Introduction
Type 2 diabetes (T2D) is a serious chronic metabolic
disorder of the 21st century and the fifth leading cause of
mortality worldwide (1,2). It is characterized by fasting
hyperglycemia, secondary to hepatic insulin resistance (IR) and
increased glucose production (3).
IR, a main characteristic of T2D, is a pathological state in which
insulin function is impaired in target tissues, including the
liver, skeletal muscle and adipose tissue. Hepatic IR is of
critical importance, as it is the primary disturbance responsible
for fasting hyperglycemia (4).
Although a number of molecular mechanisms have been suggested to
account for the development of hepatic IR (5,6),
the underlying mechanism remain unclear.
Peroxisome proliferator-activated receptor-γ (PPARγ)
coactivator-1α (PGC-1α), as a nuclear transcriptional coactivator,
regulates several important metabolic processes, including
mitochondrial biogenesis, adaptive thermogenesis, respiration,
insulin secretion and gluconeogenesis (7,8).
Previous studies performed in our laboratory, as well as others
have demonstrated that PGC-1α plays an important role in preventing
the onset and development of atherosclerosis, as well as in
diminishing neointimal formation following balloon vascular injury
(9-12). Furthermore, PGC-1α has been shown
to be involved in the pathogenesis of hepatic IR (3,13).
Gluconeogenesis is regulated by PGC-1α in the liver via the
coactivation of transcription factors, including hepatocyte nuclear
factor-4α (HNF-4α), forkhead boxO1 (FOXO1) and the glucocorticoid
receptor, to coordinate the expression of phosphoenolpyruvate
carboxykinase (PEPCK) and glucose-6-phosphatase (G6Pase) (14,15). Due to the important role of PGC-1α
in energy metabolism and insulin sensitivity, it seems to be a
candidate factor in the etiology of T2D and a drug target for
pharmacological intervention (7).
MicroRNAs (miRNAs or miRs) are endogenous non-coding
RNA molecules of 19 to 24 nucleotides in length. They play critical
roles in the negative regulation of gene expression by base pairing
to complementary sites on target mRNAs and then causing a block in
translation or triggering the degradation of the target mRNAs
(16). It has been has
systematically discovered by our group that miRNAs are stably
present in human serum/plasma and that their unique expression
patterns serve as 'fingerprints' of various diseases (17–20). Studies have demonstrated that
miRNAs play a vital role in the development of IR, a key
pathophysiological link between obesity and diabetes. For instance,
the pancreatic islet-specific miR-375 was found to regulate insulin
secretion (21). miR-103/107 have
been reported to play an important role in insulin sensitivity
(22). Ling et al found
that miR-320 was able to regulate IR (23). Moreover, muscle IR has been
reported to be related to coordinated changes in multiple miRNAs,
which indicates that miRNA detection represents a novel molecular
biomarker strategy for IR (24).
Recently, miR-696 has been shown to play an
important role in regulating PGC-1α in mouse skeletal muscle in
response to physical activity (25). In the present study, we found that
hepatic PGC-1α expression was elevated in ob/ob mice, an animal
model of insulin function deficiency, compared with normal C57BL/6
mice. On the contrary, miR-696 expression was significantly lower
in the livers of the ob/ob mice compared with the C57BL/6 mice.
Hence, we hypothesized that miR-696 may be involved in the function
of PGC-1α in the liver. The direct inhibition of PGC-1α translation
by miR-696 and the potential role of miR-696 in hepatic
gluconeogenesis were experimentally validated.
Materials and methods
Ethics statement and animal models
This study was carried out in the strict accordance
with the recommendations in the Guide for the Care and Use of
Laboratory Animals of the the National Institutes of Health. The
protocol was approved by the Institutional Review Board of Nanjing
University, Nanjing, China. Male 8-week-old C57BL/6 and ob/ob mice
were obtained from the Model Animal Research Center of Nanjing
University. The mice were sacrificed by cervical dislocation, and
the livers were removed, immediately frozen in liquid nitrogen, and
stored at −80°C for later use.
Cell culture
Primary hepatocytes were isolated from the
8-week-old male C57BL/6 mice by non-recirculating collagenase
perfusion, as previously described by Klaunig et al
(26). In brief, the livers were
initially perfused with Ca2+-free Hank's buffer and then
with collagenase type IV in Hank's buffer plus 5 mmol/l
CaCl2. The dispersed cells were resuspended and seeded
onto collagen-coated plates in DMEM supplemented with 10% fetal
bovine serum (FBS) in the presence of penicillin (100 U/ml),
streptomycin (100 mg/ml), 1 mM sodium pyruvate, 1 M dexamethasone
and 50 nM insulin. The cultures were maintained at 37°C in a
humidified atmosphere of 95% air and 5% CO2 and were
incubated for 12 h prior to the experiment. The 293T cell line was
purchased from the China Cell Culture Center. The 293T cells were
cultured in high-glucose DMEM medium (HyClone, Logan, UT, USA)
supplemented with 10% FBS (Gibco, Carlsbad, CA, USA) at 37°C in a
humidified atmosphere of 95% air and 5% CO2.
Infection with lentivirus
Recombinant lentiviruses, respectively carrying
pre-mmu-miR-696 precursor (pre-miR-696-LV), pre-non-coding sequence
(pre-NC-LV), anti-mmu-miR-696 inhibitor (anti-miR-696-LV) or
anti-non-coding sequence (anti-NC-LV), were obtained from Hanbio
(Shanghai, China). Each lentivirus contained a green fluorescent
protein (GFP) sequence so that the infection efficiency could be
monitored by fluorescence. They were individually added to
hepatocytes at 30% confluence in 6-well plates or 10-cm dishes at
an MOI of 40 together with polybrene at a final concentration of 5
µg/ml. The cells were then harvested at 3 days
post-infection for western blot analysis and reverse
transcription-quantitative PCR (RT-qPCR).
Plasmid constructs and luciferase
reporter assay
The mouse PGC-1α 3′-untranslated region (3′-UTR)
sequence (1478 bp), obtained from the GenBank database, was
amplified by PCR using a mouse genomic DNA template. The PCR
products were inserted into the XhoI/NotI sites of
the psiCHECK-2 plasmid (Promega, Madison, WI, USA) using the
following primers: forward, 5′-GCGCTCGAGGAGATGGTCAATACCTCATGGG-3′
and reverse, 5′-AATGCGGCCGCAGAAAATGTGGAAAAATATTGC-3′. Efficient
insertion was confirmed by sequencing. For the luciferase reporter
assays, the 293T cells were cultured in 6-well plates. For each
well, the cells at 70–80% confluence were co-transfected with 2
µg of the psiCHECK-2-PGC-1α 3′-UTR plasmid and 2 µg
of the pHBLV-pre-miR-696, pHBLV-anti-miR-696 or pHBLV-IRES zsgreen
plasmid using Lipofectamine 2000 (Invitrogen, Grand Island, NY,
USA) according to the manufacturer's instructions. The cells were
assayed using luciferase assay kits (Promega) at 24 h
post-transfection.
RNA isolation and RT-qPCR
Total RNA derived from the cells and liver tissues
was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA)
according to the manufacturer's instructions. RT-qPCR was carried
out using TaqMan miRNA probes (Applied Biosystems, Foster City, CA,
USA) for miRNAs and specific primers for mRNA detection,
respectively according to the manufacturer's instructions. Briefly,
for miRNAs, 1 µg of total RNA was reverse transcribed into
cDNA using AMV reverse transcriptase (Takara, Dalian, China) and
stem-loop RT primers (Applied Biosystems). Quantitative (qPCR) was
performed using a TaqMan PCR kit on an Applied Biosystems 7300
Sequence Detection system (Applied Biosystems); for mRNAs, 1
µg of RNA was reverse transcribed into cDNA with oligo
(dT)18 primers (Takara) and AMV reverse transcriptase at
42°C for 1 h, followed by 70°C for 10 min. RT-qPCR was performed
using the Applied Biosystems 7300 Sequence Detection system with
forward and reverse primers for specific genes synthesized by
Invitrogen, as well as EvaGreen (Takara); the amplification
conditions were as follows: 1 cycle of 95°C for 5 min followed by
40 cycles of 95°C for 30 sec, 60°C for 30 sec, and final 1 cycle of
72°C for 30 sec. The primers sequences used in this study are
listed in Table I. The primers
specific for miR-696 cannot be disclosed (Applied Biosystems). All
reactions were run in triplicate. After the reactions were
complete, the CT values were determined using fixed
threshold settings. miRNA expression was normalized to U6 snRNA
expression in this study. The amount of miRNA to relative to the
internal control U6 was calculated using the equation
2−ΔCT, in which ΔCT = CT miRNA −
CT U6.
 | Table IPCR primers used in this study. |
Table I
PCR primers used in this study.
| mRNA | Primer
sequences |
|---|
| Mouse U6 snRNA | Sense: |
5′-CGGGATCCGATCCGACGCCGCCATCTCTAG-3′ |
| Antisense: |
5′-CGGTCGACTAGTATATGTGCTGCCGAAGCG-3′ |
| Mouse PGC-1α | Sense: |
5′-GCGCTCGAGGAGATGGTCAATACCTCATGGG-3′ |
| Antisense: |
5′-AATGCGGCCGCAGAAAATGTGGAAAAATATTGC-3′ |
Western blot analysis
To obtain total proteins, liver tissues from the
mice or the cells (hepatocytes) were lysed in a buffer containing
50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1% Triton X-100, 0.1% SDS,
and 1 mM PMSF. The cell lysates were then centrifuged at 13,000 × g
for 5 min at 4°C and supernatants were collected for western blot
analysis. We determined the protein concentrations using the BCA
method (Pierce, Rockford, IL, USA). Equal amounts of protein (40
µg) were separated by 10% SDS-PAGE and transferred onto PVDF
membranes. The membranes were blocked for 1 h with 5% non-fat milk
in TBST, followed by overnight incubation at 4°C with primary
antibodies directed toward PGC-1α (SC-13067, 1:1000) and PEPCK
(SC-271029, 1:1000) (all from Santa Cruz Biotechnology, Santa Cruz,
CA, USA). After washing, the membranes were incubated with
corresponding secondary antibody conjugated to horseradish
peroxidase. Normalization was performed by blotting the same
samples with an antibody against GAPDH (Santa Cruz
Biotechnology).
Statistical analyses
All images of western blot analyses are
representative of at least 3 independent experiments. For each
experiment, RT-qPCR assays were performed in triplicate. The data
shown are presented as the means ± standard error for 3 independent
experiments. Differences were considered statistically significant
at P<0.05 and were assessed using the Student's t-test.
Results
Evaluation of PGC-1α and miR-696
expression in the livers of ob/ob mouse
In order to investigate the biological function of
PGC-1α in livers of mice with IR, we examined the protein level of
PGC-1α in a mouse model of genetic IR and obesity (ob/ob mice).
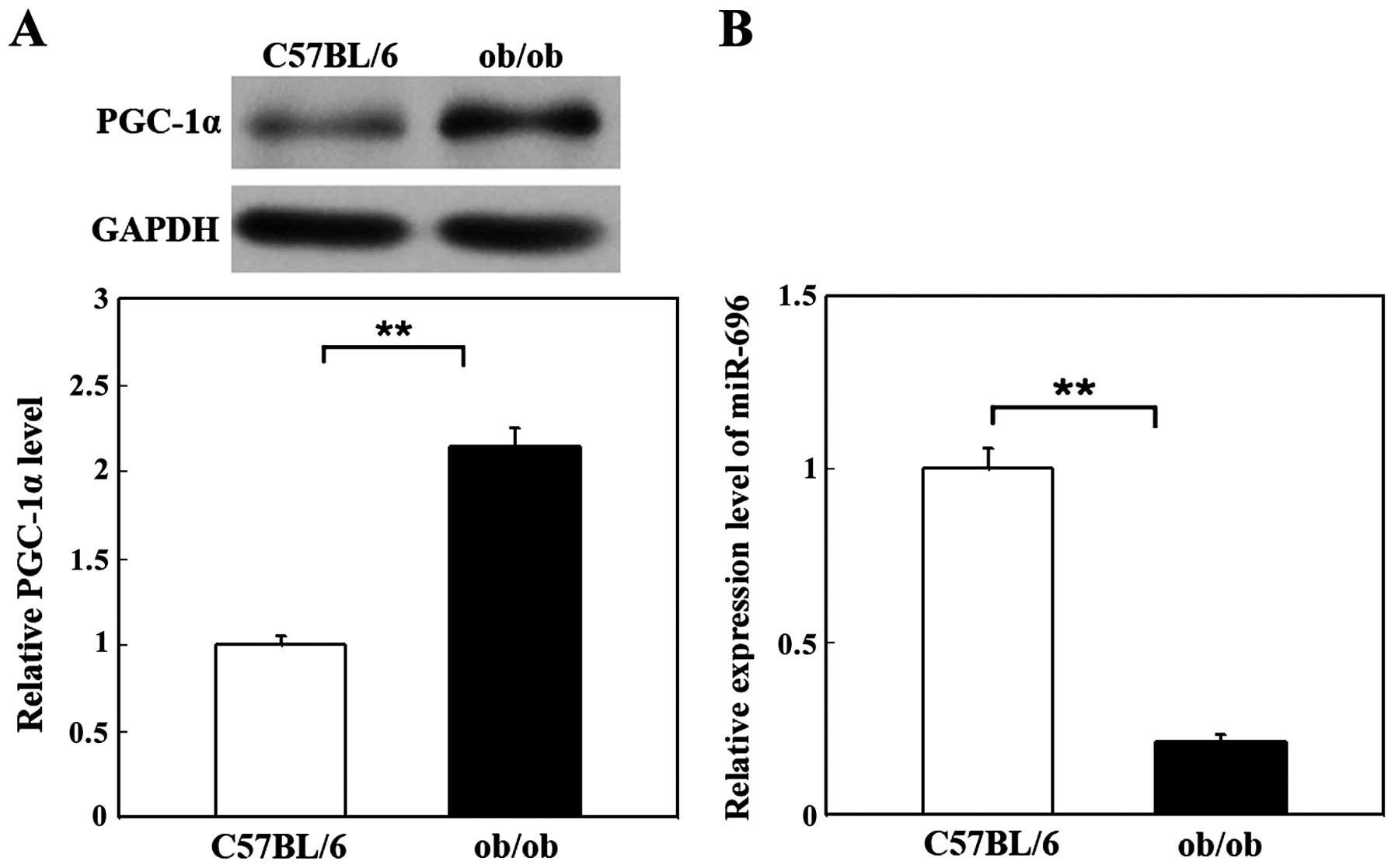
C57BL/6 mice of the same age were used as controls. As shown in
Fig. 1A, the protein expression
level of PGC-1α was significantly increased in the livers of ob/ob
mice (2.15-fold that of the normal C57BL/6 mice). The results are
representative data from 5 independent experiments. Quantification
of the protein bands was performed using Image software. These
results suggest that PGC-1α is involved in IR in the liver.
Recently, PGC-1α was deduced to be a target of
miR-696 not only by computational prediction, but also by
validating an inverse correlation between miR-696 and the PGC-1α
protein level in mouse skeletal muscle (25). In this study, in order to
investigate the correlation between miR-696 and PGC-1α in mouse
liver, we examined the level of miR-696 by RT-qPCR in the livers of
ob/ob mice and C57BL/6 mice. We found that the miR-696 level in the
livers of ob/ob mice was downregulated significantly (only
0.21-fold that of the normal controls; Fig. 1B). Taken together, these results
suggest that miR-696 negatively regulates PGC-1α expression in
mouse liver.
Regulation of PGC-1α by miR-696 in
primary hepatocytes
We further examined the correlation between miR-696
and PGC-1α by evaluating the expression of PGC-1α in primary
hepatocytes following the overexpression or knockdown of miR-696.
In our study, miR-696 overexpression/knockdown was achieved by
infecting mouse primary hepatocytes with
pre-miR-696-LV/anti-miR-696-LV. Primary hepatocytes infected by
pre-NC-LV or anti-NC-LV in the same manner were used as the
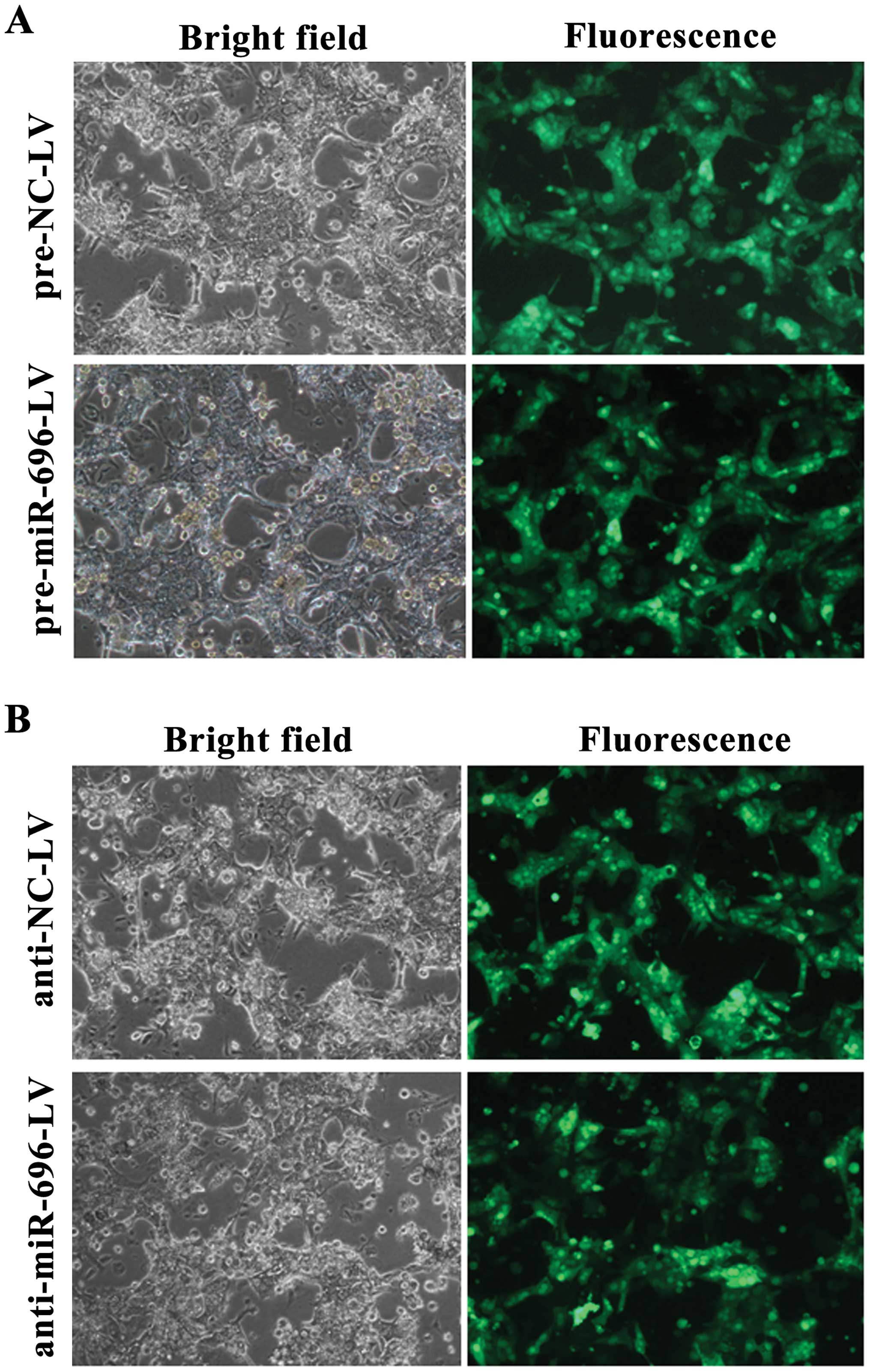
controls. The efficiency of infection at 72 h screened by the GFP
signal is shown in Fig. 2. We
also measured the efficiency of infection by RT-qPCR. As shown in
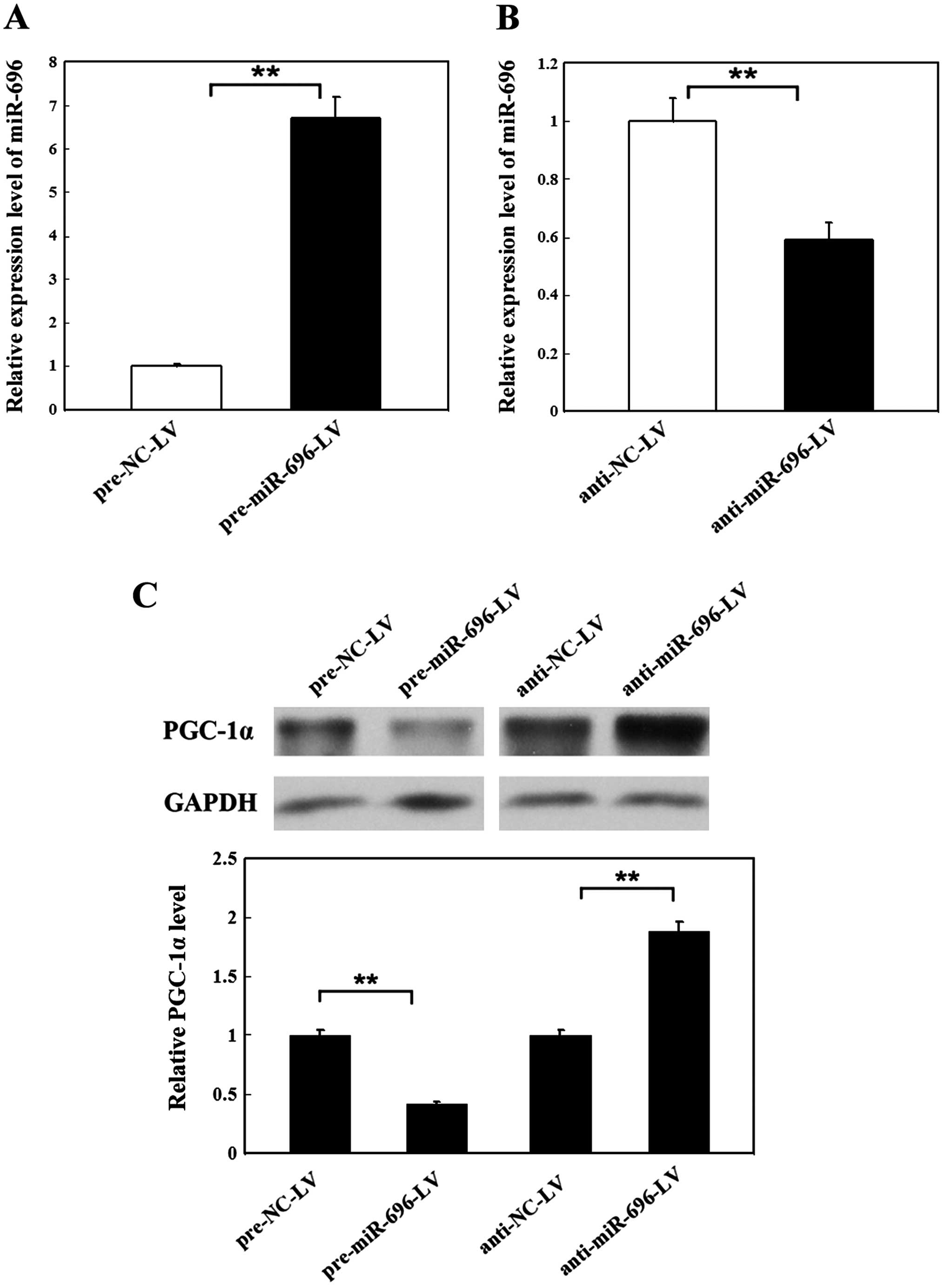
Fig. 3A, the level of miR-696 was
increased 6.7-fold in the primary hepatocytes infected by
pre-miR-696-LV compared with the controls. However, the miR-696
level was reduced by 59% after the primary hepatocytes were
infected by the anti-miR-696-LV compared with anti-NC-LV (Fig. 3B). The expression of PGC-1α was
decreased by 41% in the hepatocytes in which miR-696 was
overexpressed, and it was enhanced by 87% in the hepatocytes in
which miR-696 was knocked down (Fig.
3C). These results prove that miR-696 negatively regulates
PGC-1α in primary hepatocytes.
Identification of PGC-1α as a target of
miR-696 by luciferase binding assays
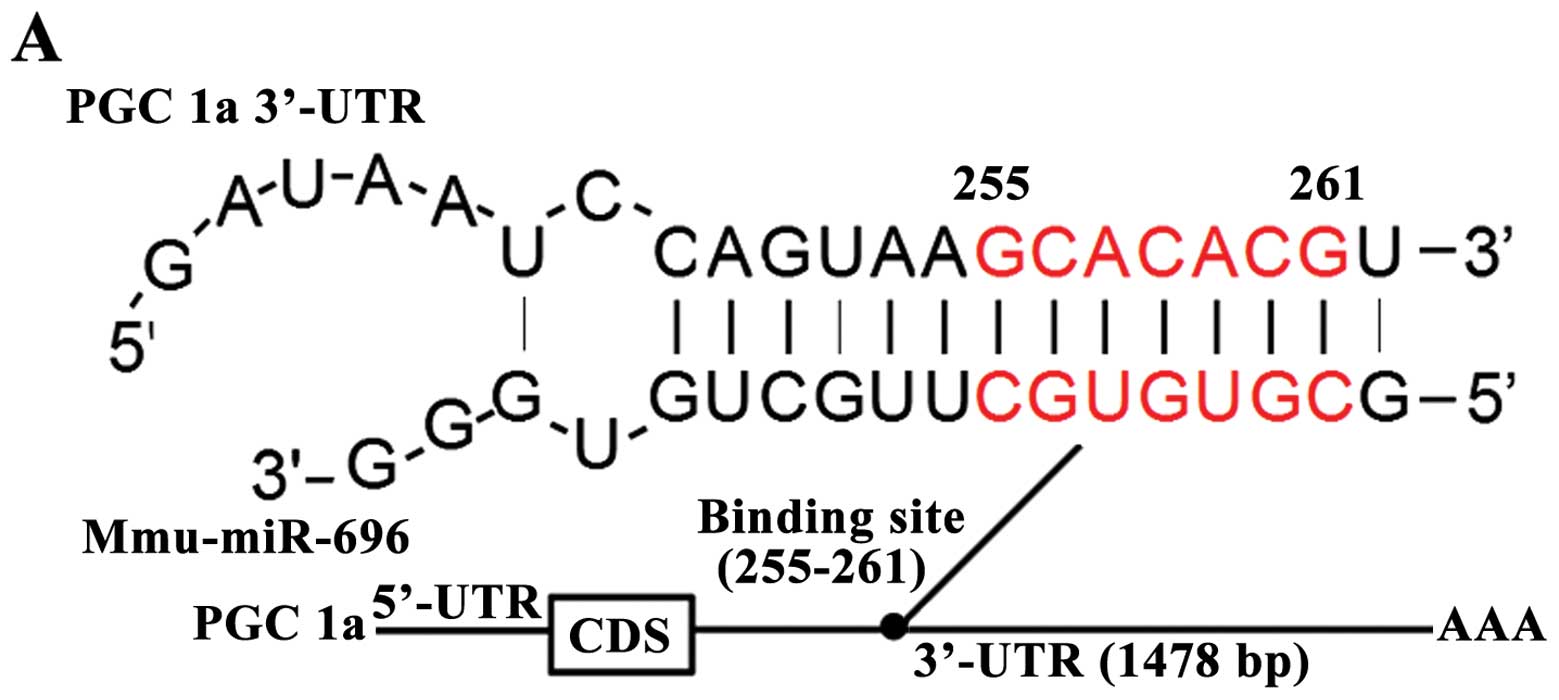
We found that there was perfect base pairing between
the 'seeds' (the core sequence that encompasses the first 2–8 bases
of the mature miRNA) and cognate targets (Fig. 4A). Thus, we then conducted a
luciferase binding assays using a psiCHECK-2 luciferase plasmid to
determine whether miR-696 suppresses PGC-1α through direct binding
to its 3′-UTR. As expected, the overexpression of miR-696 resulted
in a 62% reduction of firefly luciferase reporter activity.
However, the knockdown of miR-696 resulted in a 17% increment of
firefly luciferase reporter activity (Fig. 4B). These results unequivocally
demonstrate that miR-696 directly recognizes the 3′-UTR of the
PGC-1α transcript. Thus, the upregulation of miR-696 promotes the
suppression of PGC-1α.
Role of miR-696 targeting PGC-1α in
hepatic gluconeogenesis
PGC-1α has been reported to be an improtant
regulator of gluconeogenesis in the liver (13,14,27). It can coordinate the expression of
characteristic hepatic gluconeogenic enzymes, including PEPCK and
G6Pase through coactivating transcription factors, such as HNF-4α,
FOXO1 (14,15). We then examined the expression of
PEPCK as an indicator to examine the downstream biological
consequences of the miR-696-driven inhibition of PGC-1α expression.
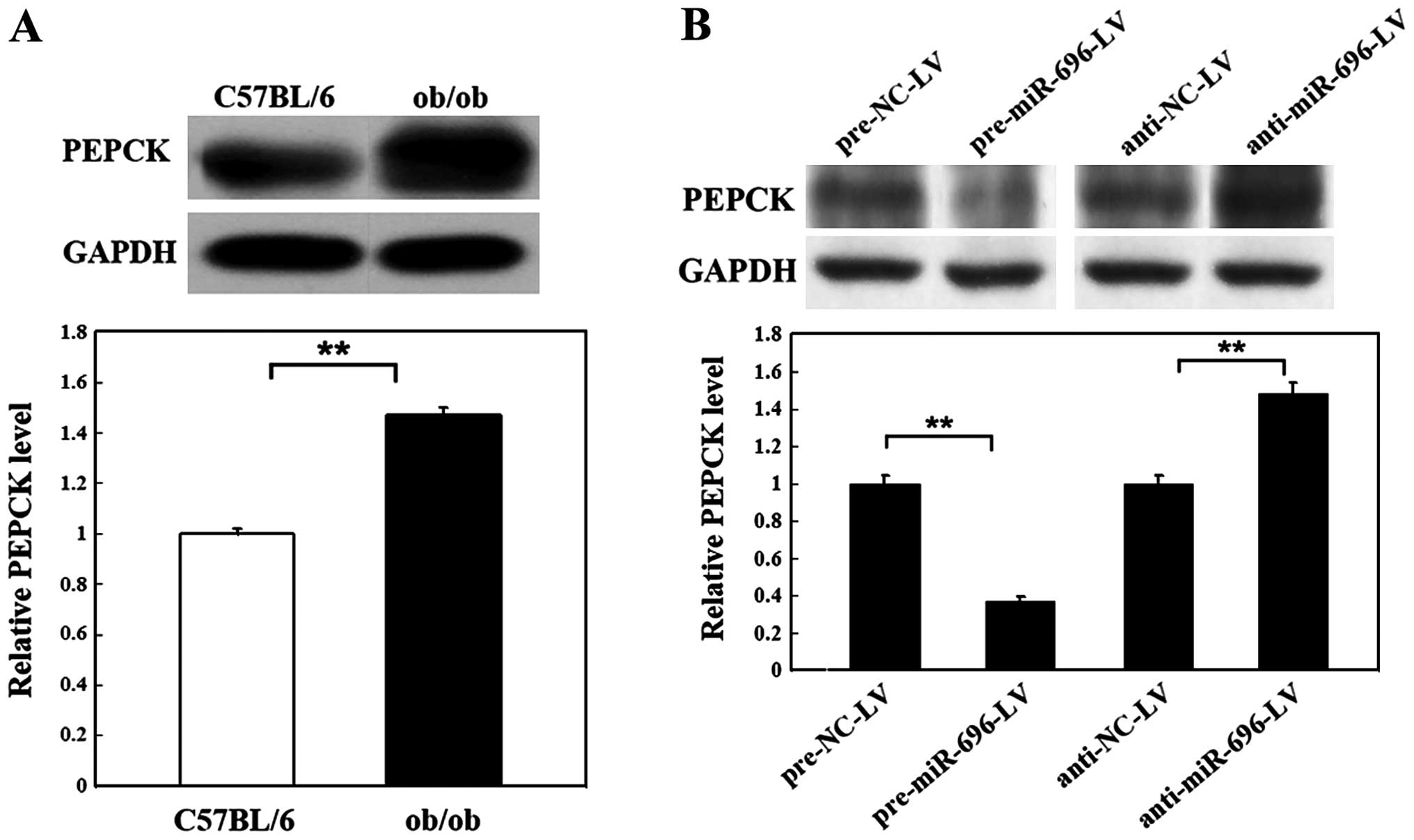
As shown in Fig. 5A, the protein
level of PEPCK was increased by 47% in the livers of ob/ob mice
compared with the normal C57BL/6 mice. We then analyzed the
molecular biological consequences of the miR-696-driven inhibition
of PGC-1α gene expression in mouse primary hepatocytes. At 72 h
following the overexpression or knockdown of miR-696, the
expression level of PEPCK was significantly inhibited by infection
with pre-miR-696-LV (only 0.37-fold that of the control). The PEPCK
level was significantly elevated by infection with anti-miR-696-LV
(1.51-fold that of the control) (Fig.
5B). These downstream effects further support the hypothesis
that miR-696 negatively regulates PGC-1α protein expression in the
mouse liver and then coordinates the expression of characteristic
hepatic gluconeogenic enzymes, including PEPCK through coactivating
transcription factors, such as HNF-4α, FOXO1, and therefore plays
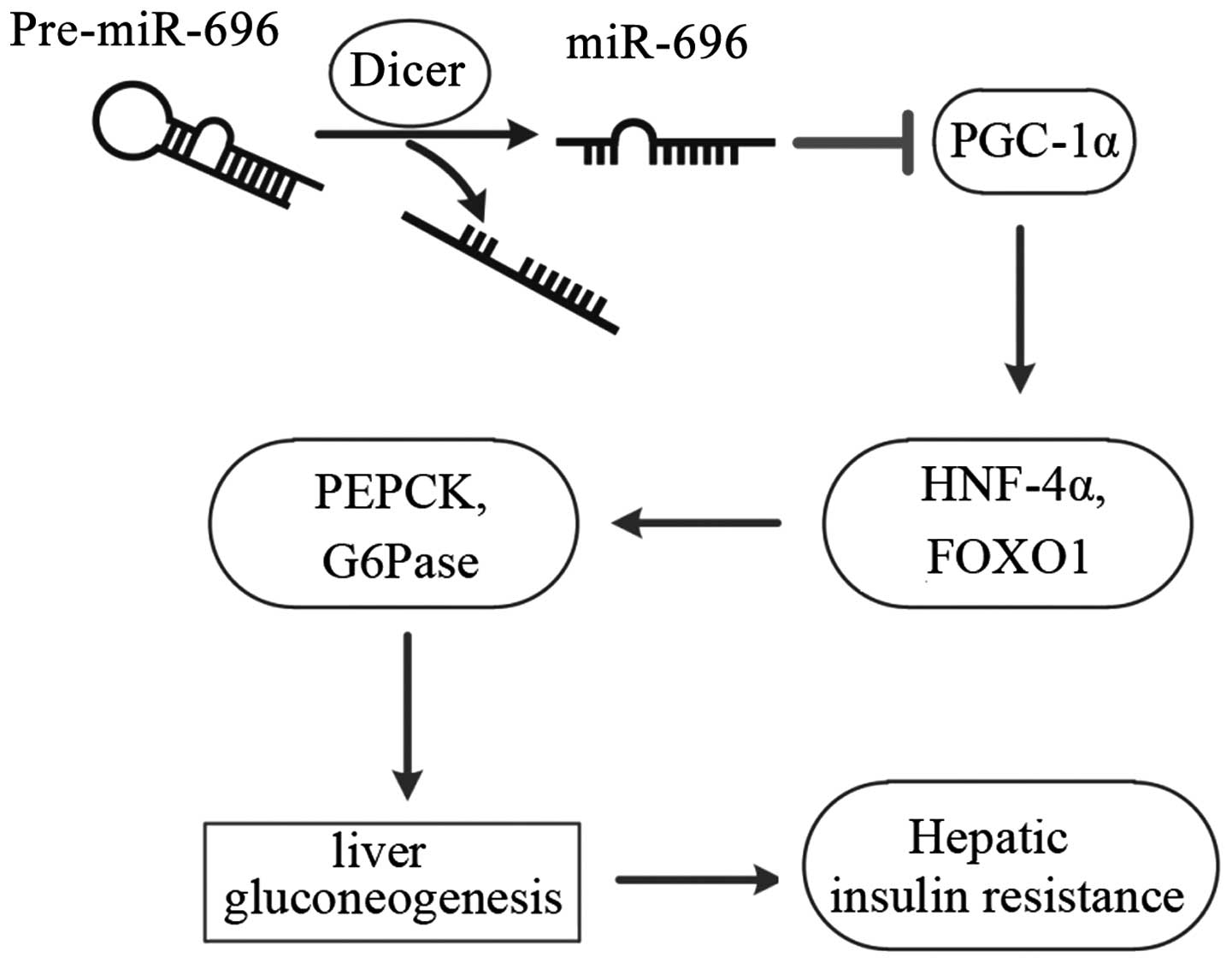
an important role in hepatic gluconeogenesis and IR (Fig. 6).
Discussion
In this study, we identified an inverse correlation
between miR-696 and PGC-1α in vivo and in vitro, and
found that PGC-1α expression was significantly decreased by the
overexpression of miR-696 through pre-miR-696-LV, whereas the
knockdown of miR-696 increased the protein level of PGC-1α. The
results of luciferase reporter assay confirmed that miR-696
directly recognizes a specific location within the 3′-UTR of PGC-1α
transcripts. The molecular biological consequences of the
miR-696-driven inhibition of PGC-1α expression were examined in the
livers of ob/ob mice and primary hepatocytes using PEPCK as an
indicator. We present convincing evidence that miR-696 plays a
significant role in hepatic gluconeogenesis and IR through the
inhibition of PGC-1α.
miRNAs are small non-coding transcripts that are
widely expressed in plants and animals to regulate gene expression
post-transcriptionally by the cleavage or translational repression
of their specific target mRNAs (28). It has been indicated by
bioinformatic studies that miRNAs have the potential to regulate
the expression of a wide spectrum of target genes (29). miRNAs have been reported to play
important roles in many diseases, including T2D (30,31). For example, the let-7 family
influence insulin sensitivity (32). Moreover, miRNAs such as miR-29,
miR-138, miR-143, miR-326 and miR-21 play an important role in
regulating glucose metabolism (33–37).
PGC-1α is known to be a potent regulator of
gluconeogenesis in the liver by coactivating transcription factors,
including HNF-4α, FOXO1. Aoi et al (25) recently reported that PGC-1α is
directly regulated by miR-696 in mouse skeletal muscle. Considering
these findings, we conducted our experiments with miR-696 to
determine its role in hepatic gluconeogenesis and IR via PGC-1α.
Our results demonstrated an inverse correlation between miR-696 and
PGC-1α in the livers of ob/ob mice compared with normal C57BL/6
mice. Additionally, an increase in miR-696 expression in primary
hepatocytes led to the downregulation of PGC-1α protein, whereas
the knockdown of miR-696 had the opposite effect on PGC-1α
expression. These observations strongly suggest that miR-696
negatively regulates the expression of PGC-1α in the liver.
The liver plays a significant role in maintaining
normal circulatory glucose levels and the gluconeogenic pathway is
a predominant contributor towards this phenomenon. PEPCK, as the
rate limiting enzyme of gluconeogenesis, catalyzes the conversion
of oxaloacetate to phosphoenolpyruvate. Taking into account that
the major downstream pathway of insulin function in the liver
culminates into the inhibition of gluconeogenic genes, we used
PEPCK as an important indicator and determined the effects of
miR-696 in hepatic gluconeogenesis on the status of PEPCK. As
compared to the control, the protein level of PEPCK was increased
by 47% in the livers of ob/ob mice compared with normal C57BL/6
mice. In primary hepatocytes, we also found that the expression of
PEPCK was suppressed by infecting mouse primary hepatocytes with
pre-miR-696-LV, and stimulated by infecting mouse primary
hepatocytes with anti-miR-696-LV. These results suggest that
miR-696 plays a vital role in hepatic gluconeogenesis and IR.
In conclusion, our in vitro and in
vivo experiments may help us to understand the importance and
mechanisms of action of miR-696 in regulating PGC-1α in hepatic
gluconeogenesis and IR, and provide hope for developing
miRNA-targeting agents for preventing and treating IR in T2D.
Acknowledgments
This study was supported by grants from the National
Natural Science Foundation of China (nos. 30570731, 30871195,
81070653, 81270907 and 81370926) to Yang Xiang.
Abbreviations:
|
T2D
|
type 2 diabetes
|
|
IR
|
insulin resistance
|
|
PPARγ
|
peroxisome proliferator-activated
receptor γ
|
|
PGC-1α
|
PPARγ coactivator-1α
|
|
miRNAs or miRs
|
microRNAs
|
|
HNF-4α
|
hepatocyte nuclear factor-4α
|
|
FOXO1
|
forkhead boxO1
|
|
PEPCK
|
phosphoenol-pyruvate carboxykinase
|
|
G6Pase
|
glucose-6-phosphatase
|
|
RT-qPCR
|
reverse transcription-quantitative
PCR
|
|
GFP
|
green fluorescent protein
|
References
|
1
|
Shaw JE, Sicree RA and Zimmet PZ: Global
estimates of the prevalence of diabetes for 2010 and 2030. Diabetes
Res Clin Pract. 87:4–14. 2010. View Article : Google Scholar
|
|
2
|
Olokoba AB, Obateru OA and Olokoba LB:
Type 2 diabetes mellitus: a review of current trends. Oman Med J.
27:269–273. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liang H, Balas B, Tantiwong P, Dube J,
Goodpaster BH, O'Doherty RM, DeFronzo RA, Richardson A, Musi N and
Ward WF: Whole body overexpression of PGC-1α has opposite effects
on hepatic and muscle insulin sensitivity. Am J Physiol Endocrinol
Metab. 296:E945–E954. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kraegen EW, Clark PW, Jenkins AB, Daley
EA, Chisholm DJ and Storlien LH: Development of muscle insulin
resistance after liver insulin resistance in high-fat-fed rats.
Diabetes. 40:1397–1403. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bajaj M and Defronzo RA: Metabolic and
molecular basis of insulin resistance. J Nucl Cardiol. 10:311–323.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yamada N: Molecular basis of metabolic
syndrome: insulin resistance. Nihon Rinsho. 64(Suppl 9): 23–29.
2006.In Japanese.
|
|
7
|
Russell AP: PGC-1alpha and exercise:
important partners in combating insulin resistance. Curr Diabetes
Rev. 1:175–181. 2005. View Article : Google Scholar
|
|
8
|
Handschin C and Spiegelman BM: Peroxisome
proliferator activated receptor gamma coactivator 1 coactivators,
energy homeostasis, and metabolism. Endocr Rev. 27:728–735. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xu W, Guo T, Zhang Y, Jiang X, Zhang Y,
Zen K, Yu B and Zhang CY: The inhibitory effect of dexamethasone on
platelet derived growth factor-induced vascular smooth muscle cell
migration through up-regulating PGC-1α expression. Exp Cell Res.
317:1083–1092. 2011. View Article : Google Scholar
|
|
10
|
Zhang Y, Liu C, Zhu L, Jiang X, Chen X, Qi
X, Liang X, Jin S, Zhang P, Li Q, et al: PGC-1alpha inhibits oleic
acid induced proliferation and migration of rat vascular smooth
muscle cells. PLoS One. 2:e11372007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jiang X, Zhang Y, Hou D, Zhu L, Xu W, Ding
L, Qi X, Sun G, Liu C, Zhang J, et al: 17beta-estradiol inhibits
oleic acid-induced rat VSMC proliferation and migration by
restoring PGC-1alpha expression. Mol Cell Endocrinol. 315:74–80.
2010. View Article : Google Scholar
|
|
12
|
Zhu L, Sun G, Zhang H, Zhang Y, Chen X,
Jiang X, Jiang X, Krauss S, Zhang J, Xiang Y and Zhang CY:
PGC-1alpha is a key regulator of glucose-induced proliferation and
migration in vascular smooth muscle cells. PLoS One. 4:e41822009.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Koo SH, Satoh H, Herzig S, Lee CH, Hedrick
S, Kulkarni R, Evans RM, Olefsky J and Montminy M: PGC-1 promotes
insulin resistance in liver through PPAR-alpha-dependent induction
of TRB-3. Nat Med. 10:530–534. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Puigserver P, Rhee J, Donovan J, Walkey
CJ, Yoon JC, Oriente F, Kitamura Y, Altomonte J, Dong H, Accili D
and Spiegelman BM: Insulin-regulated hepatic gluconeogenesis
through FOXO1-PGC-1α interaction. Nature. 423:550–555. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Rhee J, Inoue Y, Yoon JC, Puigserver P,
Fan M, Gonzalez FJ and Spiegelman BM: Regulation of hepatic fasting
response by PPARgamma coactivator-1α (PGC-1): requirement for
hepatocyte nuclear factor 4a in gluconeogenesis. Proc Natl Acad Sci
USA. 100:4012–4017. 2003. View Article : Google Scholar
|
|
16
|
Griffiths-Jones S, Grocock RJ, van Dongen
S, Bateman A and Enright AJ: miRBase: microRNA sequences, targets
and gene nomenclature. Nucleic Acids Res. 34:D140–D144. 2006.
View Article : Google Scholar :
|
|
17
|
Wang C, Yang C, Chen X, Yao B, Yang C, Zhu
C, Li L, Wang J, Li X, Shao Y, et al: Altered profile of seminal
plasma microRNAs in the molecular diagnosis of male infertility.
Clin Chem. 57:1722–1731. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu R, Zhang C, Hu Z, Li G, Wang C, Yang
C, Huang D, Chen X, Zhang H, Zhuang R, et al: A five-microRNA
signature identified from genome-wide serum microRNA expression
profiling serves as a fingerprint for gastric cancer diagnosis. Eur
J Cancer. 47:784–791. 2011. View Article : Google Scholar
|
|
19
|
Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K,
Guo J, Zhang Y, Chen J, Guo X, et al: Characterization of microRNAs
in serum: a novel class of biomarkers for diagnosis of cancer and
other diseases. Cell Res. 18:997–1006. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen X, Hu Z, Wang W, Ba Y, Ma L, Zhang C,
Wang C, Ren Z, Zhao Y, Wu S, et al: Identification of ten serum
microRNAs from a genome-wide serum microRNA expression pro?le as
novel noninvasive biomarkers for nonsmall cell lung cancer
diagnosis. Int J Cancer. 130:1620–1628. 2012. View Article : Google Scholar
|
|
21
|
Fernandez-Valverde SL, Taft RJ and Mattick
JS: MicroRNAs in β-cell biology, insulin resistance, diabetes and
its complications. Diabetes. 60:1825–1831. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Trajkovski M, Hausser J, Soutschek J, Bhat
B, Akin A, Zavolan M, Heim MH and Stoffel M: MicroRNAs 103 and 107
regulate insulin sensitivity. Nature. 474:649–653. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ling HY, Ou HS, Feng SD, Zhang XY, Tuo QH,
Chen LX, Zhu BY, Gao ZP, Tang CK, Yin WD, et al: CHANGES IN
microRNA (miR) profile and effects of miR-320 in insulin-resistant
3T3-L1 adipocytes. Clin Exp Pharmacol Physiol. 36:e32–e39. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gallagher IJ, Scheele C, Keller P, Nielsen
AR, Remenyi J, Fischer CP, Roder K, Babraj J, Wahlestedt C,
Hutvagner G, et al: Integration of microRNA changes in vivo
identifies novel molecular features of muscle insulin resistance in
type 2 diabetes. Genome Med. 2:92010. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Aoi W, Naito Y, Mizushima K, Takanami Y,
Kawai Y, Ichikawa H and Yoshikawa T: The microRNA miR 696 regulates
PGC-1{α} in mouse skeletal muscle in response to physical activity.
Am J Physiol Endocrinol Metab. 298:E799–E806. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Klaunig JE, Goldblatt PJ, Hinton DE,
Lipsky MM and Trump BF: Mouse liver cell culture. II. Primary
culture. In Vitro. 17:926–934. 1981. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yoon JC, Puigserver P, Chen G, Donovan J,
Wu Z, Rhee J, Adelmant G, Stafford J, Kahn CR, Granner DK, et al:
Control of hepatic gluconeogenesis through the transcriptional
coactivator PGC-1. Nature. 413:131–138. 2001. View Article : Google Scholar
|
|
28
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sayed D and Abdellatif M: MicroRNAs in
development and disease. Physiol Rev. 91:827–887. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Poy MN, Eliasson L, Krutzfeldt J, Kuwajima
S, Ma X, Macdonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P
and Stoffel M: A pancreatic islet-specific microRNA regulates
insulin secretion. Nature. 432:226–230. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Frost RJ and Olson EN: Control of glucose
homeostasis and insulin sensitivity by the Let-7 family of
microRNAs. Proc Natl Acad Sci USA. 108:21075–21080. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
He A, Zhu L, Gupta N, Chang Y and Fang F:
Overexpression of micro ribonucleic acid 29, highly up-regulated in
diabetic rats, leads to insulin resistance in 3T3-L1 adipocytes.
Mol Endocrinol. 21:2785–2794. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Pandey AK, Verma G, Vig S, Srivastava S,
Srivastava AK and Datta M: miR-29a levels are elevated in the db/db
mice liver and its overexpression leads to attenuation of insulin
action on PEPCK gene expression in HepG2 cells. Mol Cell
Endocrinol. 332:125–133. 2011. View Article : Google Scholar
|
|
35
|
Peschiaroli A, Giacobbe A, Formosa A,
Markert EK, Bongiorno-Borbone L, Levine AJ, Candi E, D'Alessandro
A, Zolla L, Finazzi Agrò A and Melino G: miR-143 regulates
hexokinase 2 expression in cancer cells. Oncogene. 32:797–802.
2013. View Article : Google Scholar
|
|
36
|
Kefas B, Comeau L, Erdle N, Montgomery E,
Amos S and Purow B: Pyruvate kinase M2 is a target of the
tumor-suppressive microRNA-326 and regulates the survival of glioma
cells. Neurooncol. 12:1102–1112. 2010.
|
|
37
|
Vinciguerra M, Sgroi A, Veyrat-Durebex C,
Rubbia-Brandt L, Buhler LH and Foti M: Unsaturated fatty acids
inhibit the expression of tumor suppressor phosphatase and tensin
homolog (PTEN) via microRNA-21 up-regulation in hepatocytes.
Hepatology. 49:1176–1184. 2009. View Article : Google Scholar
|