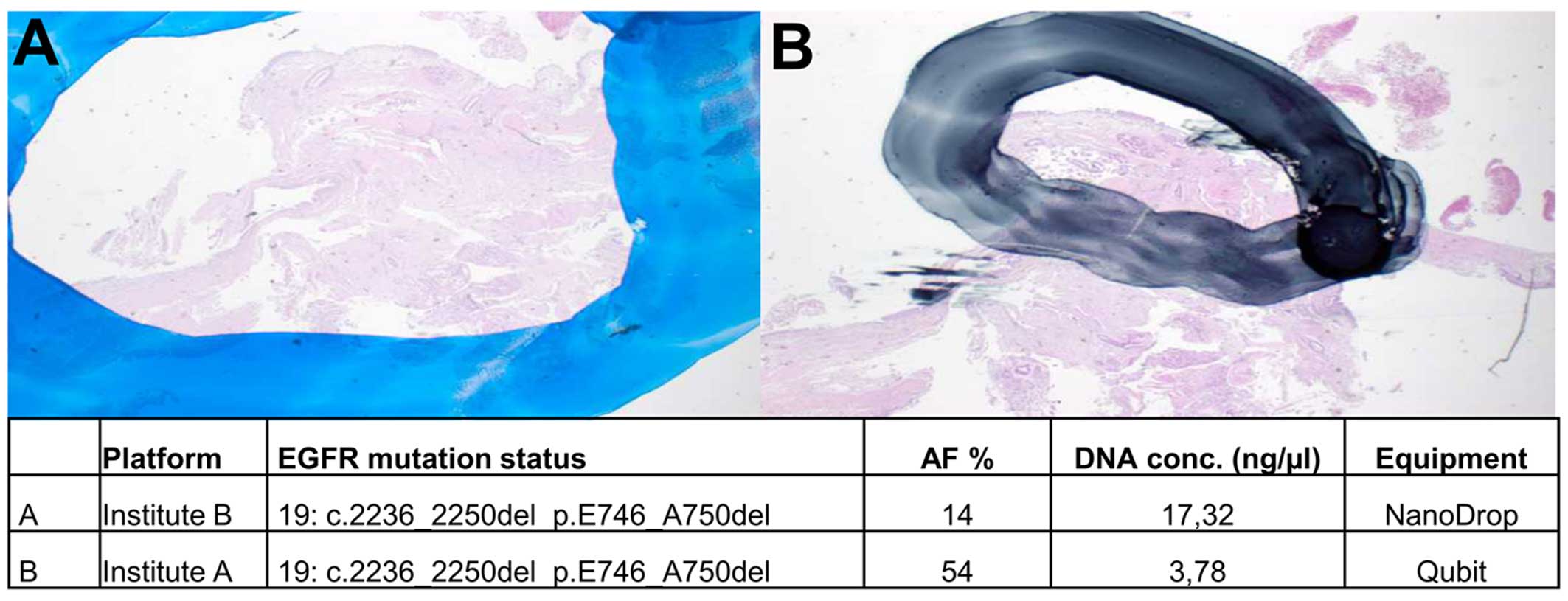
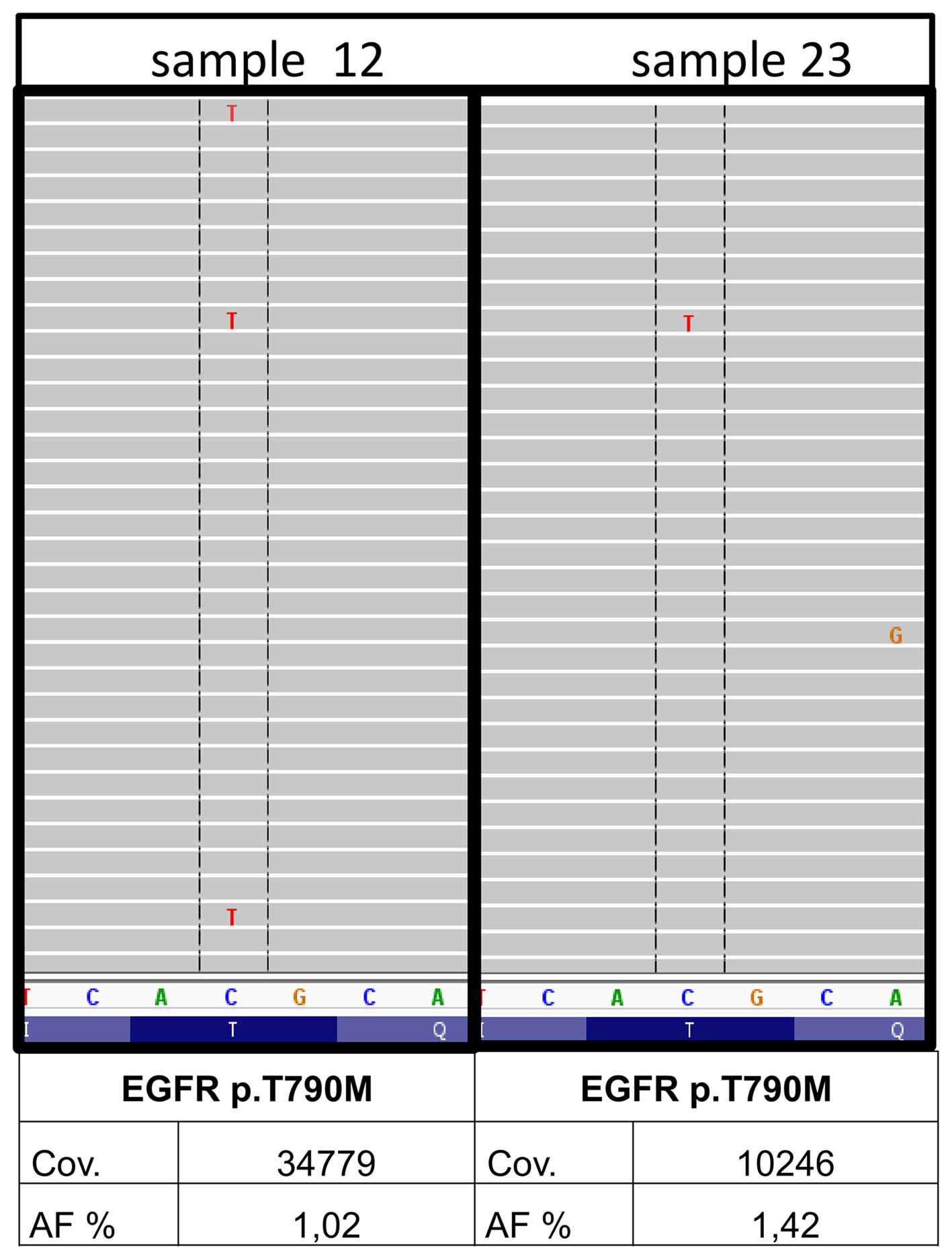
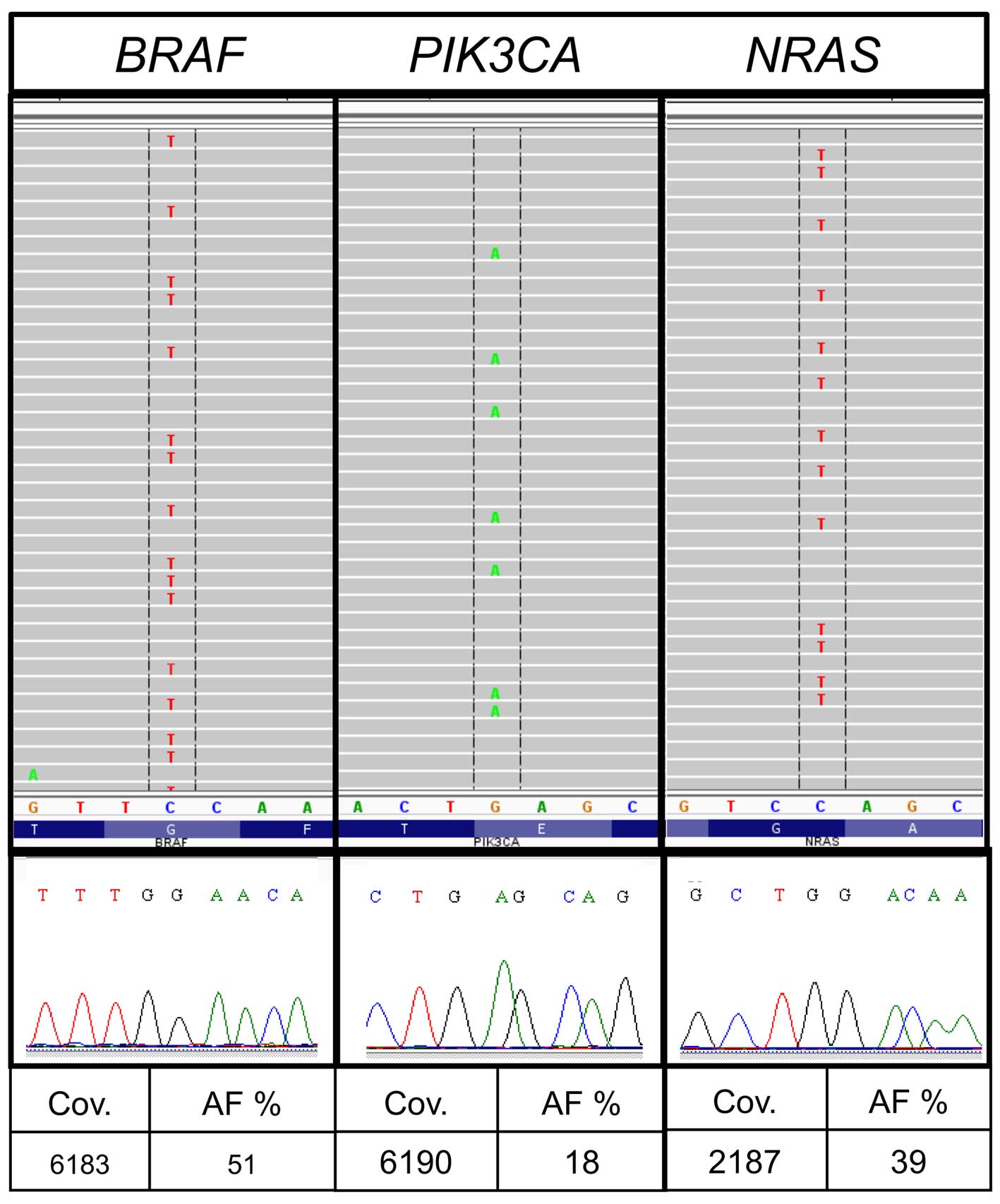
|
1
|
Endris V, Penzel R, Warth A, Muckenhuber
A, Schirmacher P, Stenzinger A and Weichert W: Molecular diagnostic
profiling of lung cancer specimens with a semiconductor-based
massive parallel sequencing approach: feasibility, costs, and
performance compared with conventional sequencing. J Mol Diagn.
15:765–775. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Clinical Lung Cancer Genome Project
(CLCGP); Network Genomic Medicine (NGM): A genomics-based
classification of human lung tumors. Sci Transl Med.
5:209ra1532013.PubMed/NCBI
|
|
3
|
Ulahannan D, Kovac MB, Mulholland PJ,
Cazier JB and Tomlinson I: Technical and implementation issues in
using next-generation sequencing of cancers in clinical practice.
Br J Cancer. 109:827–835. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hagemann IS, Devarakonda S, Lockwood CM,
Spencer DH, Guebert K, Bredemeyer AJ, Al-Kateb H, Nguyen TT,
Duncavage EJ, Cottrell CE, et al: Clinical next-generation
sequencing in patients with non-small cell lung cancer. Cancer.
121:631–639. 2015. View Article : Google Scholar
|
|
5
|
Tops BB, Normanno N, Kurth H, Amato E,
Mafficini A, Rieber N, Le Corre D, Rachiglio AM, Reiman A, Sheils
O, et al: Development of a semi-conductor sequencing-based panel
for genotyping of colon and lung cancer by the Onconetwork
consortium. BMC Cancer. 15:262015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Han JY, Kim SH, Lee YS, Lee SY, Hwang JA,
Kim JY, Yoon SJ and Lee GK: Comparison of targeted next-generation
sequencing with conventional sequencing for predicting the
responsiveness to epidermal growth factor receptor-tyrosine kinase
inhibitor (EGFR-TKI) therapy in never-smokers with lung
adenocarcinoma. Lung Cancer. 85:161–167. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
de Koning TJ, Jongbloed JD,
Sikkema-Raddatz B and Sinke RJ: Targeted next-generation sequencing
panels for monogenetic disorders in clinical diagnostics: the
opportunities and challenges. Expert Rev Mol Diagn. 15:61–70. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Meldrum C, Doyle MA and Tothill RW:
Next-generation sequencing for cancer diagnostics: A practical
perspective. Clin Biochem Rev. 32:177–195. 2011.PubMed/NCBI
|
|
9
|
Sikkema-Raddatz B, Johansson LF, de Boer
EN, Almomani R, Boven LG, van den Berg MP, van Spaendonck-Zwarts
KY, van Tintelen JP, Sijmons RH, Jongbloed JD and Sinke RJ:
Targeted next-generation sequencing can replace Sanger sequencing
in clinical diagnostics. Hum Mutat. 34:1035–1042. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Snow AN, Stence AA, Pruessner JA, Bossler
AD and Ma D: A simple and cost-effective method of DNA extraction
from small formalin-fixed paraffin-embedded tissue for molecular
oncologic testing. BMC Clin Pathol. 14:302014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Marchetti I, Iervasi G, Mazzanti CM, Lessi
F, Tomei S, Naccarato AG, Aretini P, Alberti B, Di Coscio G and
Bevilacqua G: Detection of the BRAF(V600E) mutation in fine needle
aspiration cytology of thyroid papillary microcarcinoma cells
selected by manual macrodissection: an easy tool to improve the
preoperative diagnosis. Thyroid. 22:292–298. 2012. View Article : Google Scholar
|
|
12
|
Heydt C, Fassunke J, Künstlinger H, Ihle
MA, König K, Heukamp LC, Schildhaus HU, Odenthal M, Büttner R and
Merkelbach-Bruse S: Comparison of pre-analytical FFPE sample
preparation methods and their impact on massively parallel
sequencing in routine diagnostics. PLoS One. 9:e1045662014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Peifer M, Fernández-Cuesta L, Sos ML,
George J, Seidel D, Kasper LH, Plenker D, Leenders F, Sun R, Zander
T, et al: Integrative genome analyses identify key somatic driver
mutations of small-cell lung cancer. Nat Genet. 44:1104–1110. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ihle MA, Fassunke J, König K, Grünewald I,
Schlaak M, Kreuzberg N, Tietze L, Schildhaus HU, Büttner R and
Merkelbach-Bruse S: Comparison of high resolution melting analysis,
pyrosequencing, next generation sequencing and immunohistochemistry
to conventional Sanger sequencing for the detection of p.V600E and
non-p.V600E BRAF mutations. BMC Cancer. 14:132014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang JH, Gouda-Vossos A, Dzamko N,
Halliday G and Huang Y: DNA extraction from fresh-frozen and
formalin-fixed, paraffin-embedded human brain tissue. Neurosci
Bull. 29:649–654. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Frey KG, Herrera-Galeano JE, Redden CL,
Luu TV, Servetas SL, Mateczun AJ, Mokashi VP and Bishop-Lilly KA:
Comparison of three next-generation sequencing platforms for
metagenomic sequencing and identification of pathogens in blood.
BMC Genomics. 15:962014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Loman NJ, Misra RV, Dallman TJ,
Constantinidou C, Gharbia SE, Wain J and Pallen MJ: Performance
comparison of benchtop high-throughput sequencing platforms. Nat
Biotechnol. 30:434–439. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Warth A, Penzel R, Brandt R, Sers C,
Fischer JR, Thomas M, Herth FJ, Dietel M, Schirmacher P and Bläker
H: Optimized algorithm for Sanger sequencing-based EGFR mutation
analyses in NSCLC biopsies. Virchows Arch. 460:407–414. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Moskalev EA, Stöhr R, Rieker R, Hebele S,
Fuchs F, Sirbu H, Mastitsky SE, Boltze C, König H, Agaimy A, et al:
Increased detection rates of EGFR and KRAS mutations in NSCLC
specimens with low tumour cell content by 454 deep sequencing.
Virchows Arch. 462:409–419. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hlinkova K, Babal P, Berzinec P, Majer I,
Mikle-Barathova Z, Piackova B and Ilencikova D: Evaluation of
2-year experience with EGFR mutation analysis of small diagnostic
samples. Diagn Mol Pathol. 22:70–75. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Portier BP, Kanagal-Shamanna R, Luthra R,
Singh R, Routbort MJ, Handal B, Reddy N, Barkoh BA, Zuo Z, Medeiros
LJ, et al: Quantitative assessment of mutant allele burden in solid
tumors by semiconductor-based next-generation sequencing. Am J Clin
Pathol. 141:559–572. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ausch C, Buxhofer-Ausch V, Oberkanins C,
Holzer B, Minai-Pour M, Jahn S, Dandachi N, Zeillinger R and
Kriegshäuser G: Sensitive detection of KRAS mutations in archived
formalin-fixed paraffin-embedded tissue using mutant-enriched PCR
and reverse-hybridization. J Mol Diagn. 11:508–513. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Pirker R, Herth FJ, Kerr KM, Filipits M,
Taron M, Gandara D, Hirsch FR, Grunenwald D, Popper H, Smit E, et
al: Consensus for EGFR mutation testing in non-small cell lung
cancer: results from a European workshop. J Thorac Oncol.
5:1706–1713. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dundas N, Leos NK, Mitui M, Revell P and
Rogers BB: Comparison of automated nucleic acid extraction methods
with manual extraction. J Mol Diagn. 10:311–316. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Esona MD, McDonald S, Kamili S, Kerin T,
Gautam R and Bowen MD: Comparative evaluation of commercially
available manual and automated nucleic acid extraction methods for
rotavirus RNA detection in stools. J Virol Methods. 194:242–249.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
van Eijk R, Stevens L, Morreau H and van
Wezel T: Assessment of a fully automated high-throughput DNA
extraction method from formalin-fixed, paraffin-embedded tissue for
KRAS, and BRAF somatic mutation analysis. Exp Mol Pathol.
94:121–125. 2013. View Article : Google Scholar
|
|
27
|
Do H and Dobrovic A: Sequence artifacts in
DNA from formalin-fixed tissues: Causes and strategies for
minimization. Clin Chem. 61:64–71. 2015. View Article : Google Scholar
|
|
28
|
Marchetti A, Felicioni L and Buttitta F:
Assessing EGFR mutations. N Engl J Med. 354:526–528. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wong SQ, Li J, Tan AY, Vedururu R, Pang
JM, Do H, Ellul J, Doig K, Bell A, MacArthur GA, et al: CANCER 2015
Cohort: Sequence artefacts in a prospective series of
formalin-fixed tumours tested for mutations in hotspot regions by
massively parallel sequencing. BMC Med Genomics. 7:232014.
View Article : Google Scholar
|
|
30
|
Udar N, Haigis R, Gros T, Kerry N, Barnes
B, Pokholok D, Ross M, Lucio-Eterovic AK, Zhang Q, Zenali M and
Jaeger E: A novel technique that distinguishes low-level somatic
DNA variants from FFPE-induced artifacts in solid tumors by
next-generation sequencing (NGS). International Association for the
Study of Lung Cancer. 2013.
|
|
31
|
Oxnard GR, Arcila ME, Sima CS, Riely GJ,
Chmielecki J, Kris MG, Pao W, Ladanyi M and Miller VA: Acquired
resistance to EGFR tyrosine kinase inhibitors in EGFR-mutant lung
cancer: distinct natural history of patients with tumors harboring
the T790M mutation. Clin Cancer Res. 17:1616–1622. 2011. View Article : Google Scholar :
|
|
32
|
Nguyen KS, Kobayashi S and Costa DB:
Acquired resistance to epidermal growth factor receptor tyrosine
kinase inhibitors in non-small-cell lung cancers dependent on the
epidermal growth factor receptor pathway. Clin Lung Cancer.
10:281–289. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Mok TS, Wu YL, Thongprasert S, Yang CH,
Chu DT, Saijo N, Sunpaweravong P, Han B, Margono B, Ichinose Y, et
al: Gefitinib or carboplatin-paclitaxel in pulmonary
adenocarcinoma. N Engl J Med. 361:947–957. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Rosell R, Molina MA, Costa C, Simonetti S,
Gimenez-Capitan A, Bertran-Alamillo J, Mayo C, Moran T, Mendez P,
Cardenal F, et al: Pretreatment EGFR T790M mutation and BRCA1 mRNA
expression in erlotinib-treated advanced non-small-cell lung cancer
patients with EGFR mutations. Clin Cancer Res. 17:1160–1168. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Su KY, Chen HY, Li KC, Kuo ML, Yang JC,
Chan WK, Ho BC, Chang GC, Shih JY, Yu SL and Yang PC: Pretreatment
epidermal growth factor receptor (EGFR) T790M mutation predicts
shorter EGFR tyrosine kinase inhibitor response duration in
patients with non-small-cell lung cancer. J Clin Oncol. 30:433–440.
2012. View Article : Google Scholar : PubMed/NCBI
|