Introduction
Since the discovery of radiation, practically every
industry has adopted its use in various ways. Consequently, there
has been growing concern about the biological consequences of
exposure to low-dose radiation (LDR) from medical processes or
other industrial facilities and an increased interest in genes that
are specific diagnostic markers of LDR-induced damage (1). However, the exact health risks from
exposure to LDR remain controversial and unclear (1). There are currently three models for
measuring the effects of LDR on humans. First is the linear
non-threshold (LNT) model, which is more commonly applied to
high-dose exposure, but suggests that even LDR can increase the
risk of carcinogenesis (2).
Second, unlike the LNT model, the threshold model suggests that
there is a level of radiation exposure that is harmless (3). Third is an alternative model, termed
hormesis or adaptive response, which suggests that LDR is
potentially protective and has beneficial effects (3).
Skin, which is both the largest organ and the
outermost layer of the human body, consists of two main layers, the
epidermis and dermis. Human dermal fibroblasts (HDFs) are the most
abundant cells in the dermis of the skin, and contribute to skin
firmness and elasticity through the synthesis of collagen (4,5).
Among the types of collagen, type 1 collagen is the most common
type, and is composed of two α1(I) chains and one α2(I) chain
encoded by the COL1A1 and COL1A2 genes, respectively
(5). HDFs also express matrix
metalloproteinases (MMPs), which negatively regulate collagen
levels through collagen degradation (6). The overexpression of the MMP1
gene has been observed in aged/photoaged skin in vivo
(6). Reflecting their position in
the human body, HDFs are more vulnerable than other cells to toxic
environmental agents, such as ultraviolet (UV) radiation and even
ionizing radiation. UV-induced photoaging involves the
downregulation of COL1A1 and COL1A2 and the
consequent loss of collagen, leading to fine wrinkles (4). Moreover, various studies have
indicated that skin aging induced by exposure to UV radiation is
mediated by the upregulation of MMP1 expression (4). By contrast, β-radiation has a wound
healing effect by increasing the production of collagen I
production in skin fibroblasts (7). Although the effects of UV radiation
and the underlying mechanisms responsible for UV-mediated cellular
responses have been extensively studied (4,8),
much remains unknown about the biological response of human skin to
LDR.
MicroRNAs (miRNAs or miRs) are small RNAs consisting
of 19–25 nucleotides that play essential roles in growth,
differentiation and cell death (13). A number of studies have
demonstrated that miRNAs are important regulators of cellular
responses to radiation. For example, miR-34, which is regulated by
the p53 tumor suppressor protein (9), is induced by γ-radiation in
vitro and in vivo (10,11). In addition, a recent study
demonstrated that the irradiation of human keratinocytes located in
the skin epidermis with 10 mGy and 6 Gy of γ-radiation induces a
pattern of miRNA expression that is strongly related to the
differentiation status of the irradiated cells (12). Furthermore, our previous study
demonstrated that specific miRNAs are expressed in a γ-radiation
dose-dependent manner in the human lymphoblast cell line, IM9
(13). We also identified miR-16,
miR-202, miR-303 and miR-572 as ionizing radiation-responsive
miRNAs in the IM9 cell line (14). These findings strongly suggest
that the elucidation of miRNA-based cellular mechanisms is
essential for the understanding of LDR-mediated responses in human
skin, and that certain miRNAs may be principal diagnostic markers
for LDR responses.
In the present study, we used miRNA microarray
analysis to demonstrate that LDR alters the expression profiles of
specific miRNAs in normal human dermal fibroblasts (NHDFs). We also
demonstrated that the miRNA expression profiles differ at 6 and 24
h post-irradiation. Furthermore, our results indicate that LDR has
a potential anti-aging effect through the regulation of
COL1A1 and MMP1 expression.
Materials and methods
Cell culture and irradiation
NHDFs were purchased from Lonza (Basel, Switzerland)
and maintained in Dulbecco’s modified Eagle’s medium (DMEM; Gibco;
Life Technologies, Grand Island, NY, USA) containing 10% fetal
bovine serum (FBS; Sigma-Aldrich, St. Louis, MO, USA). To evaluate
the pattern of the cell cycle and miRNA expression,
7×105 cells were seeded onto 60-mm culture plates and
cultured for 24 h. The cells were irradiated with 0.1 Gy of
γ-radiation using a Gammacell 3000 Elan irradiator
(137Cs γ-ray source; MDS Nordion, Ottawa, ON,
Canada).
Cell viability assay
The cytotoxic effects of γ-radiation (0.1 Gy) on the
NHDFs were investigated using a
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay (Sigma-Aldrich) according to the manufacturer’s instructions.
Briefly, the NHDFs were seeded and irradiated with 0.1 Gy of
γ-radiation. Following incubation for 6 or 24 h, MTT solution was
added to the cultured cells, followed by incubation at 37°C for 1
h. The medium was removed and the blue formazan crystals trapped in
the cells were dissolved in dimethyl sulfoxide (DMSO)
(Sigma-Aldrich). Cell viability was measured using an iMark plate
reader (Bio-Rad, Hercules, CA, USA) at 590 nm with a reference
filter of 620 nm. All results are presented as the mean percentage
± standard deviation (SD) of 3 independent experiments. A P-value
<0.05 as determined by the Student’s t-test was considered to
indicate a statistically significant difference.
Analysis of cell cycle by flow
cytometry
To analyze the cells in different phases of the cell
cycle, irradiated NHDFs were fixed by the addition of cold 70%
ethanol and stained by incubation with propidium iodide (PI)
staining solution [50 μg/ml PI, 0.5% Triton X-100 (both from
Sigma-Aldrich), and 100 μg/ml RNase (Qiagen, Hilden, Germany)] at
37°C for 1 h. The PI fluorescence intensity was detected using a BD
FACSCalibur flow cytometer (BD Biosciences, San Jose, CA, USA). The
mean PI fluorescence intensity was obtained from 10,000 cells using
the FLH-2 channel.
RNA purification and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was purified using TRIzol reagent (Life
Technologies, Carlsbad, CA, USA) according to the manufacturer’s
instructions. The purity and concentration of the RNA was assessed
by the ratio of absorbance at 230, 260 and 280 nm using
MaestroNano®, a micro-volume spectrophotometer
(Maestrogen, Las Vegas, NV, USA). cDNA was synthesized using a
SuperScript® III First-Strand Synthesis system for
RT-PCR (Life Technologies). Quantitative (real-time) PCR was
performed using SYBR®-Green PCR Master Mix and
SYBR®-Green RT-PCR reagents kit (Life Technologies) with
specific primers for COL1A1 and MMP1 and a Line-Gene
K RT-PCR instrument (Bioer Technology Co., Ltd., Hangzhou, China).
The CT value for each gene was normalized to glyceraldehyde
3-phosphate dehydrogenase (GAPDH). The 2−ΔΔCT
method was used to calculate the relative expression level of each
gene. The forward (F) and reverse (R) primers for human
COL1A1, MMP1 and GAPDH were 5′-AGCCAGCAGATCGAG
AACAT-3′ (COL1A1-F) and 5′-TCTTGTCCTTGGGGTT CTTG-3′
(COL1A1-R); 5′-GATGTGGAGTGCCTGATGTG-3′ (MMP1-F) and
5′-TGCTTGACCCTCAGAGACCT-3′ (MMP1-R);
5′-CGCTCTCTGCTCCTCCTGTT-3′ (GAPDH-F) and
5′-CCATGGTGTCTGAGCGATGT-3′ (GAPDH-R).
miRNA microarray analysis
RNA integrity was estimated using an Agilent 2100
Bioanalyzer® (Agilent Technologies, Santa Clara, CA,
USA). RNA samples that showed A260/280 and A260/A230 values >1.8
and an RNA integrity number >8.0 were subjected to
microRNA-based microarray analysis. Microarray analysis was
performed using a SurePrint G3 Human version 16 miRNA 8×60K
microarray kit (Agilent Technologies), according to a previously
described protocol (15).
Briefly, 50 ng of purified RNA were treated with calf intestine
alkaline phosphatase and labeled with cyanine 3-cytidine
bisphosphate. The labeled RNA was purified using a Micro Bio-Spin
P-6 column (Bio-Rad Laboratories, Hercules, CA, USA) and hybridized
with the microarray kit. The fluorescence intensity of each probe
was measured using Scanner and Feature Extraction software (Agilent
Technologies). The digitalized fluorescence intensity was analyzed
using GeneSpring GX version 11.5 software (Agilent Technologies).
The raw data were filtered using FLAG and t-tests, and applied to
the fold change analysis. The fold change analysis was conducted
based on a factor of 2.0-fold change between non-irradiated control
cells and irradiated cells.
Target prediction and bioinformatics
analysis of miRNAs
We first analyzed putative target genes of the
miRNAs using the DIANA-microT bioinformatic software tool
(http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=microT_CDS/index),
as previously described (16).
The prediction of target genes was limited by setting the threshold
value to 0.8. The putative target genes of each miRNA were further
analyzed for biological function using the Kyoto Encyclopedia of
Genes and Genomes (KEGG) pathways and Database for Annotation,
Visualization and Interrogate Discovery (DAVID, http://david.abcc.ncifcrf.gov/home.jsp)
bioinformatics resources version 6.7 according to the developer’s
protocol (17). The
‘KEGG_pathway’ category was processed by setting the threshold of
EASE score, a modified Fisher exact P-value, to 0.1, and involved
KEGG pathways displaying a value >1% (percentage of involved
target genes/total target genes in each pathway) were selected.
Results
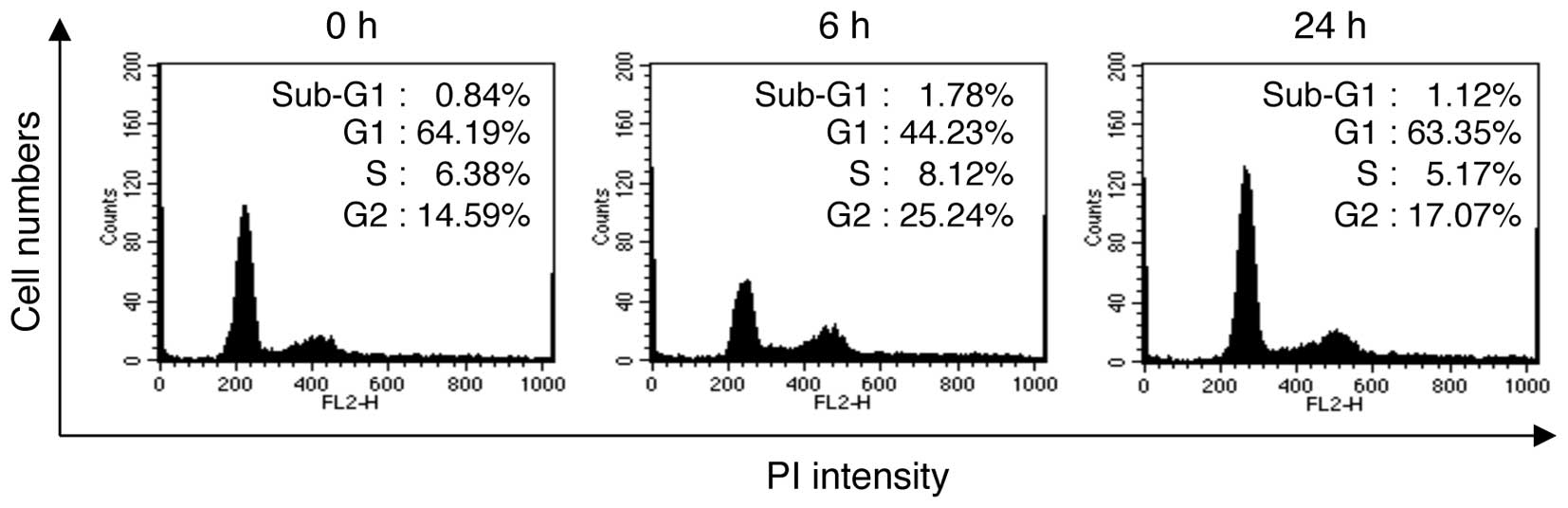
Cell cycle arrest in the G2/M phase is an
early adaptive response to LDR in NHDFs
We first determined whether LDR (0.1 Gy) affects the
viability of NHDFs. For a more precise analysis, the effects of LDR
on cell viability were analyzed at 2 different time points (6 and
24 h) after irradiation. There were no marked differences in the
viability of the cells exposed to LDR and the control cells, as
determined by MTT assay. At 6 and 24 h after irradiation, cell
viability was reduced to 92.94±3.53 and 98.04±1.82% of the control
value, respectively (data not shown). We then examined the cell
cycle patterns following the exposure of NHDFs to LDR. The cells
were irradiated with 0.1 Gy of γ-radiation and then incubated for 6
and 24 h. Following incubation, the cells were stained with PI and
the cell cycle distribution was analyzed by flow cytometry.
Exposure to LDR decreased the proportion of cells in the G1 phase
from 64.19% at 0 h to 44.23% at 6 h post-irradiation, whereas the
proportion of cells in the G2/M phase increased from 14.59 to
25.24% (Fig. 1). Of note, at 24 h
post-irradiation, the proportion of cells in the G1 phase was
63.35% and the proportion of cells in the G2/M phase was 17.07%.
These results indicate that G2/M cell cycle arrest was observed in
the NHDFs at a relatively early time point (~6 h) post-irradiation;
however, this arrest was reversed at later time points (~24 h)
post-irradiation.
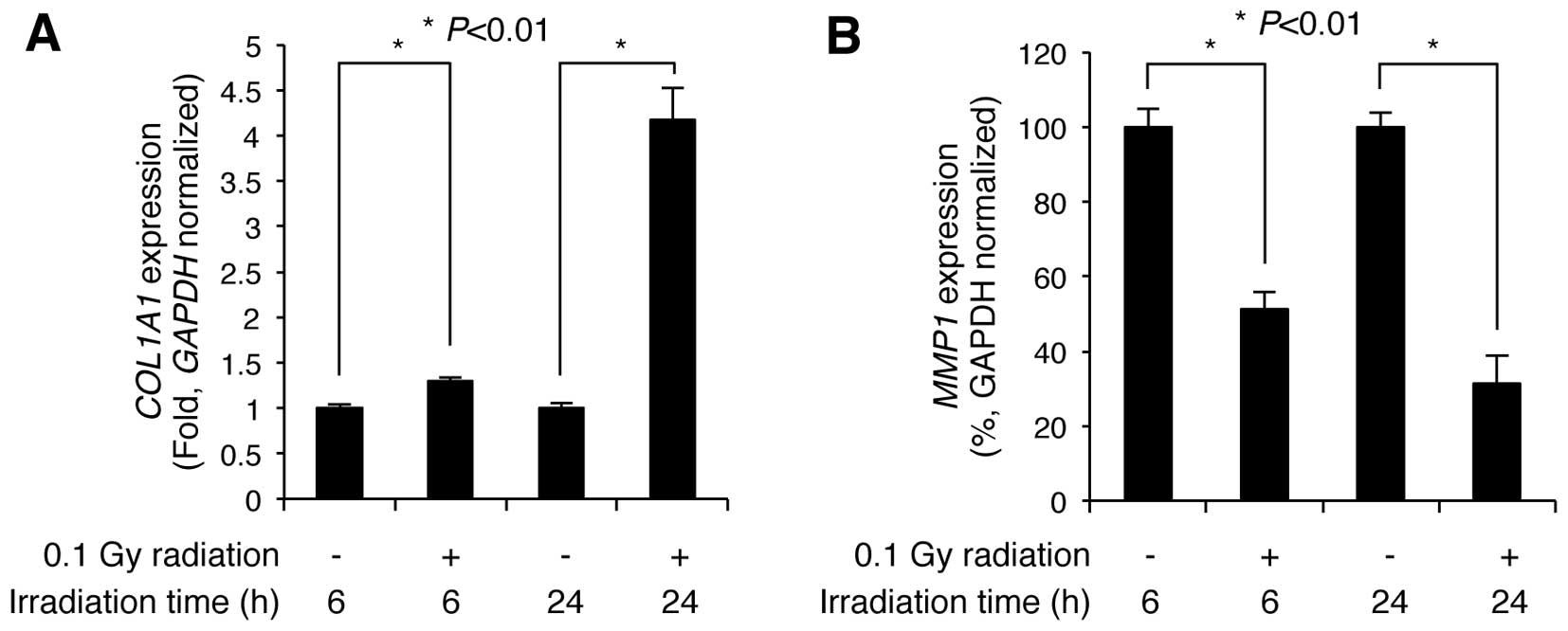
Low-dose radiation alters the expression
levels of COL1A1 and MMP1 in NHDFs
NHDFs within the dermis layer of the skin contribute
to skin firmness and elasticity by regulating the expression of
genes associated with collagen synthesis, including COL1A1
and MMP1 (5). Therefore,
we wished to examined whether LDR affects the expression levels of
these genes. The NHDFs were seeded and irradiated with 0.1 Gy of
γ-radiation. Following irradiation, the cells were incubated for an
additional 6 and 24 h, and then subjected to RT-qPCR to determine
the expression levels of COL1A1 and MMP1. Compared
with the non-irradiated control cells, COL1A1 expression was
slightly increased 1.29±0.03-fold (n=3) at 6 h post-irradiation,
but markedly increased 4.17±0.34-fold (n=3) by 24 h
post-irradiation (Fig. 2A).
Additionally, MMP1 expression significantly decreased to
51.37±4.49 and 31.33±7.44% (n=3) of the control levels at 6 and 24
h following irradiation, respectively (Fig. 2B). These results indicate that LDR
significantly induces the upregulation of COL1A1 and the
downregulation of MMP1 in NHDFs.
Specific miRNAs respond to LDR in
NHDFs
Although it is known that miRNA-mediated processes
are important in the cellular response to radiation (18), to the best of our knowledge, miRNA
responses to LDR (≥0.1 Gy) in NHDFs have not previously been
investigated. To explore the role of miRNAs in LDR-mediated
cellular responses in NHDFs, we performed miRNA microarray analysis
using a SurePrint G3 Human version 16 miRNA 8×60K microarray, which
contains 2,006 human miRNA probes. Purified RNA isolated from the
cells at 6 and 24 h post-irradiation was labeled and hybridized
onto the microarray as described in the ‘Materials and methods’.
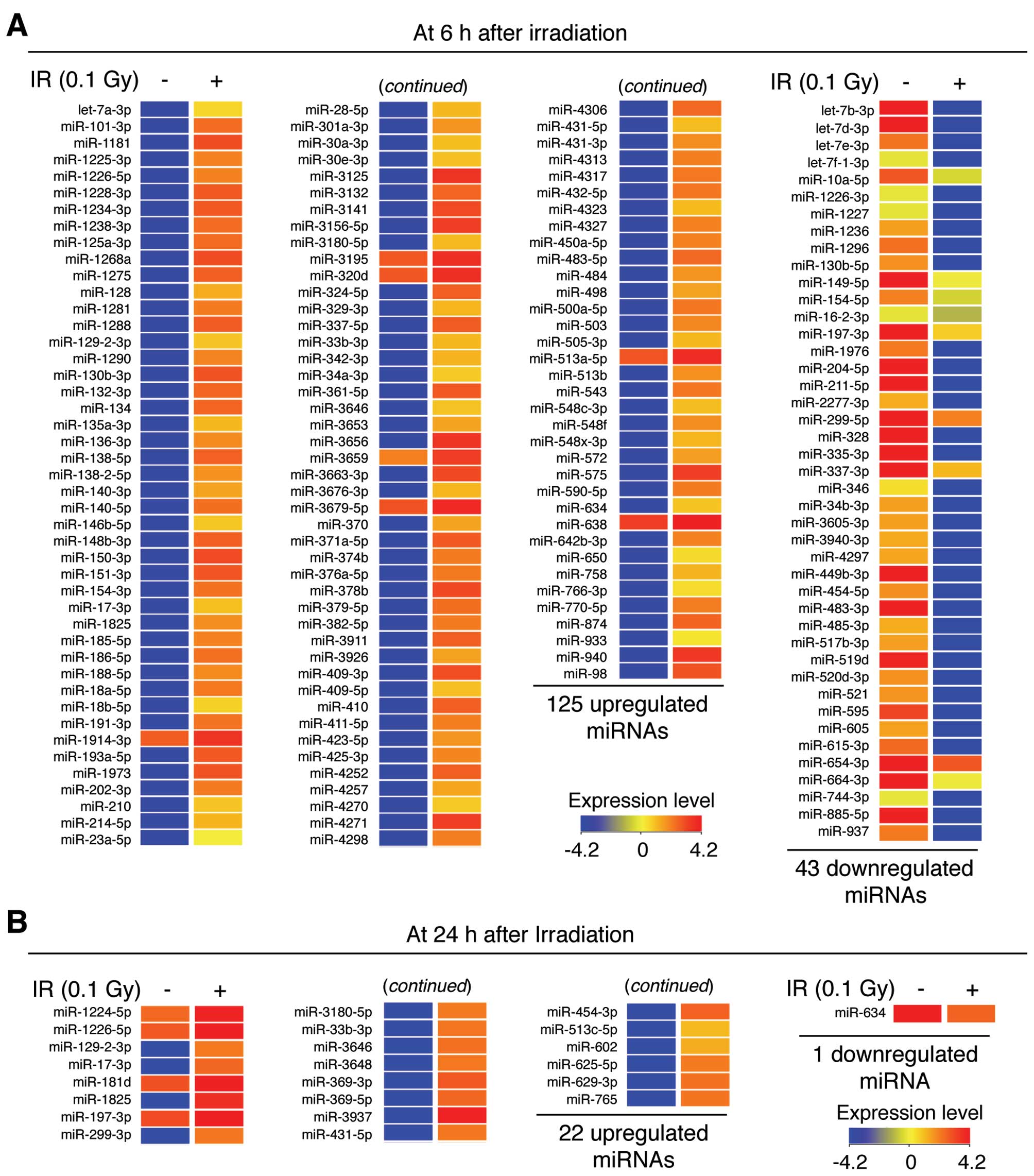
miRNAs that showed a ≥2.0-fold increase or decrease in expression
were selected using GeneSpring GX software. A number of miRNAs were
found to be significantly regulated in response to LDR in the NHDFs
(Fig. 3). At 6 h
post-irradiation, 125 miRNAs were upregulated and 43 miRNAs were
downregulated, indicating that LDR induces changes in the
expression of specific miRNAs during the early adaptive response in
NHDFs (Figs. 3A and 4A). Notably, the expression levels of
miR-3656, miR-3125 and miR-940 were significantly increased by
493.88-, 493.78- and 450.69-fold, respectively, whereas the
expression levels of miR-328, miR-885-5p and let-7d-3p were
significantly downregulated by 967.50-, 934.72- and 822.64-fold,
respectively (Table I).
Additionally, 22 miRNAs were upregulated and 1 miRNA was
downregulated at 24 h post-irradiation (Figs. 3B and 4A). Of these, miR-3937, miR-1825 and
miR-369-3p were significantly upregulated by 121.42-, 98.92- and
63.70-fold, whereas the expression level of miR-634 was
downregulated by 2.32-fold (Table
II). The complete list of differentially expressed miRNAs is
provided in Tables I and II. These data suggest that
miRNA-mediated responses are essential in the cellular
radio-adaptive response to LDR in NHDFs.
 | Table ImiRNAs that showed a >2-fold change
in expression in NHDFs at 6 h following exposure to 0.1 Gy
γ-radiation. |
Table I
miRNAs that showed a >2-fold change
in expression in NHDFs at 6 h following exposure to 0.1 Gy
γ-radiation.
| miRNA (Homo
sapiens) | Fold change |
|---|
| let-7a-3p | 42.0 |
| miR-101-3p | 193.4 |
| miR-1181 | 319.3 |
| miR-1225-3p | 139.0 |
| miR-1226-5p | 136.2 |
| miR-1228-3p | 259.7 |
| miR-1234-3p | 261.6 |
| miR-1238-3p | 193.0 |
| miR-125a-3p | 195.2 |
| miR-1268a | 320.7 |
| miR-1275 | 236.8 |
| miR-128 | 76.4 |
| miR-1281 | 153.5 |
| miR-1288 | 248.4 |
| miR-129-2-3p | 52.7 |
| miR-1290 | 135.6 |
| miR-130b-3p | 285.6 |
| miR-132-3p | 214.9 |
| miR-134 | 215.9 |
| miR-135a-3p | 64.0 |
| miR-136-3p | 121.1 |
| miR-138-5p | 223.7 |
| miR-138-2-5p | 109.3 |
| miR-140-3p | 84.9 |
| miR-140-5p | 186.7 |
| miR-146b-5p | 52.4 |
| miR-148b-3p | 233.3 |
| miR-150-3p | 333.3 |
| miR-151-3p | 277.5 |
| miR-154-3p | 180.8 |
| miR-17-3p | 57.4 |
| miR-1825 | 115.9 |
| miR-185-5p | 136.3 |
| miR-186-5p | 181.3 |
| miR-188-5p | 120.5 |
| miR-18a-5p | 160.3 |
| miR-18b-5p | 44.7 |
| miR-191-3p | 175.7 |
| miR-1914-3p | 2.0 |
| miR-193a-5p | 265.3 |
| miR-1973 | 285.5 |
| miR-202-3p | 154.2 |
| miR-210 | 53.1 |
| miR-214-5p | 67.0 |
| miR-23a-5p | 28.0 |
| miR-28-5p | 64.3 |
| miR-301a-3p | 107.8 |
| miR-30a-3p | 58.9 |
| miR-30e-3p | 56.5 |
| miR-3125 | 493.8 |
| miR-3132 | 215.1 |
| miR-3141 | 331.3 |
| miR-3156-5p | 404.0 |
| miR-3180-5p | 62.5 |
| miR-3195 | 2.1 |
| miR-320d | 2.0 |
| miR-324-5p | 229.7 |
| miR-329-3p | 65.6 |
| miR-337-5p | 249.5 |
| miR-33b-3p | 65.0 |
| miR-342-3p | 68.9 |
| miR-34a-3p | 49.1 |
| miR-361-5p | 256.2 |
| miR-3646 | 55.1 |
| miR-3653 | 84.3 |
| miR-3656 | 493.9 |
| miR-3659 | 2.8 |
| miR-3663-3p | 337.7 |
| miR-3676-3p | 65.3 |
| miR-3679-5p | 2.2 |
| miR-370 | 84.7 |
| miR-371a-5p | 251.2 |
| miR-374b | 156.3 |
| miR-376a-5p | 156.1 |
| miR-378b | 336.1 |
| miR-379-5p | 186.6 |
| miR-382-5p | 153.4 |
| miR-3911 | 229.8 |
| miR-3926 | 86.6 |
| miR-409-3p | 310.1 |
| miR-409-5p | 56.5 |
| miR-410 | 234.0 |
| miR-411-5p | 145.1 |
| miR-423-5p | 106.4 |
| miR-425-3p | 130.6 |
| miR-4252 | 234.1 |
| miR-4257 | 82.0 |
| miR-4270 | 50.3 |
| miR-4271 | 405.0 |
| miR-4298 | 145.9 |
| miR-4306 | 203.6 |
| miR-431-5p | 59.5 |
| miR-431-3p | 118.4 |
| miR-4313 | 144.6 |
| miR-4317 | 160.7 |
| miR-432-5p | 165.2 |
| miR-4323 | 61.0 |
| miR-4327 | 139.7 |
| miR-450a-5p | 140.4 |
| miR-483-5p | 202.3 |
| miR-484 | 105.4 |
| miR-498 | 86.3 |
| miR-500a-5p | 162.6 |
| miR-503 | 127.6 |
| miR-505-3p | 64.2 |
| miR-513a-5p | 2.0 |
| miR-513b | 111.6 |
| miR-543 | 166.3 |
| miR-548c-3p | 59.9 |
| miR-548f | 117.5 |
| miR-548x-3p | 66.2 |
| miR-572 | 92.5 |
| miR-575 | 401.3 |
| miR-590-5p | 153.7 |
| miR-634 | 53.6 |
| miR-638 | 2.0 |
| miR-642b-3p | 141.5 |
| miR-650 | 38.7 |
| miR-758 | 67.6 |
| miR-766-3p | 37.7 |
| miR-770-5p | 150.2 |
| miR-874 | 217.3 |
| miR-933 | 34.0 |
| miR-940 | 450.7 |
| miR-98 | 286.4 |
| let-7b-3p | −510.3 |
| let-7d-3p | −822.6 |
| let-7e-3p | −289.5 |
| let-7f-1-3p | −116.1 |
| miR-10a-5p | −4.3 |
| miR-1226-3p | −119.5 |
| miR-1227 | −114.8 |
| miR-1236 | −235.3 |
| miR-1296 | −292.4 |
| miR-130b-5p | −251.2 |
| miR-149-5p | −3.6 |
| miR-154-5p | −3.6 |
| miR-16-2-3p | −2.9 |
| miR-197-3p | −5.3 |
| miR-1976 | −282.3 |
| miR-204-5p | −690.8 |
| miR-211-5p | −695.8 |
| miR-2277-3p | −222.3 |
| miR-299-5p | −2.7 |
| miR-328 | −967.5 |
| miR-335-3p | −487.7 |
| miR-337-3p | −4.6 |
| miR-346 | −176.7 |
| miR-34b-3p | −234.2 |
| miR-3605-3p | −234.6 |
| miR-3940-3p | −225.0 |
| miR-4297 | −227.1 |
| miR-449b-3p | −652.7 |
| miR-454-5p | −253.7 |
| miR-483-3p | −746.1 |
| miR-485-3p | −223.1 |
| miR-517b-3p | −236.8 |
| miR-519d | −447.1 |
| miR-520d-3p | −251.4 |
| miR-521 | −248.1 |
| miR-595 | −366.8 |
| miR-605 | −233.4 |
| miR-615-3p | −302.1 |
| miR-654-3p | −2.4 |
| miR-664-3p | −3.7 |
| miR-744-3p | −122.9 |
| miR-885-5p | −934.7 |
| miR-937 | −282.1 |
 | Table IImiRNAs that showed a >2-fold
change in expression in NHDFs at 24 h following exposure to 0.1 Gy
γ-radiation. |
Table II
miRNAs that showed a >2-fold
change in expression in NHDFs at 24 h following exposure to 0.1 Gy
γ-radiation.
| miRNA (Homo
sapiens) | Fold change |
|---|
| miR-1224-5p | 2.2 |
| miR-1226-5p | 2.0 |
| miR-129-2-3p | 47.7 |
| miR-17-3p | 55.1 |
| miR-181d | 2.1 |
| miR-1825 | 98.9 |
| miR-197-3p | 2.2 |
| miR-299-3p | 48.3 |
| miR-3180-5p | 46.7 |
| miR-33b-3p | 49.0 |
| miR-3646 | 51.6 |
| miR-3648 | 47.7 |
| miR-369-3p | 63.7 |
| miR-369-5p | 55.1 |
| miR-3937 | 121.4 |
| miR-431-5p | 48.0 |
| miR-454-3p | 58.1 |
| miR-513c-5p | 27.1 |
| miR-602 | 30.4 |
| miR-625-5p | 46.9 |
| miR-629-3p | 50.4 |
| miR-765 | 49.3 |
| miR-634 | −2.3 |
Bioinformatics analysis of differentially
expressed miRNAs in response to LDR
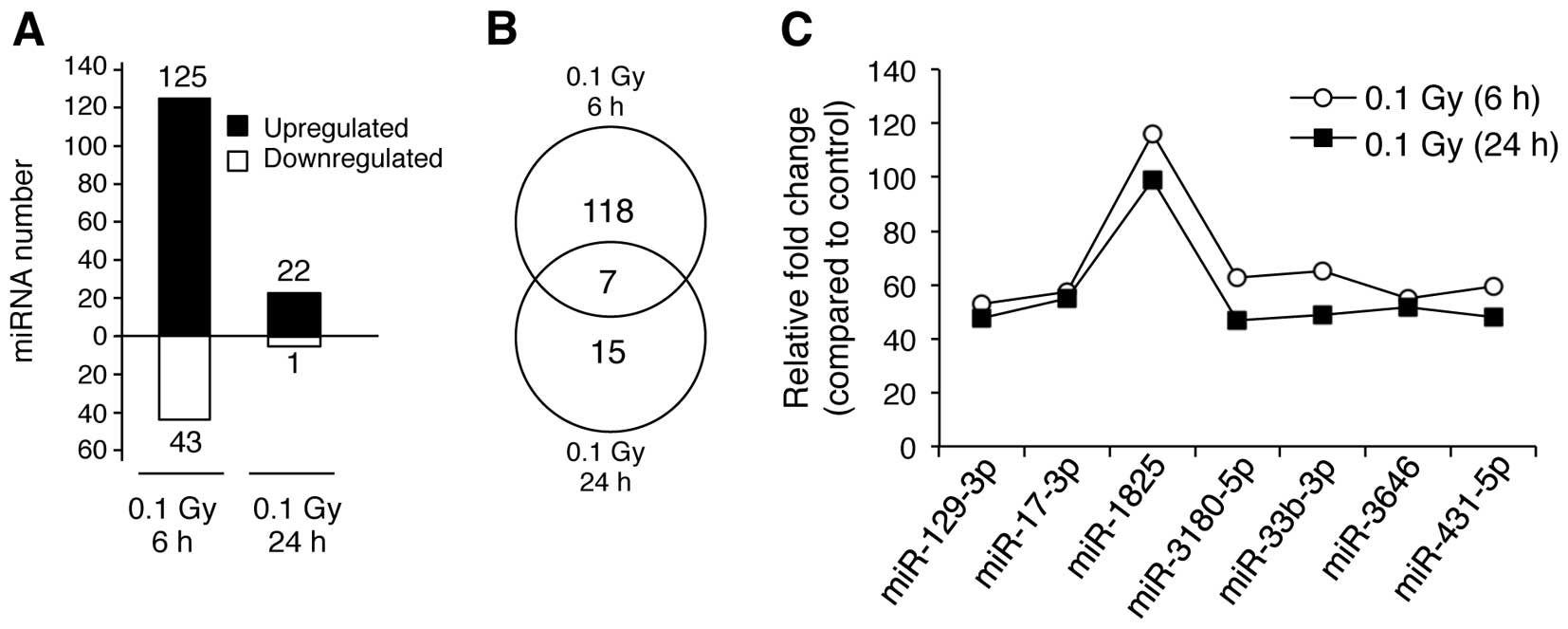
We then analyzed the biological characteristics of
the miRNAs that were upregulated in response to LDR at 6 and 24 h
(125 ‘early’ and 22 ‘late’ miRNAs) (Fig. 4A). Of the 125 miRNAs, 117 were
specifically upregulated in the early response group only (6 h
after irradiation), whereas 15 of the 22 miRNAs were specifically
upregulated in the late response group only (24 h after
irradiation), respectively (Fig.
4B). Of note, 7 miRNAs (miR-129-3p, miR-17-3p, miR-1825,
miR-3180-5p, miR-33b-3p, miR-3646 and miR-431-5p) were upregulated
in both groups (6 and 24 h post-irradiation) (Fig. 4C). Furthermore, the expression
levels of these 7 miRNAs showed only minor differences between the
late and early response groups (Fig.
4C), indicating that these are miRNAs whose expression is
generally altered in response to LDR in NHDFs.
Since miRNAs are essential regulators of almost all
cellular functions, including cell proliferation, senescence and
apoptosis through the modulation of target mRNA translation
(19), we investigated the
biological relevance of miRNA dysregulation in the LDR-induced
radio-adaptive response in NHDFs. First, we identified the
predicted target genes of each miRNA that was significantly
regulated in response to LDR in this study using the
DIANA-microT-CDS (version 5.0) web-based software tool with a
threshold value of 0.8. Following target prediction, we collected
information on the Ensembl transcript ID of the targets and
performed KEGG pathway analysis for the targets using DAVID
bioinformatics resources. To improve accuracy, the Ease Score,
which is a modified Fisher extract P-value, was fixed at 0.1 and
meaningful KEGG pathways showing a value >1% (percentage of
involved target genes/total genes involved in each pathway) were
selected. Table III shows
specific pathways for the targets of upregulated miRNAs in the
early response group (6 h post-irradiation) and the specific
pathways for the targets of the downregulated miRNAs in the early
response group are presented in Table IV. Of these, the targets of
miR-940, which was highly upregulated (450.69-fold) in the early
response group, are functionally involved in the following
cancer-related pathways: mitogen-activated protein kinase (MAPK),
WNT, Janus kinase/signal transducers and activators of
transcription (Jak-STAT), transforming growth factor-β (TGF-β) and
ErbB signaling. The targets of miR-328, which was the most highly
downregulated miRNA in the early response group, are involved in
the p53 and mammalian target of rapamycin (mTOR) signaling
pathways. Specific pathways for the targets of upregulated and
downregulated miRNAs in the late response group to LDR are shown in
Table V. The complete information
of the KEGG pathways for the miRNAs is provided in Tables III–V.
 | Table IIIFunctional annotation chart for the
top 20 miRNAs that were upregulated in the NHDFs 6 h following
exposure to 0.1 Gy γ-radiation. |
Table III
Functional annotation chart for the
top 20 miRNAs that were upregulated in the NHDFs 6 h following
exposure to 0.1 Gy γ-radiation.
| miRNA (Homo
sapiens) | Putative target
genes | KEGG pathway | Genes involved in
the term | % of involved
genes/total genes | P-value |
|---|
| miR-1181 | - | - | - | - | - |
| miR-1228-3p | 198 | - | - | - | - |
| miR-1234-3p | 19 | - | - | - | - |
| miR-1268a | 6 | - | - | - | - |
| miR-130b-3p | 473 | Endocytosis | 12 | 2.5 | 8.80E-03 |
| | TGF-β signaling
pathway | 9 | 1.9 | 1.90E-03 |
| | Insulin signaling
pathway | 9 | 1.9 | 2.50E-02 |
| |
Phosphatidylinositol signaling system | 8 | 1.7 | 3.10E-03 |
| | ErbB signaling
pathway | 6 | 1.3 | 7.90E-02 |
| | mTOR signaling
pathway | 5 | 1.1 | 4.70E-02 |
| miR-150-3p | 184 | Wnt signaling
pathway | 5 | 2.7 | 6.00E-02 |
| miR-151a-3p | 51 | Renal cell
carcinoma | 3 | 5.9 | 1.50E-02 |
| | ErbB signaling
pathway | 3 | 5.9 | 2.30E-02 |
| | Insulin signaling
pathway | 3 | 5.9 | 5.20E-02 |
| miR-193a-5p | 196 | Huntington’s
disease | 5 | 2.6 | 7.80E-02 |
| | Melanogenesis | 4 | 2 | 6.00E-02 |
| miR-1973 | 12 | - | - | - | - |
| miR-3125 | 384 | Neurotrophin
signaling pathway | 8 | 2.1 | 8.60E-03 |
| | Wnt signaling
pathway | 7 | 1.8 | 6.50E-02 |
| miR-3141 | 13 | - | - | - | - |
| miR-3156-5p | 320 | Focal adhesion | 11 | 3.4 | 2.00E-04 |
| | MAPK signaling
pathway | 11 | 3.4 | 1.90E-03 |
| | Pathways in
cancer | 9 | 2.8 | 5.60E-02 |
| | p53 signaling
pathway | 4 | 1.2 | 5.60E-02 |
| miR-3656 | 10 | - | - | - | - |
| miR-3663-3p | 305 | MAPK signaling
pathway | 12 | 3.9 | 5.90E-03 |
| miR-378b | 162 | Pathways in
cancer | 8 | 4.9 | 3.60E-02 |
| miR-409-3p | 316 | Axon guidance | 7 | 2.2 | 2.20E-02 |
| miR-409-5p | 34 | MAPK signaling
pathway | 3 | 8.8 | 6.20E-02 |
| miR-4271 | 361 | Jak-STAT signaling
pathway | 7 | 1.9 | 7.80E-02 |
| miR-575 | 241 | MAPK signaling
pathway | 8 | 3.3 | 7.70E-02 |
| | Cell cycle | 5 | 2.1 | 9.60E-02 |
| | mTOR signaling
pathway | 4 | 1.7 | 3.50E-02 |
| miR-940 | 634 | Pathways in
cancer | 23 | 3.6 | 1.50E-03 |
| | MAPK signaling
pathway | 16 | 2.5 | 3.70E-02 |
| | Wnt signaling
pathway | 11 | 1.7 | 3.20E-02 |
| | Jak-STAT signaling
pathway | 10 | 1.6 | 8.10E-02 |
| | TGF-β signaling
pathway | 9 | 1.4 | 9.00E-03 |
| | ErbB signaling
pathway | 8 | 1.3 | 2.80E-02 |
| miR-98 | 495 | MAPK signaling
pathway | 19 | 3.8 | 1.70E-04 |
| | Pathways in
cancer | 19 | 3.8 | 2.00E-03 |
| | Wnt signaling
pathway | 10 | 2 | 1.70E-02 |
| | Jak-STAT signaling
pathway | 9 | 1.8 | 4.90E-02 |
| | p53 signaling
pathway | 8 | 1.6 | 1.80E-03 |
| | Insulin signaling
pathway | 8 | 1.6 | 6.30E-02 |
| | TGF-β signaling
pathway | 7 | 1.4 | 2.60E-02 |
| | Apoptosis | 6 | 1.2 | 7.70E-02 |
 | Table IVFunctional annotation chart for the
top 20 miRNAs that were downregulated in the NHDFs 6 h following
exposure to 0.1 Gy γ-radiation. |
Table IV
Functional annotation chart for the
top 20 miRNAs that were downregulated in the NHDFs 6 h following
exposure to 0.1 Gy γ-radiation.
| miRNA (Homo
sapiens) | Putative target
genes | KEGG pathway | Genes involved in
the term | % of involved
genes/total genes | P-value |
|---|
| let-7b-3p | 610 | Pathways in
cancer | 23 | 3.8 | 6.80E-04 |
| | MAPK signaling
pathway | 18 | 3 | 4.90E-03 |
| | Wnt signaling
pathway | 12 | 2 | 8.70E-03 |
| | TGF-β signaling
pathway | 9 | 1.5 | 6.40E-03 |
| let-7d-3p | - | - | - | - | - |
| let-7e-3p | 2 | - | - | - | - |
| miR-1296 | 91 | - | - | - | - |
| miR-130b-5p | 361 | Ubiquitin-mediated
proteolysis | 6 | 1.7 | 6.10E-02 |
| miR-1976 | 429 | Drug
metabolism | 5 | 1.2 | 5.90E-03 |
| | Pyrimidine
metabolism | 5 | 1.2 | 7.90E-02 |
| miR-204-5p | 178 | Jak-STAT signaling
pathway | 5 | 2.8 | 5.00E-02 |
| | Huntington’s
disease | 5 | 2.8 | 7.80E-02 |
| | Melanogenesis | 4 | 2.2 | 6.00E-02 |
| miR-211-5p | 178 | Jak-STAT signaling
pathway | 5 | 2.8 | 5.40E-02 |
| | Melanogenesis | 4 | 2.2 | 6.30E-02 |
| miR-328 | 103 | p53 signaling
pathway | 4 | 3.9 | 4.30E-03 |
| | mTOR signaling
pathway | 3 | 2.9 | 2.60E-02 |
| miR-335-3p | 2,294 | Pathways in
cancer | 56 | 2.4 | 1.20E-03 |
| | MAPK signaling
pathway | 39 | 1.7 | 7.20E-02 |
| | Calcium signaling
pathway | 33 | 1.4 | 3.70E-03 |
| | Wnt signaling
pathway | 28 | 1.2 | 9.20E-03 |
| | Insulin signaling
pathway | 22 | 1 | 7.60E-02 |
| | TGF-β signaling
pathway | 21 | 0.9 | 1.20E-03 |
| | ErbB signaling
pathway | 21 | 0.9 | 1.20E-03 |
| | p53 signaling
pathway | 14 | 0.6 | 3.70E-02 |
| miR-449-3p | 305 | Pathways in
cancer | 10 | 3.3 | 9.50E-02 |
| | Neurotrophin
signaling pathway | 7 | 2.3 | 1.70E-02 |
| miR-454-5p | 33 | - | - | - | - |
| miR-483-3p | 155 | RNA
degradation | 5 | 3.2 | 7.10E-04 |
| | Purine
metabolism | 4 | 2.6 | 9.90E-02 |
| miR-519d | 739 | MAPK signaling
pathway | 23 | 3.1 | 3.20E-04 |
| | Insulin signaling
pathway | 12 | 1.6 | 1.10E-02 |
| | Chemokine signaling
pathway | 12 | 1.6 | 8.80E-02 |
| | TGF-β signaling
pathway | 10 | 1.4 | 4.70E-03 |
| | ErbB signaling
pathway | 9 | 1.2 | 1.50E-02 |
| miR-520d-3p | 647 | MAPK signaling
pathway | 19 | 2.9 | 3.30E-04 |
| | Regulation of actin
cytoskeleton | 15 | 2.3 | 2.20E-03 |
| | Pathways in
cancer | 15 | 2.3 | 6.60E-02 |
| | Insulin signaling
pathway | 11 | 1.7 | 3.80E-03 |
| | TGF-β signaling
pathway | 8 | 1.2 | 9.60E-03 |
| | ErbB signaling
pathway | 8 | 1.2 | 9.60E-03 |
| miR-521 | 6 | - | - | - | - |
| miR-595 | 35 | - | - | - | - |
| miR-615-3p | 25 | - | - | - | - |
| miR-885-5p | 280 | Tight junction | 7 | 2.5 | 8.00E-03 |
| | RNA
degradation | 5 | 1.8 | 6.40E-03 |
| miR-937 | 5 | - | - | - | - |
 | Table VFunctional annotation chart for the
top 20 upregulated and the single downregulated miRNA in NHDFs 24 h
following exposure to 0.1 Gy γ-radiation. |
Table V
Functional annotation chart for the
top 20 upregulated and the single downregulated miRNA in NHDFs 24 h
following exposure to 0.1 Gy γ-radiation.
| miRNA (Homo
sapiens) | Putative target
genes | KEGG pathway | Genes involved in
the term | % of involved
genes/total genes | P-value |
|---|
| miR-1224-5p | 263 | Axon guidance | 5 | 1.9 | 5.00E-02 |
| miR-129-2-3p | 181 | Valine, leucine and
isoleucine degradation | 3 | 1.7 | 5.30E-02 |
| miR-17-3p | 384 | MAPK signaling
pathway | 12 | 3.1 | 7.60E-03 |
| | Insulin signaling
pathway | 7 | 1.8 | 3.30E-02 |
| | mTOR signaling
pathway | 6 | 1.6 | 2.20E-03 |
| | TGF-β signaling
pathway | 5 | 1.3 | 7.00E-02 |
| miR-1825 | 390 | Pathways in
cancer | 13 | 3.3 | 1.30E-02 |
| | MAPK signaling
pathway | 10 | 2.6 | 4.80E-02 |
| miR-197-3p | 422 | Ubiquitin mediated
proteolysis | 8 | 1.9 | 2.60E-02 |
| miR-299-3p | 238 | Purine
metabolism | 5 | 2.1 | 6.20E-02 |
| miR-3180-5p | 629 | Axon guidance | 11 | 1.7 | 6.20E-03 |
| | Wnt signaling
pathway | 10 | 1.6 | 4.30E-02 |
| miR-33b-3p | 147 | - | - | - | - |
| miR-3646 | 656 | Ubiquitin mediated
proteolysis | 14 | 2.1 | 4.70E-04 |
| | Wnt signaling
pathway | 12 | 1.8 | 9.90E-03 |
| |
Phosphatidylinositol signaling system | 6 | 0.9 | 9.30E-02 |
| miR-3648 | 3 | - | - | - | - |
| miR-369-3p | 1,171 | Pathways in
cancer | 34 | 2.9 | 1.10E-03 |
| | MAPK signaling
pathway | 23 | 2 | 5.80E-02 |
| | Jak-STAT signaling
pathway | 16 | 1.4 | 3.40E-02 |
| | TGF-β signaling
pathway | 15 | 1.3 | 4.10E-04 |
| | ErbB signaling
pathway | 12 | 1 | 1.10E-02 |
| | p53 signaling
pathway | 9 | 0.8 | 4.20E-02 |
| miR-369-5p | 2 | - | - | - | - |
| miR-3937 | 11 | - | - | - | - |
| miR-431-5p | 307 |
Aldosterone-regulated sodium
reabsorption | 4 | 1.3 | 2.00E-02 |
| miR-454-3p | 546 | Endocytosis | 16 | 2.9 | 2.80E-04 |
| | Insulin signaling
pathway | 9 | 1.6 | 4.40E-02 |
| |
Phosphatidylinositol signaling system | 8 | 1.5 | 5.60E-03 |
| | TGF-β signaling
pathway | 7 | 1.3 | 4.10E-02 |
| | mTOR signaling
pathway | 5 | 0.9 | 6.50E-02 |
| | Inositol phosphate
metabolism | 5 | 0.9 | 7.20E-02 |
| miR-513c-5p | 160 | Tight junction | 4 | 2.5 | 4.00E-02 |
| |
Phosphatidylinositol signaling system | 3 | 1.9 | 6.60E-02 |
| miR-602 | 22 | - | - | - | - |
| miR-625-5p | 357 | Tight junction | 8 | 2.2 | 9.20E-03 |
| | Ubiquitin mediated
proteolysis | 8 | 2.2 | 1.00E-02 |
| | Apoptosis | 7 | 2 | 4.20E-03 |
| miR-629-3p | 563 | PPAR signaling
pathway | 6 | 1.1 | 3.20E-02 |
| | Riboflavin
metabolism | 3 | 0.5 | 6.20E-02 |
| miR-765 | 696 | Endocytosis | 11 | 1.6 | 9.50E-02 |
| | MAPK signaling
pathway | 18 | 3 | 4.90E-03 |
| | Wnt signaling
pathway | 12 | 2 | 8.70E-03 |
| | TGF-β signaling
pathway | 9 | 1.5 | 6.40E-03 |
| miR-634a | 240 | Ubiquitin-mediated
proteolysis | 6 | 2.5 | 5.60E-02 |
Discussion
The present study demonstrates that LDR (0.1 Gy)
induces distinct cellular responses in NHDFs. A MTT-based cell
viability assay demonstrated a slight decrease in cell viability at
6 h after irradiation, but this decrease was reversed by 24 h after
irradiation. Similar results were obtained by FACS analysis and PI
staining; at 6 h after irradiation the proportion of irradiated
cells which had accumulated in the G2/M phase was markedly higher
than that of non-irradiated cells; however, this value had returned
close to the value of the control at 24 h. These data indicate that
NHDFs have distinct radio-adaptive responses to LDR at different
time points post-irradiation. The early radio-adaptive response to
LDR appears to activate cell cycle arrest mechanisms, but does not
fully block cell cycle progression. By contrast, the late
radio-adaptive response to LDR may activate radioresistance
mechanisms. A slight decrease in cell viability and a partial
increase in the G2/M cell population were only evident at 6 h
post-irradiation; by 24 h post-irradiation, the decrease in cell
viability and G2/M cell cycle arrest were reversed and had returned
close to the control levels. A similar result was reported in a
previous study on human glioblastoma T98G and U373MG cell lines, in
which the percentage of cells in the G2/M phase of the cell cycle
following exposure to different doses of γ-radiation (0.2–2 Gy)
increased as early as 8 h following exposure to radiation, although
this accumulation was clearly transient and its duration was
proportional to the exposure dose (20). In addition, in our previous study,
we demonstrated similar results in the human B lymphoblastic cell
line, IM9, which showed a marked increase in the proportion of
cells in the G2/M phase within 6 h after exposure to 1 Gy of
γ-radiation (14). Collectively,
our data indicate that the radio-adaptive response of human skin
fibroblasts to 0.1 Gy of γ-radiation involves dual mechanisms
depending on the time point following exposure.
We also found that exposure to 0.1 Gy of γ-radiation
had a potential anti-aging effect on NHDFs. At 24 h after exposure
to LDR, there was a marked increase in the expression of
COL1A1 and a decrease in the expression of MMP1.
However, the anti-aging effects of LDR must be considered in
relation to its safety as the overproduction of collagen is one of
the etiological factors of the skin disease, scleroderma (21). A similar result was previously
reported using single doses of β-radiation in human Tenon’s capsule
fibroblasts (hTfs) (7). A
growth-arresting dose (1,000 cGy) of β-radiation increases the
production of collagen I and therefore has a wound healing effect
(7). However, various aspects of
this response, including collagen structure and skin inflammation
following exposure to LDR, have not yet been investigated, and
further in vitro and in vivo experiments are required
to fully clarify this issue.
In the present study, data from a MTT-based cell
viability assay and a FACS-based cell cycle analysis assay revealed
that the early radio-adaptive response of NHDFs involving a
decrease in cell viability and G2/M phase arrest was reversed to
near-control levels by 24 h post-irradiation. Although these
results suggest that cellular conditions at 24 h after irradiation
are similar to those of control (non-irradiated) cells, our
additional data clearly indicate that these cell conditions differ
from each other. Firstly, RT-qPCR demonstrated that the
intracellular levels of COL1A1 and MMP1 transcripts
were markedly altered 24 h following exposure to 0.1 Gy of
γ-radiation. Secondly, miRNA microarray analysis revealed that
microRNA expression patterns differed significantly between the
control cells and the irradiated cells at 24 h post-irradiation.
Thirdly, the expression levels of the altered miRNAs were markedly
different in the irradiated cells from those in the control cells.
Notably, the expression levels of 16 miRNAs, including miR-3937 and
miR-1825, were significantly increased by >40-fold at 24 h
post-irradiation. These results indicated that, although cell
viability and cell cycle distribution were reversed, returning to
values close to the control values, the intercellular conditions at
24 h post-irradiation differed from those of the non-irradiated
control cells and the irradiated cells presumably had distinct
cellular properties.
Using miRNA microarray analysis, we revealed the
existence of specific miRNAs in NHDFs with early and late
radio-adaptive responses following exposure to 0.1 Gy γ-radiation.
Among the early response miRNAs, the biological functions of
miR-3656 and miR-3125, which were the most highly upregulated
(493.88- and 493.78-fold, respectively), to the best of our
knowledge, have not been previously studied. Our bioinformatics
analysis revealed that these two miRNAs are functionally related to
the Wnt signaling pathway. A recent study demonstrated that LDR
(0.3 Gy) stimulates Wnt/β-catenin signaling to induce neural stem
cell proliferation (22).
Therefore, these 2 miRNAs may be valuable novel targets for
radio-adaptive response mechanisms. In addition, among the miRNAs
that showed an altered expression in the present study, 7 miRNAs
(miR-129-3p, miR-17-3p, miR-1825, miR-3180-5p, miR-33b-3p, miR-3646
and miR-431-5p) were upregulated at both 6 and 24 h following
exposure to LDR and are potential LDR-specific biomarkers in human
skin. Moreover, the expression levels of these miRNAs were highly
upregulated by >40-fold and were consistent between the 2
post-irradiation time points. These data indicate that these miRNAs
are constantly expressed following exposure to LDR, independent of
the post-irradiation time point.
In conclusion, to the best of our knowledge, we
provide the first evidence that LDR (0.1 Gy) induces dual
radio-adaptive responses in NHDFs. We also demonstrated that LDR
induced potential anti-aging effects through the regulation of the
expression of collagen synthesis-relating genes. Furthermore, LDR
induced changes in the expression profiles of specific miRNAs, and
some of the deregulated miRNAs were specific to the early or late
radio-adaptive response. Although further studies are required to
validate the deregulated miRNAs, our results provide novel
information on miRNA-mediated adaptive responses to LDR in
NHDFs.
Acknowledgements
Dr Seunghee Bae was supported by the KU Research
Professor Program of Konkuk University, and this study was
supported by grant no. 20131610101840 from the Ministry of Trade,
Industry and Energy of Republic of Korea.
References
|
1
|
Nguyen PK and Wu JC: Radiation exposure
from imaging tests: is there an increased cancer risk? Expert Rev
Cardiovasc Ther. 9:177–183. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cleaver JE: Biology and genetics in the
biological effects of ionizing radiation (BEIR VII) report. Health
Physics. 89:S32. 2005.
|
|
3
|
Feinendegen LE: Evidence for beneficial
low level radiation effects and radiation hormesis. Br J Radiol.
78:3–7. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sjerobabski-Masnec I and Situm M: Skin
aging. Acta Clin Croat. 49:515–518. 2010.
|
|
5
|
Gelse K, Pöschl E and Aigner T: Collagens
- structure, function, and biosynthesis. Adv Drug Deliv Rev.
55:1531–1546. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Quan T, Qin Z, Xia W, Shao Y, Voorhees JJ
and Fisher GJ: Matrix-degrading metalloproteinases in photoaging. J
Investig Dermatol Symp Proc. 14:20–24. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Constable PH, Crowston JG, Occleston NL
and Khaw PT: The effects of single doses of beta radiation on the
wound healing behaviour of human Tenon’s capsule fibroblasts. Br J
Ophthalmol. 88:169–173. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rastogi RP, Richa, Kumar A, Tyagi MB and
Sinha RP: Molecular mechanisms of ultraviolet radiation-induced DNA
damage and repair. J Nucleic Acids. 2010:5929802010. View Article : Google Scholar
|
|
9
|
Chang TC, Wentzel EA, Kent OA, et al:
Transactivation of miR-34a by p53 broadly influences gene
expression and promotes apoptosis. Mol Cell. 26:745–752. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
He L, He X, Lim LP, et al: A microRNA
component of the p53 tumour suppressor network. Nature.
447:1130–1134. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu C, Zhou C, Gao F, et al: MiR-34a in
age and tissue related radio-sensitivity and serum miR-34a as a
novel indicator of radiation injury. Int J Biol Sci. 7:221–233.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Joly-Tonetti N, Viñuelas J, Gandrillon O
and Lamartine J: Differential miRNA expression profiles in
proliferating or differentiated keratinocytes in response to gamma
irradiation. BMC Genomics. 14:1842013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cha HJ, Seong KM, Bae S, et al:
Identification of specific microRNAs responding to low and high
dose γ-irradiation in the human lymphoblast line IM9. Oncol Rep.
22:863–868. 2009.PubMed/NCBI
|
|
14
|
Cha HJ, Shin S, Yoo H, et al:
Identification of ionizing radiation-responsive microRNAs in the
IM9 human B lymphoblastic cell line. Int J Oncol. 34:1661–1668.
2009.PubMed/NCBI
|
|
15
|
Cha HJ, Lee KS, Lee GT, et al: Altered
miRNA expression profiles are involved in the protective effects of
troxerutin against ultraviolet B radiation in normal human dermal
fibroblasts. Int J Mol Med. 33:957–963. 2014.PubMed/NCBI
|
|
16
|
Paraskevopoulou MD, Georgakilas G,
Kostoulas N, et al: DIANA-microT web server v5.0: service
integration into miRNA functional analysis workflows. Nucleic Acids
Res. 41:W169–W173. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kraemer A, Anastasov N, Angermeier M,
Winkler K, Atkinson MJ and Moertl S: MicroRNA-mediated processes
are essential for the cellular radiation response. Radiat Res.
176:575–586. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Huang Y, Shen XJ, Zhou Q, Wang SP, Tang SM
and Zhang GZ: Biological functions of microRNAs: a review. J
Physiol Biochem. 67:129–139. 2011. View Article : Google Scholar
|
|
20
|
Fernet M, Mégnin-Chanet F, Hall J and
Favaudon V: Control of the G2/M checkpoints after exposure to low
doses of ionising radiation: implications for
hyper-radiosensitivity. DNA Repair (Amst). 9:48–57. 2010.
View Article : Google Scholar
|
|
21
|
Varga J and Abraham D: Systemic sclerosis:
a prototypic multisystem fibrotic disorder. J Clin Invest.
117:557–567. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wei LC, Ding YX, Liu YH, et al: Low-dose
radiation stimulates Wnt/β-catenin signaling, neural stem cell
proliferation and neurogenesis of the mouse hippocampus in vitro
and in vivo. Curr Alzheimer Res. 9:278–289. 2012. View Article : Google Scholar : PubMed/NCBI
|