Introduction
Colorectal cancer (CRC) has become a major clinical
problem and ranks as the third most prevalent cancer and the fourth
leading cause of cancer-associated death worldwide (1). This is due to the limitations of
chemotherapy as a result of drug resistance and organ toxicities
(2). To overcome these serious
limitations, the design of novel approaches targeting
cancer-related signaling pathways is key to improving treatment
outcomes in colon cancer.
The Notch signaling pathway plays a vital role in
several cellular processes. Notch signaling is considered to play
an oncogenic role in the pathogenesis of CRC (3,4).
Previous studies have also revealed that Notch signaling is
deregulated in several types of human tumors, including colon
cancer (5,6), which implies that screening various
chemotherapeutic agents for their anti-colon cancer potency by
targeting the Notch pathway may be extremely valuable. In humans,
the Notch signaling pathway has four receptors (Notch-1, -2, -3,
and -4) and five ligands (Jagged-1, Jagged-2, Delta-1, Delta-3 and
Delta-4) (7). The interactions
between these molecules determines the fate of the cell (8). Notch receptor-ligand interactions
lead to proteolytic cleavage of Notch by γ-secretase and other
proteases, which causes the release of the Notch intracellular
domain (NICD) from the plasma membrane and initiates its subsequent
translocation into the nucleus. In the nucleus, the NICD binds to
one of three cofactors, CBF-1/Suppressor of Hairless/Lag-1 (CSL),
mastermind-like (MAML)-1 and p300/CBP, to generate a complex that
acts as a transcriptional coactivator (3,9).
This complex then activates the transcription of target genes, such
as Hes-1 and Hey-1 (10,11). Thus, chemotherapeutic agents that
more specifically block the Notch signaling pathway to inhibit the
proteolytic cleavage activity of γ-secretase would be an essential
development in cancer therapy.
Niclosamide, an anthelminthic drug, has been used in
the treatment of tapeworm infections for approximately five decades
(12). Several studies have
independently reported that niclosamide is active against cancer
cells, including leukemia (13),
colon cancer (14), glioblastoma
(15) and breast cancer (16), although its precise mechanism of
antitumor action remains only partially understood. Due to this
lack of understanding, we performed this study in order to
determine whether niclosamide triggers the inactivation of the
Notch pathway, which in turn may contribute to the increased
expression of members of the microRNA (miRNA or miR)-200 family
that are suppressed in colon cancer.
Mounting evidence indicates that cross-talk occurs
between miRNAs and the Notch signaling pathway during
carcinogenesis (17). miRNAs are
a class of small, evolutionary conserved, non-coding RNAs that bind
to the 3′ untranslated region of messenger RNAs (mRNAs) in order to
regulate gene expression (18–20). miRNAs are involved in various
regulatory cellular functions including cell proliferation,
differentiation and apoptosis (21). They regulate the Notch signaling
pathway and Notch-related genes and affect various types of cancer
(20). Moreover, some miRNAs have
been reported to be oncomirs, which act either as oncogenes or as
oncosuppressors (22). This
indicates that further understanding of the Notch signaling-miRNA
circuit may be critical for the discovery of novel chemotherapeutic
agents for the treatment of colon cancer.
Five members of the miR-200 family, miR-200a,
miR-200b, miR-200c, miR-141 and miR-429, which are located on
chromosomes 1 and 12 in humans, have been identified (20,23). miRNA-200 family members play a
vital role in suppressing cancer metastasis by inhibiting the
epithelial-mesenchymal transition (EMT). Decreased levels of
miR-200c and miR-141 are associated with the increased expression
of the transcription factor ZEB-1 (20), which in turn activates the Notch
signaling pathway by targeting Notch ligand Jagged-1 and
mastermind-like co-activators MAML-2 and -3 (24). Through suppression of the
miRNA-200 family, ZEB-1 upregulates the Notch signaling pathway in
tumor cells (24). ZEB-1 is an
EMT activator, and its expression promotes tumor metastasis by
inducing EMT activation (25). It
has previously been shown that overexpression of the miRNA-200
family may inhibit EMT through increased E-cadherin expression
targeting ZEB1 and ZEB2 (23).
Low levels of miR-141 and miR-200c affect stem cell properties and
drug resistance in pancreatic adenocarcinoma and basal breast
cancer types (24). Furthermore,
it has been shown that re-expression of miR-141 and miR-429 hinders
Jagged-1 in metastatic prostate cancer cells, which suggests a
novel means of controlling the fate of the Notch pathway (26). A study by Wang et al
revealed that transfection of miR-200b in Rink-1 cells (pancreatic
cell line) inactivated the Notch signaling pathway directly by
decreasing the levels of Jagged-1/2 and those of their target genes
Hes-1, Hey-2 and Bcl-2, which led to the inhibition of cell growth
(27). Taken together, these
findings show the connection among Notch signaling, miRNA-200 and
ZEB in cancer. Nevertheless, a more in-depth investigation is
warranted in order to understand how the miR-200 family directly
regulates the Notch signaling pathway.
Collectively, these findings suggest that
pharmacologic inactivation of Notch signaling with niclosamide may
have potential therapeutic applications in the treatment of colon
cancer. Herein, we report for the first time, to the best of our
knowledge, a novel mechanism by which niclosamide inhibits colon
cancer progression through upregulating the tumor suppressor
miRNA-200 family and suppressing the Notch pathway. This strategy
may be of therapeutic value in colon cancer and provide the basis
for the development of specific inhibitors.
Materials and methods
Cell lines and cell culture
Human colon cancer cell lines LoVo and SW620 were
purchased from the Cell Bank of the Shanghai Institutes for
Biological Sciences, Chinese Academy of Sciences (Shanghai, China).
All LoVo, SW620 and HCT116 (China Infrastructure of Cell Line
Resources, Beijing, China) cells were grown in RPMI-1640 medium
supplemented with 10% heat-inactivated fetal bovine serum (FBS) and
1% penicillin and streptomycin. The cell lines were maintained in a
humidified incubator at 37°C with an atmosphere of 5%
CO2.
Reagents and antibodies
Niclosamide was obtained from Sigma-Aldrich (St.
Louis, MO, USA).
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT;
also known as thiazolyl blue and methylthiazolyldiphenyl
tetrazolium bromide) was purchased from Sigma-Aldrich. The Annexin
V-FITC Apoptosis Detection kit was purchased from Vazyme Biotech
(Nanjing, China). Dimethyl sulfoxide (DMSO) was obtained from
BioSharp (Hefei, China). The primary antibodies, rabbit polyclonal
anti-Notch1 (ab27526), rabbit polyclonal anti-Notch2 (ab8926),
rabbit polyclonal to anti-Notch3 (ab23426) and rabbit polyclonal
anti-Hey1 (ab22614) were purchased from Abcam (Cambridge, UK).
Mouse anti-β-actin monoclonal antibody (TA-09) was purchased from
ZSGB-BIO (Beijing, China). The secondary antibodies,
peroxidase-conjugated AffiniPure goat anti-rabbit IgG (ZB-2301) and
peroxidase-conjugated AffiniPure goat anti-mouse IgG (ZB-2305),
were purchased from ZSGB-BIO.
Cell proliferation assay
For the cell proliferation assay, HCT116, LoVo and
SW620 cells (2–3×103) were seeded in 96-well plates (one
plate for each day) and incubated for 24 h to allow the cells to
attach to the bottom of the wells. The cells were then treated with
various concentrations of niclosamide or the corresponding volume
of DMSO for 24, 48 and 72 h. To determine cell proliferation, MTT
was added at a final concentration of 0.5 mg/ml and incubated for 3
h at 37°C and 5% CO2 in a humidified incubator.
Crystallized MTT was resolved by 1:1 DMSO and isopropanol, and the
absorption of each well was measured at 570 nm using a microplate
spectrophotometer (xMark: BioRad, Berkeley, CA, USA). In addition,
the morphology of the cells was observed under an inverted phase
contrast microscope, and images were captured with the microscope
(1X71; Olympus, Tokyo, Japan).
Wound-healing assay
For the wound-healing assay, HCT116, LoVo and SW620
cells were grown to confluence in 6-well plates. A perpendicular
scratch wound was generated with a 200 µl pipette tip, and
the wells were washed twice with phosphate-buffered saline (PBS) to
remove the detached cells; the cells were then incubated with 1
µM niclosamide or DMSO for 48 h. Images of each well were
captured at 0 and 48 h. Wound healing was quantified by measuring
the distance between the wound edges using Image-Pro Plus 6.0
software.
Transwell (Boyden chamber) migration
assay
A Boyden chamber (8 mm pore size; Corning, Inc.,
Acton, MA, USA) was used to perform the assay. A total of
2.5×105 HCT116, LoVo or SW620 cells in 200 µl
serum free medium were added to the top chamber and treated with
different concentrations of niclosamide. Then, 600 µl medium
with 20% FBS was added to the bottom chamber to act as the
chemoattractant. The cells were allowed to migrate for 24 h.
Non-migrated cells in the top chamber were removed. The migrated
cells were fixed in 4% paraformaldehyde and stained with 0.5%
crystal violet (Biosharp, Hefei, China). The migrated cells were
counted and images were captured under an inverted microscope.
Apoptotic assay
To examine the apoptosis inducing effect of
niclosamide, an Annexin V-FITC apoptosis detection kit (Vazyme
Biotech) was used. The cells, HCT116 and SW620 (4×105
cells/well) were seeded in a 6-well plate overnight and treated
with niclosamide at the indicated concentrations. After the 48-h
treatments, the cells were harvested and washed with cold PBS
twice. Apoptosis levels were studied using the apoptosis detection
kit according to the manufacturer's instructions by flow cytometry.
The data were then analyzed using FlowJo software.
Western blot analysis
For total protein extraction, HCT116, LoVo and SW620
cells (4×105) were seeded overnight in 6-well plates and
treated with niclosamide. After 48 h, the cells were lysed with a
total protein extraction kit (KeyGen, Nanjing, China). To assess
the total protein concentration, a BCA protein assay was performed
using a BCA protein assay kit (Beyotime Biotechnology, Shanghai,
China) according to the manufacturer's instructions. The lysates of
equal protein concentrations were then separated by sodium dodecyl
sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and
transferred to nitrocellulose membranes using a wet transfer
system. The membranes were incubated in blocking solution
containing 5% nonfat dry milk in Tris-buffered saline containing
0.1% v/v Tween-20 (TBST) for 1 h at room temperature. The membranes
were incubated overnight at 4°C with rabbit anti-human Notch1
polyclonal antibodies (dilution, 1:500), rabbit anti-human Notch2
polyclonal antibodies (dilution, 1:1,000), rabbit anti-human Notch3
polyclonal antibodies (dilution, 1:500), rabbit anti-human Hey1
polyclonal antibodies (dilution, 1:200) and mouse anti-β-actin
polyclonal antibodies (dilution, 1:500), followed by incubation for
1 h at room temperature with HRP-conjugated anti-rabbit antibody
(dilution, 1:5,000) or anti-mouse IgG antibody (dilution, 1:2,000).
The antibody protein complexes were visualized with an
electrochemical luminescence reagent (ECL system; Pierce; Thermo
Fisher Scientific, Rockford, IL, USA) according to the
manufacturer's instructions, and subsequent exposure to ImageQuant
LAS 500 (GE Healthcare Life Sciences, Shanghai, China) for 30 sec
to 10 min.
miRNA expression analysis
To determine the effect of niclosamide on the
expression of the miR-200 family (miR-200a, miR-200b, miR-200c,
miR-141 and miR-429) in colon cancer cells, the cells were seeded
overnight in 6-well plates and treated with niclosamide. After 48
h, total RNA from the cultured cells was extracted using TRIzol
reagent (Takara Biotech, Beijing, China) according to the
manufacturer's instructions. To detect mature miRNA expression,
reverse transcription-quantitative polymerase chain reaction
(RT-qPCR) was performed. The extracted RNA was reverse transcribed,
and cDNA was synthesized using an miRcute miRNA First-Strand cDNA
synthesis kit (Tiangen Biotech, Beijing, China) according to the
manufacturer's instructions. qPCR was then conducted with an
miRcute miRNA qPCR detection kit (SYBR-Green I) (Tiangen Biotech).
U6 was used as an endogenous control. The following PCR cycling
conditions were used: 95°C for 30 sec followed by 40 cycles of 95°C
for 5 sec and 60°C for 30 sec. In addition, melting curves were
used to estimate non-specific amplification. The relative
expression level was calculated using the ΔΔCt method. The primer
sequences used in this study are presented in Table I.
 | Table IList of miRNAs and mature miRNA
sequences used in this study. |
Table I
List of miRNAs and mature miRNA
sequences used in this study.
| miRNA | Mature miRNA
sequence |
|---|
|
hsa-miR-200a-3p-F |
GCGCTAACACTGTCTGGTAACGATGT |
|
hsa-miR-200b-3p-F |
GCCGTAATACTGCCTGGTAATGATGA |
|
hsa-miR-200c-3p-F |
CGTAATACTGCCGGGTAATGATGGA |
|
hsa-miR-141-3p-F |
GCGGTAACACTGTCTGGTAAAGATGG |
|
hsa-miR-429-3p-F |
GCGGTAATACTGTCTGGTAAAACCGT |
| U6-F | CTCGCTTCGGCACA |
| U6-R |
AACGCTTCACGAATTTGCGT |
Statistical analysis
All statistical analyses were performed using
GraphPad Prism 5 software. A one-way analysis of variance (ANOVA)
was used for comparisons among the groups, and a two-way ANOVA was
used to compare two independent variables among the groups,
followed by Tukey's test to compare all pairs of columns. The data
are shown as the means ± SE; P<0.05 was considered to indicate a
significant difference.
Results
Niclosamide affects the growth and
morphology of colon cancer cells
To determine whether niclosamide exerts direct
effects on colon cancer cells, we performed MTT assays to examine
the inhibitory effect of niclosamide on the proliferation of
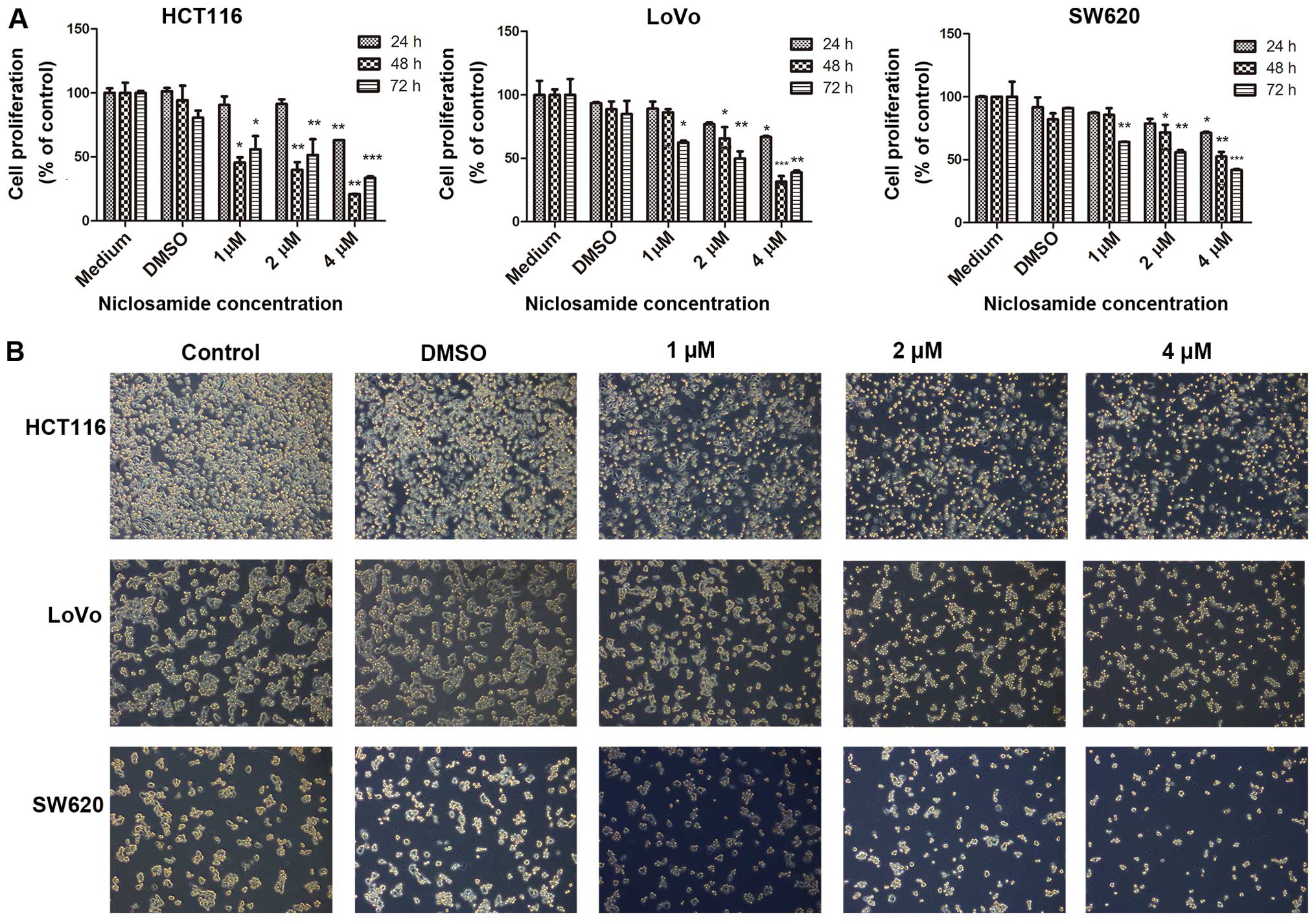
different colon cancer cell lines. As shown in Fig. 1A, progressive inhibition of the
growth of the HCT116, LoVo and SW620 cell lines occurred following
treatment with niclosamide at concentrations of 1, 2 and 4
µM for different time periods (24, 48 and 72 h). As shown in
Fig. 1A, there was no significant
difference in the viability of cells treated with 0.1% DMSO and the
control cells grown with medium only; therefore, we used both as
the controls for our subsequent experiments. Niclosamide
significantly suppressed the proliferation of colon cancer cells
after 24 h at a dose of 4 µM, which was followed by further
significant suppression over the next 48 and 72 h in a dose- and
time-dependent manner. The analysis of cell morphology revealed
that niclosamide treatment resulted in a dose-dependent change in
cellular morphology, with a stretched morphology at lower
niclosamide concentrations and a rounded morphology at higher
niclosamide concentrations (Fig.
1B).
Niclosamide induces the apoptosis of
colon cancer cells
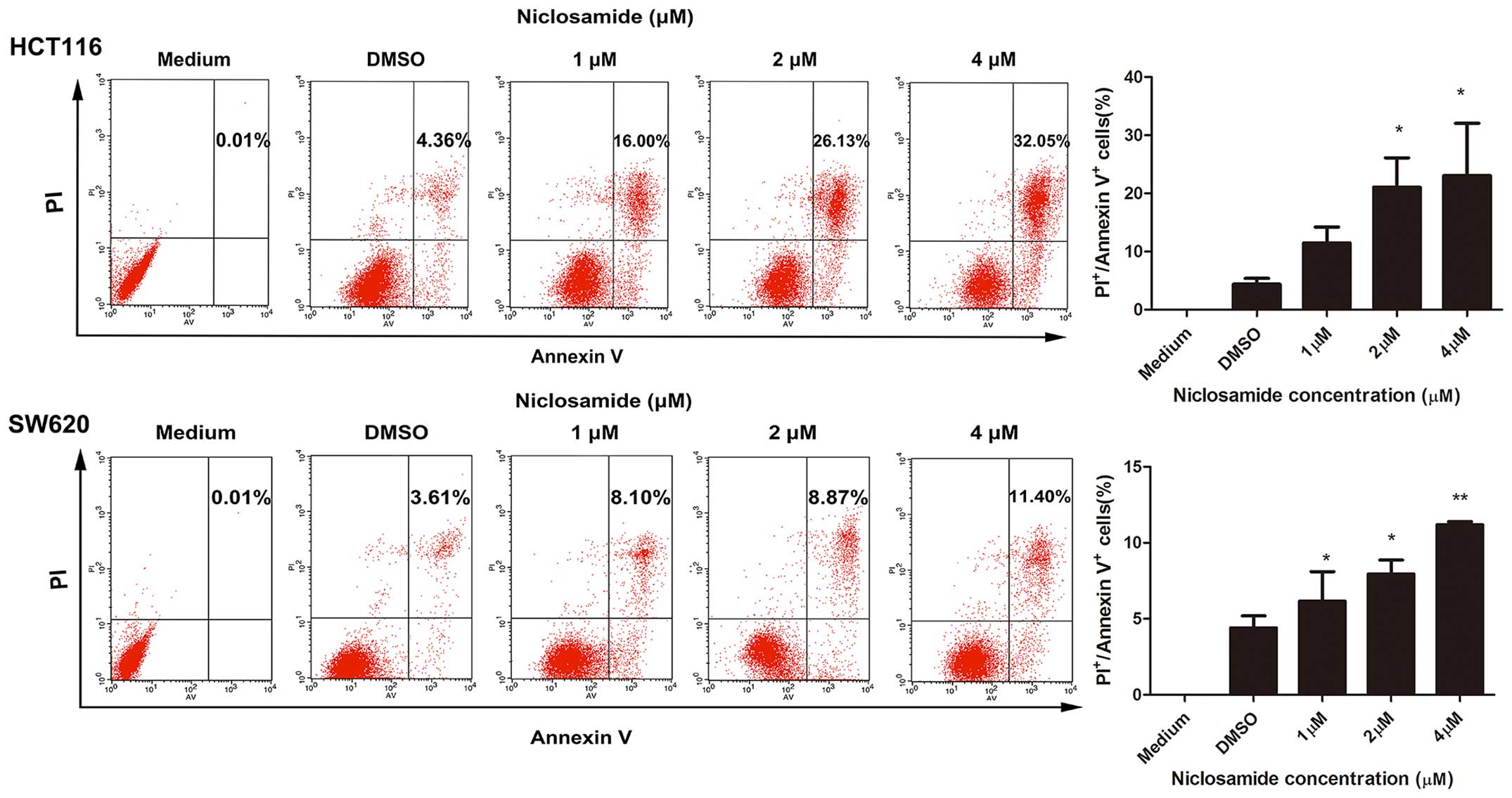
It has been reported that niclosamide induces the
apoptosis of breast cancer cells (16). To determine whether niclosamide
induced the apoptosis of colon cancer cells, we performed an
apoptosis assay using the Annexin V-FITC apoptosis detection kit.
HCT116 and SW620 cells were treated with 1, 2 and 4 µM
niclosamide, with medium and DMSO serving as controls, for 48 h.
Following treatment, the apoptosis-inducing effect of niclosamide
was clearly observed in both cell lines (Fig. 2). The induction of apoptosis was
dose-dependent. Specifically, niclosamide at concentrations of 1, 2
and 4 µM induced apoptosis rates of 16±2.763, 26.13±5.095
and 32.05±9.015%, respectively, in the HCT116 cells. Similarly,
niclosamide at concentrations of 1, 2 and 4 µM triggered
apoptosis at rates of 8.10±1.945, 8.87±0.9300 and 11.40±0.2000%,
respectively, in the SW620 cells. These results clearly indicate
that niclosamide induces the apoptosis of colon cancer cells.
Niclosamide decreases the migration of
colon cancer cells
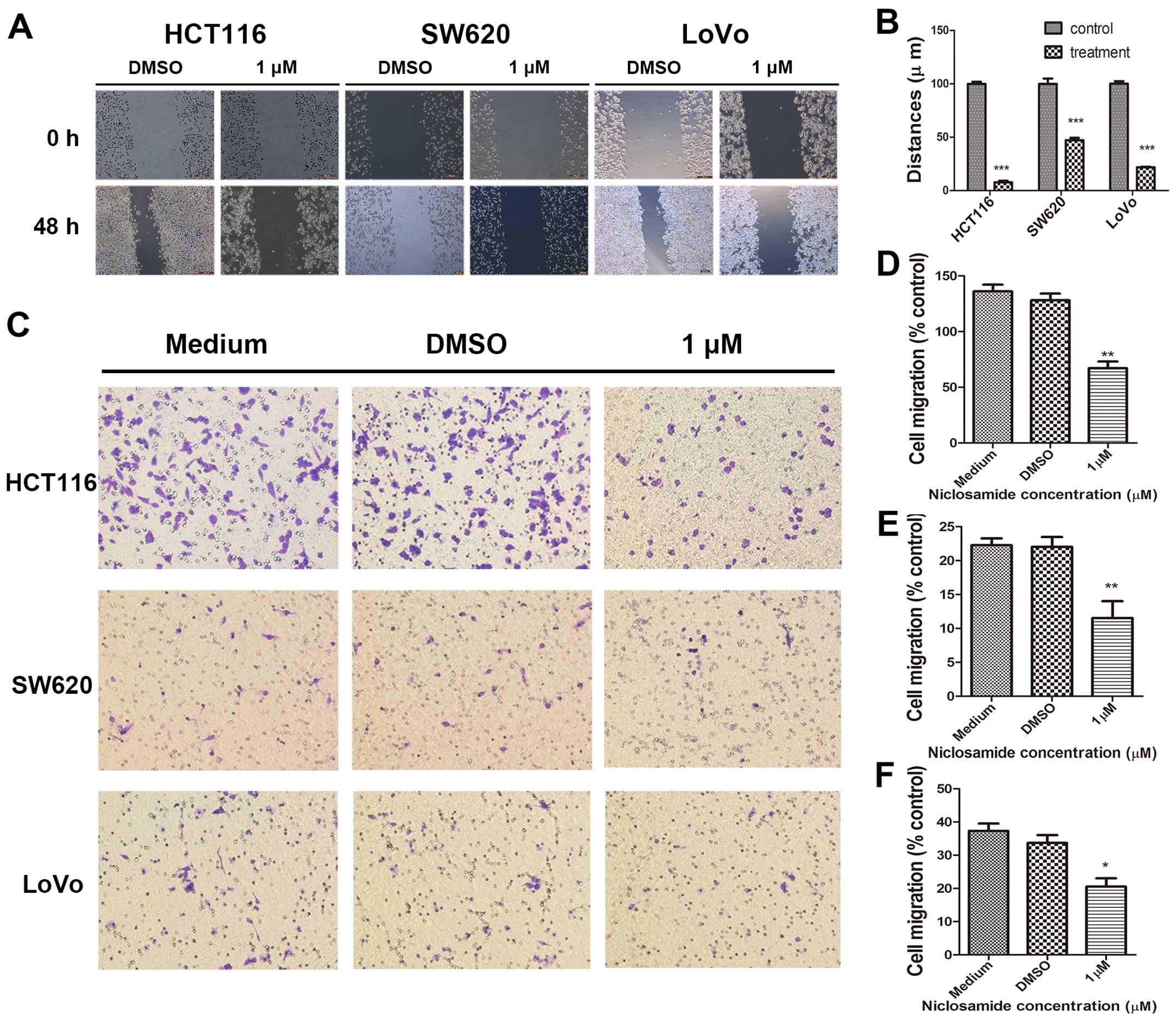
Tumor cell migration remains one of the key steps in
successful cancer metastasis. Therefore, to determine whether
niclosamide inhibits colon cancer cell migration, we conducted
wound-healing assays on HCT116, SW620 and LoVo cells. As shown in
Fig. 3A, niclosamide treatment at
a concentration of 1 µM, significantly decreased the wound
healing capacity of HCT116, SW620 and LoVo cells compared with the
control cells after 48 h. In the HCT116 cells, niclosamide
treatment decreased wound closure by 92.337±1.356% compared with
the control. Niclosamide treatment inhibited wound closure by
52.85±2.129% in SW620 cells. Similarly, niclosamide treatment in
LoVo cells decreased their wound healing ability by 78.19±0.4531%
(Fig. 3B). To confirm these
results, we also performed a Transwell migration assay and found
that niclosamide inhibited the migration of the HCT116, SW620 and
LoVo cells after 24 h (Fig. 3C).
At a 1 µM concentration, niclosamide significantly decreased
migration by 49.26±4.412% in the HCT116 cells compared with the
control (Fig. 3D). In addition,
niclosamide decreased migration by 54.49±6.696% in the SW620 cells
(Fig. 3E). Furthermore, in the
LoVo cells, migration was inhibited by 54.49±6.696% with 1
µM niclosamide (Fig. 3F).
These results show that niclosamide suppresses the migration of
colon cancer cells.
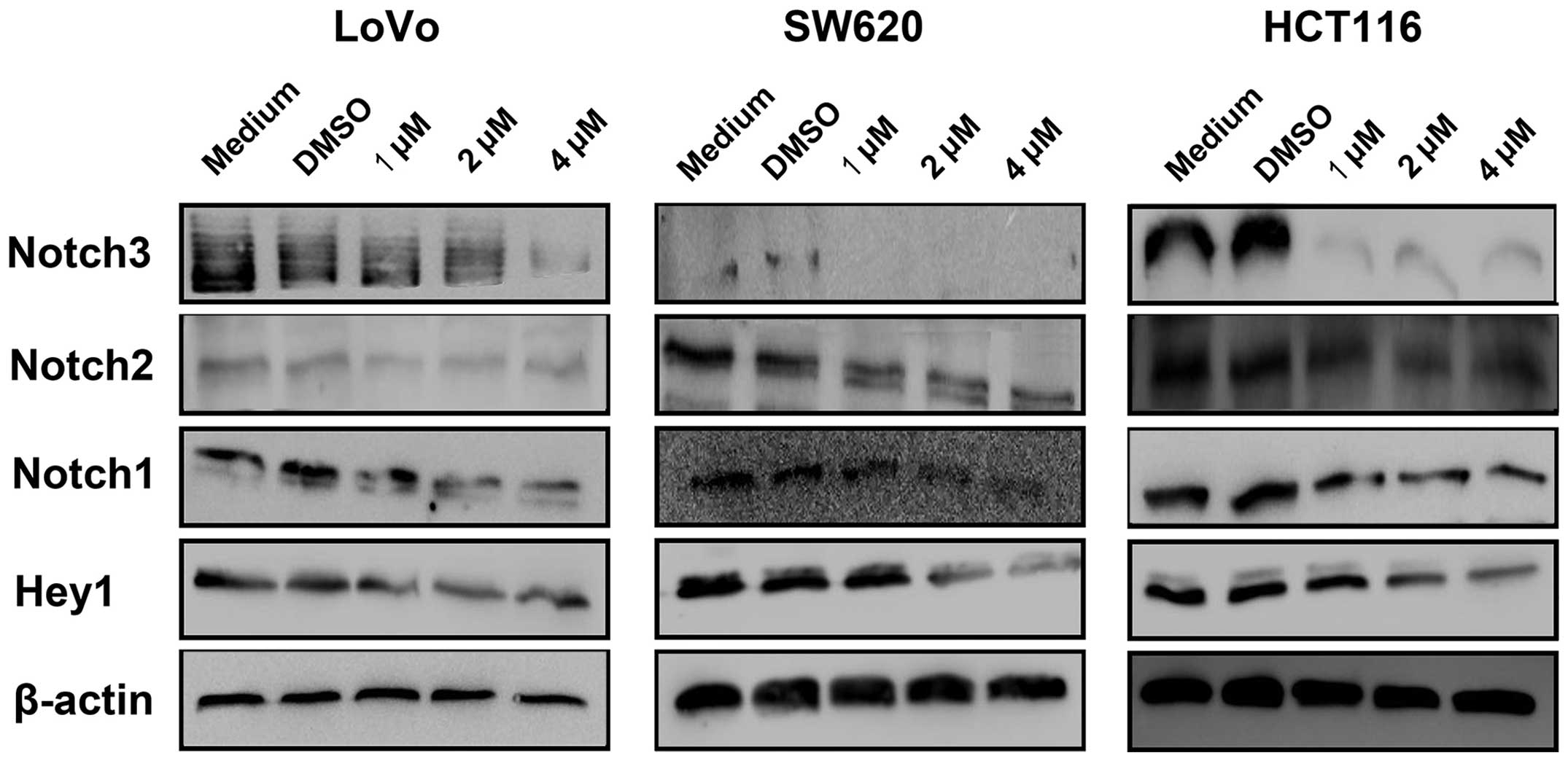
Niclosamide downregulates the Notch
signaling pathway
Previous studies have indicated that the Notch
pathway regulates cell proliferation and migration and induces
apoptosis during cancer development (28,29). In this study, we examined whether
niclosamide affected the Notch pathway to inhibit the
proliferation, decrease the migration and induce the apoptosis of
colon cancer cells. The expression of Notch1, Notch2, Notch3 and
the downstream target gene Hey-1 in HCT116, SW620 and LoVo cells
was assessed using western blot analysis following the 48-h
treatment of colon cancer cells with different niclosamide
concentrations (1, 2 and 4 µM). Western blot analysis
demonstrated that niclosamide treatment resulted in reduced levels
of the Notch receptors and Hey1 in the colon cancer cells (Fig. 4).
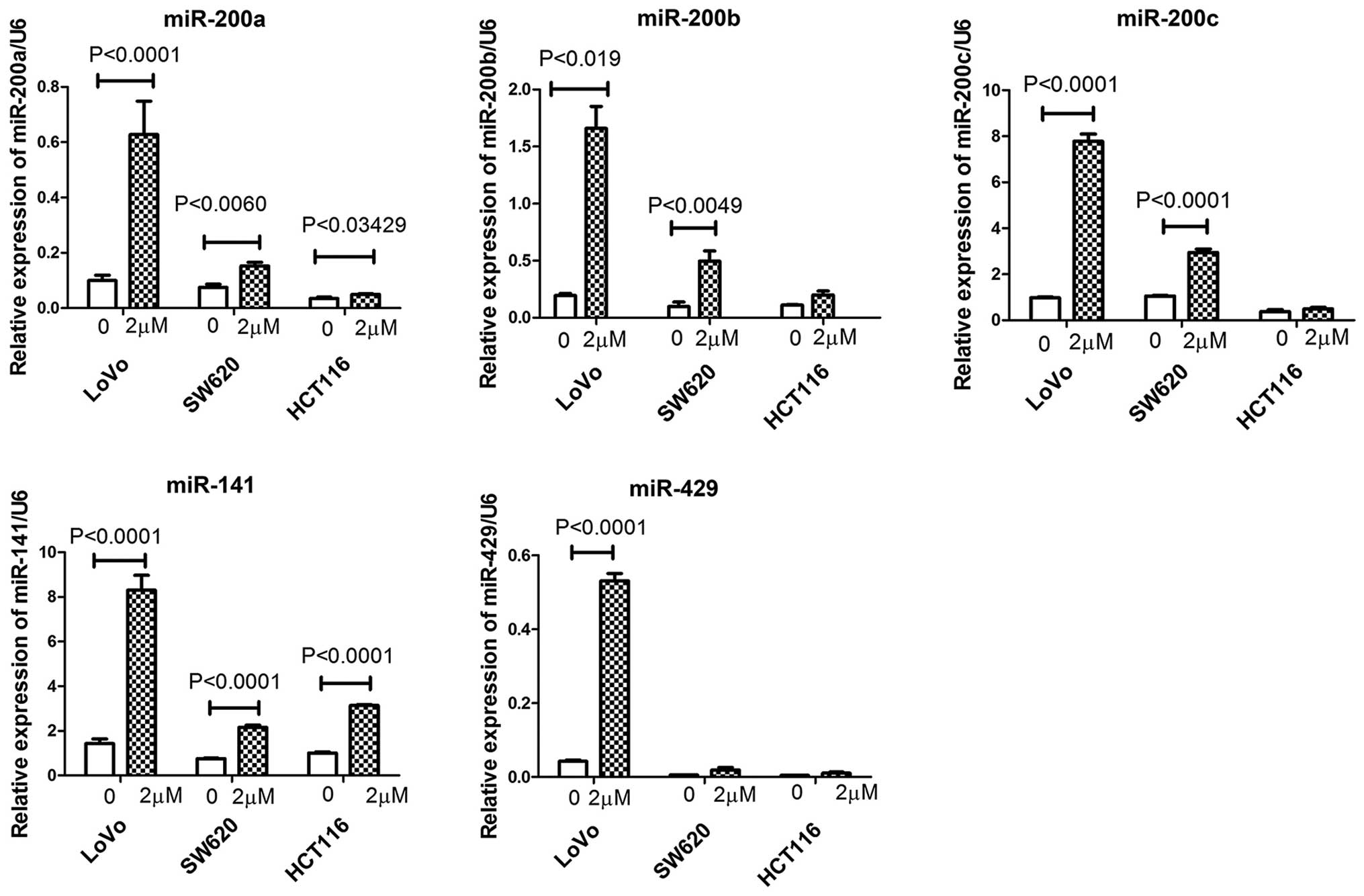
Effect of niclosamide on the expression
of miRNAs in colon cancer cells
Previous studies have shown that the miR-200 family
plays a role in the carcinogenesis of colon cancer (30) and that cross-talk occurs between
miRNAs and Notch signaling pathways during the development and
progression of tumors (17).
Therefore, we investigated whether niclosamide treatment regulated
the expression of the miR-200 family (miR-200a, miR-200b, miR-200c,
miR-141 and miR-429) in colon cancer cells using RT-qPCR. Following
a 48-h treatment of the colon cancer cells with niclosamide at a
concentration of 2 µM, we found that niclosamide treatment
increased the relative expression of miR-200a and miR-141 in the
HCT116, SW620 and LoVo colon cancer cells (Fig. 5). Niclosamide increased the
relative expression of miR-200b and miR-200c in the LoVo and SW620
cells but not in the HCT116 cells. Notably, niclosamide treatment
increased the relative expression of miR-429 only in the LoVo cells
but not in the HCT116 and SW620 cells. These data indicate that
niclosamide differentially regulates the expression of these
tumor-associated miRNAs in colon cancer cells.
Discussion
CRC has become a major problem and is one of the
most frequent causes of cancer-related death worldwide (1). It remains incurable, which indicates
the need for novel therapeutic strategies. For nearly five decades,
niclosamide has been used for the treatment of intestinal parasite
infections in humans (31).
Notably, a number of studies have shown that it also has potential
anticancer activity in both in vitro and in vivo
experiments through the inactivation of multiple signaling pathways
(13,38); however, the underlying molecular
mechanisms remain poorly understood. In the present study, we aimed
to explore the potential anticancer activity of niclosamide in
three colon cancer cell lines (LoVo, SW620 and HCT116) and to
elucidate the underlying molecular mechanism responsible for the
suppressive effects of niclosamide in colon cancer. Under normal
conditions, the activated Notch signaling pathway plays a vital
role in controlling the apoptosis, differentiation and
proliferation of cells (32).
Accumulating evidence indicates that inappropriate activation of
the Notch signaling plays a critical role in the pathogenesis of
cancer (33). Recent studies have
revealed that Notch receptors and their ligands are aberrantly
activated in many types of human cancers, including colon cancer
(34,35). Thus, considerable effort has been
made to block this pathway with an increasing range of
pharmacologic agents, primarily through the inhibition of Notch
cleavage (36). Therefore, the
downregulation of the Notch signaling pathway may be a novel
approach for the treatment of colon cancer.
Multiple studies by different groups have shown that
niclosamide inhibits several molecules and signaling pathways,
including the nuclear factor-κB (NF-κB) signaling pathway (13), the mTOR-signaling pathway
(37), the Wnt/β-catenin
signaling pathway (38), the
signal transducers and activators of the transcription 3 (Stat3)
signaling pathway (16), and the
Notch signaling pathway (39).
However, the effects of niclosamide on colon cancer cells and the
mechanisms responsible for these effects are not completely
understood. In this study, we observed that colon cancer cells
exposed to niclosamide treatment experienced growth inhibition and
apoptosis induction, as demonstrated by the MTT assay and Annexin
V-FITC apoptosis detection assay, respectively, which suggests that
this treatment has therapeutic implications for the management of
colon cancer. To explore the mechanism of increased apoptosis
induced by niclosamide, we investigated the activity of the Notch
pathway, which plays a key role in inhibiting the apoptotic
response. We demonstrated for the first time, to the best of our
knowledge, that niclosamide downregulates the expression of the
Notch receptors (Notch1, Notch2 and Notch3) as well as the
downstream target gene Hey1, which is likely to inhibit the
development of colon cancer. Thus, we hypothesized that the
possible novel mechanism responsible for the triggering of
apoptosis by niclosamide may be due to the inhibition of the Notch
gene expression. Notch signaling genes are functionally active and
highly expressed, and suppress apoptosis and promote cell growth in
colon cancer (5,40). Moreover, the results of the
Transwell assay showed that niclosamide, at a low concentration of
1 µM, inhibited the migration of colon cancer cells in
vitro, which suggests that it may be a possible candidate for
treating colon cancer metastasis. As the activation of Notch
signaling may affect the upregulation of various other signaling
pathways, such as the NF-κB, PI3K/AKT, c-MYC and EGFR pathways
(41), an effective and in-depth
mechanistic study of the effects of niclosamide on these pathways
through Notch signaling may be crucial for the design of novel
therapeutic approaches for the treatment of colon cancer. In this
regard, our findings may serve as a base for further study of the
anti-colon cancer effects of niclosamide due to the inactivation of
Notch signaling affecting multiple target genes in these
pathways.
miRNAs are key players that function as endogenous
post-transcriptional gene controllers to mediate protein synthesis
or mRNA stability (18,19). It has been demonstrated that a
large number of miRNAs are associated with tumor growth and
progression by regulating the expression and transcription of many
tumor-associated genes (22,42,43). Several studies have suggested that
miR-200 serves as a potential tumor suppressor, primarily by
repressing the acquisition of the EMT phenotype during tumor
development and progression (44,45). The decreased expression of miR-200
has been observed in many tumors, such as pancreatic, breast and
prostate cancer, which is associated with tumor invasion and
metastasis (46). For the first
time, to the best of our knowledge, this study demonstrated that
the miR-200 family was downregulated in colon cancer cells and that
inactivation of the Notch pathway by niclosamide led to increased
expression of the miR-200 family (miR-200a, miR-200b, miR-200c,
miR-141 and miR-429). This suggests that the Notch pathway is
involved in the regulation of the miR-200 family, and that
niclosamide may induce the re-expression of these miRNAs, which
would be highly valuable in attenuating the aggressiveness of tumor
cells.
In conclusion, these experiments provide mechanistic
evidence that the Notch pathway is inhibited in colon cancer cells
in response to niclosamide treatment. The downregulation of Notch
signaling by niclosamide is accompanied by the upregulation of
tumor suppressor miRNAs (miR-200a, miR-200b, miR-200c, miR-141 and
miR-429). Therefore, we propose that the inhibition of Notch
signaling is a novel strategy that may be used to impede the
induction of cancer survival mechanisms in colon cancer. However,
further in-depth, preclinical experiments using appropriate animal
models are warranted.
Acknowledgments
The present study was supported by grants from the
Chinese National Science Foundation Projects (nos. 81372669,
31270867 and 31470800), the Chinese State Key Program in Basic
Research (no. 2012CB822103), the Science and Technology Planning
Project of Liaoning province, China (no. 2012225020), and the
Project of Chinese Ministry of Health (no. W2012RQ23).
References
|
1
|
Brenner H, Kloor M and Pox CP: Colorectal
cancer. Lancet. 383:1490–1502. 2014. View Article : Google Scholar
|
|
2
|
Miyamoto S, Nakanishi M and Rosenberg DW:
Suppression of colon carcinogenesis by targeting Notch signaling.
Carcinogenesis. 34:2415–2423. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Katoh M and Katoh M: Notch signaling in
gastrointestinal tract (Review). Int J Oncol. 30:247–251. 2007.
|
|
4
|
Prasetyanti PR, Zimberlin CD, Bots M,
Vermeulen L, Melo FS and Medema JP: Regulation of stem cell
self-renewal and differentiation by Wnt and Notch are conserved
throughout the adenoma-carcinoma sequence in the colon. Mol Cancer.
12:1262013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Reedijk M, Odorcic S, Zhang H, Chetty R,
Tennert C, Dickson BC, Lockwood G, Gallinger S and Egan SE:
Activation of Notch signaling in human colon adenocarcinoma. Int J
Oncol. 33:1223–1229. 2008.PubMed/NCBI
|
|
6
|
Zhang Y, Li B, Ji ZZ and Zheng PS: Notch1
regulates the growth of human colon cancers. Cancer. 116:5207–5218.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mumm JS and Kopan R: Notch signaling: from
the outside in. Dev Biol. 228:151–165. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li JL and Harris AL: Notch signaling from
tumor cells: a new mechanism of angiogenesis. Cancer Cell. 8:1–3.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Iso T, Kedes L and Hamamori Y: HES and
HERP families: multiple effectors of the Notch signaling pathway. J
Cell Physiol. 194:237–255. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bhanot U, Köhntop R, Hasel C and Möller P:
Evidence of Notch pathway activation in the ectatic ducts of
chronic pancreatitis. J Pathol. 214:312–319. 2008. View Article : Google Scholar
|
|
11
|
Ehebauer M, Hayward P and Arias AM: Notch,
a universal arbiter of cell fate decisions. Science. 314:1414–1415.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Garin JP, Despeignes J and Billerau M:
Present treatment of taeniasis with niclosamide. Lyon Med.
212:1581–1588. 1964.In French. PubMed/NCBI
|
|
13
|
Jin Y, Lu Z, Ding K, Li J, Du X, Chen C,
Sun X, Wu Y, Zhou J and Pan J: Antineoplastic mechanisms of
niclosamide in acute myelogenous leukemia stem cells: inactivation
of the NF-kappaB pathway and generation of reactive oxygen species.
Cancer Res. 70:2516–2527. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sack U, Walther W, Scudiero D, Selby M,
Kobelt D, Lemm M, Fichtner I, Schlag PM, Shoemaker RH and Stein U:
Novel effect of antihelminthic niclosamide on S100A4-mediated
metastatic progression in colon cancer. J Natl Cancer Inst.
103:1018–1036. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wieland A, Trageser D, Gogolok S, Reinartz
R, Höfer H, Keller M, Leinhaas A, Schelle R, Normann S, Klaas L, et
al: Anticancer effects of niclosamide in human glioblastoma. Clin
Cancer Res. 19:4124–4136. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ye T, Xiong Y, Yan Y, Xia Y, Song X, Liu
L, Li D, Wang N, Zhang L and Zhu Y: et al The anthelmintic drug
niclosamide induces apoptosis, impairs metastasis and reduces
immunosuppressive cells in breast cancer model. PLoS One.
9:e858872014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang Z, Li Y, Kong D, Ahmad A, Banerjee S
and Sarkar FH: Cross-talk between miRNA and Notch signaling
pathways in tumor development and progression. Cancer Lett.
292:141–148. 2010. View Article : Google Scholar
|
|
18
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Denli AM, Tops BB, Plasterk RH, Ketting RF
and Hannon GJ: Processing of primary microRNAs by the
Microprocessor complex. Nature. 432:231–235. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Prokopi M, Kousparou CA and Epenetos AA:
The secret role of microRNAs in cancer stem cell development and
potential therapy: a Notch-pathway approach. Front Oncol.
4:3892015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Srinivasan S, Selvan ST, Archunan G,
Gulyas B and Padmanabhan P: MicroRNAs -the next generation
therapeutic targets in human diseases. Theranostics. 3:930–942.
2013. View Article : Google Scholar
|
|
22
|
Esquela-Kerscher A and Slack FJ: Oncomirs
- microRNAs with a role in cancer. Nat Rev Cancer. 6:259–269. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Korpal M and Kang Y: The emerging role of
miR-200 family of microRNAs in epithelial-mesenchymal transition
and cancer metastasis. RNA Biol. 5:115–119. 2008. View Article : Google Scholar
|
|
24
|
Brabletz S, Bajdak K, Meidhof S, Burk U,
Niedermann G, Firat E, Wellner U, Dimmler A, Faller G, Schubert J
and Brabletz T: The ZEB1/miR-200 feedback loop controls Notch
signalling in cancer cells. EMBO J. 30:770–782. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Burk U, Schubert J, Wellner U, Schmalhofer
O, Vincan E, Spaderna S and Brabletz T: A reciprocal repression
between ZEB1 and members of the miR-200 family promotes EMT and
invasion in cancer cells. EMBO Rep. 9:582–589. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Vallejo DM, Caparros E and Dominguez M:
Targeting Notch signalling by the conserved miR-8/200 microRNA
family in development and cancer cells. EMBO J. 30:756–769. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang Z, Banerjee S, Ahmad A, Li Y, Azmi
AS, Gunn JR, Kong D, Bao B, Ali S, Gao J, et al: Activated K-ras
and INK4a/Arf deficiency cooperate during the development of
pancreatic cancer by activation of Notch and NF-κB signaling
pathways. PLoS One. 6:e205372011. View Article : Google Scholar
|
|
28
|
Ferrari-Toninelli G, Bonini SA, Uberti D,
Buizza L, Bettinsoli P, Poliani PL, Facchetti F and Memo M:
Targeting Notch pathway induces growth inhibition and
differentiation of neuroblastoma cells. Neuro Oncol. 12:1231–1243.
2010.PubMed/NCBI
|
|
29
|
Rasul S, Balasubramanian R, Filipović A,
Slade MJ, Yagüe E and Coombes RC: Inhibition of gamma-secretase
induces G2/M arrest and triggers apoptosis in breast cancer cells.
Br J Cancer. 100:1879–1888. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Senfter D, Holzner S, Kalipciyan M,
Staribacher A, Walzl A, Huttary N, Krieger S, Brenner S, Jäger W,
Krupitza G, et al: Loss of miR-200 family in 5-fluorouracil
resistant colon cancer drives lymphendothelial invasiveness in
vitro. Hum Mol Genet. 24:3689–3698. 2015.PubMed/NCBI
|
|
31
|
Li Y, Li PK, Roberts MJ, Arend RC, Samant
RS and Buchsbaum DJ: Multi-targeted therapy of cancer by
niclosamide: a new application for an old drug. Cancer Lett.
349:8–14. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Pastò A, Serafin V, Pilotto G, Lago C,
Bellio C, Trusolino L, Bertotti A, Hoey T, Plateroti M, Esposito G,
et al: NOTCH3 signaling regulates MUSASHI-1 expression in
metastatic colorectal cancer cells. Cancer Res. 74:2106–2118. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ranganathan P, Weaver KL and Capobianco
AJ: Notch signalling in solid tumours: a little bit of everything
but not all the time. Nat Rev Cancer. 11:338–351. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Dai Y, Wilson G, Huang B, Peng M, Teng G,
Zhang D, Zhang R, Ebert MP, Chen J, Wong BC, et al: Silencing of
Jagged1 inhibits cell growth and invasion in colorectal cancer.
Cell Death Dis. 5:e11702014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ozawa T, Kazama S, Akiyoshi T, Murono K,
Yoneyama S, Tanaka T, Tanaka J, Kiyomatsu T, Kawai K, Nozawa H, et
al: Nuclear Notch3 expression is associated with tumor recurrence
in patients with stage II and III colorectal cancer. Ann Surg
Oncol. 21:2650–2658. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Shih IeM and Wang TL: Notch signaling,
gamma-secretase inhibitors, and cancer therapy. Cancer Res.
67:1879–1882. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Balgi AD, Fonseca BD, Donohue E, Tsang TC,
Lajoie P, Proud CG, Nabi IR and Roberge M: Screen for chemical
modulators of autophagy reveals novel therapeutic inhibitors of
mTORC1 signaling. PLoS One. 4:e71242009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Osada T, Chen M, Yang XY, Spasojevic I,
Vandeusen JB, Hsu D, Clary BM, Clay TM, Chen W, Morse MA and Lyerly
HK: Antihelminth compound niclosamide downregulates Wnt signaling
and elicits antitumor responses in tumors with activating APC
mutations. Cancer Res. 71:4172–4182. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang AM, Ku HH, Liang YC, Chen YC, Hwu YM
and Yeh TS: The autonomous notch signal pathway is activated by
baicalin and baicalein but is suppressed by niclosamide in K562
cells. J Cell Biochem. 106:682–692. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Fernández-Majada V, Aguilera C, Villanueva
A, Vilardell F, Robert-Moreno A, Aytés A, Real FX, Capella G, Mayo
MW, Espinosa L and Bigas A: Nuclear IKK activity leads to
dysregulated notch-dependent gene expression in colorectal cancer.
Proc Natl Acad Sci USA. 104:276–281. 2007. View Article : Google Scholar :
|
|
41
|
Qiao L and Wong BC: Role of Notch
signaling in colorectal cancer. Carcinogenesis. 30:1979–1986. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Jones KB, Salah Z, Del Mare S, Galasso M,
Gaudio E, Nuovo GJ, Lovat F, LeBlanc K, Palatini J, Randall RL, et
al: miRNA signatures associate with pathogenesis and progression of
osteosarcoma. Cancer Res. 72:1865–1877. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kobayashi E, Hornicek FJ and Duan Z:
MicroRNA involvement in osteosarcoma. Sarcoma. 2012:3597392012.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kong D, Li Y, Wang Z, Banerjee S, Ahmad A,
Kim HR and Sarkar FH: miR-200 regulates PDGF-D-mediated
epithelial-mesenchymal transition, adhesion, and invasion of
prostate cancer cells. Stem Cells. 27:1712–1721. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Li Y, VandenBoom TG II, Kong D, Wang Z,
Ali S, Philip PA and Sarkar FH: Up-regulation of miR-200 and let-7
by natural agents leads to the reversal of
epithelial-to-mesenchymal transition in gemcitabine-resistant
pancreatic cancer cells. Cancer Res. 69:6704–6712. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhang B, Pan X, Cobb GP and Anderson TA:
microRNAs as oncogenes and tumor suppressors. Dev Biol. 302:1–12.
2007. View Article : Google Scholar
|