Introduction
Human embryonic stem (hES) cells have been widely
used in regenerative medicine due to their capacity to
differentiate into various cell types both in vitro and
in vivo (1,2). However, transplant rejection and the
unstable epigenetic state of hES cells from human embryos limit
their use in research and therapy. Human parthenogenetic embryonic
stem (hPES) cells, the genetic materials of which are derived
entirely from a single oocyte, are considered to be a possible
means to resolve the issue of immune rejection (3), and several hPES cell lines have been
generated (4–8). These stem cell lines have exhibited
infinite proliferation, self-renewal and differentiation
properties, similar to embryonic stem cell lines in
vitro.
The completely undifferentiated status and the
original genetic characteristics of hES cells are important for
clinical and research trials (9).
It has been previously reported that the evolution and selection of
hES cell clones cultured in vitro causes genetic and
epigenetic changes, which alters the behavior and fate of these hES
cells (10–13). The genetic and epigenetic
stabilities of hES cells are crucial for their use in regenerative
medicine. Epigenetic changes include DNA methylation, histone
modifications, genomic imprinting and X chromosome inactivation
(XCI).
XCI involves one of the X chromosomes in cells of a
female mammal and is crucial for embryo formation and cell biology
(14). To date, hES cells have
been shown to have 3 different XCI statuses. With status I in the
hES cells, both X chromosomes are activated in the undifferentiated
stage, and XCI occurs randomly following differentiation, which is
close to what occurs in mouse embryonic stem cells (15–17). With status II, XCI has already
occurred in undifferentiated hES cells, and approximately 20–70% of
hES cells can be found with X-inactive specific transcript (XIST)
clouds accumulated on a specific chromosome (11,18). Finally, with status III, XCI has
occurred without XIST RNA expression (11). Certain studies have demonstrated
that the hES cell XCI states are related to the culture conditions
used and spontaneous differentiation potential (19). However, the XCI statuses of hPES
cell lines have not been thoroughly investigated to date.
Thus, in the present study, we assessed the statuses
of hPES cell lines following prolonged passaging in culture in
vitro. We focused on the XCI status of hPES cell lines (hPES-1
and hPES-2) under long-term culture conditions (>50 passages)
in vitro. We found that hPES cells also had 3 XCI statuses,
although different XCI statuses could be found within the same cell
line. These differences in XCI status in hPES cells may be related
to their X chromosome instability. Furthermore, low expression
levels of X-linked genes were detected in the hPES cells that were
related to their XIST RNA expression levels. The XCI status is
related to the genetic characteristics or strains of embryonic stem
cells that are cultured in vitro and the freezing conditions
used. Our findings suggest that it is essential to assess the XCI
status of hES cells and to consider this as one of the indicators
used for evaluating the quality of hES cells.
Materials and methods
Ethics statement
Our protocols were approved by the Ethics Committee
of the First Affiliated Hospital of Sun Yat-Sen University. Donors
voluntarily donated experimental materials with no financial
compensation and written informed consent was obtained.
Derivation and culture of hES, human
foreskin fibroblasts (HFFs) and human endometrial stromal cells
(hESCs)
Three hES lines were analyzed in this study,
including human biparental embryonic stem cell line-1 [hBES-1,
passage (P)12], hPES cell line-1 (hPES-1, P10) and hPES cell line-2
(hPES-2, P10). The hBES-1 cells were from a cHES1 cell line that
was derived and propagated in our embryonic stem cell laboratory,
as previously described (20).
The hPES-1 and hPES-2 cells were from hPES1 and hPES2 cell lines
that were also derived and propagated in our laboratory, as
previously described (6).
Culture, cryopreservation and warming methods for undifferentiated
hESCs and embryoid body (EB) formation were as previously
described, and the origins and detailed characterizations of the
pluripotency of these cell lines were verified (6,20,21). The derivation and culture of hESCs
and HFFs were as previously described (22,23).
Spontaneous hESC differentiation was induced as
previously described (6,20). hPES-1 and hPES-2 cells at P60 and
hPES-2′ cells at P70 were removed from the dishes using 1 mg/ml of
collagenase IV (cat. no. 17104-019; Invitrogen/Gibco, Grand Island,
NY, USA) and cultured under suspension conditions. Spontaneous EBs
were grown in medium that included 80% Knockout-DMEM (Cat. no.
10829-018), supplemented with 20% serum replacement, 0.1 mM
2-mercaptoethanal and 1% non-essential amino acids (Cat. no.
11140-050) (all from Invitrogen/Gibco). After 14 days, all
differentiated samples from EBs were collected.
Karyotyping
The hBES-1 (P19) colonies, hESCs and HFFs were
incubated with 0.2 μg/ml of colchicine (Invitrogen/Gibco) at
37°C for 3 h. The cells were collected, trypsinized, washed with
phosphate-buffered saline (PBS) (Cat. no. 10010-049;
Invitrogen/Gibco) and then incubated with 0.075 M potassium
chloride at 37°C for 10 min. These cells were fixed with
methanol:glacial acetic acid (1:3) 3 times and then dropped onto
glass slides. Chromosome spreads were Giemsa-banded and
photographed. Karyotypes were assessed using normal G-banding
procedures and 50 metaphase II spreads were examined for each
sample. A normal karyotype showed normal chromosome numbers and
G-banding patterns in the spreads examined.
Array-based comparative genomic
hybridization (aCGH)
The hBES-1 (P42), hPES-1 (P70), hPES-2 (P20) and
hPES-2 (P55) cell colonies were lysed, after which genomic DNA was
amplified using a SurePlex DNA Amplification system (BlueGnome
Ltd., Cambridge, UK), according to the manufacturer’s instructions.
Whole genome amplification (WGA) products were processed as
previously described (24)
according to the BlueGnome protocol (available at: http://www.cytochip.com/). Briefly, WGA products were
fluorescently labeled and competitively hybridized to 24sure
V3/24sure + arrays with matched control SureRef reference DNA
(male/female) (both from BlueGnome Ltd.) in an array CGH experiment
format. A laser scanner, InnoScanw 710 AL (Innopsys, Carbonne,
France), was used to excite the hybridized fluorophores of a
Fluorescent Labelling system (BlueGnome Ltd.), and used to read and
store the resulting hybridization images. The scanned images were
then analyzed and quantified using algorithm fixed settings with
BlueFuse Multi software (BlueGnome Ltd.), a software package that
automatically performed the steps of grid placement,
quantification, normalization and post-processing.
Flow cytometry for hESC phenotyping
The hBES-1 (P70) and hPES-2 (P70) cell colonies were
digested with 0.25% Trypsin-EDTA (Cat. no. 15400-054;
Invitrogen/Gibco) to prepare single cells. The hESCs
(1−2×106) were suspended in 0.5 ml of Dulbecco’s
phosphate-buffered saline (DPBS; Cat. no. 14190-144;
Invitrogen/Gibco). Subsequenlty, 2–3 ml of 70% ethanol were added
to each tube, mixed and incubated at 4°C for 30 min. Cell
suspensions were washed, centrifuged and resuspended in 400
μl of DPBS. Cell suspensions were filtered, 1 mg/ml of
propidium iodide (Asegene, Guangdong, China) was added followed by
incubation for 30 min. Cell suspensions were analyzed using a
FACSCalibur Flow Cytometer with CellQuest software
(Becton-Dickinson, Bergen County, NJ, USA).
DNA fluorescence in situ hybridization
(FISH)
For DNA FISH analysis, the hBES-1, hPES-1 and hPES-2
cells were dropped onto wet slides and dried at room temperature
overnight. They were then fixed with 0.1% Tween-20 and 0.01 N HCl
and dehydrated with an ethanol series at concentrations of 70, 85
and 100%. CEPX/CEPY (Cytocell, Banbury, Oxfordshire, UK) was used
for hybridization for 2 h. Finally, the cells were stained with
4′,6-diamidino-2-phenylindole (DAPI; Cytocell) for 5 min. The cells
were examined under a fluorescence microscope (Leica DMIRE 2; Leica
Microsystems GmbH, Wetzlar, Germany). At least 20 cells were
examined for each experiment.
RNA FISH
The hESCs, hBES-1 (P20 and P40), hPES-1 (P20 and
P40), hPES-2 (P20 and P40) and hPES-2′ (P70) cells were used for
RNA FISH analysis. XIST RNA-FISH (Biosearch Technologies, Novato,
CA, USA) was carried out according to the protocol provided by
Stellaris FISH Probes (Biosearch Technologies; https://www.biosearchtech.com/display.aspx?catid=224%2c318),
as previously described (15).
The XIST probe sequences are listed in Table I. hES cell colonies and hESCs were
cultured on Millicell EZ slides (Cat. no. PEZGS0496; Millipore,
Billerica, MA, USA). Stained colonies were examined under a
fluorescence microscope (Leica DMIRE 2; Leica Microsystems
GmbH).
 | Table IXIST probe sequences. |
Table I
XIST probe sequences.
| Probe sequence
(5′→3′) | Probe sequence
name |
|---|
|
gaattgcagcgctttaagaactgaagg | Human
XIST-RNAFISHprobe_1 |
|
gagagagtaagaaatatggctgcagca | Human
XIST-RNAFISHprobe_2 |
|
gacgtgtcaagaagacactaggagaaa |
HumanXIST-RNAFISHprobe_3 |
|
gaagggaatcagcaggtatccgatacc | Human
XIST-RNAFISHprobe_4 |
|
gatattccagagagtgcaacaacccac | Human
XIST-RNAFISHprobe_5 |
|
cttagcttaactgcagagtcattctct | Human
XIST-RNAFISHprobe_6 |
|
ccgagttatgcggcaagtctaaaatgg | Human
XIST-RNAFISHprobe_7 |
|
tgcctgacctgctatcatccatcttgc | Human
XIST-RNAFISHprobe_8 |
|
ttagctcatgcaatgcacatgacttcc | Human
XIST-RNAFISHprobe_9 |
|
cgatacaacaatcacgcaaagctccta | Human
XIST-RNAFISHprobe_10 |
|
ccgcaatgtcaaaatcgccattttaag | Human
XIST-RNAFISHprobe_11 |
|
cattttggacaacctaacaaagcacag | Human
XIST-RNAFISHprobe_12 |
|
acttgaacactgcgacagaactggatc | Human
XIST-RNAFISHprobe_13 |
|
catcttttcctgtgtgaccgcacatgt | Human
XIST-RNAFISHprobe_14 |
|
catgttttacactgcggcaagaccttc | Human
XIST-RNAFISHprobe_15 |
|
catatgacaacgcctgccatattgtcc | Human
XIST-RNAFISHprobe_16 |
|
gatgtccacgtgacaaaagccatgata | Human
XIST-RNAFISHprobe_17 |
|
ctctaattggctgtgatcaattccacc | Human
XIST-RNAFISHprobe_18 |
|
gtgtgtcatcagtctaattccatcttc | Human
XIST-RNAFISHprobe_19 |
|
gtgttcctcttgaggaaggcaggaatt | Human
XIST-RNAFISHprobe_20 |
|
tcagtactgaagatcagcaatgccaag | Human
XIST-RNAFISHprobe_21 |
|
cagagtgctgtctaatccaatgggtag | Human
XIST-RNAFISHprobe_22 |
|
cgactggtagtcttcatgattaatggg | Human
XIST-RNAFISHprobe_23 |
|
ctctaagaatgagtcagtcccactgct | Human
XIST-RNAFISHprobe_24 |
|
aaggtggtaggtagttcacactatcta | Human
XIST-RNAFISHprobe_25 |
|
aaggaaacttgggtagtcagaactcag | Human
XIST-RNAFISHprobe_26 |
|
attgtagcgtgcaaataggatacagag | Human
XIST-RNAFISHprobe_27 |
|
ctagtacagaggtcttgagtagtaagg | Human
XIST-RNAFISHprobe_28 |
|
cactgctgaacactagggaagtgagtg | Human
XIST-RNAFISHprobe_29 |
|
ctagtgcaaaggtcttgactagaggtc |
HumanXIST-RNAFISHprobe_30 |
|
tagcactcctgctgctttgccaaggag | Human
XIST-RNAFISHprobe_31 |
|
gcagtataagagaagaagcactagcta | Human
XIST-RNAFISHprobe_32 |
|
agcgggattctactctaacataggggc | Human
XIST-RNAFISHprobe_33 |
|
caagagagtgaattcaggctagttaga | Human
XIST-RNAFISHprobe_34 |
|
tacttccagctgggatgtaaatacagt | Human
XIST-RNAFISHprobe_35 |
|
caattacatgccatctacagttcgaag | Human
XIST-RNAFISHprobe_36 |
|
gataggtcagaaacccaagtctaattg | Human
XIST-RNAFISHprobe_37 |
|
ggccttaggtgtcaccaaccatgctgt | Human
XIST-RNAFISHprobe_38 |
|
ctagtgcatagcaacctcgacaaatac | Human
XIST-RNAFISHprobe_39 |
|
cagtgtgcgattacgcacataaatgtc | Human
XIST-RNAFISHprobe_40 |
|
gagagtaggaccttattcacatggaat | Human
XIST-RNAFISHprobe_41 |
Immunofluorescence staining
The hESCSs, hBES-1 (P20 and P40), hPES-1 (P20 and
P40), hPES-2 (P20 and P40), and hPES-2′ (P70) cells were used for
immunofluorescence staining. The hES cell colonies and hESCs were
cultured on Millicell EZ slides (Millipore) and fixed with 4%
paraformaldehyde (Cat. no. P6148) for 20–30 min, treated with 0.5%
Triton X-100 (Cat. no. X100) for 20 min and then blocked with 10%
goat serum (Cat. no. G9023) (all from Sigma, St. Louis, MO, USA)
for 1 h. The cells were then incubated with primary antibodies at
4°C overnight. The primary antibodies included mouse anti-histone
H3 trimethyl K27 (H3K27me3; 1:100; Cat. no. ab6147) and rabbit
anti-histone H3 acetyl K9 (H3K9ac; 1:200; Cat. no. ab61231) (both
from Abcam, Cambridge, UK). The cells were then rinsed 3 times with
PBS and incubated at 37°C for 60 min with goat anti-mouse IgM R-PE
(1:200; Cat. no. 488800A; Invitrogen, Carlsbad, CA, USA) or goat
anti-rabbit IgG FITC (1:200; Cat. no. A24532; Invitrogen), and
finally stained with DAPI for 5 min. The stained colonies were
examined under a fluorescence microscope (Leica DMIRE 2; Leica
Microsystems GmbH).
Real-time polymerase chain reaction
(PCR)
XIST expression (probe XIST ID: Hs01079824_m1;
Ambion, Austin, TX, USA) was assessed in the hESCs and HFFs, the
hBES-1, hPES-1 and hPES-2 cells at P20, P40 and P60, the hPES-2′
undifferentiated cells at P50, P60 and P70, as well as in EBs of
hPES-1 (P60), hPES-2 (P60) and hPES-2′ (P70) cells. X-linked gene
expression was assessed in the hBES-1, hPES-1 and hPES-2 cells at
P20, P40 and P60, and in the hPES-2′ undifferentiated cells at P50,
P60 and P70. Real-time PCR was carried out using TaqMan gene
expression Cells-to CT kits (Cat. no. 4399002; Ambion) as described
in the TaqMan gene expression Cells-to CT kit protocol (http://www.lifetechnologies.com/order/catalog/product/4399002?ICID=search-product).
The target X-linked genes were alpha thalassemia/mental
retardation, X-Linked (ATRX; assay ID: Hs00230877_m1) and
cysteine-rich hydrophobic domain 1 (CHIC1; assay ID: Hs01371424_m1)
(both from Ambion). Relative XIST gene and other X-linked gene
expression levels were calculated using the 2−∆∆CT
method following normalization to GAPDH (assay ID: Hs02758991_m1;
Ambion) expression levels.
Pluripotent characterizations of hPES-2
cells
To exclude the possibility that XIST expression in
undifferentiated hPES-2 cells occurred due to their differentiation
in culture, we assessed the pluripotent characterizations of the
hPES2 cells. EBs were used to assess the differentiation capability
of the hPES-2 cells in vitro using specific
immunofluorescence staining.
Alkaline phosphatase (AP) activity was assessed by
histochemical staining. The hPES-2 cell colonies on a mouse
embryonic fibroblast (MEF) feeder layer were fixed with 4%
paraformaldehyde for 20–30 min, treated with 0.5% Triton X-100 for
10 min and then stained with BCIP/NBT (Beyotime, Haimen, Jiangsu,
China) for 5–13 min prior to examination.
Briefly, for immunofluorescence staining, the hPES-2
cell colonies were incubated with the following primary antibodies
against stage-specific embryonic antigens: rat anti-human
stage-specific embryonic antigen (SSEA)3 monoclonal antibody (Cat.
no. LV1528429), rat anti-human SSEA4 monoclonal antibody (Cat. no.
LV1488380), mouse anti-human tumor-rejection antigen (TRA)-1-60
monoclonal antibody (Cat. no. LV1541028), mouse anti-human TRA-1-81
monoclonal antibody (Cat. no. LV1580855); mouse anti-human
octamer-binding transcription factor 4 (OCT-4) monoclonal antibody
(Cat. no. MAB4419A4) and mouse anti-human Nanog homeobox (NANOG)
monoclonal antibody (Cat. no. MABD24A4) (all from Millipore) (all
were used at 1:100). The secondary antibodies were as follows: goat
anti-rat IgM 488 (Cat. no. 549138), goat anti-mouse IgM R-PE (Cat.
no. 488800A) (both from Invitrogen), goat anti-rat IgG, FITC (Cat.
no. CW0167) and goat anti-mouse IgG, FITC (Cat. no. CW0113) (both
from CWBIO, Beijing, China) (all were used at 1:200).
Gene expression levels in the pluripotent hPES2
cells were assessed by real-time PCR. The primers used for OCT-4,
REX1 [also referred to as zinc-finger protein-42 (ZFP42)], SRY (sex
determining region Y)-box 2 (SOX2), NANOG, Lin-28 homolog A (LIN28)
and nucleophosmin (NPM1) are listed in Table II. β-actin was used as a control.
PCR products were size-fractionated by 1% agarose gel
electrophoresis and were visualized by ethidium bromide staining.
Final analysis was made using an image analyzer (Bio-Rad, Hercules,
CA, USA).
 | Table IIPrimers for pluripotency genes. |
Table II
Primers for pluripotency genes.
| Gene | Forward
(5′-3′) | Reverse
(5′-3′) | Annealing
temperature (°C) | Product size
(bp) |
|---|
| OCT-4 |
GACAACAATGAGAACCTTCAGGAGA |
TTCTGGCGCCGGTTACAGAACCA | 55 | 218 |
| NANOG |
CAGAAGGCCTCAGCACCTAC |
CTGTTCCAGGCCTGATTGTT | 55 | 216 |
| REX-1 |
GCGTACGCAAATTAAAGTCCAGA |
CAGCATCCTAAACAGCTCGCAGAAT | 58 | 306 |
| SOX-2 |
CCCCCGGCGGCAATAGCA |
TCGGCGCCGGGGAGATACAT | 58 | 448 |
| LIN28 |
AGTAAGCTGCACATGGAAGG |
ATTGTGGCTCAATTCTGTGC | 58 | 420 |
| NPM1 |
TGGTGCAAAGGATGAGTTGC |
GTCATCATCTTCATCAGCAGC | 58 | 343 |
| β-actin |
CGGATGTCCACGTCACACTT |
GTTGCTATCCAGGCTGTGGT | 55 | 469 |
Statistical analysis
The results from real-time PCR for the expression
levels of the different genes in the different cell lines were
compared by ANOVA. Pearson correlation coeffi-cients were
determined to assess possible associations between XIST RNA and
X-linked gene expression levels. A P-value of <0.05 was
considered to indicate a statistically significant difference.
Results
Karyotype instability in hPES cell
lines
The hBES-1, hPES-1 and hPES-2 are hES cell lines
that have been strictly validated. The hBES-1 cell line is the
parent source of hES cell lines, and hPES-1 and hPES-2 cells are
well known parthenogenetic embryonic stem cell lines (6,20).
The hBES-1, hPES-1 and hPES-2 cells had stabilized at >70
generations when cultured under identical culture conditions. The
hBES-1, hPES-1 and hPES-2 cells retained the unique morphological
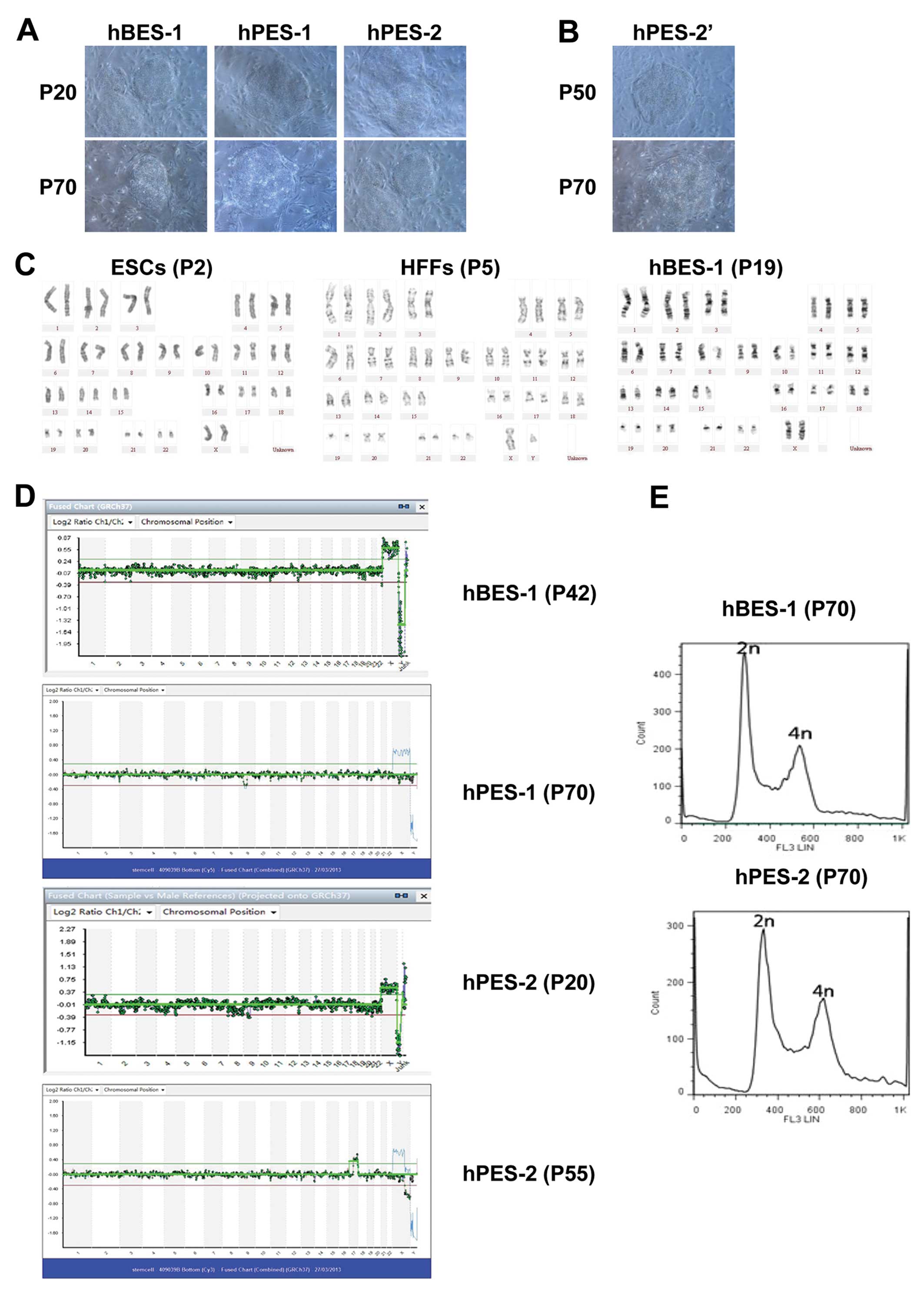
and growth characteristics of hESCs (Fig. 1A and B).
We found that the chromosome karyotypes of hPES-2
cells changed during their long-term culture in vitro. At
P20 (early generations), the results from aCGH for the hPES-2 cells
were 46,XX. At P55, X chromosome microdeletions were observed in
the hPES-2 cells, and the aCGH results were 46,XX,del(x)
(q22.3;q28),+(17)(q121.31;q25.3)
(Fig. 1C–E). In our previous
study [Mai et al (6)], we
reported X chromosome microdeletions in the same hPES cell lines.
These phenomena indicated that the hPES-2 cell chromosomes were
unstable and that their karyotypes could change during long-term
culture. These phenomena did not occur in the hBES-1 and hPES-1
cell lines (Table III).
 | Table IIIKaryotypes of human embryonic stem
cells. |
Table III
Karyotypes of human embryonic stem
cells.
| Chromosome
karyotyping | aCGH | FACS |
|---|
| hBES-1 | 46,XX (P19) | 46,XX (P42) | Diploid (P70) |
| hPES-1 | 46,XX (P40)
[6[ | 46,XX (P70) | N/A |
| hPES-2 |
46,XX,del(X)(q22;q24), del(1)(q21;q25)
(P57) [6[ | 46,XX
(P20)
46,XX,del(X)(q22.3;q28),+(17)(q121.31;q25.3) (P55) | Diploid (P70) |
Three XCI statuses identified in hPES
cells
XIST RNA and H3K27me3 accumulation on X chromosomes
has been reported to be a sign of XCI, accompanied by the loss of
H3K9ac expression at H3K27me3 accumulation sites (25–31). In this study, we used XIST RNA,
H3K27me3 and H3K9ac as indicators of XCI.
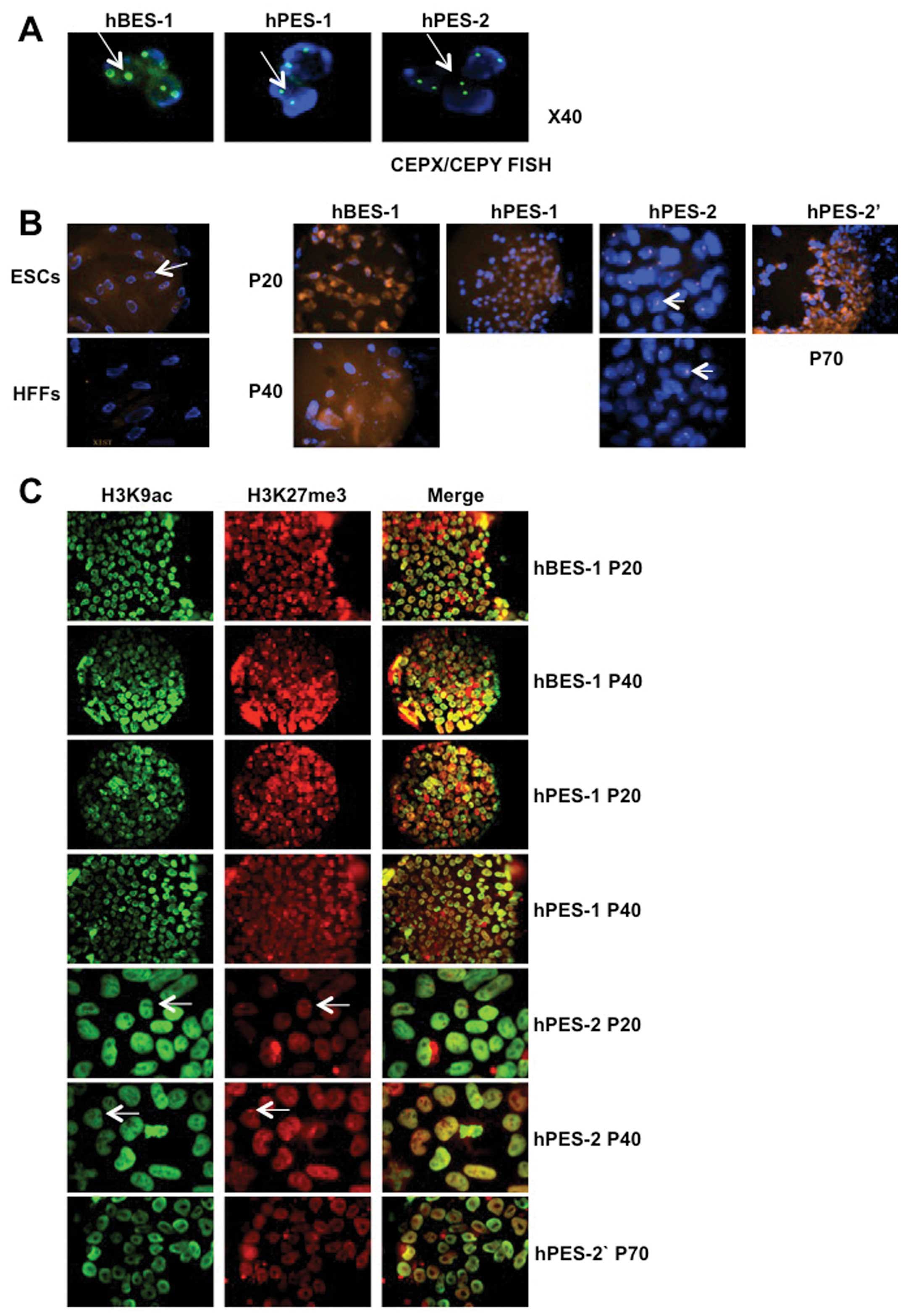
We assessed the XCI status of hBES-1, hPES-1 and
hPES-2 cell clones at P20 and P40, and found that XIST RNA and
H3K27me3 did not accumulate in the hBES-1 and hPES-1 cells, which
suggested that early-passage hBES-1 and hPES-1 cells did not
undergo XCI. Subsequently, we further verified the X chromosome
contents in the hBES-1, hPES-1 and hPES-2 cells using CEPX/CEPY
FISH. These 3 hES cell lines all contained 2 X chromosomes
(Fig. 2A). However, XIST RNA and
H3K27me3 accumulation and H3K9ac loss were observed in the hPES-2
cells at P20, which suggested that XCI had been activated during
early passage and maintained in the hPES-2 cells at P40 (Fig. 2B and C, Table IV). A previous study suggested
that XCI in mouse embryonic stem cells indicated cell
differentiation (32). However,
in this study, the hPES-2 cells still expressed high levels of
pluripotency genes and proteins and formed EBs in vitro,
which suggested that the hPES-2 cells did not undergo
differentiation (Fig. 3).
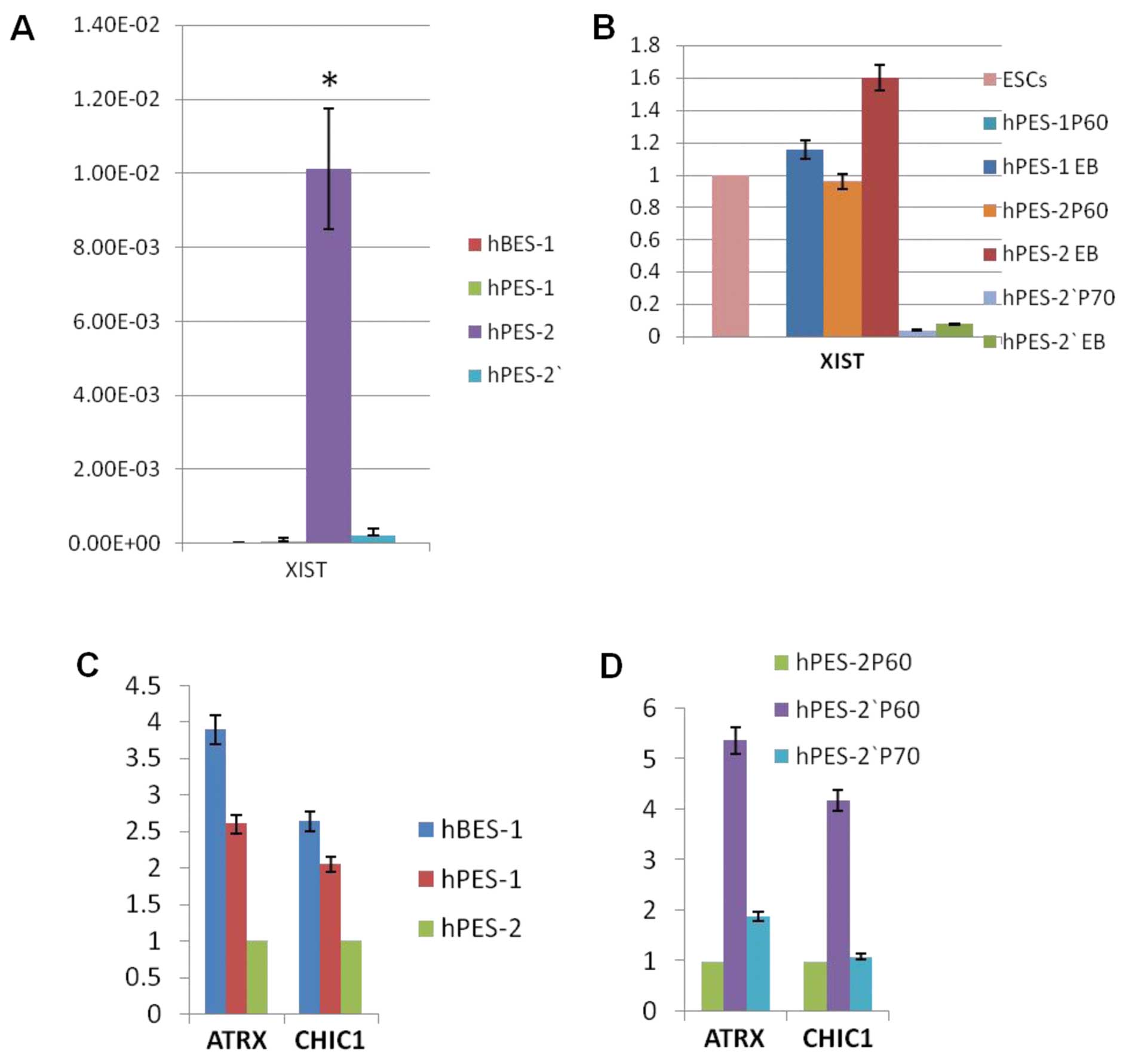
Real-time PCR results also showed that XIST RNA expression was high
in hPES-2 cells, but was very low or negative in hBES-1 and
hPES-1cells (P<0.001) (Fig.
4A).
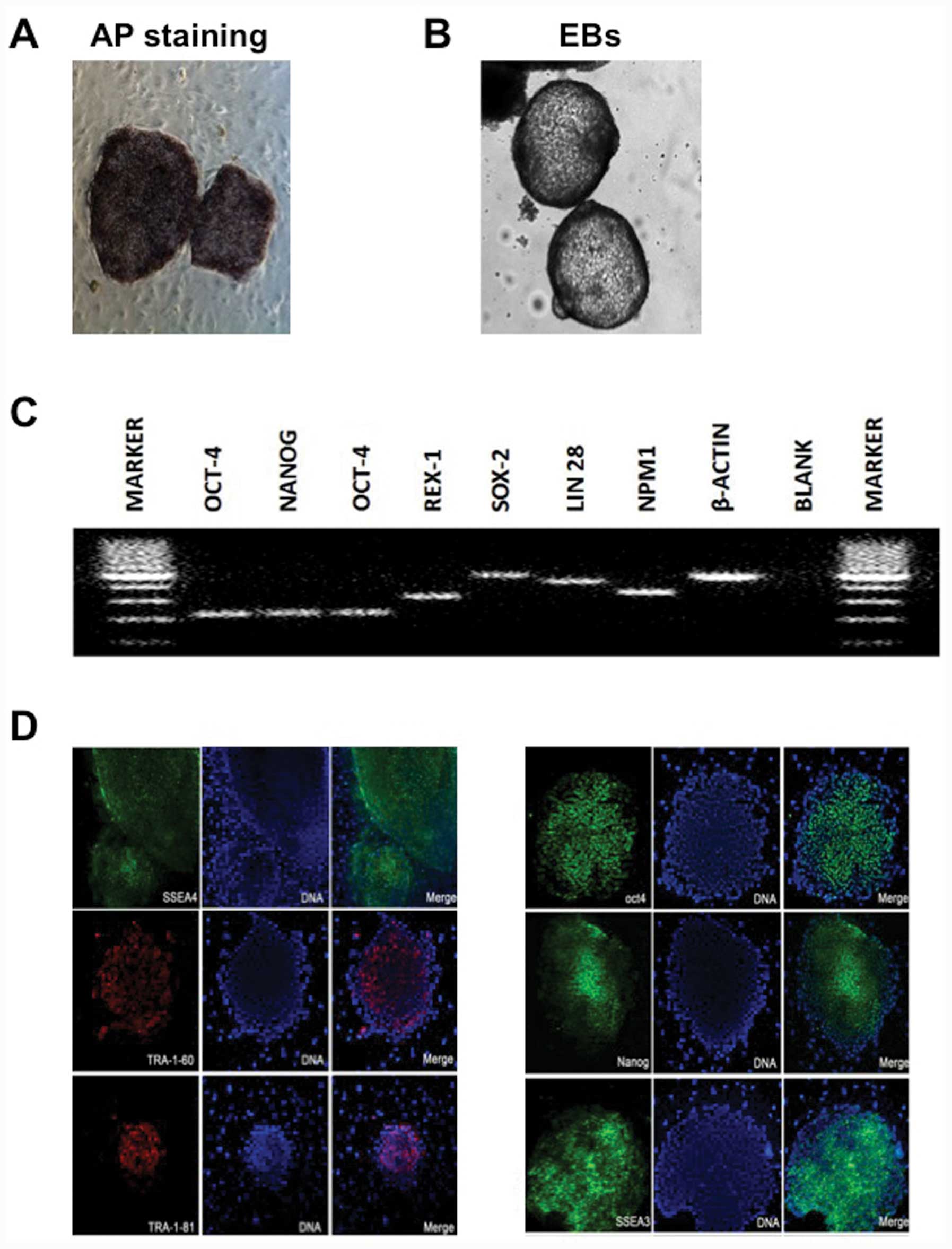 | Figure 3Pluripotentiality of human
parthenogenetic embryonic stem cell line-2 (hPES-2). (A) Alkaline
phosphatase (AP) staining, (B) embryoid bodies (EBs), (C)
pluripotency gene expression, including octamer-binding
transcription factor 4 (OCT-4), REX1 [also referred to as
zinc-finger protein-42 (Zfp42)], SRY (sex determining region Y)-box
2 (SOX2), Nanog homeobox (NANOG), Lin-28 homolog A (LIN28) and
nucleophosmin (β-actin was used as a control), amplified by
real-time PCR, and (D) pluripotency immunofluorescent markers,
including stage-specific embryonic antigen (SSEA)3, SSEA4,
tumor-rejection antigen (TRA)-1-60, TRA-1-81, OCT-4 and NANOG. |
 | Table IVXIST RNA and H3K27me3 statuses of hES
cell lines. |
Table IV
XIST RNA and H3K27me3 statuses of hES
cell lines.
| XIST RNA
| H3K27me3
|
|---|
| P20 | P40 | P20 | P40 |
|---|
| hBES-1 | − | − | − | − |
| hPES-1 | − | − | − | − |
| hPES-2 | + | + | + | + |
|
| P70 | P70 |
|
| hPES-2′ | − | − |
To validate the XCI status in the hPES-2 cells,
hPES-2 cells at P45 (subclones of hPES-2 cells; designated as
hPES-2′ cells) that had been stored in liquid nitrogen were
recovered. These cells grew normally and passed to P70. However,
these cells differed from the hPES-2 cells, as the XIST RNA
expression levels in the hPES-2′ cells were very low (P<0.001;
Fig. 4A); XIST RNA and H3K27me3
did not accumulate on X chromosomes (Fig. 2B and C, Table IV). In EB forming assays, the
hPES-1 and hPES-2 EBs had a higher XIST RNA expression, whereas the
hPES-2′ EB XIST RNA expression was extremely low (Fig. 4B).
All of these results indicated that there were
different XCI statuses in the hPES cells: i) pre-XCI status: hPES-1
cells that did not express XIST RNA, and re-expressed XIST RNA
following differentiation; ii) XCI status: hPES-2 cells expressed
XIST RNA and this expression was sustained following
differentiation; and iii) quiescent XCI status: low XIST RNA
expression in hPES-2′ cells, which remained low following
differentiation.
X-linked gene expression levels in hPES
cells
ATRX and CHIC1 are X-linked genes that are silenced
during XCI. As expected, the ATRX and CHIC1 expression levels were
very low in the hPES-2 cells, and higher in the hBES-1 and hPES-1
cells. The ATRX and CHIC1 expression levels in the hPES-2′ cells at
P60 were higher than those in the hPES-2 cells at P60, but were
decreased in the hPES-2′ cells at P70 and were as low as in those
in the hPES-2 cells at P60, suggesting that a low XIST RNA
expression in the hPES-2′ cells at P70 had occurred (Fig. 4C–D).
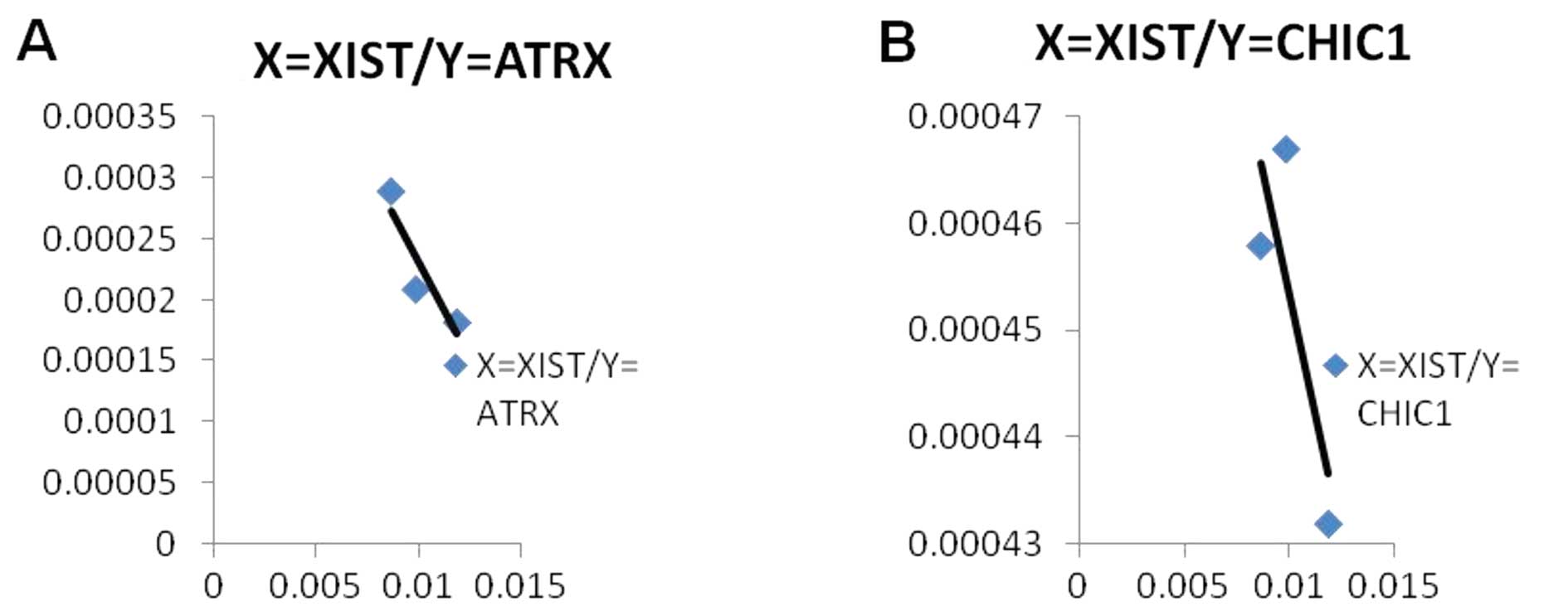
The results from a correlation analysis suggested
that there was a tendency for a negative correlation between
X-linked genes and XIST RNA expression levels, although this was
not statistically significant (P>0.05; n=3). An increased sample
size may have provided a significant result. However, this still
indicated that XIST RNA mediated the silencing of these X-linked
genes (Fig. 5).
Discussion
Using hPES cells may be a means to resolve human
embryo stem cell transplantation immune rejection issues (33,34), although the security of their
application remains a concern. Studies have found that human and
murine parthenogenetic embryonic stem cells have genetic
instabilities and epigenetic abnormalities (33,35). Thus, it is essential to understand
the genetic and epigenetic characteristics of hPES.
In the current study, we found that some hPES cell
lines had chromosome karyotype instabilities. The hPES-1 cells had
a stable karyotype following serial passage, whereas the hPES-2
cells lost some X chromosome fragments after 50 passages. Previous
studies have demonstrated that parthenogenetic embryonic stem cells
often undergo changes associated with karyotype abnormalities
(33). Liu el al (36) also reported X chromosome losses in
hPES cells. These, as previously suggested, the genetic
characteristics of hPES cells would not be stable following
long-term culture (37).
To date, 3 XCI statuses have been reported for hES
cells. With status I in hES cells, 2 X chromosomes are both
activated in the undifferentiated stage, and XCI occurs randomly
following differentiation, similar to what occurs in mouse
embryonic stem cells (15–17).
With status II, XCI has already occurred in undifferentiated hES
cells, and approximately 20–70% of hES cells will have XIST clouds
accumulated on specific chromosomes (11,18). With status III, XCI has occurred
without XIST RNA expression (11). The XCI status may be associated
with the genetic characteristics of the cells themselves, although
it may also be affected by the culture conditions used. Some
investigators have assumed that XCI in hES cells moved from state I
to a transition state II to III, as an adaptation to the
environmental conditions in vitro (11,18). Lengner et al (19) found that blastocyst-derived hES
cells did not have XCI when cultured under low oxygen
concentrations (5%); this status was associated with the
pluripotency of stem cells and was affected by the oxygen
concentration and atmospheric pressure. These data indicate that
the culture conditions used, the methods used to establish cell
lines, as well as other factors affect the XCI status of hES
cells.
In the present study, we also found 3 XCI statuses
that respectively matched those of hES cells: i) pre-XCI status,
which was similar to hES status I: hPES-1 cells did not express
XIST RNA, but did express X-linked genes, and re-expressed XIST RNA
following differentiation, hPES-1 cells for example; ii) XCI
status, which was similar to hES status II: hPES-2 cells expressed
XIST RNA and expressed X-linked genes at a very low level, and XIST
RNA expression was sustained following differentiation, hPES-2
cells for example; and iii) quiescent XCI status, which was similar
to hES status III: low XIST RNA and X-linked gene expression in
hPES-2′ cells, and XIST RNA expression remained at a low level
following differentiation, hPES-2′ cells for example. The results
observed for the hPES-2′ cells may have been due to their
development from hPES-2.
The hES cells used in this study all underwent
freezing and thawing and long-term culture in vitro. A
previous study suggested that the freezing process and the in
vitro culture conditions may alter the epigenetic state of stem
cells (19). In this study,
unstable XCI was found in the hPES-2 cells, and stable XCI (XCI I)
was found in the hPES-1 and hBES-1 cells, which was similar to the
results from the study by Liu et al (38). Unstable XCI may have been
associated with chromosome instability in the hPES-2 cells.
Unstable chromosomes rendered this cell line susceptible to
environmental conditions and the freezing process used, which may
have been the result of a environmental adaptations.
There is no paternal genetic material in
parthenogenetic embryonic stem cells, and the 2 X chromosomes are
all from the mother. The XCI status of hPES cells that lack
paternal genetic material seems to be similar to that of embryonic
stem cells. Liu et al (36) found that early-passage hPES cells
did not exhibit XCI, but that XIST RNA expression and the XCI
status emerged slowly during the course of long-term culture in
vitro. Others have found that different hPES cells lines have
different XCI states, which was similar to the situation with hES
cells (38). To the best of our
understanding, the XCI status of hES cells and the factors that
affect it are more significant.
In conclusion, our data demonstrated that the
chromosome karyotypes of some hPES cell lines exhibited
instabilities. Similar to the hES cells, the hPES cells had 3 XCI
statuses. Unstable XCI of hPES-2 cells may be related to chromosome
instability. Unstable chromosomes render this cell line susceptible
to environmental conditions and the freezing process used, which
may be the result of an environmental adaptation. XCI plays
important roles in sustaining embryo formation and cell biological
activity (14), and an abnormal
XCI can result in some diseases (39). Thus, it is essential to routinely
assess the XCI status of hES cells, including hPES cells prior to
their use in clinical applications.
Acknowledgments
We are very grateful to Professor Mai Qingyun and
Professor Li Tao for their assitance in the derivation and
characterization of the hBES-1, hPES-1 and hPES-2 cell lines. This
study was supported in part by grants from the National Natural
Science Foundation of China (81270750); the National 973 program
(2012CB947604); Guangdong Provincial Key Laboratory of Reproductive
Medicine (2012A06140003).
References
|
1
|
Lindvall O and Kokaia Z: Stem cells for
the treatment of neurological disorders. Nature. 441:1094–1096.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Vats A, Bielby RC, Tolley NS, Nerem R and
Polak JM: Stem cells. Lancet. 366:592–602. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Szabó P and Mann JR: Expression and
methylation of imprinted genes during in vitro differentiation of
mouse parthenogenetic and androgenetic embryonic stem cell lines.
Development. 120:1651–1660. 1994.PubMed/NCBI
|
|
4
|
Lin G, OuYang Q, Zhou X, et al: A highly
homozygous and parthenogenetic human embryonic stem cell line
derived from a one-pronuclear oocyte following in vitro
fertilization procedure. Cell Res. 17:999–1007. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Revazova ES1, Turovets NA, Kochetkova OD,
Kindarova LB, Kuzmichev LN, Janus JD and Pryzhkova MV:
Patient-specific stem cell lines derived from human parthenogenetic
blastocysts. Cloning and Stem Cells. 9:432–449. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mai Q, Yu Y, Li T, et al: Derivation of
human embryonic stem cell lines from parthenogenetic blastocysts.
Cell Res. 17:1008–1019. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Revazova ES, Turovets NA, Kochetkova OD,
et al: HLA homozygous stem cell lines derived from human
parthenogenetic blastocysts. Cloning Stem Cells. 10:11–24. 2008.
View Article : Google Scholar
|
|
8
|
Lu Z, Zhu W, Yu Y, et al: Derivation and
long-term culture of human parthenogenetic embryonic stem cells
using human foreskin feeders. J Assist Reprod Genet. 27:285–291.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhao Q, Wang J, Zhang Y, Kou Z, Liu S and
Gao S: Generation of histocompatible androgenetic embryonic stem
cells using spermatogenic cells. Stem Cells. 28:229–239. 2010.
|
|
10
|
Maitra A, Arking DE, Shivapurkar N, et al:
Genomic alterations in cultured human embryonic stem cells. Nat
Genet. 37:1099–1103. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Silva SS, Rowntree RK, Mekhoubad S and Lee
JT: X-chromosome inactivation and epigenetic fluidity in human
embryonic stem cells. Proc Nat Acad Sci USA. 105:4820–4825. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Allegrucci C, Thurston A, Lucas E and
Young L: Epigenetics and the germline. Reproduction. 129:137–149.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Allegrucci C, Denning C, Priddle H and
Young L: Stem-cell consequences of embryo epigenetic defects.
Lancet. 364:206–208. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tomkins DJ, McDonald HL, Farrell SA and
Brown CJ: Lack of expression of XIST from a small ring X chromosome
containing the XIST locus in a girl with short stature, facial
dysmorphism and developmental delay. Eur J Hum Genet. 10:44–51.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sugawara O, Takagi N and Sasaki M:
Correlation between X-chromosome inactivation and cell
differentiation in female preimplantation mouse embryos. Cytogenet
Cell Genet. 39:210–219. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hall LL, Byron M, Butler J, et al:
X-inactivation reveals epigenetic anomalies in most hESC but
identifies sublines that initiate as expected. J Cell Physiol.
216:445–452. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dhara SK and Benvenisty N: Gene trap as a
tool for genome annotation and analysis of X chromosome
inactivation in human embryonic stem cells. Nucleic Acids Res.
32:3995–4002. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shen Y, Matsuno Y, Fouse SD, et al:
X-inactivation in female human embryonic stem cells is in a
nonrandom pattern and prone to epigenetic alterations. Proc Nat
Acad Sci USA. 105:4709–4714. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lengner CJ, Gimelbrant AA, Erwin JA, et
al: Derivation of pre-X inactivation human embryonic stem cells
under physiological oxygen concentrations. Cell. 141:872–883. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li T, Zhou CQ, Mai QY and Zhuang GL:
Establishment of human embryonic stem cell line from gamete donors.
Chin Med J (Engl). 118:116–122. 2005.
|
|
21
|
Li T, Mai Q, Gao J and Zhou C:
Cryopreservation of human embryonic stem cells with a new bulk
vitrification method. Biol Reprod. 82:848–853. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ryan IP, Schriock ED and Taylor RN:
Isolation, characterization, and comparison of human endometrial
and endometriosis cells in vitro. J Clin Endocrinol Metab.
78:642–649. 1994.PubMed/NCBI
|
|
23
|
Qin XY, Sone H, Kojima Y, et al:
Individual variation of the genetic response to bisphenol a in
human foreskin fibroblast cells derived from cryptorchidism and
hypospadias patients. PLoS One. 7:e527562012. View Article : Google Scholar
|
|
24
|
Fiorentino F1, Spizzichino L, Bono S, et
al: PGD for reciprocal and Robertsonian translocations using array
comparative genomic hybridization. Hum Reprod. 26:1925–1935. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Brockdorff N, Ashworth A, Kay GF, et al:
Conservation of position and exclusive expression of mouse Xist
from the inactive X chromosome. Nature. 351:329–331. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Brown CJ, Ballabio A, Rupert JL,
Lafreniere RG, Grompe M, Tonlorenzi R and Willard HF: A gene from
the region of the human X inactivation centre is expressed
exclusively from the inactive X chromosome. Nature. 349:38–44.
1991. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Penny GD, Kay GF, Sheardown SA, Rastan S
and Brockdorff N: Requirement for Xist in X chromosome
inactivation. Nature. 379:131–137. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Heard E and Disteche CM: Dosage
compensation in mammals: fine-tuning the expression of the X
chromosome. Genes Dev. 20:1848–1867. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Okamoto I, Otte AP, Allis CD, Reinberg D
and Heard E: Epigenetic dynamics of imprinted X inactivation during
early mouse development. Science. 303:644–649. 2004. View Article : Google Scholar
|
|
30
|
Costanzi C and Pehrson JR: Histone
macroH2A1 is concentrated in the inactive X chromosome of female
mammals. Nature. 393:599–601. 1998. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chadwick BP and Willard HF: Cell
cycle-dependent localization of macroH2A in chromatin of the
inactive X chromosome. J Cell Biol. 157:1113–1123. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Navarro P, Chambers I, Karwacki-Neisius V,
Chureau C, Morey C, Rougeulle C and Avner P: Molecular coupling of
Xist regulation and pluripotency. Science. 321:1693–1695. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kim K, Lerou P, Yabuuchi A, et al:
Histocompatible embryonic stem cells by parthenogenesis. Science.
315:482–486. 2007. View Article : Google Scholar
|
|
34
|
Taylor CJ, Bolton EM, Pocock S, Sharples
LD, Pedersen RA and Bradley JA: Banking on human embryonic stem
cells: estimating the number of donor cell lines needed for HLA
matching. Lancet. 366:2019–2025. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Jiang H, Sun B, Wang W, et al: Activation
of paternally expressed imprinted genes in newly derived
germline-competent mouse parthenogenetic embryonic stem cell lines.
Cell Res. 17:792–803. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liu W, Yin Y, Jiang Y, et al: Genetic and
epigenetic X-chromosome variations in a parthenogenetic human
embryonic stem cell line. J Assist Reprod Genet. 28:303–313. 2011.
View Article : Google Scholar :
|
|
37
|
Allegrucci C and Young LE: Differences
between human embryonic stem cell lines. Hum Reprod Update.
13:103–120. 2007. View Article : Google Scholar
|
|
38
|
Liu W, Guo L, He W, Li Q and Sun X: Higher
copy number variation and diverse X chromosome inactivation in
parthenote-derived human embryonic stem cells. J Reprod Dev.
58:642–648. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ganesan S, Richardson AL, Wang ZC, et al:
Abnormalities of the inactive X chromosome are a common feature of
BRCA1 mutant and sporadic basal-like breast cancer. Cold Spring
Harb Symp Quant Biol. 70:93–97. 2005. View Article : Google Scholar
|