detected from the transfected cells. This result indicated that the
9-bp sequence around D1 from nt 25531 to nt 25539, which is exactly
in the 5′ element, is necessary for the activity of the splice
donor site D1. Similarly, the 20-bp sequence just upstream of the
splice site A1 at nt 26597 from nt 26577 to nt 26596, which
includes the 3′ element, is important for splice acceptor site A1,
as mutation in this region resulted in the loss of A1-related
transcripts (transcripts II and III). However, the determinant
regulatory region that is necessary for activity of splice acceptor
site A2 at nt 26633 is located not in the 3′ element but in the
sequence from nt 26641 to nt 26650, as the plasmid containing a
mutant sequence in this region (pCMV-UL21.5-mut-A2-3) led to the
loss of A2-related transcripts (transcript IV).
Overall, in addition to the UL21.5 transcript, four
novel low-abundance 3′-coterminal spliced transcripts were
identified to transcribe from the UL21.5 gene locus during
late-phase infection. Three splice donor sites and three splice
acceptor sites found in this region were validated to be functional
splice sites by an in vitro expression system. Determinant
regulatory regions of one splice donor site and two splice acceptor
sites were identified in corresponding regions by mutation
experiments. Alteration of the donor or acceptor splice sites may
result in the loss of their related transcripts.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (30672248, 30901625, 81171580
and 81171581) and the Specialized Research Fund for the Doctoral
Program of Higher Education (20112104110012) and the Outstanding
Scientific Fund of Shengjing Hospital.
References
|
1
|
Mocarski E, Shenk T and Pass RF:
Cytomegaloviruses. Fields Virology. Knipe DM, Howley PM, Griffin
DE, Lamb RA, Martin MA, Roizman B and Straus SE: 5th edition.
Lippincott-Raven Publishers; Philadelphia, PA: pp. 2702–2772.
2007
|
|
2
|
Mocarski ES: Cytomegaloviruses and their
replication. Fields virology. Fields BN, Knipe DM and Howley PM: 2.
3rd edition. Lippincott-Raven Publishers; Philadelphia, PA: pp.
2447–2492. 1996
|
|
3
|
Du G, Dutta N, Lashmit P and Stinski MF:
Alternative splicing of the human cytomegalovirus major
immediate-early genes affects infectious-virus replication and
control of cellular cyclin-dependent kinase. J Virol. 85:804–817.
2011. View Article : Google Scholar :
|
|
4
|
Griffiths PD, Cope AV, Hassan-Walker AF
and Emery VC: Diagnostic approaches to cytomegalovirus infection in
bone marrow and organ transplantation. Transpl Infect Dis.
1:179–186. 1999. View Article : Google Scholar
|
|
5
|
Chee MS, Bankier AT, Beck S, et al:
Analysis of the protein-coding content of the sequence of human
cytomegalovirus strain AD169. Curr Top Microbiol Immunol.
154:125–169. 1990.PubMed/NCBI
|
|
6
|
Davison AJ, Dolan A, Akter P, et al: The
human cytomegalovirus genome revisited: comparison with the
chimpanzee cytomegalovirus genome. J Gen Virol. 84:17–28. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Dolan A, Cunningham C, Hector RD, et al:
Genetic content of wild-type human cytomegalovirus. J Gen Virol.
85:1301–1312. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dunn W, Trang P, Zhong Q, Yang E, van
Belle C and Liu F: Human cytomegalovirus expresses novel microRNAs
during productive viral infection. Cell Microbiol. 7:1684–1695.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Murphy E, Rigoutsos I, Shibuya T and Shenk
TE: Reevaluation of human cytomegalovirus coding potential. Proc
Natl Acad Sci USA. 100:13585–13590. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Murphy E, Yu D, Grimwood J, et al: Coding
potential of laboratory and clinical strains of human
cytomegalovirus. Proc Natl Acad Sci USA. 100:14976–14981. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ma Y, Wang N, Li M, et al: Human CMV
transcripts: an overview. Future Microbiol. 7:577–593. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Maniatis T and Reed R: An extensive
network of coupling among gene expression machines. Nature.
416:499–506. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sandri-Goldin RM: Viral regulation of mRNA
export. J Virol. 78:4389–4396. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Awasthi S, Isler JA and Alwine JC:
Analysis of splice variants of the immediate-early 1 region of
human cytomegalovirus. J Virol. 78:8191–8200. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shirakata M, Terauchi M, Ablikim M, et al:
Novel immediate-early protein IE19 of human cytomegalovirus
activates the origin recognition complex I promoter in a
cooperative manner with IE72. J Virol. 76:3158–3167. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gatherer D, Seirafian S, Cunningham C, et
al: High-resolution human cytomegalovirus transcriptome. Proc Natl
Acad Sci USA. 108:19755–19760. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bresnahan WA and Shenk T: A subset of
viral transcripts packaged within human cytomegalovirus particles.
Science. 288:2373–2376. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Rawlinson WD and Barrell BG: Spliced
transcripts of human cytomegalovirus. J Virol. 67:5502–5513.
1993.PubMed/NCBI
|
|
19
|
Dai W, Wade B and Thomas S: Human
cytomegalovirus encodes a highly specific RANTES decoy receptor.
Proc Natl Acad Sci USA. 101:16642–16647. 2004. View Article : Google Scholar
|
|
20
|
Ma YP, Ruan Q, Ji YH, et al: Novel
transcripts of human cytomegalovirus clinical strain found by cDNA
library screening. Genet Mol Res. 10:566–575. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sun Z, Ren G, Ma Y, et al: Transcription
pattern of UL131A-128 mRNA in clinical strains of human
cytomegalovirus. J Biosci. 35:365–370. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Qi Y, Ma Y, He R, et al: Characterization
of 3′ termini of human cytomegalovirus UL138–UL145 transcripts in a
clinical strain. Microbiol Immunol. 55:95–99. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Adam BL, Jervey TY, Kohler CP, et al: The
human cytomegalovirus UL98 gene transcription unit overlaps with
the pp28 true late gene (UL99) and encodes a 58-kilodalton early
protein. J Virol. 69:5304–5310. 1995.PubMed/NCBI
|
|
24
|
Stenberg RM, Witte PR and Stinski MF:
Multiple spliced and unspliced transcripts from human
cytomegalovirus immediate-early region 2 and evidence for a common
initiation site within immediate-early region 1. J Virol.
56:665–675. 1985.PubMed/NCBI
|
|
25
|
Terhune SS, Schröer J and Shenk T: RNAs
are packaged into human cytomegalovirus virions in proportion to
their intracellular concentration. J Virol. 78:10390–10398. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bishop J, Morton J, Rosbach M and
Richardson M: Three abundance classes in HeLa cell messenger RNA.
Nature. 250:199–204. 1974. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang SM, Fears SC, Zhang L, Chen JJ and
Rowley JD: Screening polydA/dT)− cDNAs for gene
identification. Proc Natl Acad Sci USA. 97:4162–4167. 2000.
View Article : Google Scholar
|
|
28
|
Kapranov P, Cawley SE, Drenkow J, et al:
Large-scale transcriptional activity in chromosomes 21 and 22.
Science. 296:916–919. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kerry JA, Sehgal A, Barlow SW, et al:
Isolation and characterization of a low-abundance splice variant
from the human cytomegalovirus major immediate-early gene region. J
Virol. 69:3868–3872. 1995.PubMed/NCBI
|
|
30
|
Hassan-Walker AF, Cope AV, Griffiths PD
and Emery VC: Transcription of the human cytomegalovirus natural
killer decoy gene, UL18, in vitro and in vivo. J Gen Virol.
79:2113–2116. 1998.PubMed/NCBI
|
|
31
|
Adair R, Liebisch GW and Colberg-Poley AM:
Complex alternative processing of human cytomegalovirus UL37
pre-mRNA. J Gen Virol. 84:3353–3358. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tenney DJ and Colberg-Poley AM: RNA
analysis and isolation of cDNAs derived from the human
cytomegalovirus immediate-early region at 0.24 map units.
Intervirology. 31:203–214. 1990.PubMed/NCBI
|
|
33
|
Scalzo AA, Forbes CA, Smith LM and Loh LC:
Transcriptional analysis of human cytomegalovirus and rat
cytomegalovirus homologues of the M73/M73.5 spliced gene family.
Arch Virol. 154:65–75. 2009. View Article : Google Scholar
|
|
34
|
Kotenko SV, Saccani S, Izotova LS,
Mirochnitchenko OV and Pestka S: Human cytomegalovirus harbors its
own unique IL-10 homolog (cmvIL-10). Proc Natl Acad Sci USA.
97:1695–1700. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lockridge KM, Zhou SS, Kravitz RH, et al:
Primate cytomegalovirus encode and express an IL-10-like protein.
Virology. 268:272–280. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kondo K, Xu J and Mocarski ES: Human
cytomegalovirus latent gene expression in granulocyte-macrophage
progenitors in culture and in seropositive individuals. Proc Natl
Acad Sci USA. 93:11137–11142. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hutchinson NI, Sondermeyer RT and Tocci
MJ: Organization and expression of the major genes from the long
inverted repeat of the human cytomegalovirus genome. Virology.
155:160–171. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Greenaway PJ and Wilkinson GW: Nucleotide
sequence of the most abundantly transcribed early gene of human
cytomegalovirus strain AD169. Virus Res. 7:17–31. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Demarchi JM: Human cytomegalovirus DNA:
restriction enzyme cleavage maps and map locations for
immediate-early, early, and late RNAs. Virology. 114:23–38. 1981.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Black DL: Mechanisms of alternative
pre-messenger RNA splicing. Annu Rev Biochem. 72:291–336. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Hastings ML and Krainer AR: Pre-mRNA
splicing in the new millennium. Curr Opin Cell Biol. 13:302–309.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Schwartz SH, Silva J, Burstein D, Pupko T,
Eyras E and Ast G: Large-scale comparative analysis of splicing
signals and their corresponding splicing factors in eukaryotes.
Genome Res. 18:88–103. 2008. View Article : Google Scholar :
|
|
43
|
Cartegni L, Chew SL and Krainer AR:
Listening to silence and understanding nonsense: exonic mutations
that affect splicing. Nat Rev Genet. 3:285–298. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Reed R and Maniatis T: Intron sequences
involved in lariat formation during pre-mRNA splicing. Cell.
41:95–105. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Ruskin B and Green MR: Role of the 3′
splice site consensus sequence in mammalian pre-mRNA splicing.
Nature. 317:732–734. 1985. View Article : Google Scholar : PubMed/NCBI
|