Introduction
Visceral adipose tissue contributes to the
pathophysiology of metabolic syndrome, which includes conditions
such as insulin resistance, hypertension, hyperlipidemia and
hyperuricemia (1,2). The basic strategy to treat metabolic
syndrome is diet and exercise to prevent patients from
cardiovascular disease and diabetes mellitus (3). However, visceral obesity negates
efforts to cut down on the energy supply and metabolize excess
energy (1). Medication or surgery
is considered when lifestyle alteration fails, although the
indication for bariatric surgery is limited to severely obese cases
and those complicated with organ failure (4). Thus, the majority of metabolic
syndrome cases with no response to diet and exercise are treated
with medication.
Metformin is an oral biguanide drug that was
introduced into clinical practice in the 1950s for the treatment of
type 2 diabetes (5). Although
metformin has been reported to suppress lipogenesis in a murine
preadipocyte cell line, in contrast to thiazolidine, which induces
the maturation of preadipocytes (6,7),
the effect of metformin on human visceral adipose tissue has not
been clearly demonstrated.
MicroRNAs (miRNAs or miRs) have been suggested as
therapeutic targets in metabolic syndrome due to their role in the
maturation of fat cells and the differentiation of adipocytes from
mesenchymal stromal stem cells (8–10).
The aim of the present study was to determine
whether metformin suppresses the differentiation of human
preadipocytes and to identify miRNAs associated with the regulation
of lipid metabolism in these cells.
Materials and methods
Reagents
Primary antibodies for peroxisome
proliferator-activated receptor γ (PPARγ) (cat. no. ab27649) and
CCAAT-enhancer-binding protein α (C/EBPα) (cat. no. ab40761) were
supplied by Abcam (Cambridge, UK). The primary antibody for β-actin
(cat. no. AC-15) was obtained from Sigma Aldrich (St. Louis, MO,
USA). The primers and probes used in RT-qPCR for PPARγ (Assay ID:
Hs01115513_m1), C/EBPα (assay ID: Hs00269972_s1) and β-actin (assay
ID: Hs01060665_g1) were supplied by Life technologies (Carlsbad,
CA, USA).
Cell line and culture
Poietics™ human visceral preadipocytes (HPrAD-vis)
were purchased from Lonza (Walkersville, MD, USA). Cells were
plated and subcultured according to the manufacturer's
instructions. Briefly, the cells were suspended in growth media at
1×105 cells/ml, and 1.5 ml of the cell suspension was
seeded in 6-well dishes. The cells were incubated for 24 h. For the
treated group, 1.5 ml of differentiation media (containing insulin,
dexamethasone, indomethacin and isobutylmethylxanthine) including 2
or 10 mM metformin was added to the growth media in each well,
yielding final concentrations of metformin of 1 and 5 mM,
respectively. As a control, 1.5 ml of differentiation media without
metformin was added to the control cells in growth media. Cells or
media were harvested at the indicated time points.
Oil Red O staining
Adipogenic differentiation of the cells was assessed
with Oil Red O staining as previously described (11). The cells were washed with PBS and
fixed in 4% paraformaldehyde for 10 min. The cells were then washed
with 3% isopropanol, followed by staining with newly filtered Oil
Red O staining solution for 10 min. After washing with distilled
water, the cells were destained in 100% isopropanol for 15 min. The
stained area was measured using Image J (NIH, Bethesda, MD,
USA).
Enzyme-linked immunosorbent assay for
adiponectin in culture media
The adiponectin concentration in the culture media
was measured using an enzyme-linked immunosorbent assay with human
Adiponectin/Acrp30 (R&D systems, Minneapolis, MN, USA)
(12). The assay was performed
according to the manufacturer's protocol.
Cell proliferation assay
Cell proliferation was evaluated using a WST-8
assay, as previously described (13,14). Briefly, cell growth was measured
with 10% WST-8 (Dojindo Laboratories, Tokyo, Japan) in a microplate
reader according to the absorbance at 450 nm at the indicated time
point.
RT-qPCR
Reverse-transcription and qPCR were performed to
measure changes in messenger RNA (mRNA) expression following
metformin treatment. Taqman® Gene Expression Assays
(Life Technologies) were used to determine the expression level of
mRNAs, and β-actin was used as an internal control. mRNAs were
reverse transcribed using the Taqman® RNA Reverse
Transcription kit (Life Technologies) according to the
manufacturer's protocol. Briefly, total RNA was extracted using the
RNeasy Mini kit (Qiagen, Venlo, The Netherlands) and diluted to 1.0
ng/µl. Reverse transcription was performed in a 20 µl
reaction volume consisting of 10 µl of RNA, 2 µl of
10X RT random primers and 8 µl of reverse transcription
master mix. As a result, 0.5 ng/µl of cDNA was produced per
tube. The PCR reaction was performed in a final volume of 20
µl, consisting of 2 µl of cDNA (0.5 ng), 1 µl
of 20X qPCR assay mix, 7 µl of nuclease-free water and 10
µl of Taqman® Fast Universal Master mix according
to the manufacturer's protocol. The cDNA was amplified and
quantified using StepOnePlus™ (Life Technologies). The mRNA
expression levels were standardized to β-actin.
Western blot analysis
The cell lysates were processed as previously
described (15). All the steps
were carried out at 4°C. Protein concentrations were measured using
a spectrophotometer Nano Drop 2000 (Thermo Fisher Scientific Inc.,
Wilmington, DE). The samples were electrophoresed using 10%
SDS-PAGE gels, and the proteins were transferred to nitrocellulose
membranes. The membranes were then incubated with primary
antibodies after being blocked and incubated with horseradish
peroxidase (HRP)-conjugated secondary antibodies. Immunoreactive
proteins were visualized with an enhanced chemiluminescence
detection system (PerkinElmer, San Jose, CA, USA) on X-ray film, as
previously described (13,14).
miRNA array analysis
Total RNA of cultured cells was extracted using the
miRNeasy Mini kit (Qiagen) as previously described (13,14,16–21). The samples were labeled using a
miRCURY Hy3 Power Labeling kit (Exiqon, Vedbaek, Denmark) and
hybridized onto a human miRNA Oligo chip, version 14.0 (Toray
Industries Inc., Tokyo, Japan). Scanning was conducted using a
3D-Gene Scanner 3000 (Toray Industries Inc.). 3D-Gene extraction
software (ver. 1.2, Toray Industries Inc.) was used to read the raw
intensity of the image. The raw data were analyzed with
GeneSpringGX ver. 10.0 (Agilent Technologies, Santa Clara, CA, USA)
and quantile normalized (22).
The fold-changes in miRNA expression level between the treated and
control groups were calculated. Hierarchical clustering was
accomplished using the furthest neighbor method and Pearson's
product-moment correlation coefficients as a metric.
Results
Metformin suppresses the differentiation
of preadipocytes with no proliferative effect on the cells
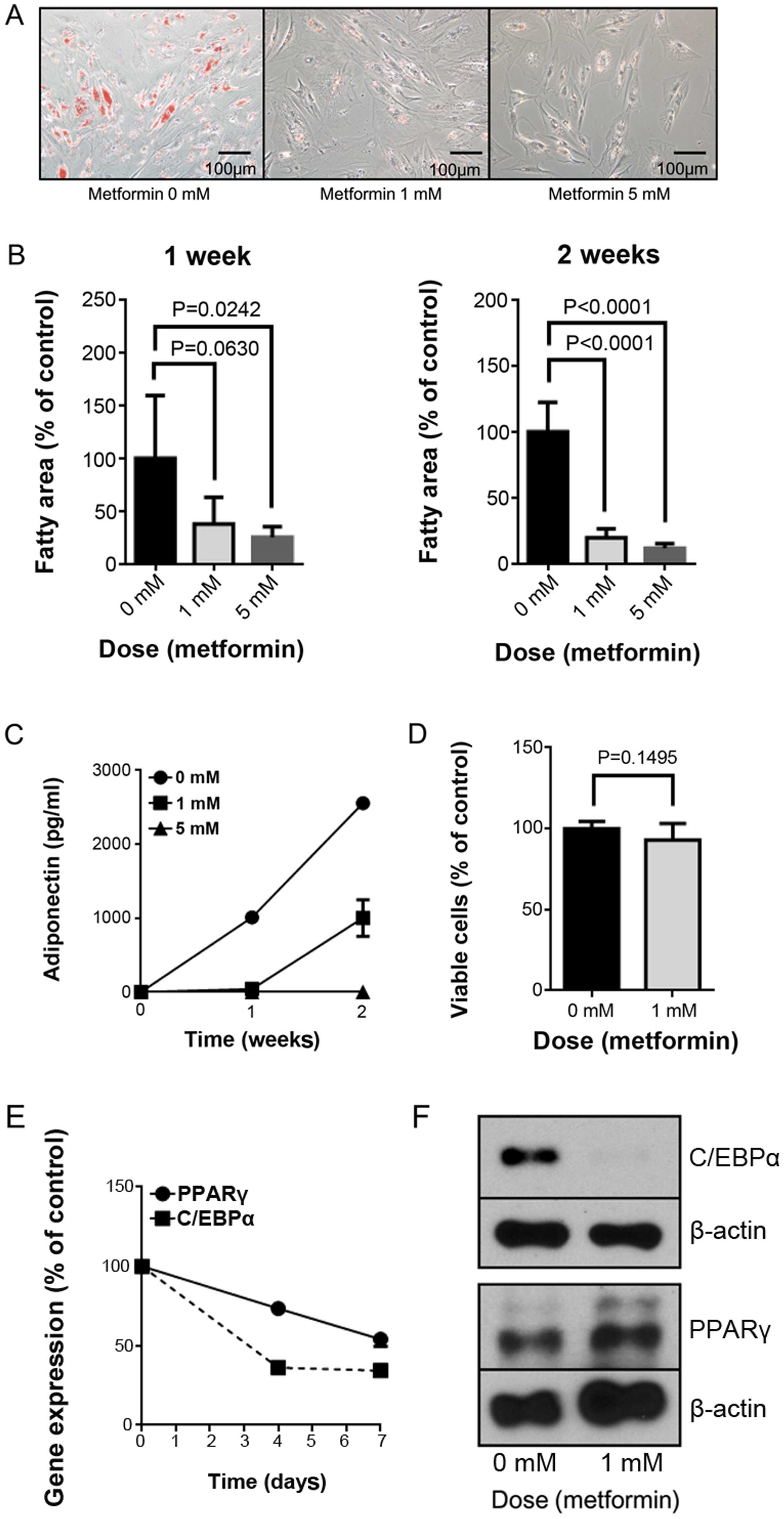
We studied the effect of metformin on the maturation
of HPrAD-vis cells through morphological analysis with Oil Red O
staining. Metformin (1 and 5 mM) significantly reduced lipid
droplet accumulation in preadipocytes, as shown in Fig. 1A and B.
We also analyzed adiponectin secretion by measuring
the concentration in the culture media. As shown in Fig. 1C, metformin decreased the
adiponectin concentration on days 7 and 14 of culture. However,
according to the WST-8 assay, metformin did not have any
proliferative or growth-inhibitory effect on the cells.
We then examined whether the expression levels of
key genes involved in adipocyte differentiation (PPARγ and C EBPα)
were altered by metformin. The mRNA expression of PPARγ and C/EBPα
was downregulated by metformin treatment on day 7 (Fig. 1D). Downregulation of C/EBPα
translation was confirmed by western blot analysis (Fig. 1E). These experiments were repeated
three times, and identical results were obtained.
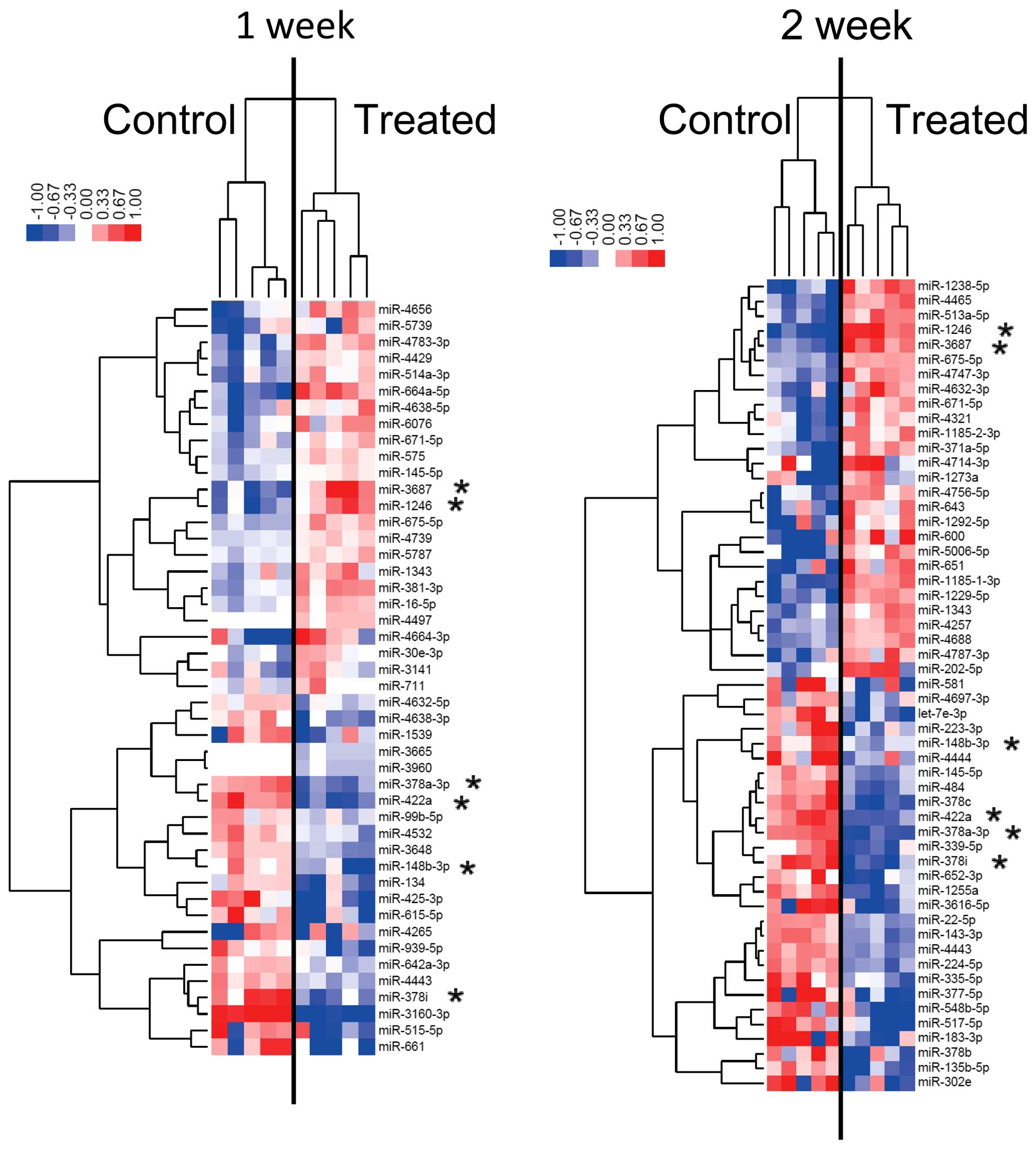
Differences in miRNA expression in
HPrAD-vis cells cultured in vitro with and without metformin
Using custom microarrays, we analyzed the expression
of human miRNAs in HPrAD-vis cells treated with or without
metformin. As shown in Table I,
for HPrAD-vis cells treated with or without 5 mM metformin, 6
miRNAs were significantly upregulated after 1 week of metformin
treatment, while 13 miRNAs were downregulated. After 2 weeks of 5
mM metformin treatment, 27 miRNAs were significantly upregulated
and 6 miRNAs were downregulated compared to the control (Table II). As shown in Tables I and II, 2 upregulated miRNAs (miR-3687 and
miR-1246), 4 downregulated miRNAs including miR-378 family members
were observed in the cells cultured for 1 and 2 weeks with
metformin. The expression level of miR-1246 increased in a
time-dependent manner.
 | Table IStatistical results and chromosomal
locations of miRNAs in HPrAD-vis cells treated with 5 mM metformin
for 1 week compared to control cells (P<0.05). |
Table I
Statistical results and chromosomal
locations of miRNAs in HPrAD-vis cells treated with 5 mM metformin
for 1 week compared to control cells (P<0.05).
| Name | Fold-change
(treated/control) | SD | P-value | Location |
|---|
| miR-664a-5p | 2.57 | 0.406 | 0.0090 | 1 |
| miR-3687a | 2.34 | 0.910 | 0.0229 | 21 |
| miR-4664-3p | 2.30 | 0.904 | 0.0153 | 8 |
| miR-1246a | 1.78 | 0.363 | 0.0107 | 2 |
| miR-4783-3p | 1.74 | 0.183 | 0.0010 | 2 |
| miR-4656 | 1.58 | 0.266 | 0.0019 | 7 |
| miR-1539 | 0.64 | 0.362 | 0.0136 | 18 |
| miR-148b-3pa | 0.64 | 0.198 | 0.0109 | 12 |
| miR-4638-3p | 0.63 | 0.165 | 0.0170 | 5 |
| miR-515-5p | 0.62 | 0.325 | 0.0205 | 19 |
| miR-134 | 0.61 | 0.273 | 0.0313 | 14 |
| miR-939-5p | 0.60 | 0.239 | 0.0360 | 8 |
| miR-378a-3pa | 0.54 | 0.069 | 0.0004 | 5 |
| miR-378ia | 0.49 | 0.090 | 0.0115 | 22 |
| miR-422aa | 0.45 | 0.066 | 0.0068 | 15 |
| miR-425-3p | 0.42 | 0.185 | 0.0420 | 3 |
| miR-661 | 0.42 | 0.307 | 0.0244 | 8 |
| miR-615-5p | 0.40 | 0.233 | 0.0481 | 12 |
| miR-3160-3p | 0.21 | 0.019 | 0.0037 | 11 |
 | Table IIStatistical results and chromosomal
locations of miRNAs in HPrAD-vis cells treated with 5 mM metformin
for 2 weeks compared to control cells (P<0.05). |
Table II
Statistical results and chromosomal
locations of miRNAs in HPrAD-vis cells treated with 5 mM metformin
for 2 weeks compared to control cells (P<0.05).
| Name | Fold-change
(treated/control) | SD | P-value | Location |
|---|
| miR-1246a | 3.22 | 0.450 | 0.0006 | 2 |
| miR-600 | 2.58 | 1.002 | 0.0052 | 9 |
| miR-1238-5p | 2.41 | 0.441 | 0.0017 | 19 |
| miR-3687a | 2.28 | 0.423 | 0.0034 | 21 |
| miR-1185-1-3p | 2.28 | 0.233 | 0.0012 | 14 |
| miR-1229-5p | 2.08 | 0.160 | 0.0017 | 5 |
| miR-671-5p | 2.03 | 0.314 | 0.0209 | 7 |
| miR-371a-5p | 2.02 | 0.241 | 0.0225 | 19q13 |
| miR-5006-5p | 1.99 | 0.306 | 0.0339 | 13 |
| miR-651 | 1.97 | 0.497 | 0.0448 | X |
| miR-4465 | 1.93 | 0.222 | 0.0079 | 6 |
| miR-4756-5p | 1.91 | 0.208 | 0.0352 | 20 |
| miR-4632-3p | 1.88 | 0.584 | 0.0251 | 1 |
| miR-643 | 1.87 | 0.397 | 0.0319 | 19 |
| miR-513a-5p | 1.85 | 0.259 | 0.0039 | X |
| miR-1292-5p | 1.85 | 0.416 | 0.0155 | 20 |
| miR-4714-3p | 1.77 | 0.693 | 0.0408 | 15 |
| miR-1185-2-3p | 1.75 | 0.271 | 0.0359 | 14 |
| miR-1273a | 1.72 | 0.369 | 0.0236 | 8q22.2 |
| miR-202-5p | 1.71 | 0.437 | 0.0495 | 10 |
| miR-4747-3p | 1.66 | 0.239 | 0.0134 | 19 |
| miR-4787-3p | 1.63 | 0.356 | 0.0431 | 3 |
| miR-4321 | 1.62 | 0.267 | 0.0262 | 19 |
| miR-1343 | 1.60 | 0.233 | 0.0008 | 11 |
| miR-4257 | 1.58 | 0.149 | 0.0014 | 1 |
| miR-4688 | 1.57 | 0.152 | 0.0014 | 11 |
| miR-675-5p | 1.51 | 0.041 | 0.0003 | 11 |
| miR-148b-3pa | 0.65 | 0.064 | 0.0285 | 12 |
| miR-22-5p | 0.64 | 0.049 | 0.0007 | 17 |
| miR-145-5p | 0.64 | 0.067 | 0.0014 | 5 |
| miR-4697-3p | 0.61 | 0.308 | 0.0115 | 11q25 |
| miR-581 | 0.59 | 0.281 | 0.0168 | 5 |
| miR-4443 | 0.59 | 0.045 | 0.0005 | 3 |
| miR-224-5p | 0.59 | 0.065 | 0.0004 | X |
| miR-143-3p | 0.58 | 0.064 | 0.0015 | 5 |
| miR-335-5p | 0.56 | 0.101 | 0.0166 | 7 |
| miR-484 | 0.55 | 0.084 | 0.0030 | 16 |
| miR-339-5p | 0.54 | 0.133 | 0.0070 | 7 |
| miR-223-3p | 0.52 | 0.149 | 0.0311 | X |
| miR-652-3p | 0.48 | 0.246 | 0.0226 | X |
| miR-1255a | 0.47 | 0.156 | 0.0074 | 4 |
| miR-422aa | 0.46 | 0.048 | 0.0020 | 15 |
| miR-378b | 0.45 | 0.370 | 0.0192 | 3 |
| miR-135b-5p | 0.43 | 0.239 | 0.0201 | 1 |
| miR-548b-5p | 0.43 | 0.252 | 0.0045 | 6 |
| miR-302e | 0.42 | 0.250 | 0.0209 | 11p15 |
| miR-378a-3pa | 0.41 | 0.044 | 0.0001 | 5 |
| miR-3616-5p | 0.41 | 0.229 | 0.0208 | 20 |
| let-7e-3p | 0.40 | 0.208 | 0.0055 | 19 |
| miR-4444 | 0.40 | 0.118 | 0.0455 | 2 |
| miR-378c | 0.40 | 0.091 | 0.0055 | 10 |
| miR-377-5p | 0.38 | 0.203 | 0.0325 | 14 |
| miR-378i | 0.37 | 0.120 | 0.0014 | 22 |
| miR-517-5p | 0.36 | 0.274 | 0.0225 | 19 |
| miR-183-3p | 0.34 | 0.180 | 0.0222 | 7 |
Unsupervised hierarchical clustering analysis, using
Pearson's correlations, showed that HPrAD-vis cells treated in
vitro for 1 or 2 weeks with metformin clustered separately from
the untreated cells (Fig. 2). The
subset of 19 miRNAs in HPrAD-vis cells treated for 1 week with
metformin and the 33 miRNAs detected in HPrAD-vis cells treated for
2 weeks were found to exhibit >1.5-fold alterations in the
expression levels between the metformin-treated and control groups.
These microarray data are registered at the NCBI Gene expression
Omnibus (GEO) under accession number GSE55665.
Discussion
In the present study, metformin suppressed lipid
accumulation and adiponectin secretion with no suppression of cell
growth. In addition, the expression levels of PPARγ and C/EBPα in
HPrAD-vis cells were altered following treatment, indicating that
metformin suppressed the differentiation of human preadipocytes
(6,23,24). Additionally, the miRNA profiles of
metformin-treated preadipocytes and control cells showed
differential clustering. In particular, the expression of miR-1246
and miR-3687 increased following metformin administration, while
the expression of miR-378 family members was decreased.
Differentiation of preadipocytes is associated with
the accumulation of lipid droplets in the cells. Preadipocytes
differentiate in mature fatty cells when they are exposed to an
excess energy supply, such as through overnutrition (7). Mature adipocytes secrete several
types of adipokines, including adiponectin (23). Additionally, the differentiation
of adipocytes from mesenchymal stromal stem cells is characterized
by alterations in genes such as PPARγ and C/EBP (25). Indeed, PPARγ and C/EBPα are
representative of the genes upregulated in preadipocytes during the
differentiation process from preadipocytes to mature adipocytes
(24).
In a previous report, metformin was shown to
suppress lipid droplet accumulation in murine preadipocytes
(6). Similarly, in the present
study, human preadipocytes showed reduced lipid accumulation
without a decrease in cell numbers. Furthermore, the secretion of
adiponectin, the mRNA expression of PPARγ and C/EBPα and the
protein expression of C/EBP in human preadipocytes decreased under
metformin administration, similar to previous findings on murine
preadipocytes (6). Collectively,
our results suggest that metformin inhibits the differentiation of
human preadipocytes.
Adipocyte differentiation is regulated via miRNAs;
these small non-coding RNAs target the mRNAs of genes involved in
each step of the differentiation from mesenchymal stromal stem
cells to mature adipocytes (26).
miRNAs are evolutionarily endogenous non-coding RNAs that have been
identified as post-transcriptional regulators of gene expression.
miRNAs bind mainly to the 3′ untranslated regions (UTRs) of target
mRNAs, resulting in mRNA degradation or the blockade of mRNA
translation (27,28). Thus, miRNAs play crucial roles in
the differentiation, maturation and intracellular metabolism of
cells and the effects of various drugs, as shown in the findings of
the present study (13,14,16–21). Several miRNAs have been shown to
contribute to lipid metabolism (8), with roles for miRNAs reported for
lipid synthesis, metabolism, transportation and storage.
In the present study, upregulation of miR-1246 and
miR-3687 and downregulation of the miR-378 family were found to be
involved in the differentiation of preadipocytes.
Mice genetically lacking miR-378 family members are
known to be resistant to high-fat diet-induced obesity (10). In addition, miR-378 family members
were found to be downregulated in murine preadipocytes, the
differentiation of which was suppressed by metformin treatment
(29). Therefore, metformin may
exert its inhibitory effect on visceral adipocytes via
downregulation of the miR-378 family.
Our data further suggest that the upregulation of
miR-1246 and miR-3687 and downregulation of miR-422a are associated
with the effect of metformin on the inhibition of preadipocyte
differentiation. To the best of our knowledge, this report is the
first to describe the association between three miRNAs above and
lipid metabolism. Notably, mice and rats lack miR-1246, miR-3687 or
miR-422a. Thus, the murine in vitro model was not used to
examine the expression levels of these miRNAs following metformin
treatment. The significance of miR-1246, miR-3687 and miR-422a in
adipogenesis and the maturation of preadipocytes thus remains to be
investigated.
In a previous study, miR-137 was shown to play a
role in the differentiation of preadipocytes (30). However, our results showed that
miR-137 expression was not altered during the differentiation of
preadipocyte cells. Thus, the role of miR-137 in preadipocyte
maturation should be investigated further in future studies.
In conclusion, metformin suppresses the maturation
of human preadipocytes in vitro and altered the miRNA
profile of cells. In particular, our results identify miR-1246,
miR-3687 and miR-422a as promising therapeutic targets for the
treatment of visceral obesity.
Abbreviations:
|
ELISA
|
enzyme linked immunosorbent assay
|
|
PCR
|
polymerase chain reaction
|
|
SDS-PAGE
|
sodium dodecyl sulfate-polyacrylamide
gel electrophoresis
|
References
|
1
|
Kaur J: A comprehensive review on
metabolic syndrome. Cardiol Res Pract. 2014:9431622014.PubMed/NCBI
|
|
2
|
Al-Daghri NM, Al-Attas OS, Alokail MS,
Alkharfy KM, Charalampidis P, Livadas S, Kollias A, Sabico SL and
Chrousos GP: Visceral adiposity index is highly associated with
adiponectin values and glycaemic disturbances. Eur J Clin Invest.
43:183–189. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Simmons RK, Alberti KG, Gale EA, Colagiuri
S, Tuomilehto J, Qiao Q, Ramachandran A, Tajima N, Brajkovich
Mirchov I, et al: The metabolic syndrome: Useful concept or
clinical tool? Report of a WHO Expert Consultation. Diabetologia.
53:600–605. 2010. View Article : Google Scholar
|
|
4
|
O'Brien PE, MacDonald L, Anderson M,
Brennan L and Brown WA: Long-term outcomes after bariatric surgery:
Fifteen-year follow-up of adjustable gastric banding and a
systematic review of the bariatric surgical literature. Ann Surg.
257:87–94. 2013. View Article : Google Scholar
|
|
5
|
Inzucchi SE, Bergenstal RM, Buse JB,
Diamant M, Ferrannini E, Nauck M, Peters AL, Tsapas A, Wender R and
Matthews DR: Management of hyperglycaemia in type 2 diabetes: A
patient-centered approach. Position statement of the American
Diabetes Association (ADA) and the European Association for the
Study of Diabetes (EASD). Diabetologia. 55:1577–1596. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Alexandre KB, Smit AM, Gray IP and
Crowther NJ: Metformin inhibits intracellular lipid accumulation in
the murine pre-adipocyte cell line, 3T3-L1. Diabetes Obes Metab.
10:688–690. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liang J, Fu M, Ciociola E, Chandalia M and
Abate N: Role of ENPP1 on adipocyte maturation. PLoS One.
2:e8822007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fernández-Hernando C, Ramírez CM, Goedeke
L and Suárez Y: MicroRNAs in metabolic disease. Arterioscler Thromb
Vasc Biol. 33:178–185. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gao Y, Xue J, Li X, Jia Y and Hu J:
Metformin regulates osteoblast and adipocyte differentiation of rat
mesenchymal stem cells. J Pharm Pharmacol. 60:1695–1700. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Carrer M, Liu N, Grueter CE, Williams AH,
Frisard MI, Hulver MW, Bassel-Duby R and Olson EN: Control of
mitochondrial metabolism and systemic energy homeostasis by
microRNAs 378 and 378*. Proc Natl Acad Sci USA. 109:15330–15335.
2012. View Article : Google Scholar
|
|
11
|
Wilson B, Liotta LA and Petricoiniii E:
Dynamic protein pathway activation mapping of adipose-derived stem
cell differentiation implicates novel regulators of adipocyte
differentiation. Mol Cell Proteomics. 12:2522–2535. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Berg AH, Combs TP and Scherer PE:
ACRP30/adiponectin: An adipokine regulating glucose and lipid
metabolism. Trends Endocrinol Metab. 13:84–89. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kato K, Gong J, Iwama H, Kitanaka A, Tani
J, Miyoshi H, Nomura K, Mimura S, Kobayashi M, Aritomo Y, et al:
The anti-diabetic drug metformin inhibits gastric cancer cell
proliferation in vitro and in vivo. Mol Cancer Ther. 11:549–560.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fujita K, Iwama H, Sakamoto T, Okura R,
Kobayashi K, Takano J, Katsura A, Tatsuta M, Maeda E, Mimura S, et
al: Galectin-9 suppresses the growth of hepatocellular carcinoma
via apoptosis in vitro and in vivo. Int J Oncol. 46:2419–2430.
2015.PubMed/NCBI
|
|
15
|
Masaki T, Okada M, Tokuda M, Shiratori Y,
Hatase O, Shirai M, Nishioka M and Omata M: Reduced C-terminal Src
kinase (Csk) activities in hepatocellular carcinoma. Hepatology.
29:379–384. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Miyoshi H, Kato K, Iwama H, Maeda E,
Sakamoto T, Fujita K, Toyota Y, Tani J, Nomura T, Mimura S, et al:
Effect of the anti-diabetic drug metformin in hepatocellular
carcinoma in vitro and in vivo. Int J Oncol. 45:322–332.
2014.PubMed/NCBI
|
|
17
|
Kobayashi M, Kato K, Iwama H, Fujihara S,
Nishiyama N, Mimura S, Toyota Y, Nomura T, Nomura K, Tani J, et al:
Antitumor effect of metformin in esophageal cancer: In vitro study.
Int J Oncol. 42:517–524. 2013.
|
|
18
|
Fujimori T, Kato K, Fujihara S, Iwama H,
Yamashita T, Kobayashi K, Kamada H, Morishita A, Kobara H, Mori H,
et al: Antitumor effect of metformin on cholangiocarcinoma: In
vitro and in vivo studies. Oncol Rep. 34:2987–2996. 2015.PubMed/NCBI
|
|
19
|
Fujihara S, Kato K, Morishita A, Iwama H,
Nishioka T, Chiyo T, Nishiyama N, Miyoshi H, Kobayashi M, Kobara H,
et al: Antidiabetic drug metformin inhibits esophageal
adenocarcinoma cell proliferation in vitro and in vivo. Int J
Oncol. 46:2172–2180. 2015.PubMed/NCBI
|
|
20
|
Kato K, Iwama H, Yamashita T, Kobayashi K,
Fujihara S, Fujimori T, Kamada H, Kobara H and Masaki T: The
anti-diabetic drug metformin inhibits pancreatic cancer cell
proliferation in vitro and in vivo: Study of the microRNAs
associated with the antitumor effect of metformin. Oncol Rep.
35:1582–1592. 2016.
|
|
21
|
Fujita K, Kobara H, Mori H, Fujihara S,
Chiyo T, Matsunaga T, Nishiyama N, Ayaki M, Yachida T, Morishita A,
et al: Differences in miRNA expression profiles between GIST and
leiomyoma in human samples acquired by submucosal tunneling biopsy.
Endosc Int Open. 3:E665–E671. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bolstad BM, Irizarry RA, Astrand M and
Speed TP: A comparison of normalization methods for high density
oligonucleotide array data based on variance and bias.
Bioinformatics. 19:185–193. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Nigro E, Scudiero O, Monaco ML, Palmieri
A, Mazzarella G, Costagliola C, Bianco A and Daniele A: New insight
into adiponectin role in obesity and obesity-related diseases.
BioMed Res Int. 2014:6589132014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wu Z, Rosen ED, Brun R, Hauser S, Adelmant
G, Troy AE, McKeon C, Darlington GJ and Spiegelman BM:
Cross-regulation of C/EBP alpha and PPAR gamma controls the
transcriptional pathway of adipogenesis and insulin sensitivity.
Mol Cell. 3:151–158. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nakagami H: The mechanism of white and
brown adipocyte differentiation. Diabetes Metab J. 37:85–90. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Son YH, Ka S, Kim AY and Kim JB:
Regulation of adipocyte differentiation via MicroRNAs. Endocrinol
Metab (Seoul). 29:122–135. 2014. View Article : Google Scholar
|
|
27
|
Callegari E, Elamin BK, Sabbioni S,
Gramantieri L and Negrini M: Role of microRNAs in hepatocellular
carcinoma: A clinical perspective. Onco Targets Ther. 6:1167–1178.
2013.PubMed/NCBI
|
|
28
|
Kerr TA, Korenblat KM and Davidson NO:
MicroRNAs and liver disease. Transl Res. 157:241–252. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gerin I, Bommer GT, McCoin CS, Sousa KM,
Krishnan V and MacDougald OA: Roles for miRNA-378/378* in adipocyte
gene expression and lipogenesis. Am J Physiol Endocrinol Metab.
299:E198–E206. 2010.PubMed/NCBI
|
|
30
|
Shin KK, Kim YS, Kim JY, Bae YC and Jung
JS: miR-137 controls proliferation and differentiation of human
adipose tissue stromal cells. Cell Physiol Biochem. 33:758–768.
2014. View Article : Google Scholar : PubMed/NCBI
|