Introduction
Mesenchymal stem cells (MSCs) were first derived
from bone marrow and are characterized by their self-renewal
ability and their capacity to develop into various mesenchymal
tissue cells (1–3). Much of this differentiation process
depends on the ability of the MSCs to proliferate and differentiate
under the influence of various growth factors and cytokines
(4–7). For example, the role of growth
factors in bone repair is widely recognized, particularly with
regard to bone morphogenetic protein (BMP), fibroblast growth
factor (FGF), platelet-derived growth factor (PDGF), vascular
endothelial growth factor (VEGF), insulin-like growth factor-I
(IGF-I) and transforming growth factor-β (TGF-β) (8,9).
In a recent study of ours, we demonstrated that PDGF-induced
phosphoinositide 3-kinase (PI3K)-mediated signaling promoted the
TGF-β-induced osteogenic differentiation of MSCs in a
TGF-β-activated extracellular signal-regulated kinase (ERK)
kinase-dependent manner (10). In
a another recent study, we demonstrated that MSCs-secreted protein,
scrapie responsive gene-1 (SCRG1), and its receptor bone marrow
stromal cell antigen-1 (BST1), which played important roles in the
maintenance of stemness and in the suppression of the osteogenic
differentiation of MSCs (11).
VEGF, an important growth factor for bone repair,
regulates numerous cellular events associated with angiogenesis and
vasculogenesis, such as tissue remodeling during embryonic
development and in adults (12).
The mammalian VEGF signaling pathway consists of 5 glycoprotein
ligands from the VEGF family (VEGF-A, -B, -C, -D and placental
growth factor), 3 transmembrane receptors [VEGF receptor (VEGFR)1,
VEGFR2 and VEGFR3] and 2 co-receptors (neuropilin-1 and -2)
(13–22). VEGF-A binding to VEGFR2 is
believed to be the key signaling pathway mediating angiogenesis
(14,23). VEGF-A enhances proliferation and
survival, promotes cell migration, increases vascular permeability,
and alters gene expression in endothelial cells (13,14). VEGF-B binding to VEGFR1 promotes
the survival of endothelial cells, pericytes, and smooth muscle
cells and upregulates the expression of prosurvival genes (24). VEGF-C and VEGF-D bind to the
receptors, VEGFR2 and VEGFR3 (22). VEGF-C expression has been shown to
be associated with advanced metastasis in colorectal cancer
(25) and to play a role in
lymphangiogenesis and/or metastasis to lymph nodes in multiple
types of cancer, including colorectal (26) and breast cancer (27,28). VEGF-D is also involved in
lymphangiogenesis and lymphatic metastasis (29,30). On the other hand, in contrast to
the well-studied VEGF signaling in endothelial cells, the VEGF
signaling pathways in cells involved in bone repair, such as MSCs
and osteoblasts, remains less well known (31). Osteoblasts express VEGFR1, VEGFR2
and the co-receptor, neuropilin (32). The expression of VEGF and its
receptors in differentiating osteoblasts has been detected in
cultured cells (32,33), and an in vitro cell culture
study suggested a role for VEGFR2 in both osteoblast
differentiation and survival (34).
In a recent study of ours, we established 3 MSC
lines (SG-2, SG-3, and SG-5) derived from the bone marrow of green
fluorescent protein (GFP)-transgenic mice (35). These cell lines clearly expressed
the mouse MSC markers, stem cells antigen-1 (Sca-1) and CD44, and
the SG-2 and SG-5 cells retained their potential for osteogenic and
adipogenic differentiation. In addition, we examined the reactions
of the TGF-β superfamily in these MSC lines. The analysis of
cytokine and cytokine receptor expression in these MSC lines
revealed that BMP receptor 1B was most strongly expressed in the
SG-3 cells, which underwent osteogenesis in response to BMP. TGF-β
receptor II was more strongly expressed in the SG-3 and SG-5 cells.
However, we unexpectedly noted that the phosphorylation of Smad2, a
major transcription factor, was induced by TGF-β1 in the SG-2
cells, but not in the SG-3 or SG-5 cells. These findings
demonstrated the establishment of TGF-β-responsive SG-2 MSCs,
BMP-responsive SG-3 MSCs, and TGF-β/BMP-non-responsive SG-5
MSCs.
In the present study, we focused on membrane
proteins that are expressed specifically in SG-2 cells in order to
facilitate the sorting and identification of the MSCs. VEGFR3, the
gene product of FMS-like tyrosine kinase 4 (Flt4), was
strongly expressed only in the SG-2 cells, but not in the SG-3 and
SG-5 cells. Our findings demonstrate the role of VEGF-C, a specific
ligand of VEGFR3, in the regenerative ability of the mouse MSC
line, TGF-β-responsive SG-2 cells.
Materials and methods
Mouse MSC lines
In a recent study of ours, we described the
establishment process and culture method for all MSC lines derived
from the bone marrow of GFP-transgenic mice: TGF-β-responsive SG-2,
BMP-responsive SG-3 and TGF-β/BMP-non-responsive SG-5 cells
(35). These cell lines were
cultured in Dulbecco's modified Eagle's medium (DMEM;
Sigma-Aldrich, St. Louis, MO, USA) supplemented with 10% fetal
bovine serum (FBS; HyClone, GE Healthcare Life Sciences, Logan, UT,
USA, Logan, UT, USA) at 37°C under hypoxic conditions (5%
O2, 5% CO2 and 90% N2).
DNA microarray analysis
Whole genome expression was analyzed for the
bone-marrow derived SG-2, SG-3 and SG-5 MSC lines. Total RNA was
extracted using ISOGEN reagent (Nippon Gene Co., Ltd., Tokyo,
Japan). Filgen, Inc. (Nagoya, Japan) performed the DNA microarray
analyses, including reverse transcription labeling, microarray
hybridization, scanning and raw data analyses. For hybridization, 3
GeneChip Mouse Gene 2.0 ST arrays (Affymetrix, Santa Clara, CA,
USA) were used. These analyses were conducted by the Research
Institute of Bio-System Informatics (Tohoku Chemical Co., Ltd.,
Morioka, Japan).
Flow cytometry
Almost confluent SG-2, SG-3 and SG-5 cells
(1.0×105) were suspended in ice-cold phosphate-buffered
saline (PBS) containing 0.5% FBS and 2 mM EDTA. The cells were
incubated with phycoerythrin (PE)-conjugated anti-mouse VEGFR3
(CD310) antibody (1:10, clone AFL4, #130-102-216; Miltenyi Biotec,
Bergisch Gladbach, Germany) for 1 h at 4°C in the dark. Acquisition
was performed with an EPICS XL ADC system (Beckman Coulter, Inc.,
Brea, CA, USA).
Western blot analysis
The SG-2, SG-3 and SG-5 cells were serum-starved
overnight and stimulated with 10 ng/ml VEGF-C (R&D Systems,
Inc., Minneapolis, MN, USA) for 1 h at 37°C under hypoxic
conditions. The cells were washed twice with ice-cold PBS and then
lysed in RIPA buffer (50 mM Tris-HCl, pH 7.2, 150 mM NaCl, 1%
NP-40, 0.5% sodium deoxycholate and 0.1% SDS) containing protease
and phosphatase inhibitor cocktails (Sigma-Aldrich). Protein
content was measured using BCA reagent (Pierce/Thermo Fisher
Scientific, Waltham, MA, USA). Samples containing equal amounts of
protein were separated using 12.5% SDS-polyacrylamide gel
electrophoresis and transferred to a polyvinylidene difluoride
membrane (Merck Millipore, Darmstadt, Germany). After blocking with
5% non-fat dry milk in T-TBS (50 mM Tris-HCl, pH 7.2, 150 mM NaCl
and 0.1% Tween-20), the membrane was incubated with primary
anti-Akt (#9272), anti-phospho-Akt (Ser473) [phosphorylated
(p-)Akt; #9271], anti-p44/42 mitogen-activated protein kinase
(MAPK; ERK1/2; #9102), anti-phospho-p44/42 MAPK (Thr202/Tyr204)
(p-ERK1/2; #9101), anti-p38 MAPK (p38; #9212), anti-phospho-p38
MAPK (T180/Y182) (p-p38; #9211), anti-stress-activated protein
kinase/c-Jun N-terminal kinase (SAPK/JNK) (JNK; #9252) and
anti-phospho-SAPK/JNK (Thr183/Tyr185) (p-JNK; #9251) antibodies
(all from Cell Signaling Technology, Danvers, MA, USA), and
anti-β-actin (clone C4; Santa Cruz Biotechnology, Dallas, TX, USA)
antibody as a loading control for normalization. The blots were
incubated with an alkaline phosphatase-conjugated secondary
antibody and developed using the BCIP/NBT membrane phosphatase
substrate system (Kirkegaard & Perry Laboratories,
Gaithersburg, MD, USA).
Cell proliferation assay
Cell proliferation was analyzed using a colorimetric
assay for cleavage of the tetrazolium salt WST-1 (Roche
Diagnostics, Basel, Switzerland) by mitochondrial dehydrogenases in
viable cells. The measured absorbance of the dye directly
correlates with the number of metabolically active cells in the
culture. The cells were cultured in 96-well plates (Nunc; Thermo
Fisher Scientific) in growth medium with/without 10 ng/ml VEGF-C
under hypoxic conditions. After 5 days, the cells were incubated
for a further 1 h at 37°C with 100 µl medium containing 10
µl WST-1 reagent. The samples were shaken for 1 min, and
absorbance was measured at 450 nm using an MPR-A4i microplate
reader (Tosoh Corp., Tokyo, Japan).
Transwell migration assay
The migration assay was performed as reported
previously using Transwell cell culture inserts (BD Biosciences,
Franklin Lakes, NJ, USA) that were 6.5 mm in diameter with
8-µm pore filters (11).
The cells (5.0×104) were suspended in 350 µl
serum-free DMEM containing 0.1% BSA (Sigma-Aldrich) and seeded into
the upper well; 600 µl normal growth medium with/without 10
ng/ml VEGF-C was placed in the lower well of the Transwell plate.
Following incubation for 15 h under hypoxic conditions, the cells
that had not migrated from the upper side of the filter were
scraped off with a cotton swab, and the filters were stained with
the three-step stain set (Diff-Quik; Sysmex, Kobe, Japan). The
number of cells that had migrated to the lower side of the filter
was counted under a light microscope in 5 high-power fields (×400
magnification; Olympus IX70; Olympus Corp., Tokyo, Japan). The
experiment was performed in triplicate.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
The SG-2, SG-3 and SG-5 cells were stimulated with
10 ng/ml VEGF-C (R&D Systems) or 5.0 ng/ml TGF-β (Calbiochem,
Merck Millipore). After 48 h, total RNA from each cell was isolated
using ISOGEN reagent (Nippon Gene Co., Ltd.) according to the
manufacturer's instructions. First-strand cDNA was synthesized from
total RNA using the PrimeScript RT Reagent kit (Takara Bio, Otsu,
Japan). RT-qPCR was performed on a Thermal Cycler Dice Real-Time
system with SYBR Premix Ex Taq II (both from Takara Bio) and
specific oligonucleotide primers (Table I) using a two-step cycle procedure
(denaturation at 95°C for 5 sec, annealing and extension at 60°C
for 30 sec) for 40 cycles. For each test run, cDNA derived from 50
ng total RNA as a template and 0.4 µM primer pair was used.
mRNA expression was normalized to glyceraldehyde 3-phosphate
dehydrogenase (Gapdh), and the relative amounts of each mRNA
in each sample were calculated using the ΔΔCq method. The relative
mRNA expression levels are expressed as fold increase or decrease
relative to the control.
 | Table IPrimer sequences. |
Table I
Primer sequences.
| Gene name | Symbol | Primer sequence
(5′→3′) |
|---|
| Prospero homeobox
1 | Prox1 | Forward:
CGCTTAGCATTGCTGTTGCTG |
| | Reverse:
GAGCCATTCCTGGGTGATGTC |
| Lymphatic vessel
endothelial hyaluronan receptor 1 | Lyve1 | Forward:
GAGCCATTCAAAGTACCAGGTCCTAA |
| | Reverse:
ACATGTGCCTGGTTCCAAAG |
| Runt-related
transcription factor 2 | Runx2 | Forward:
GACGTGCCCAGGCGTATTTC |
| | Reverse:
AAGGTGGCTGGGTAGTGCATTC |
| Alkaline
phosphatase, liver/bone/kidney | Alpl | Forward:
ACACCTTGACTGTGGTTACTGCTGA |
| | Reverse:
CCTTGTAGCCAGGCCCGTTA |
| Integrin-binding
sialoprotein | Ibsp | Forward:
AGAACAATCCGTGCCACTCACTC |
| | Reverse:
AGTAGCGTGGCCGGTACTTAAAGA |
| Bone
gamma-carboxyglutamate (gla) protein | Bglap | Forward:
CGGCCCTGAGTCTGACAAA |
| | Reverse:
TCTGTAGGCGGTCTTTAAGCCATA |
| Glyceraldehyde
3-phosphate dehydrogenase | Gapdh | Forward:
TGTGTCCGTCGTGGATCTGA |
| | Reverse:
TTGCTGTTGAAGTCGCAGGAG |
Statistical analysis
All experiments were repeated at least 3 times.
Representative images or data are shown. The numerical data are
presented as the means ± standard deviation (SD). Differences
between averages and percentages between control and tests were
statistically analyzed using paired two-tailed Student's t-tests. A
P-value <0.05 was considered to indicate a statistically
significant difference.
Results
Higher expression of VEGFR3 in SG-2
cells
To identify the genes that modulate the regenerative
ability of MSCs, we used DNA microarrays to characterize the
specific gene expression profiles observed in the TGF-β-responsive
SG-2 cells. We identified 105 genes that were ≥10-fold more
strongly expressed in the SG-2 cells compared to the SG-3 and SG-5
cells (Table II). Among these
genes, we focused on VEGFR3, the Flt4 gene product, since,
as a cell surface antigen, it is useful for identifying and sorting
MSCs from various tissues. The gene expression level of Flt4
in the SG-2 cells was more than 16.5- and 32.0-fold higher than
that in the SG-3 and SG-5 cells, respectively. These results were
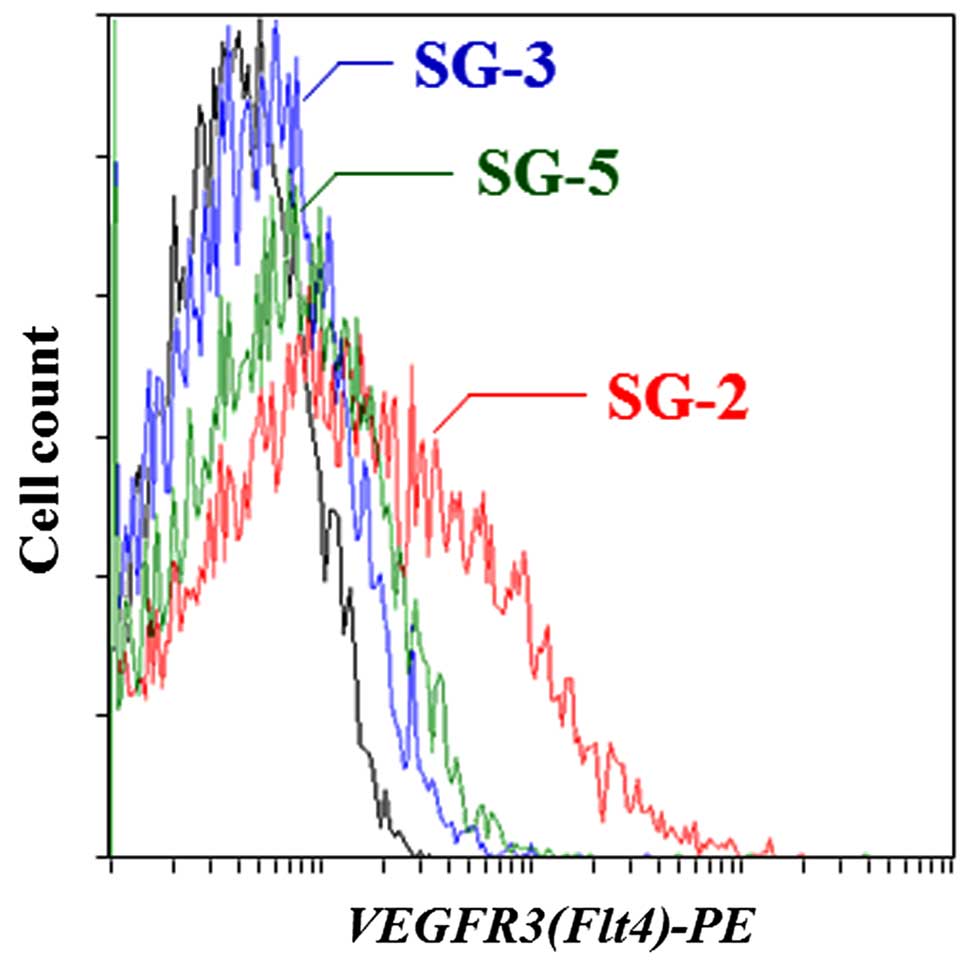
further confirmed by flow cytometry (Fig. 1) and indicated that the SG-2 cells
expressed higher levels of VEGFR3 on the cell surface than the SG-3
and SG-5 cells.
 | Table IIGenes expressed ≥10-fold stronger in
the SG-2 cells compared to the SG-3 and SG-5 cells. |
Table II
Genes expressed ≥10-fold stronger in
the SG-2 cells compared to the SG-3 and SG-5 cells.
| Symbol | Gene name | Fold change in SG-2
|
|---|
| vs. SG-3 | vs. SG-5 |
|---|
| Thrb | Thyroid hormone
receptor beta | 75.7 | 58.4 |
| Grhl2 | Grainyhead-like 2
(Drosophila) | 72.9 | 56.5 |
|
Olfr1497 | Olfactory receptor
1497 | 69.6 | 53.1 |
|
Arhgap15 | Rho GTPase
activating protein 15 | 66.5 | 51.4 |
| Hoxd11 | Homeobox D11 | 57.5 | 44.1 |
| Cfc1 | Cripto, FRL-1,
cryptic family 1 | 56.2 | 43.8 |
| Kcnq5 | Potassium
voltage-gated channel, subfamily Q, member 5 | 53.5 | 41.5 |
| Scgb2b7 | Secretoglobin,
family 2B, member 7 | 50.6 | 39.0 |
| Fn3krp | Fructosamine 3
kinase related protein | 55.4 | 36.2 |
| Tcp10a | T-complex protein
10a | 43.5 | 41.6 |
|
Serpina6 | Serine (or
cysteine) peptidase inhibitor, clade A, member 6 | 46.0 | 35.0 |
| Cym | Chymosin | 40.4 | 30.3 |
|
Ppp1r3fos | Protein phosphatase
1, regulatory subunit 3F, opposite strand | 39.2 | 30.6 |
| Syn3 | Synapsin III | 69.7 | 22.7 |
| Lypd6 | LY6/PLAUR domain
containing 6 | 38.3 | 30.4 |
| Olfr772 | Olfactory receptor
772 | 38.6 | 29.7 |
| Olfr2 | Olfactory receptor
2 | 38.4 | 29.4 |
| Zfp92 | Zinc finger protein
92 | 26.5 | 43.1 |
| Fam81b | Family with
sequence similarity 81, member B | 38.1 | 28.8 |
| Ssxb1 | Synovial sarcoma, X
member B, breakpoint 1 | 37.4 | 28.6 |
| Vmn2r25 | Vomeronasal 2,
receptor 25 | 36.7 | 28.7 |
| Olfr368 | Olfactory receptor
368 | 35.9 | 28.2 |
| Arhgef4 | Rho guanine
nucleotide exchange factor (GEF) 4 | 31.8 | 30.9 |
| Il21 | Interleukin 21 | 35.7 | 27.8 |
| Fpr-rs3 | Formyl peptide
receptor, related sequence 3 | 34.8 | 27.1 |
| Cldn13 | Claudin 13 | 34.4 | 27.0 |
| Trim43c | Tripartite
motif-containing 43C | 35.1 | 26.6 |
| Caskin1 | CASK interacting
protein 1 | 34.4 | 26.3 |
| Commd7 | COMM domain
containing 7 | 34.1 | 25.3 |
| Prom2 | Prominin 2 | 33.5 | 25.4 |
|
St6galnac3 | ST6
(α-N-acetyl-neuraminyl-2,3-β-galactosyl-1,3)-N-acetyl-galactosaminide
α-2,6-sialyltransferase 3 | 32.6 | 25.8 |
| Slco6c1 | Solute carrier
organic anion transporter family, member 6c1 | 30.4 | 23.6 |
| Nhlrc4 | NHL repeat
containing 4 | 29.8 | 23.7 |
| Chrd | Chordin | 21.6 | 33.8 |
| Olfr117 | Olfactory receptor
117 | 31.0 | 22.5 |
|
Igkv4-53 | Immunoglobulin κ
variable 4-53 | 26.3 | 25.3 |
| Ctag2 | Cancer/testis
antigen 2 | 28.8 | 22.9 |
| F2rl3 | Coagulation factor
II (thrombin) receptor-like 3 | 28.9 | 22.6 |
| Kynu | Kynureninase
(L-kynurenine hydrolase) | 28.4 | 21.2 |
| Spata22 | Spermatogenesis
associated 22 | 27.2 | 21.2 |
|
Vmn1r191 | Vomeronasal 1
receptor 191 | 27.1 | 21.2 |
| Ctnnd1 | Catenin (cadherin
associated protein), delta 1 | 26.0 | 20.5 |
|
Vmn1r234 | Vomeronasal 1
receptor 234 | 26.2 | 19.8 |
| Zfp174 | Zinc finger protein
174 | 26.4 | 19.5 |
| Dgat2l6 | Diacylglycerol
O-acyltransferase 2-like 6 | 25.7 | 19.7 |
|
Nudt12os | Nudix (nucleoside
diphosphate linked moiety X)-type motif 12, opposite strand | 25.3 | 19.6 |
| Zap70 | Zeta-chain (TCR)
associated protein kinase | 25.3 | 19.1 |
| Flt4 | FMS-like tyrosine
kinase 4 | 16.5 | 32.0 |
| Hes3 | Hairy and enhancer
of split 3 (Drosophila) | 24.8 | 19.3 |
| Sema6d | Sema domain,
transmembrane domain (TM), and cytoplasmic domain, (semaphorin)
6D | 15.8 | 33.9 |
| Slc17a4 | Solute carrier
family 17 (sodium phosphate), member 4 | 16.9 | 29.5 |
| Tbx2 | T-box 2 | 24.0 | 18.1 |
| Wdr95 | WD40 repeat domain
95 | 22.7 | 17.6 |
| Fer1l5 | Fer-1-like 5 (C.
elegans) | 22.4 | 17.8 |
| Gbx2 | Gastrulation brain
homeobox 2 | 15.0 | 29.1 |
| Cd33 | CD33 antigen | 22.4 | 17.6 |
| Tdrd5 | Tudor domain
containing 5 | 22.8 | 17.0 |
| Lix1 | Limb expression 1
homolog (chicken) | 21.4 | 16.7 |
| Lgals4 | Lectin, galactose
binding, soluble 4 | 21.5 | 16.2 |
| Pip5k1b |
Phosphatidylinositol-4-phosphate 5-kinase,
type 1β | 21.4 | 15.9 |
| Gsdma2 | Gasdermin A2 | 21.3 | 15.9 |
|
Ifi27l2b | Interferon,
α-inducible protein 27 like 2 beta | 16.0 | 19.8 |
| Zcchc18 | Zinc finger, CCHC
domain containing 18 | 17.2 | 17.8 |
| Olfr384 | Olfactory receptor
384 | 19.5 | 15.5 |
|
Olfr1123 | Olfactory receptor
1123 | 11.4 | 33.8 |
| Myo18b | Myosin XVIIIb | 11.8 | 29.6 |
| Ss18 | Synovial sarcoma
translocation, Chromosome 18 | 23.4 | 13.0 |
|
Plxna4os1 | Plexin A4, opposite
strand 1 | 19.5 | 14.4 |
| Fam115e | Family with
sequence similarity 115, member E | 19.1 | 14.5 |
| Fbxl5 | F-box and
leucine-rich repeat protein 5 | 13.7 | 20.1 |
| Megf10 | Multiple
EGF-like-domains 10 | 18.5 | 14.3 |
| Robo3 | Roundabout homolog
3 (Drosophila) | 18.3 | 14.3 |
|
Igkv4-91 | Immunoglobulin
kappa chain variable 4-91 | 18.3 | 14.2 |
| Elavl3 | ELAV (embryonic
lethal, abnormal vision, Drosophila)-like 3 (Hu antigen
C) | 15.7 | 16.1 |
| Slc17a8 | Solute carrier
family 17 (sodium-dependent inorganic phosphate cotransporter),
member 8 | 11.7 | 24.4 |
| Gpr39 | G protein-coupled
receptor 39 | 15.7 | 15.8 |
|
Catsper1 | Cation channel,
sperm associated 1 | 18.3 | 13.6 |
| Zscan4a | Zinc finger and
SCAN domain containing 4A | 17.4 | 13.8 |
|
Ceacam14 | Carcinoembryonic
antigen-related cell adhesion molecule 14 | 17.8 | 13.3 |
| Acnat1 | Acyl-coenzyme A
amino acid N-acyltransferase 1 | 12.4 | 17.8 |
|
Arhgap26 | Rho GTPase
activating protein 26 | 17.4 | 12.6 |
|
Olfr1270 | Olfactory receptor
1270 | 16.6 | 13.0 |
| Lrrc25 | Leucine rich repeat
containing 25 | 11.4 | 19.1 |
| Capsl |
Calcyphosine-like | 16.5 | 12.5 |
| Gnl2 | Guanine nucleotide
binding protein-like 2 (nucleolar) | 16.1 | 12.8 |
| Ctnna3 | Catenin (cadherin
associated protein), alpha 3 | 16.4 | 12.6 |
| Itm2a | Integral membrane
protein 2A | 16.4 | 12.4 |
| Sema5b | Sema domain, seven
thrombospondin repeats (type 1 and type 1-like), transmembrane
domain (TM) and short cytoplasmic domain, (semaphorin) 5B | 20.5 | 10.6 |
| Il17c | Interleukin
17C | 15.7 | 12.2 |
| Chrna9 | Cholinergic
receptor, nicotinic, alpha polypeptide 9 | 15.4 | 12.1 |
| Acap1 | ArfGAP with
coiled-coil, ankyrin repeat and PH domains 1 | 10.8 | 17.0 |
| Spint5 | Serine protease
inhibitor, Kunitz type 5 | 15.1 | 11.4 |
| Rab5b | RAB5B, member RAS
oncogene family | 12.5 | 13.3 |
|
Olfr1122 | Olfactory receptor
1122 | 10.5 | 16.4 |
| Grm8 | Glutamate receptor,
metabotropic 8 | 17.6 | 10.1 |
|
Igh-VJ558 | Immunoglobulin
heavy chain (J558 family) | 14.4 | 11.2 |
| Ptprz1 | Protein tyrosine
phosphatase, receptor type Z, polypeptide 1 | 14.4 | 11.1 |
| Cd300lh | CD300 antigen-like
family member H | 15.2 | 10.4 |
| Mcoln1 | Mucolipin 1 | 11.7 | 12.8 |
| Olfr649 | Olfactory receptor
649 | 13.9 | 10.9 |
| Tlr8 | Toll-like receptor
8 | 13.9 | 10.7 |
| Mb21d1 | Mab-21 domain
containing 1 | 10.8 | 13.2 |
|
Itgb1bp2 | Integrin beta 1
binding protein 2 | 13.3 | 10.5 |
| Defb28 | Defensin beta
28 | 13.3 | 10.4 |
| Cypt2 | Cysteine-rich
perinuclear theca 2 | 13.4 | 10.3 |
Promotion of the migratory ability and
proliferative activity of SG-2 cells by VEGF-C
Subsequently, we examined the effects of VEGF-C, a
specific ligand of VEGFR3, on the MSC lines (SG-2, SG-3 and SG-5).
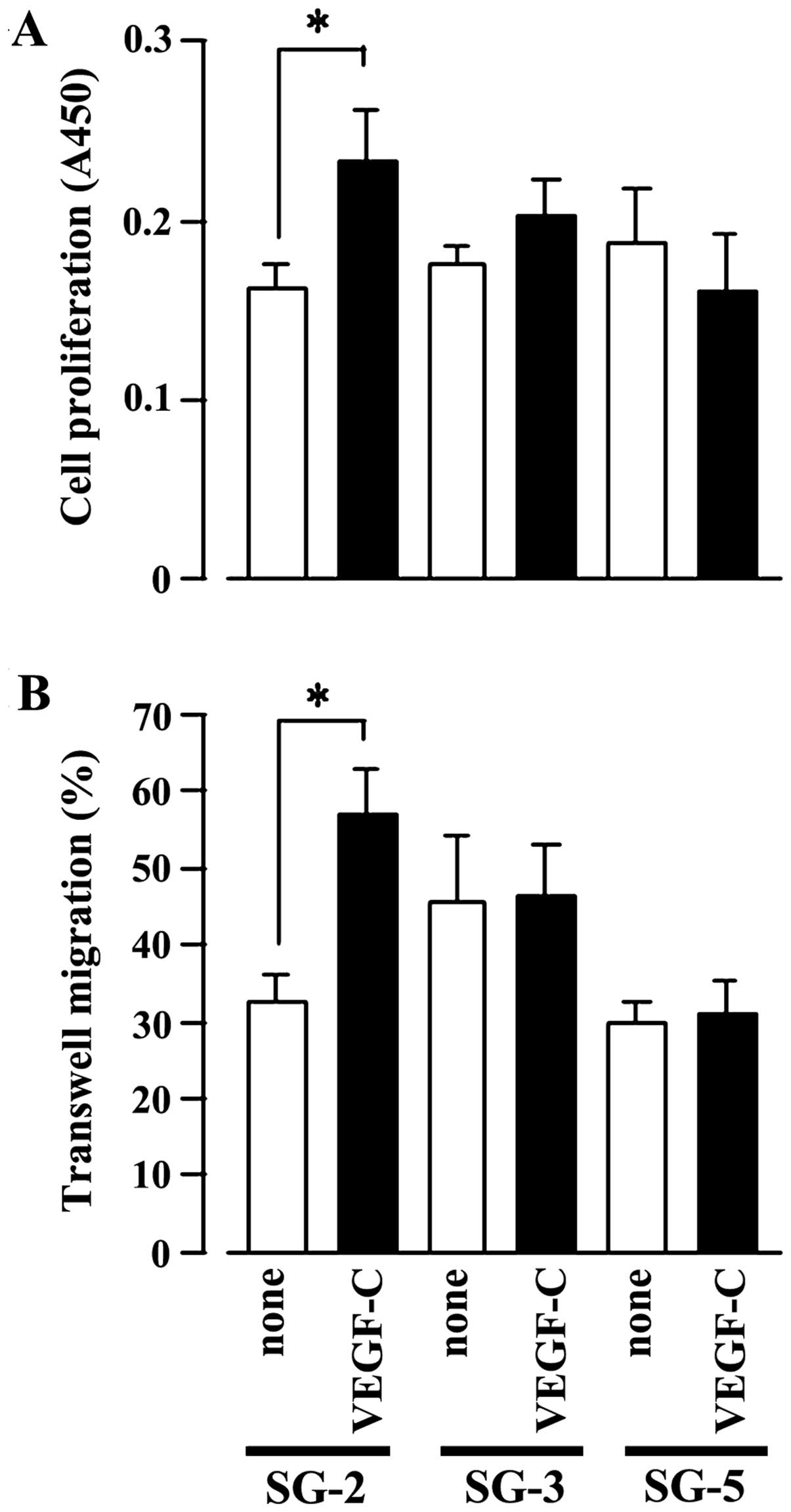
Indeed, VEGF-C significantly stimulated SG-2 cell proliferation
(Fig. 2A) and cell migration
(Fig. 2B), but had no effect on
the SG-3 or SG-5 cells. These results strongly suggest that VEGF-C
specifically promotes the proliferative activity and migratory
ability of the SG-2 cells through VEGFR3.
Phosphorylation of ERK1/2 in SG-2 cells
by stimulation with VEGF-C
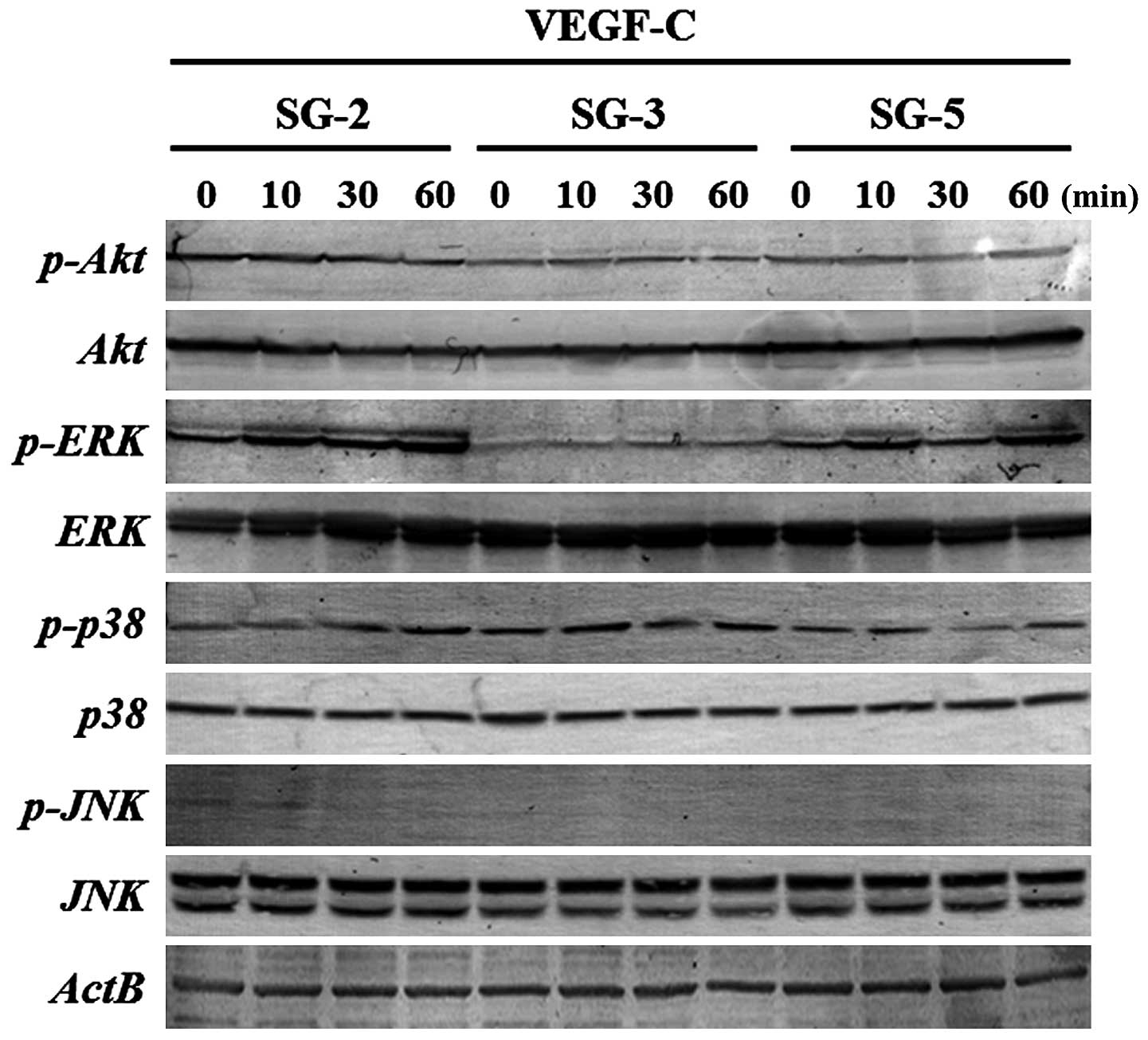
To clarify the signaling pathways activated by
VEGF-C in SG-2 cells, we evaluated the phosphorylation status of
molecules in the PI3K/Akt- and MAPK-mediated pathways. ERK1/2
phosphorylation was markedly upregulated in the SG-2 cells upon
VEGF-C stimulation, whereas that in the SG-3 and SG-5 cells was
unaffected (Fig. 3). These
results suggest that VEGF-C enhances the proliferative activity and
migratory ability of the MSCs through the ERK1/2 pathway in
Flt4-positive SG-2 cells.
Increase in the expression of lymphatic
endothelial differentiation marker genes following stimulation of
SG-2 cells with VEGF-C
The VEGF-C gene encodes a ligand for VEGFR3 that is
expressed mainly in lymphatic endothelia (18). Furthermore, the VEGF-C/VEGFR3
pathway was the first critical pathway described for the
development of the lymphatic vascular tree (36). As the SG-2 cells respond to both
TGF-β and VEGF-C, we examined the effects of TGF-β or VEGF-C
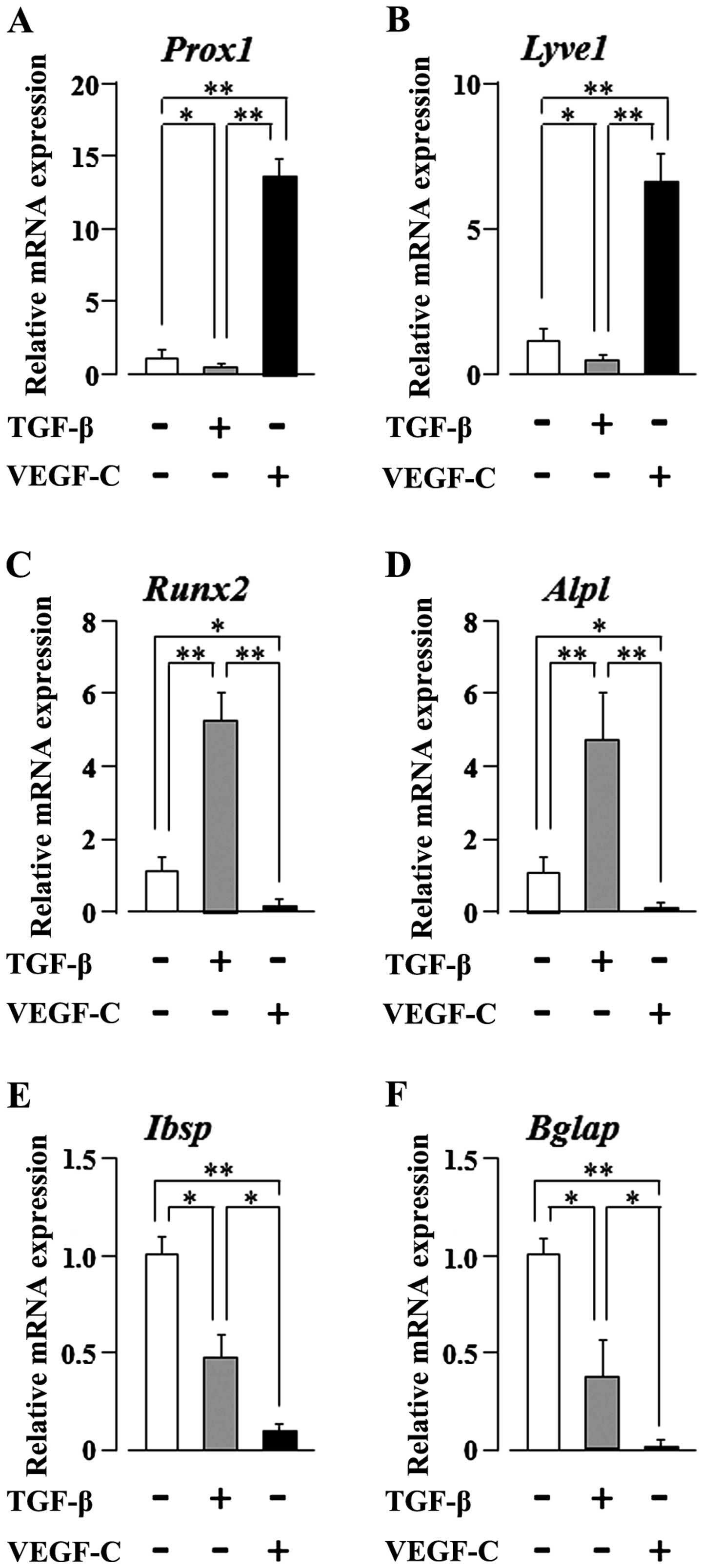
stimulation on their multi-differentiation potential. VEGF-C
clearly and significantly increased the mRNA expression levels of
the lymphatic endothelial cell markers, prospero homeobox 1
(Prox1) and lymphatic vessel endothelial hyaluronan receptor
1 (Lyve1) (p<0.01), in the SG-2 cells, whereas the levels
were clearly and significantly decreased following stimulation of
the cells with TGF-β (p<0.05; Fig.
4A and B). We previously reported that TGF-β promotes the
osteogenic differentiation of bone marrow-derived MSCs (10). Therefore, in this study, we
further investigated the mRNA expression of osteogenic
differentiation markers in the SG-2 cells following TGF-β or VEGF-C
stimulation. TGF-β clearly and significantly increased the
expression of the early-stage osteogenic differentiation marker
genes, Runt-related transcription factor 2 (Runx2) and
alkaline phosphatase, liver/bone/kidney (Alpl) (p<0.01),
in the SG-2 cells; by contrast, VEGF-C clearly and significantly
decreased the expression of these early-stage osteogenic
differentiation markers (p<0.05; Fig. 4C and D). Of note, TGF-β
unexpectedly decreased the expression of the late-stage osteogenic
differentiation markers, integrin-binding sialoprotein
(Ibsp) and bone gamma-carboxyglutamate (Gla) protein
(Bglap) (p<0.05; Fig. 4E
and F). On the other hand, as expected, VEGF-C suppressed the
expression of these late-stage differentiation markers (p<0.01;
Fig. 4E and F). These results
suggest that VEGF-C and TGF-β reciprocally regulate the commitment
of MSCs to differentiate into lymphatic endothelial or osteoblastic
phenotypes, respectively. On the other hand, both TGF-β and VEGF-C
appear to suppress the final maturation of osteoblastic MSC
differentiation during late-stage osteogenesis.
Discussion
As demonstrated in our previous study, the
TGF-β-responsive, Flt4-positive SG-2 MSC line retained both
osteogenic and adipogenic differentiation potentials (35). Herein, we focused on SG-2-specific
membrane protein expression and identified high expression levels
of VEGFR3, the Flt4 gene product (Table II and Fig. 1). Furthermore, we found that the
VEGFR3-specific ligand, VEGF-C, significantly increased the
proliferative activity and migratory ability of the SG-2 cells
(Fig. 2). VEGF potently promotes
angiogenesis and is indispensable for vascular development
(37,38), and the tyrosine kinase receptor,
VEGFR2, is the primary transmitter of VEGF signals in endothelial
cells (39,40). The binding of VEGF-A to VEGFR2
activates downstream signaling, including the MAPK pathways
(41,42). Other VEGF family members and other
signaling mediators affect and overlap with the function of VEGF-A
(22,43,44). VEGFR3 is activated by the VEGF
homologues, VEGF-C and VEGF-D, which, when fully proteolytically
processed, also stimulate VEGFR2 and induce the formation and
activation of VEGFR2-VEGFR3 heterodimers (36,45,46). Since in this study VEGF-C
stimulation induced ERK1/2 phosphorylation in the SG-2 cells, the
promotion of the migratory ability and proliferative activity of
Flt4-positive MSCs appears to depend on the activation of
the MAPK cascade (Fig. 3).
The VEGF-C/VEGFR3 pathway was the first critical
pathway described for the development of the lymphatic vascular
tree (36). It has been
demonstrated that VEGFR3 expression starts during mouse embryonic
day 8.5 in developing blood vessels, and VEGFR3-deficient embryos
die at midgestation from defects in the remodeling of primary
vascular networks (47). In adult
tissues, VEGFR3 expression occurs mainly in lymphatic endothelial
cells (47–50), and VEGFR3-positive lymphatic
vessels appear concurrently with blood vessels during wound
healing, but regress rapidly (51). However, VEGFR3-expressing
endothelial cells may also be found in the fenestrated capillaries
of several adult organs, including the bone marrow, splenic and
hepatic sinusoids, kidney glomeruli and endocrine glands (50). Notably, in human cancer,
VEGFR3-expressing vascular endothelial cells are detected in
angiogenic capillaries (52,53), and the inactivation of VEGFR3
signaling with blocking antibodies in nude mice has been shown to
suppress tumor growth by inhibiting angiogenesis (54).
MSCs have been reported to home towards hypoxic
micro-environments in vivo, and hypoxic tumor cells
specifically recruit MSCs by activating survival pathways that
facilitate tumor progression (55). Based on these findings, the
results of our study suggest that Flt4-positive MSCs play an
important role in tumor angiogenesis and lymphatic vessel
formation. Previous studies have demonstrated the VEGF-mediated
differentiation of lymphatic endothelial cells from bone
marrow-derived MSCs (56,57).
In this study, our results indicated that
stimulation with VEGF-C increased the expression of lymphatic
endothelial cell marker genes in Flt4-positive SG-2 cells
(Fig. 4A and B). More
interestingly, VEGF-C suppressed the expression of osteogenic
differentiation marker genes (Fig. 4C
and D). On the other hand, TGF-β suppressed the lymphatic
endothelial commitment of SC-2 cells (Fig. 4A and B). Thus, we concluded that
VEGF-C and TGF-β reciprocally regulate MSC commitment to
differentiation into lymphatic endothelial or osteoblastic
phenotypes, respectively. However, TGF-β and VEGF-C both seem to
suppress the maturation of osteoblastic MSC differentiation at the
late stage of the osteogenic process, suggesting that additional
cellular signals must be necessary for the progression of
osteoblastic differentiation of some types of MSCs. In addition,
VEGF-C positively regulated the migration and proliferation of the
Flt4-positive SG-2 cells (Fig.
2). The migratory ability and the proliferative activity are
necessary conditions of MSCs, suggesting the novel possibility that
VEGF-C plays an important role in determining MSC
characteristics.
Our findings provide new insight into the molecular
mechanisms underlying the regenerative activity of MSCs. In future
studies, we aim to determine whether these results are reproducible
in vivo by transplanting GFP-expressing SG-2 cells into
suitable animal experimental models to facilitate their
discrimination from the surrounding donor cells.
Acknowledgments
The present study was supported in part by JSPS
KAKENHI grant nos. 25463053 to N.C., 25893221 and 15K20633 to S.S.,
26462932 to H.K. and 26670852 to A.I.; a Grant-in-Aid for Strategic
Medical Science Research Centre from the Ministry of Education,
Culture, Sports, Science and Technology of Japan, 2010–2014; and a
grant from the Keiryokai Research Foundation grant no. 120 to N.C.
and S.S., 2013.
References
|
1
|
Prockop DJ: Marrow stromal cells as stem
cells for nonhematopoietic tissues. Science. 276:71–74. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pittenger MF, Mackay AM, Beck SC, Jaiswal
RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S and
Marshak DR: Multilineage potential of adult human mesenchymal stem
cells. Science. 284:143–147. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Docheva D, Popov C, Mutschler W and
Schieker M: Human mesenchymal stem cells in contact with their
environment: surface characteristics and the integrin system. J
Cell Mol Med. 11:21–38. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Vidane AS, Zomer HD, Oliveira BM,
Guimarães CF, Fernandes CB, Perecin F, Silva LA, Miglino MA,
Meirelles FV and Ambrósio CE: Reproductive stem cell
differentiation: extracellular matrix, tissue microenvironment, and
growth factors direct the mesenchymal stem cell lineage commitment.
Reprod Sci. 20:1137–1143. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen BY, Wang X, Chen LW and Luo ZJ:
Molecular targeting regulation of proliferation and differentiation
of the bone marrow-derived mesenchymal stem cells or mesenchymal
stromal cells. Curr Drug Targets. 13:561–571. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Soleymaninejadian E, Pramanik K and
Samadian E: Immunomodulatory properties of mesenchymal stem cells:
cytokines and factors. Am J Reprod Immunol. 67:1–8. 2012.
View Article : Google Scholar
|
|
7
|
Chau JF, Leong WF and Li B: Signaling
pathways governing osteoblast proliferation, differentiation and
function. Histol Histopathol. 24:1593–1606. 2009.PubMed/NCBI
|
|
8
|
Pountos I, Georgouli T, Henshaw K, Bird H,
Jones E and Giannoudis PV: The effect of bone morphogenetic
protein-2, bone morphogenetic protein-7, parathyroid hormone, and
platelet-derived growth factor on the proliferation and osteogenic
differentiation of mesenchymal stem cells derived from osteoporotic
bone. J Orthop Trauma. 24:552–556. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hughes FJ, Turner W, Belibasakis G and
Martuscelli G: Effects of growth factors and cytokines on
osteoblast differentiation. Periodontol. 2000 41:48–72. 2006.
View Article : Google Scholar
|
|
10
|
Yokota J, Chosa N, Sawada S, Okubo N,
Takahashi N, Hasegawa T, Kondo H and Ishisaki A: PDGF-induced
PI3K-mediated signaling enhances the TGF-β-induced osteogenic
differentiation of human mesenchymal stem cells in a
TGF-β-activated MEK-dependent manner. Int J Mol Med. 33:534–542.
2014.PubMed/NCBI
|
|
11
|
Aomatsu E, Takahashi N, Sawada S, Okubo N,
Hasegawa T, Taira M, Miura H, Ishisaki A and Chosa N: Novel
SCRG1/BST1 axis regulates self-renewal, migration, and osteogenic
differentiation potential in mesenchymal stem cells. Sci Rep.
4(3652)2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ferrara N: Vascular endothelial growth
factor: basic science and clinical progress. Endocr Rev.
25:581–611. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ellis LM and Hicklin DJ: VEGF-targeted
therapy: mechanisms of anti-tumour activity. Nat Rev Cancer.
8:579–591. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dvorak HF: Vascular permeability
factor/vascular endothelial growth factor: a critical cytokine in
tumor angiogenesis and a potential target for diagnosis and
therapy. J Clin Oncol. 20:4368–4380. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Orlandini M, Marconcini L, Ferruzzi R and
Oliviero S: Identification of a c-fos-induced gene that is related
to the platelet-derived growth factor/vascular endothelial growth
factor family. Proc Natl Acad Sci USA. 93:11675–11680. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Roche PA and Cresswell P: Proteolysis of
the class II-associated invariant chain generates a peptide binding
site in intracellular HLA-DR molecules. Proc Natl Acad Sci USA.
88:3150–3154. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Olofsson B, Pajusola K, Kaipainen A, von
Euler G, Joukov V, Saksela O, Orpana A, Pettersson RF, Alitalo K
and Eriksson U: Vascular endothelial growth factor B, a novel
growth factor for endothelial cells. Proc Natl Acad Sci USA.
93:2576–2581. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Joukov V, Pajusola K, Kaipainen A, Chilov
D, Lahtinen I, Kukk E, Saksela O, Kalkkinen N and Alitalo K: A
novel vascular endothelial growth factor, VEGF-C, is a ligand for
the Flt4 (VEGFR-3) and KDR (VEGFR-2) receptor tyrosine kinases.
EMBO J. 15(1751)1996.PubMed/NCBI
|
|
19
|
Soker S, Takashima S, Miao HQ, Neufeld G
and Klagsbrun M: Neuropilin-1 is expressed by endothelial and tumor
cells as an isoform-specific receptor for vascular endothelial
growth factor. Cell. 92:735–745. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Migdal M, Huppertz B, Tessler S, Comforti
A, Shibuya M, Reich R, Baumann H and Neufeld G: Neuropilin-1 is a
placenta growth factor-2 receptor. J Biol Chem. 273:22272–22278.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Neufeld G, Kessler O and Herzog Y: The
interaction of Neuropilin-1 and Neuropilin-2 with tyrosine-kinase
receptors for VEGF. Adv Exp Med Biol. 515:81–90. 2002. View Article : Google Scholar
|
|
22
|
Cao Y: Positive and negative modulation of
angiogenesis by VEGFR1 ligands. Sci Signal. 2:re12009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kerbel RS: Tumor angiogenesis. N Engl J
Med. 358:2039–2049. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang F, Tang Z, Hou X, Lennartsson J, Li
Y, Koch AW, Scotney P, Lee C, Arjunan P, Dong L, et al: VEGF-B is
dispensable for blood vessel growth but critical for their
survival, and VEGF-B targeting inhibits pathological angiogenesis.
Proc Natl Acad Sci USA. 106:6152–6157. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hanrahan V, Currie MJ, Gunningham SP,
Morrin HR, Scott PA, Robinson BA and Fox SB: The angiogenic switch
for vascular endothelial growth factor (VEGF)-A, VEGF-B, VEGF-C,
and VEGF-D in the adenoma-carcinoma sequence during colorectal
cancer progression. J Pathol. 200:183–194. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang TB, Chen ZG, Wei XQ, Wei B and Dong
WG: Serum vascular endothelial growth factor-C and
lymphoangiogenesis are associated with the lymph node metastasis
and prognosis of patients with colorectal cancer. ANZ J Surg.
81:694–699. 2011. View Article : Google Scholar
|
|
27
|
Skobe M, Hawighorst T, Jackson DG, Prevo
R, Janes L, Velasco P, Riccardi L, Alitalo K, Claffey K and Detmar
M: Induction of tumor lymphangiogenesis by VEGF-C promotes breast
cancer metastasis. Nat Med. 7:192–198. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wu QW, She HQ, Liang J, Huang YF, Yang QM,
Yang QL and Zhang ZM: Expression and clinical significance of
extracellular matrix protein 1 and vascular endothelial growth
factor-C in lymphatic metastasis of human breast cancer. BMC
Cancer. 12(47)2012. View Article : Google Scholar
|
|
29
|
Karnezis T, Shayan R, Caesar C, Roufail S,
Harris NC, Ardipradja K, Zhang YF, Williams SP, Farnsworth RH, Chai
MG, et al: VEGF-D promotes tumor metastasis by regulating
prostaglandins produced by the collecting lymphatic endothelium.
Cancer Cell. 21:181–195. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Stacker SA, Caesar C, Baldwin ME, Thornton
GE, Williams RA, Prevo R, Jackson DG, Nishikawa S, Kubo H and Achen
MG: VEGF-D promotes the metastatic spread of tumor cells via the
lymphatics. Nat Med. 7:186–191. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu Y and Olsen BR: Distinct VEGF
functions during bone development and homeostasis. Arch Immunol
Ther Exp (Warsz). 62:363–368. 2014. View Article : Google Scholar
|
|
32
|
Deckers MM, Karperien M, van der Bent C,
Yamashita T, Papapoulos SE and Löwik CW: Expression of vascular
endothelial growth factors and their receptors during osteoblast
differentiation. Endocrinology. 141:1667–1674. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zelzer E, McLean W, Ng YS, Fukai N,
Reginato AM, Lovejoy S, D'Amore PA and Olsen BR: Skeletal defects
in VEGF(120/120) mice reveal multiple roles for VEGF in
skeletogenesis. Development. 129:1893–1904. 2002.PubMed/NCBI
|
|
34
|
Alonso V, de Gortázar AR, Ardura JA,
Andrade-Zapata I, Alvarez-Arroyo MV and Esbrit P: Parathyroid
hormone-related protein (107-139) increases human osteoblastic cell
survival by activation of vascular endothelial growth factor
receptor-2. J Cell Physiol. 217:717–727. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sawada S, Chosa N, Takizawa N, Yokota J,
Igarashi Y, Tomoda K, Kondo H, Yaegashi T and Ishisaki A:
Establishment of mesenchymal stem cell lines derived from the bone
marrow of GFP-transgenic mice exhibiting diversity in intracellular
TGF-β and BMP signaling. Mol Med Rep. 13:2023–2031. 2016.
|
|
36
|
Tammela T and Alitalo K:
Lymphangiogenesis: molecular mechanisms and future promise. Cell.
140:460–476. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ferrara N, Carver-Moore K, Chen H, Dowd M,
Lu L, O'Shea KS, Powell-Braxton L, Hillan KJ and Moore MW:
Heterozygous embryonic lethality induced by targeted inactivation
of the VEGF gene. Nature. 380:439–442. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Carmeliet P, Mackman N, Moons L, Luther T,
Gressens P, Van Vlaenderen I, Demunck H, Kasper M, Breier G, Evrard
P, et al: Role of tissue factor in embryonic blood vessel
development. Nature. 383:73–75. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Shalaby F, Rossant J, Yamaguchi TP,
Gertsenstein M, Wu XF, Breitman ML and Schuh AC: Failure of
blood-island formation and vasculogenesis in Flk-1-deficient mice.
Nature. 376:62–66. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Gille H, Kowalski J, Li B, LeCouter J,
Moffat B, Zioncheck TF, Pelletier N and Ferrara N: Analysis of
biological effects and signaling properties of Flt-1 (VEGFR-1) and
KDR (VEGFR-2). A reassessment using novel receptor-specific
vascular endothelial growth factor mutants. J Biol Chem.
276:3222–3230. 2001. View Article : Google Scholar
|
|
41
|
Takahashi T, Ueno H and Shibuya M: VEGF
activates protein kinase C-dependent, but Ras-independent
Raf-MEK-MAP kinase pathway for DNA synthesis in primary endothelial
cells. Oncogene. 18:2221–2230. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Meadows KN, Bryant P and Pumiglia K:
Vascular endothelial growth factor induction of the angiogenic
phenotype requires Ras activation. J Biol Chem. 276:49289–49298.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Huang H, Bhat A, Woodnutt G and Lappe R:
Targeting the ANGPT-TIE2 pathway in malignancy. Nat Rev Cancer.
10:575–585. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Weis SM and Cheresh DA: αV integrins in
angiogenesis and cancer. Cold Spring Harb Perspect Med.
1:a0064782011. View Article : Google Scholar
|
|
45
|
Dixelius J, Makinen T, Wirzenius M,
Karkkainen MJ, Wernstedt C, Alitalo K and Claesson-Welsh L:
Ligand-induced vascular endothelial growth factor receptor-3
(VEGFR-3) hetero-dimerization with VEGFR-2 in primary lymphatic
endothelial cells regulates tyrosine phosphorylation sites. J Biol
Chem. 278:40973–40979. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Nilsson I, Bahram F, Li X, Gualandi L,
Koch S, Jarvius M, Söderberg O, Anisimov A, Kholová I, Pytowski B,
et al: VEGF receptor 2/-3 heterodimers detected in situ by
proximity ligation on angiogenic sprouts. EMBO J. 29:1377–1388.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Dumont DJ, Jussila L, Taipale J,
Lymboussaki A, Mustonen T, Pajusola K, Breitman M and Alitalo K:
Cardiovascular failure in mouse embryos deficient in VEGF
receptor-3. Science. 282:946–949. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Kaipainen A, Korhonen J, Mustonen T, van
Hinsbergh VW, Fang GH, Dumont D, Breitman M and Alitalo K:
Expression of the fms-like tyrosine kinase 4 gene becomes
restricted to lymphatic endothelium during development. Proc Natl
Acad Sci USA. 92:3566–3570. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Jussila L, Valtola R, Partanen TA, Salven
P, Heikkilä P, Matikainen MT, Renkonen R, Kaipainen A, Detmar M,
Tschachler E, et al: Lymphatic endothelium and Kaposi's sarcoma
spindle cells detected by antibodies against the vascular
endothelial growth factor receptor-3. Cancer Res. 58:1599–1604.
1998.PubMed/NCBI
|
|
50
|
Partanen TA, Arola J, Saaristo A, Jussila
L, Ora A, Miettinen M, Stacker SA, Achen MG and Alitalo K: VEGF-C
and VEGF-D expression in neuroendocrine cells and their receptor,
VEGFR-3, in fenestrated blood vessels in human tissues. FASEB J.
14:2087–2096. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Paavonen K, Puolakkainen P, Jussila L,
Jahkola T and Alitalo K: Vascular endothelial growth factor
receptor-3 in lymphangio-genesis in wound healing. Am J Pathol.
156:1499–1504. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Valtola R, Salven P, Heikkilä P, Taipale
J, Joensuu H, Rehn M, Pihlajaniemi T, Weich H, deWaal R and Alitalo
K: VEGFR-3 and its ligand VEGF-C are associated with angiogenesis
in breast cancer. Am J Pathol. 154:1381–1390. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Partanen TA, Alitalo K and Miettinen M:
Lack of lymphatic vascular specificity of vascular endothelial
growth factor receptor 3 in 185 vascular tumors. Cancer.
86:2406–2412. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Kubo H, Fujiwara T, Jussila L, Hashi H,
Ogawa M, Shimizu K, Awane M, Sakai Y, Takabayashi A, Alitalo K, et
al: Involvement of vascular endothelial growth factor receptor-3 in
maintenance of integrity of endothelial cell lining during tumor
angiogenesis. Blood. 96:546–553. 2000.PubMed/NCBI
|
|
55
|
Rattigan Y, Hsu JM, Mishra PJ, Glod J and
Banerjee D: Interleukin 6 mediated recruitment of mesenchymal stem
cells to the hypoxic tumor milieu. Exp Cell Res. 316:3417–3424.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Wei L, Liu Y, Chen G, Fang Y, Song X, Dong
P, Gao J, Liu R, Ding Z, Bi Y and Liu Z: Differentiation of
lymphatic endothelial cells from bone marrow mesenchymal stem cells
with VEGFs. Lymphology. 45:177–187. 2012.
|
|
57
|
Zhan J, Li Y, Yu J, Zhao Y, Cao W, Ma J,
Sun X, Sun L, Qian H, Zhu W and Xu W: Culture medium of bone
marrow-derived human mesenchymal stem cells effects lymphatic
endothelial cells and tumor lymph vessel formation. Oncol Lett.
9:1221–1226. 2015.PubMed/NCBI
|