Introduction
The four Dengue viruses (DENV 1–4) are enveloped,
positive strand RNA viruses, and cause a broad spectrum of clinical
manifestations (1). The DENV, as
well as West Nile virus (WNV), Japanese encephalitis virus (JEV)
and Hepatitis C virus (HCV) belongs to the family
Flaviviridae, which follow the similar replication
mechanisms (2). The DENV RNA is
translated into a single poly-protein precursor that is
subsequently cleaved into three structural proteins and seven
non-structural proteins. The DENV NS3 and NS5 are directly involved
in viral RNA replication, with NS3 acting as an RNA helicase and
NS5 acting as the RNA-dependent RNA polymerase (2). In addition, DENV is dependent on
host factors to establish successful infection and accomplish viral
replication (2).
Several RNA binding proteins have been reported to
be involved in the replication of DENV RNA. Yocupicio-Monroy et
al (3,4) reported that the La, calreticulin,
and protein disulfide isomerase proteins from a human monocytic
cell line bind to the 3′ untranslated region (UTR) of DENV-4, and
La protein interacts with the viral 3′ UTRs and is redistributed in
DENV-infected cells. Another study by De Nova-Ocampo et al
(5) showed that the La protein,
translation elongation factor-1α (EF-1α), and polypyrimidine
tract-binding protein (PTB) interact with the genomic 3′ UTR of
DENV-4 in which the CS1-L-SL region is responsible for forming
specific and stable complexes with cellular proteins. Additionally,
different regions located in the DENV 3′ UTR (+) have been shown to
interact with several other proteins such as poly(A)-binding
protein, NF90, p100, Y box-binding protein-1, HNRNPs A1, A2/B2 and
Q (6–9). Ward et al (10) reported that one of the P-body
protein DDX6 bound to DENV RNA during infection and this
interaction was mediated by the DB1 and DB2 structures in the 3′
UTR, whereas the stress granule (SG) proteins G3BP1, Caprin1 and
USP10 bound to the variable region (VR) in the 3′ UTR. Considering
that those proteins colocalized with DENV replication sites and
bound to different sites on 3′ UTR, they suggested that the DENV-2
RNA was guided to a site for assembly of P-body and SG proteins
which could regulate whether the viral RNA undergoes translation,
replication or packaging. Further identification of host factors
interacting with DENV RNA may improve our understanding of the DENV
amplification cycle.
The LSm1-7 complex is known to be an important
component of the P-body, and acts as the highly conserved
RNA-binding protein to regulate the fate of cellular mRNAs
(11,12). The LSm1-7 complex is also involved
in the replication of several viruses. A screen for host factors
required for Brome mosaic virus (BMV) gene expression identified
that LSm1-7 complex plays an essential role in translation and in
the translation-replication transit of the BMV genome (13,14). Pérez-Vilaró et al (15) and Scheller et al (16) showed that the LSm1-7 complex is
required for efficient translation of the HCV genome and that these
requirements are functionally linked to the 5′ and 3′ UTRs, which
are known to play key roles in the regulation of HCV translation
and replication.
Given the conserved function of the LSm1-7 complex
in the mRNA processing and the common mechanism of all (+) RNA
viruses replication, we hypothesized that the LSm1-7 complex may be
used by DENV RNA to replicate in human cells. In the present study,
we showed that LSm1 can interact with DENV RNA specifically. By
measuring DENV RNA amplification and infectious virus particle
production, our results indicated that the LSm1-7 complex is
necessary for DENV replication.
Materials and methods
Cell culture and cytoplasmic extract
A549 and Vero-E6 cells were maintained in RPMI-1640
(Invitrogen, Carlsbad, CA, USA) supplemented with 10% FBS (Hyclone
Laboratories Inc., Logan, UT, USA), 10 U/ml penicillin and 10
μg/ml streptomycin (Pen/Strep; Gibco-Invitrogen, Carlsbad,
CA, USA) at 37°C in a 5% CO2 atmosphere. C6/36 (Aedes
albopictus) cells were cultured at 28°C in RPMI-1640
supplemented with 10% FBS.
For preparation of the cytoplasmic extract (S-100),
A549 cells were adapted for growth on a large scale. Cell extracts
from A549 cells were prepared using the procedure as described by
Paranjape and Harris (9).
DENV-2 amplification and infection
DENV-2 was amplified in C6/36 cells as previously
described (17). For DENV-2
infection, the cells were incubated with the virus (MOI=1) for 2 h
at 37°C with occasional rocking. After 2 h, the cells were rinsed,
overlaid with complete medium, and incubated at various time
points.
Immunofluorescence assay and confocal
microscopy
A549 cells were seeded onto 8-well coverslips
(LabTek, Bloomington, IN, USA) and infected with DENV-2 for 24 h.
After being fixed and permeabilized, the cells were incubated with
blocking buffer [PBS containing 3% bovine serum albumin (BSA)] for
1 h. Primary antibodies were diluted in blocking buffer (1:300 for
anti-LSm1 and anti-Dcp1B, Proteintech Group, Chicago, IL, USA;
1:200 for anti-dsRNA, English and Scientific Consulting Kft.,
Szirák, Hungary) and incubated with cells at 4°C overnight. The
cells were then washed and incubated for 1 h with fluorescently
labelled Alexa Fluor 488 goat anti-rabbit and Alexa Fluor 594 goat
anti-mouse antibodies (Invitrogen). Coverslips were mounted in
ProLong Gold antifade reagent containing DAPI (Invitrogen), and the
cells were visualized under a laser confocal microscope LSM 510
(Carl Zeiss, Inc., Thornwood, NY, USA). The images were captured
and analyzed by using ZEN software (Carl Zeiss, Inc.).
RNA interference
The siRNAs, target LSm1 (siLSm1:
5′-CAAACUUAGUGCUACAUCATT-3′) and the scrambled sequence as a
negative control (siNC: 5′-UUCUCCGAACGUGUCACGUTT-3′) were
transfected into A549 cells with 33 and 25 nM of each siRNA for the
first and second transient transfections, respectively. The effects
of LSm1 knockdown were detected 48 h post-secondary transfection by
immunoblotting LSm1 protein and mRNA levels. For detection of the
effect of LSm1 knockdown on DENV replication, siRNA-transfected
A549 cells were infected with DENV-2. At the indicated time points,
the supernatant and cell RNAs were collected for infection of
Vero-E6 cells and relative reverse transcription quantitative
polymerase chain reaction (RT-qPCR) analysis, respectively.
Relative RT-qPCR analysis
LSm1 gene expression and viral RNA synthesis were
analyzed by relative quantitative RT-PCR (primers: LSm1-F,
5′-GACTTGGAAAAGGAGAGTGACA-3′; LSm1-R, 5′-GGGCAAAAGATTAGTACTCATC-3′;
DENV-NS5-F, 5′-ACAAGTAGAACAACCTGGTCCAT-3′; DENV-NS5-R:
5′-GCCGCACCATTGGTCTTCTC-3′). The housekeeping gene β-actin
(β-actin-F, 5′-TGACGGGGTCACCCACACT G-3′; β-actin-R,
5′-AAGCTGTAGCCGCGCTCGGT-3′) was used as an internal control. Total
RNA was isolated by using RNAiso and total cDNA was synthesized by
using PrimeScript™ RT Master Mix (both from Takara Bio, Inc.,
Shiga, Japan). Each cDNA was amplified with the above primers and
SYBR Premix Ex Taq™ II (Takara Bio, Inc.) for 40 cycles and data
were analyzed by using Mx3005P System Software (Stratagene, La
Jolla, CA, USA). Relative gene expression refers to the magnitude
of the signal generated for the NS5 gene of DENV from siLSm1- and
siNC-treated cells using the β-actin gene as the internal
control.
Western blotting
The samples were separated by SDS-PAGE followed by
immunoblotting analysis using a rabbit poly-clonal anti-LSm1 or a
mouse anti-β-actin mAb (both from Proteintech Group).
RNA immunoprecipitation
The interaction of LSm1 with DENV viral RNA during
infection was studied by using RNA immunoprecipitation assay.
Briefly, the infected cells were collected and cross-linked with
0.1% formaldehyde. The cells were then disrupted by sonication in
RIP assay buffer (50 mM Tris-HCl, pH 7.5, 1% Nonidet P-40, 0.5%
sodium deoxycholate, 0.05% SDS, 1 mM EDTA and 150 mM NaCl, 0.2 mM
PMSF, 1X protease inhibitors and 40 units RNase inhibitor), and the
resulting lysates were precleared with protein A-agarose (Santa
Cruz Biotechnology, Inc., Santa Cruz, CA, USA). Immunoprecipitation
was carried out using anti-LSm1 monoclonal antibody or normal
rabbit IgG (as a negative control). Immune complexes were
precipitated with protein A-agarose. Viral RNA immunoprecipitation
was isolated using an RNeasy mini kit (Qiagen). RT-PCR was
conducted using primers 3′ UTR-F (5′-AAGGCAAAACTAACATGA AAC-3′) and
3′ UTR-R (5′-AGAACCTGTTGATTCAACAGC-3′).
RNA pull-down assay
To create DNA templates for the in vitro
transcription of DENV 3′ UTR, plasmid FL-D2 (18) containing the full-length DENV-2
genome was used as the template to amplify 454 nt of the DENV 3′
UTR. PCR was performed with primers 3′ UTR-F (5′-TAATACGACTCACT
ATAGGGAAGGCAAAACTAACATGAAAC-3′), including the T7 promoter and 3′
UTR-R. PCR templates were used for T7 RiboMAX Express Large Scale
RNA Production System (Promega, Madison, WI, USA) to generate DENV
3′ UTR RNA according to the manufacturer’s instructions. To
generate DENV RNA 3′ UTR-Bio, the Pierce™ RNA 3′ End Biotinylation
kit (Thermo Scientific, Waltham, MA USA) was used to attach a
single biotinylated nucleotide to the 3′ terminus of a 3′ UTR RNA
strand.
The pull-down assay was then performed. Briefly, 25
μg of DENV RNA 3′ UTR-Bio in RNA binding buffer (150 mM
NaCl, 50 mM Hepes-HCl pH 7.5, 0.5% NP-40) was heated to 95°C for 5
min, cooled to room temperature, combined with paramagnetic
streptavidin beads (Dynabeads C1, Dynabeads® MyOne™
Streptavidin C1, Invitrogen) and incubated on a rotation wheel at
4°C for 30 min. The beads were then washed three times with RNA
washing buffer (250 mM NaCl, 50 mM Hepes-HCl pH 7.5, 0.5% NP-40 and
10 mM MgCl2) prior to incubation with 400 μg of
S-100 fraction containing 40 units RNase inhibitor (Takara Bio,
Inc.) and 20 μg yeast tRNA (Invitrogen) by agitation for 30
min at 4°C. After incubation beads were washed another three times
with RNA wash buffer, fractions from the beads were boiled for 5
min in 0.1% SDS for subsequent western blotting.
Flow cytometric analysis
Vero-E6 cells were infected with DENV-2 containing
supernatant from LSm1 knockdown cells or siNC-treated cells
infected with DENV-2 in 6-well culture plates. Thirty-six hours
post-infection, the cells were collected and treated in 100
μl of Cytofix/Cytoperm buffer (Becton-Dickinson) and then
washed according to the manufacturer’s instructions. The cells were
resuspended with the DENV antibody 4G2 conjugated to fluorescein
isothiocyanate (FITC) (A190-108F; Cambridge Bioscience) and
incubated for 1 h at 4°C, followed by two rounds of washing, and
then resuspended in 300 μl of PBS. Flow cytometry was
carried out with a BD LSR II instrument, with data analysis using
FlowJo software (Tree Star, Inc., Ashland, OR, USA).
Results
The host factor LSm1 interacts with DENV
RNA in vitro
LSm1 has been shown to be involved in the
propagation of the positive strand RNA virus BMV and HCV. Following
infection with these viruses, LSm1 interacts directly with viral
RNA genomes via the 5′ and 3′ UTRs. To investigate whether LSm1 was
present in the ribonucleoprotein (RNP) complex formed between the
DENV-2 RNA and host factors, an RNA pull-down assay was performed.
The paramagnetic streptavidin beads-based RNA pull-down assay using
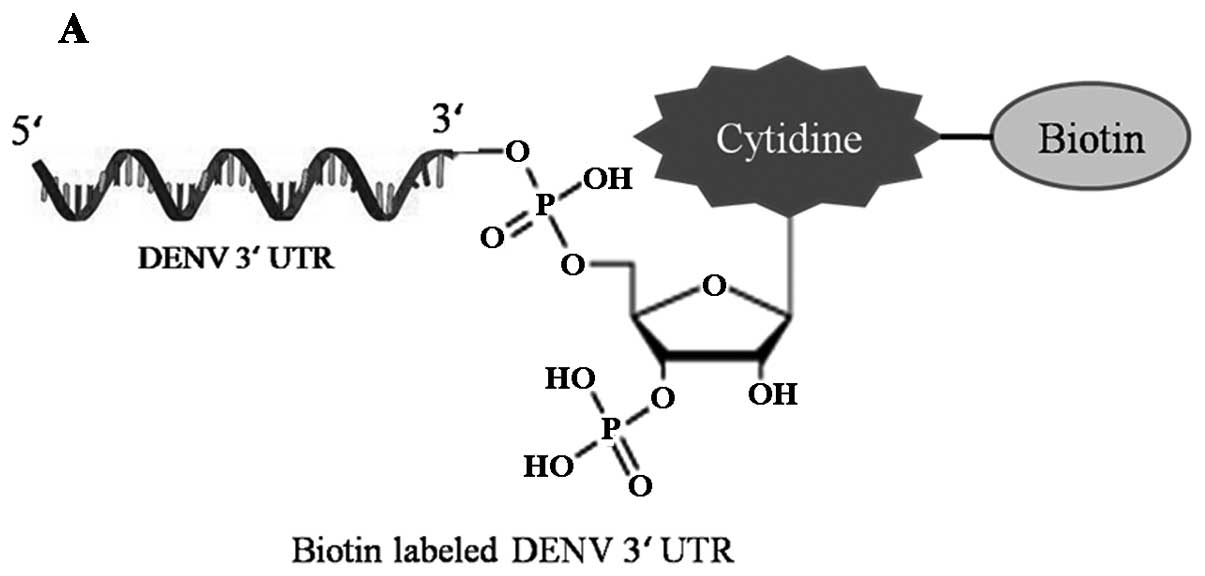
biotinylated DENV RNA 3′ UTR (Fig.
1A) was performed. Purified DENV 3′ UTR-Bio RNA was bound to
beads and incubated with A549 cell S-100 fractions, and the
interacting proteins were analyzed by western blotting. DENV 3′
UTR-Bio RNA but not beads alone was able to precipitate LSm1
present in cell extracts (Fig.
1B, left). As indicated from this result LSm1 can interact with
DENV RNA specifically in vitro.
LSm1 associates with DENV RNA in infected
cells and colocalizes with the DENV replication complex
To determine whether LSm1 associates with DENV RNA
during a virus infection, a viral RNA immunoprecipitation assay was
performed using anti-LSm1 antibody. A549 cells were infected with
DENV-2 at an MOI of 1. Cells were harvested 24 h post-inoculation
(p.i.) and treated with 0.1% formaldehyde to cross-link protein RNA
complexes. After the cell lysates were incubated with anti-LSm1
antibody, the RNA-LSm1-7 complexes were isolated after high
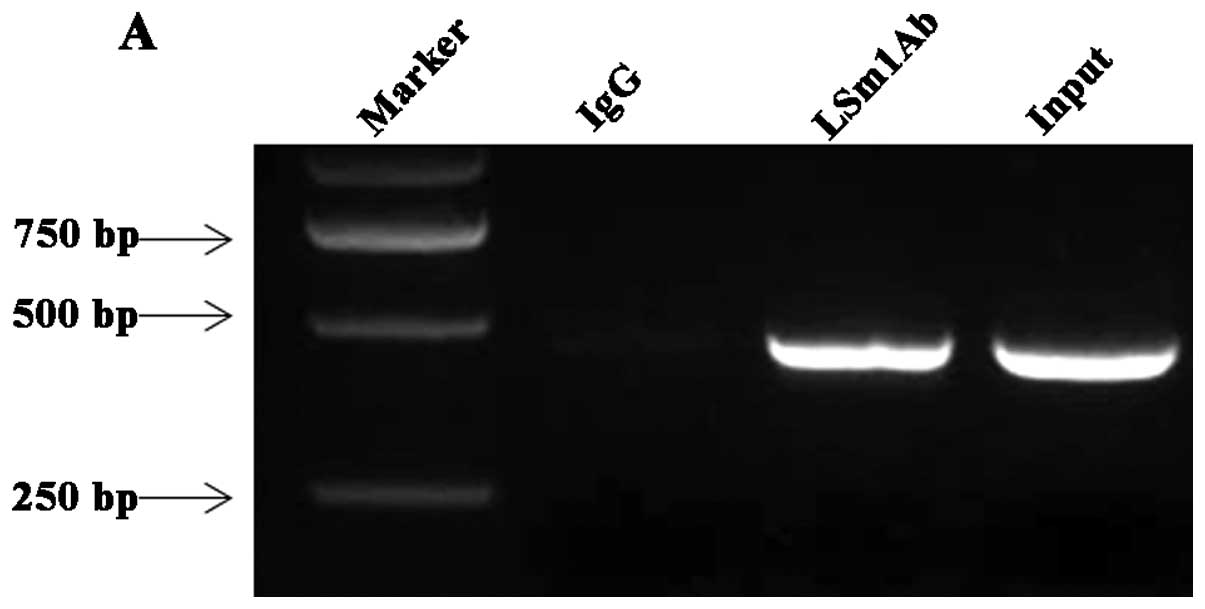
stringency washing and RNA was extracted. RT-PCR was conducted with
the primers targeted to DENV 3′ UTR. As shown in Fig. 2A, DENV RNA bound to LSm1 was
detected 24 h p.i. Normal rabbit IgG precipitation controls run in
parallel were negative. These results suggested that LSm1 interacts
with DENV RNA during the course of DENV-2 infection in
vivo.
To determine the potential role of LSm1 in the DENV
life cycle, we examined the subcellular localization of LSm1 during
DENV-2 infection using confocal laser scanning microscopy. To
detect DENV RNA, we used an antibody specific to dsRNA that was
used previously as a marker for sites of DENV replication (19). A549 cells were infected with
DENV-2 at an MOI of 1. After 24 h, the cells were fixed,
permeabilized and stained with anti-dsRNA and anti-LSm1 antibody.
For comparison, uninfected cells were stained with the same
antibodies. Nuclei were stained with DAPI, and the coverslips were
analyzed under a confocal laser scanning microscope. In the
uninfected cells, we observed detectable LSm1 exclusively. In the
infected cells, LSm1 localized to sites of DENV replication were
indicated by dsRNA signal (Fig.
2B).
LSm1 depletion inhibits the replication
of DENV RNA and release of infectious particles
LSm1 has been previously involved in HCV replication
and translational silencing. To determine whether LSm1 has a
potential role in DENV-2 replication, we established silencing
conditions for LSm1 in A549 cells followed by DENV-2 infection and
analyzed the levels of viral RNA in the infected cells and
infectious viral particles in the supernatant. The results showed
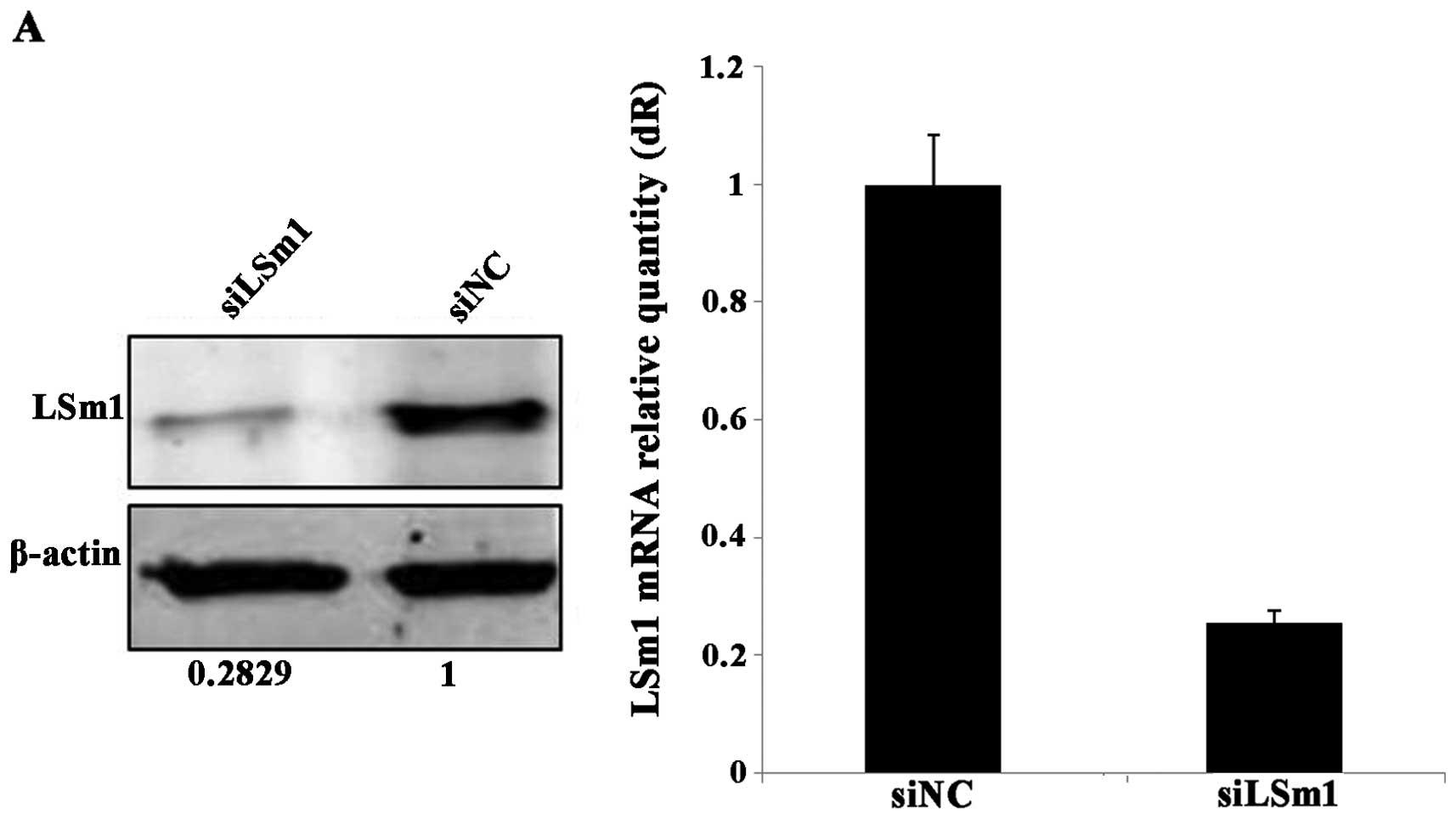
that targeting LSm1 siRNA-mediated silencing specifically reduced
71.71% of the LSm1 protein and 74.33% of LSm1 mRNA when using the
non-targeting siRNA siNC as a reference (Fig. 3A). At the time of maximal
silencing, A549 cells were infected with DENV-2. After 36 h,
intracellular DENV RNAs were quantified by RT-qPCR, whereas cell
supernatants were collected for the detection of infectious
particles. Intracellular DENV RNA levels were significantly reduced
by 59.43% compared to the siNC control while downregulating the
LSm1 protein 71.71% (Fig. 3B).
Moreover, DENV-2 production from siLsm1-transfected cells was also
reduced by 36.24% compared to the siNC control (Fig. 3C). Decrease of viral RNA
accumulation and particle production in LSm1 downregulation of
cells indicated LSm1 is invovled in early stages of the viral life
cycle, such as translation and replication.
Replication of DENV RNA in vivo is
associated with P-body
P-bodies are distinct foci within the cytoplasm of
the eukaryotic cell and play fundamental roles in mRNA translation,
RNA silencing, and RNA degradation as well as viral infection
processes (20–22). In the present study, we examined
whether the positive regulating role of LSm1 in DENV RNA
replication was associated with P-body. In the
5′-3′-deadenylation-dependent mRNA decay pathway, mRNA exit from
translation and shortening of the poly(A) tail is followed by
decapping via the Dcp1/Dcp2 decapping enzyme (23). This decapping process is not
necessary for the HCV life cycle and Dcp protein is a commonly used
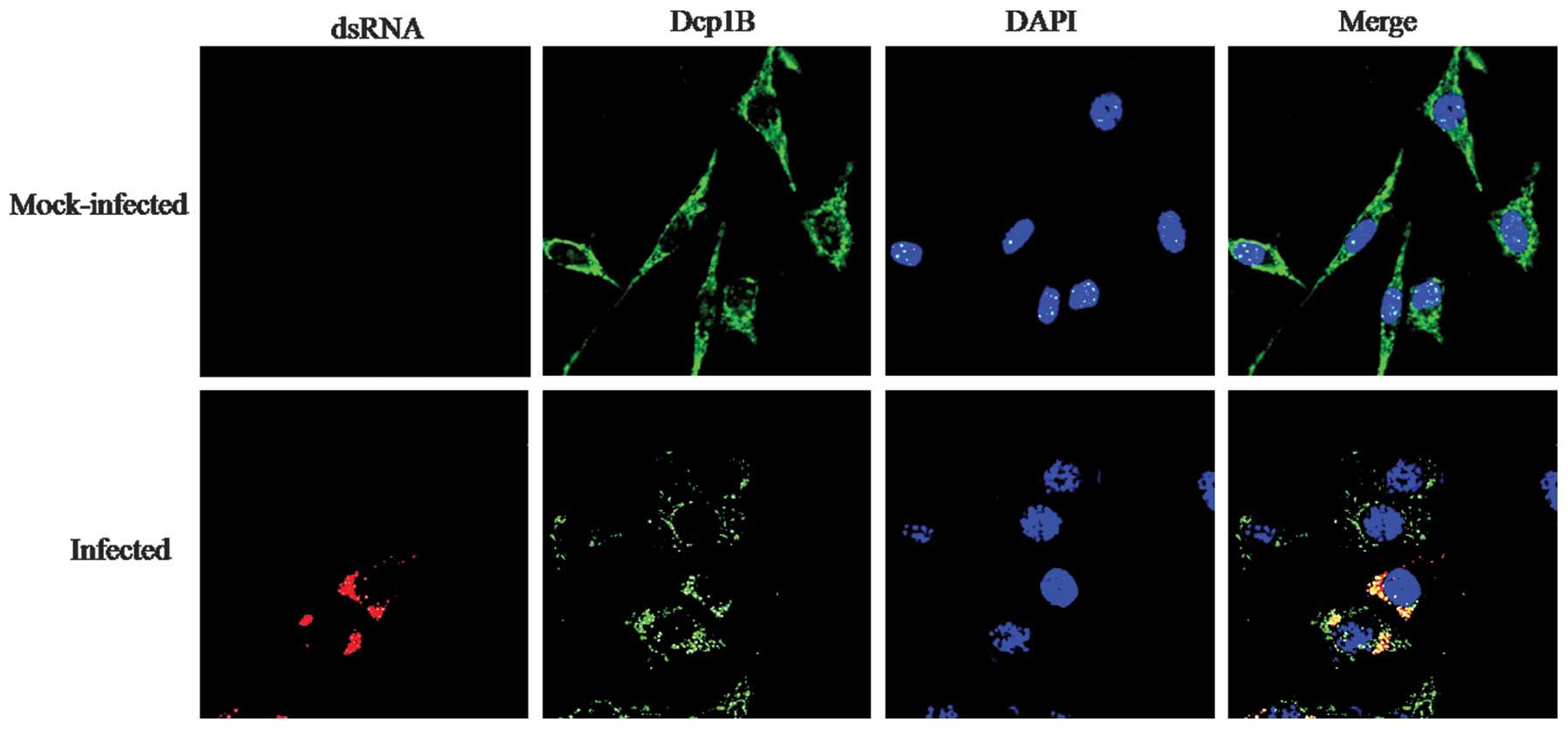
marker to detect P-bodies (16,24,25). We examined the subcellular
localization of Dcp1B during DENV-2 infection. A549 cells were
infected with DENV-2, and the cells were fixed, permeabilized and
stained with anti-dsRNA and anti-Dcp1B antibody. The nuclei were
stained with DAPI, and the coverslips were analyzed under a
confocal laser scanning microscope. In the uninfected cells, we
observed detectable Dcp1B exclusively. In the infected cells, Dcp1B
colocalized with dsRNA signal (Fig.
4). This result showed that DENV RNA replicates reside within
the P-bodies.
Discussion
Flavivirus 3′ UTRs are known to have important
regulatory significance for viral RNA replication and translation
(26). Host factors binding 3′
UTR-mediated RNA-protein interaction also play important roles in
the flavivirus RNA replication. Several cell proteins interacting
with DENV 3′ UTR have been reported (5–10,27). The LSm1-7 complexes have been
identified to bind HCV 5′ and 3′ UTR, and are required for
efficient HCV RNA translation and intracellular HCV RNA
accumulation (15,16,25,28). By using two independent
methodologies, we showed that LSm1 can bind 3′ UTR of DENV-2 and as
the positive regulator for viral RNA replication.
The Sm and Sm-like (LSm) proteins constitute a
conserved family whose members function in multiple aspects of RNA
metabolism. The LSm1-7 complex is localized in the cytoplasm and
plays a key role in the regulation of the fate of cellular mRNAs
from translation to degradation in the
5′-3′-deadenylation-dependent mRNA decay pathway (11,12). In the present study, our data
clearly demonstrate that LSm1-7 complexes are required for
efficient DENV-2 propagation in human cells in culture.
Downregulation of the LSm1-7 complexes significantly reduced DENV
RNA replication and infectious viral particle release. It is known
that the function of LSm1-7 complex as activator of the decapping
of cellular mRNAs seems to involve their binding to short oligo(A)
tracts at the 3′ end of deadenylated cellular mRNAs (29). This interaction then inhibits
trimming of the 3′ end while simultaneously promoting decapping and
subsequent degradation. However, the role of the LSm1-7 complexes
on virus life cycles may be different as DENV 3′ end does not
contain the poly(A) tract. Accordingly, binding of the LSm1-7
complex to 3′ UTR may facilitate rearrangement in the viral RNP
structure required for the 5′-3′ interaction. Long-range RNA-RNA
interactions between the 5′ and 3′ ends, which circularize the DENV
genome have been proposed to be required for RNA replication.
Notably, the LSm1-7 complex is a core component of
P-body. P-bodies are highly dynamic cytoplasmic granules conserved
among eukaryotes, which are sites at which non-translating mRNAs
accumulate for different fates such as degradation, storage or
returning to translation (20).
It is known that several proteins within P-bodies or SGs can
enhance or limit viral infection. Moreover, some viral RNAs and
proteins, as well as host antiviral defense proteins, accumulate in
P-bodies and/or SGs (30).
Additionally, the LSm1-7 complex is thought to play a role in mRNP
remodeling in P-body and this determines whether mRNAs within the
P-body are to be translated, stored or degraded (31). By using confocal laser scanning
microscopy, we observed the colocalization of the DENV dsRNA and
Dcp1B, which indicates that the replication process occurs at the
site of P-body. Together with the results that colocali zation of
the DENV dsRNA and LSm1-7 complex occurs in the perinuclear area,
it is possible that the LSm1-7 complex binds to the DENV 3′ UTR,
and this RNP acts as a platform to recruit P-body components such
as DDX6 and PatL1, and then form a P-body-like foci where
replication occurs.
The interactions between the LSm1-7 complex and DENV
RNA may facilitate rearrangements in the viral RNP structure and
composition, recruiting proteins such as the LSm1-7 complex, DDX6
and PatL1 from the cellular mRNA repression/decay machinery and,
instead of promoting decay, may promote viral RNA translation and
subsequent transfer to replication. DDX6 has been shown to be
involved in HCV and DENV RNA replication, and function as a
positive regulator in the HCV and DENV viral cycle associated with
the viral 3′ UTR (10,15,25,32). This view is consistent with
another suggestion made for the regulation of DENV RNA. Ward et
al (10) have shown that DDX6
interacts with DENV-2 RNA in vivo and binds to the DB2 and
DB1 structures in the DENV-2 3′ UTR. The interaction between DDX6
and the DB structures required the formation of a potential
pseudoknot structure, suggesting that the complex tertiary
structures in the DENV 3′ UTR would be required for DDX6 function.
Although the sites of DENV replication and translation have yet to
be separated, it is possible that the LSm1-7 complex together with
DDX6, key components of P-body, act to sort viral RNAs throughout
the entire process, from translation and replication to the sites
of viral packaging.
The results of the present study have demonstrated
that the LSm1-7 complex, a cellular protein binding to the DENV RNA
3 ′ UTR acts as a positive regulator of DENV replication. It is
likely that the LSm1-7 complex functions directly or indirectly as
an RBP rearranging the second structure of viral RNA to promote the
formation of a complex suitable for replication.
Acknowledgments
This study was supported by the National Natural
Science Foundation of China (NSFC 31370193).
References
|
1
|
Wiwanitkit V: Dengue fever: diagnosis and
treatment. Expert Rev Anti Infect Ther. 8:841–845. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rodenhuis-Zybert IA, Wilschut J and Smit
JM: Dengue virus life cycle: viral and host factors modulating
infectivity. Cell Mol Life Sci. 67:2773–2786. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yocupicio-Monroy M, Padmanabhan R, Medina
F and del Angel RM: Mosquito La protein binds to the 3′
untranslated region of the positive and negative polarity dengue
virus RNAs and relocates to the cytoplasm of infected cells.
Virology. 357:29–40. 2007. View Article : Google Scholar
|
|
4
|
Yocupicio-Monroy RM, Medina F, Reyes-del
Valle J and del Angel RM: Cellular proteins from human monocytes
bind to dengue 4 virus minus-strand 3′ untranslated region RNA. J
Virol. 77:3067–3076. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
De Nova-Ocampo M, Villegas-Sepúlveda N and
del Angel RM: Translation elongation factor-1alpha, La, and PTB
interact with the 3′ untranslated region of dengue 4 virus RNA.
Virology. 295:337–347. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Polacek C, Friebe P and Harris E:
Poly(A)-binding protein binds to the non-polyadenylated 3′
untranslated region of dengue virus and modulates translation
efficiency. J Gen Virol. 90:687–692. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gomila RC, Martin GW and Gehrke L: NF90
binds the dengue virus RNA 3′ terminus and is a positive regulator
of dengue virus replication. PLoS One. 6:e166872011. View Article : Google Scholar
|
|
8
|
Lei Y, Huang Y, Zhang H, Yu L, Zhang M and
Dayton A: Functional interaction between cellular p100 and the
dengue virus 3′ UTR. J Gen Virol. 92:796–806. 2011. View Article : Google Scholar
|
|
9
|
Paranjape SM and Harris E: Y box-binding
protein-1 binds to the dengue virus 3′-untranslated region and
mediates antiviral effects. J Biol Chem. 282:30497–30508. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ward AM, Bidet K, Yinglin A, Ler SG, Hogue
K, Blackstock W, Gunaratne J and Garcia-Blanco MA: Quantitative
mass spectrometry of DENV-2 RNA-interacting proteins reveals that
the DEAD-box RNA helicase DDX6 binds the DB1 and DB2 3′ UTR
structures. RNA Biol. 8:1173–1186. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Beggs JD: Lsm proteins and RNA processing.
Biochem Soc Trans. 33:433–438. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Khusial P, Plaag R and Zieve GW: LSm
proteins form heptameric rings that bind to RNA via repeating
motifs. Trends Biochem Sci. 30:522–528. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Diez J, Ishikawa M, Kaido M and Ahlquist
P: Identification and characterization of a host protein required
for efficient template selection in viral RNA replication. Proc
Natl Acad Sci USA. 97:3913–3918. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Galao RP, Chari A, Alves-Rodrigues I,
Lobão D, Mas A, Kambach C, Fischer U and Díez J: LSm1-7 complexes
bind to specific sites in viral RNA genomes and regulate their
translation and replication. RNA. 16:817–827. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pérez-Vilaró G, Scheller N, Saludes V and
Díez J: Hepatitis C virus infection alters P-body composition but
is independent of P-body granules. J Virol. 86:8740–8749. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Scheller N, Mina LB, Galao RP, Chari A,
Giménez-Barcons M, Noueiry A, Fischer U, Meyerhans A and Díez J:
Translation and replication of hepatitis C virus genomic RNA
depends on ancient cellular proteins that control mRNA fates. Proc
Natl Acad Sci USA. 106:13517–13522. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pang X, Zhang M and Dayton AI: Development
of Dengue virus type 2 replicons capable of prolonged expression in
host cells. BMC Microbiol. 1:182001. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Polo S, Ketner G, Levis R and Falgout B:
Infectious RNA transcripts from full-length dengue virus type 2
cDNA clones made in yeast. J Virol. 71:5366–5374. 1997.PubMed/NCBI
|
|
19
|
Emara MM and Brinton MA: Interaction of
TIA-1/TIAR with West Nile and dengue virus products in infected
cells interferes with stress granule formation and processing body
assembly. Proc Natl Acad Sci USA. 104:9041–9046. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Eulalio A, Behm-Ansmant I, Schweizer D and
Izaurralde E: P-body formation is a consequence, not the cause, of
RNA- mediated gene silencing. Mol Cell Biol. 27:3970–3981. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jakymiw A, Pauley KM, Li S, Ikeda K, Lian
S, Eystathioy T, Satoh M, Fritzler MJ and Chan EK: The role of
GW/P-bodies in RNA processing and silencing. J Cell Sci.
120:1317–1323. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chan EK, Yao B and Fritzler MJ:
Reflections on ten years of history of, and future prospects for,
GW182 and GW/P body research. Adv Exp Med Biol. 768:261–270. 2013.
View Article : Google Scholar
|
|
23
|
Coller J and Parker R: Eukaryotic mRNA
decapping. Annu Rev Biochem. 73:861–890. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yu SF, Lujan P, Jackson DL, Emerman M and
Linial ML: The DEAD-box RNA helicase DDX6 is required for efficient
encapsidation of a retroviral genome. PLoS Pathog. 7:e10023032011.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ariumi Y, Kuroki M, Kushima Y, Osugi K,
Hijikata M, Maki M, Ikeda M and Kato N: Hepatitis C virus hijacks
P-body and stress granule components around lipid droplets. J
Virol. 85:6882–6892. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Men R, Bray M, Clark D, Chanock RM and Lai
CJ: Dengue type 4 virus mutants containing deletions in the 3′
noncoding region of the RNA genome: analysis of growth restriction
in cell culture and altered viremia pattern and immunogenicity in
rhesus monkeys. J Virol. 70:3930–3937. 1996.PubMed/NCBI
|
|
27
|
Agis-Juárez RA, Galván I, Medina F,
Daikoku T, Padmanabhan R, Ludert JE and del Angel RM:
Polypyrimidine tract-binding protein is relocated to the cytoplasm
and is required during dengue virus infection in Vero cells. J Gen
Virol. 90:2893–2901. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Roberts AP, Doidge R, Tarr AW and Jopling
CL: 2013. The P body protein LSm1 contributes to stimulation of
hepatitis C virus translation, but not replication, by
microRNA-122. Nucleic Acids Res. 42:1257–1269. 2014. View Article : Google Scholar :
|
|
29
|
Chowdhury A, Mukhopadhyay J and Tharun S:
The decapping activator Lsm1p-7p-Pat1p complex has the intrinsic
ability to distinguish between oligoadenylated and polyadenylated
RNAs. RNA. 13:998–1016. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Beckham CJ and Parker R: P bodies, stress
granules, and viral life cycles. Cell Host Microbe. 3:206–212.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Parker R and Sheth U: P bodies and the
control of mRNA translation and degradation. Mol Cell. 25:635–646.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Huys A, Thibault PA and Wilson JA:
Modulation of hepatitis C virus RNA accumulation and translation by
DDX6 and miR-122 are mediated by separate mechanisms. PLoS One.
8:e674372013. View Article : Google Scholar : PubMed/NCBI
|